Abstract
Vaccination and SIV challenge of macaque species is the best animal model for evaluating candidate HIV vaccines in pre-clinical studies. As such, robust assays optimized for use in nonhuman primates are necessary for reliable ex vivo measurement of immune responses and identification of potential immune correlates of protection. We optimized and qualified an 8-color intracellular cytokine staining assay for the measurement of IFNγ, IL-2, and TNF from viable CD4 and CD8 T cells from cryopreserved rhesus macaque PBMC stimulated with peptides. After optimization, five laboratories tested assay performance using the same reagents and PBMC samples; similar results were obtained despite the use of flow cytometers with different configurations. The 8-color assay was then subjected to a pre-qualification study to quantify specificity and precision. These data were used to set positivity thresholds and to design the qualification protocol. Upon completion of the qualification study, the assay was shown to be highly reproducible with low inter-aliquot, inter-day, and inter-operator variability according to the qualification criteria with an overall variability of 20–40% for each outcome measurement. Thus, the 8-color ICS assay was formally qualified according to the ICH guidelines Q2 (R1) for specificity and precision indicating that it is considered a standardized/robust assay acceptable for use in pre-clinical trial immunogenicity testing.
Keywords: Flow cytometry, Immune assays, Phenotype, Memory, Antigen-specific
1. Introduction
Since HIV was identified as the etiological cause of AIDS 28 years ago, the virus has infected approximately 60 million people worldwide and caused 25 million deaths. In order to curb the spread of HIV and eventually eliminate the AIDS pandemic, the production of an effective prophylactic vaccine is paramount. Results released in 2009 from the RV144 clinical trial in Thailand are promising (Rerks-Ngarm et al., 2009). In this study, prime-boost vaccination was shown to be moderately effective, reducing the risk of HIV infection by 31%. The vaccine regimen induced production of non-neutralizing antibodies as well as CD4 T cell responses. In addition, previous studies provide evidence suggesting vaccine-induced CD8 T cell responses decrease mortality post infection (Letvin et al., 2006; Mattapallil et al., 2006). Taken together, the data suggest that efficacious vaccines may require elicitation of cellular responses from both CD4 and CD8 T cells, in addition to humoral responses. As such, the current focus is on improving vaccine efficacy and the nonhuman primate (NHP) is the animal model of choice for this purpose.
Simian immunodeficiency virus (SIV) infection in the Indian rhesus macaque is comparable, immunologically and pathologically, to HIV infection in humans. The nucleotide sequence of SIV is similar to HIV-1, and SIV infection in the rhesus model leads to an immunodeficiency syndrome that closely resembles AIDS in humans, characterized by early viremia, loss of adaptive immunity, CD4 T cell depletion and subsequent death of infected animals (Gardner, 1989; McClure et al., 1989). In order to define the potential role and correlates of cellular responses to the protection of a vaccine, assays for measuring immune responses to vaccination must be adapted and optimized for use in NHP.
IFNγ ELISpot is the most commonly used method for measuring vaccine-induced Ag-specific T cell responses. However, this method does not easily distinguish which cell type is producing the cytokine, therefore providing limited information. Previous data has demonstrated that examination of IFNγ alone does not adequately identify the magnitude of a vaccine-induced immune response (De Rosa et al., 2004; Sun et al., 2008) and a number of studies have demonstrated that positive outcomes are associated with T cells that produce multiple effector cytokines (i.e., T cell quality) in addition to the magnitude of this response (De Rosa et al., 2004; Betts et al., 2006; Darrah et al., 2007). Intracellular cytokine staining (ICS) is a comprehensive and informative method for evaluating antigen-specific T cell responses since it provides a quantitative assessment of the type (CD4 vs CD8) and phenotype of responsive cells that defines the magnitude as well as the quality of the response.
The International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) establishes guidelines for the validation of analytical procedures that are included as part of registration applications that are submitted within the EC, Japan, and USA. As such, assays used to measure T cell responses from vaccines in clinical trials should be validated according the specifications in Guideline Q2(R1), Validation of Analytical Procedures: Text and Methodology (http://www.ich.org/products/guidelines/quality/article/quality-guidelines.html, 2005). This document describes in detail the characteristics that should be considered for assay validation: 1) accuracy 2) precision (repeatability, intermediate precision, and reproducibility) 3) specificity 4) linearity 5) range and 6) robustness. Two other parameters, detection limit and quantitation limit may also be considered under some circumstances. An IFNγ ELISpot assay was validated (Russell et al., 2003) and an ICS assay for human samples was qualified and partially validated (Horton et al., 2007) according to these guidelines.
Qualification is often the initial stage of validation and includes tests of precision and specificity, at a minimum, while validation also includes tests of accuracy, linearity, range, and robustness. When an assay is qualified, it means there is evidence that the assay performs within the prospectively established acceptance ranges for each parameter evaluated. These acceptance ranges are established based upon the intended use of the assay, and by meeting these criteria the assay is deemed “fit for purpose” (Ritter et al., 2004). Full assay validation is not required for pre-clinical studies in nonhuman primates (NHP); however, since data from NHP studies are used to support vaccine selection for clinical studies in humans, it is important to demonstrate that the assays used in NHP studies perform as expected.
In this report, we describe the optimization and qualification of an eight color ICS assay designed to measure cytokine responses from frozen rhesus macaque PBMC. The panel, which was previously described (Foulds et al., 2012), measures IFNγ, IL-2, and TNF production from CD4 and CD8 T cells, and has an 11-color variant that provides for the additional identification of memory cell subsets based on the expression of CD28, CD45RA, and CCR7. After development and optimization in one laboratory, the assay was evaluated in a comparative study by five different laboratories in the U.S. and Europe to examine reproducibility between laboratories. Once the panel and assay were approved, we began the process to formally qualify the assay. A pre-qualification study was performed first to establish the parameters by which the assay would be evaluated during the qualification. Then, we performed a formal qualification study to assess the specificity and precision of the assay according to these parameters. For precision studies, repeatability describes precision under the same operating conditions over a short period of time (intra-assay precision) and intermediate precision describes intra-laboratory variation (inter-day, inter-operator, etc.). The ICS assay described here was shown to have low inter-vial (intra-assay), inter-day, and inter-operator variability according to ICH guidelines Q2 (R1) and was formally qualified for the measurement of immune responses from NHP in pre-clinical studies.
2. Methods
2.1. Animals
Specificity experiments were performed on cryopreserved PBMC isolated by Ficoll centrifugation of 20 ml of whole blood from healthy SIV-uninfected (SIV−) colony-bred rhesus macaques (Macaca mulatta). Precision experiments were performed using cryopreserved PBMC and splenocytes from immunized and SIVmac251-infected (SIV+) rhesus macaques, respectively. Immunized animals received 3 doses of recombinant DNA and one dose of recombinant Ad5 encoding SIV Gag, Env, and Pol antigens via a prime/boost regimen (Mattapallil et al., 2006). These animals were apheresed at different time points post-immunization in order to obtain sufficient PBMC from a single time point for use in the pre-qualification and qualification studies. Splenocytes were obtained from euthanized SIV+ animals infected with SIVmac251 for other studies. All animals were housed at Bioqual, Inc. (Rockville, MD) in accordance with American Association for Accreditation of Laboratory Animal Care guidelines. All animal studies were approved by the Vaccine Research Center (NIH) Institutional Animal Care and Use Committee.
2.2. Cell preparation and antigenic stimulation
Cryopreserved PBMC and splenocytes were thawed in a 37C water bath until only a small pea-sized piece of ice was left. The cells were then transferred to pre-warmed R10 [RPMI 1640 (BioWhittaker, Walkersville, MD), 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin G, 100 μg/ml streptomycin] with 50 U/ml Benzonase (Novagen, Madison, WI) and washed. Cells were resuspended at 1–2 million cells/ml in R10 in 15 ml conical tubes and “rested” overnight in a 37C/5% CO2 incubator with the caps loosened one turn and the tubes slightly slanted. In vitro stimulations were performed the following morning. In brief, cells were transferred to a 96 well v-bottom plate at 1 to 3 million cells/well and stimulated with peptide pools (15-mers overlapping by 11 amino acids spanning SIVmac239 Env, Gag, and Pol; provided by the NIH AIDS Research & Reference Reagent Program, Germantown, MD) at a final concentration of 2 μg/ml, in the presence of Brefeldin A at a final concentration of 10 μg/ml, for 6 h. Negative controls received an equal concentration of DMSO (the peptide diluent). At the end of the incubation, the plate was transferred to 4C overnight. This overnight storage was shown not to affect the assay.
2.3. Intracellular staining procedure
Staining for cell surface and intracellular molecules was performed as described (Foulds et al., 2012). The following monoclonal antibodies were used: CD4-QD605 (clone MT477; conjugated in-house) (Chattopadhyay et al., 2007), CD8-Pacific Blue (clone RPA-T8; BD Biosciences), CD69-ECD (clone TP1.55.3; Beckman Coulter), CD3-Cy7APC (clone SP34.2; BD Biosciences), IFNγ-APC (clone B27; BD Biosciences), IL-2-PE (clone MQ1-17 H12; BD Biosciences), and TNF-FITC (clone Mab11; BD Biosciences). Aqua LIVE/DEAD kit (Invitrogen, Carlsbad, CA) was used to exclude dead cells. The final composition of the panel is shown in Table 1. All antibodies were titrated to determine the optimal dilution. Samples were acquired on an LSR II flow cytometer and analyzed using FlowJo version 9.3 (Treestar, Inc., Ashland, OR).
Table 1.
8-color ICS panel for Rhesus macaques.
| Marker | Dye | Staining step |
|---|---|---|
| Dead cells | Aqua blue | Pre-surface |
| CD4 | QD605 | Surface |
| CD3 | Cy7-APC | Intracellular |
| CD8 | Pacific blue | Intracellular |
| CD69 | ECD | Intracellular |
| IFNγ | APC | Intracellular |
| IL-2 | PE | Intracellular |
| TNF | FITC | Intracellular |
2.4. Statistical analysis
For Figs. 1 and 2, Prism (GraphPad Software, La Jolla, CA) was used to perform Wilcoxon matched-pairs signed rank tests across experimental conditions. The same software was used to determine coefficients of variation for Fig. 3. For Fig. 4 and Tables 2, 3, 4, 6, 7 and Supplementary Table 1 analyses were performed on the 6 marginal cytokine responses (IFNγ, IL-2, and TNF in CD4 and CD8 T cells) and all descriptive and inferential statistical analyses were performed using SAS, StatXact, and/or R statistical software. For Fig. 6, analyses were computed using JMP software (SAS, Cary, NC).
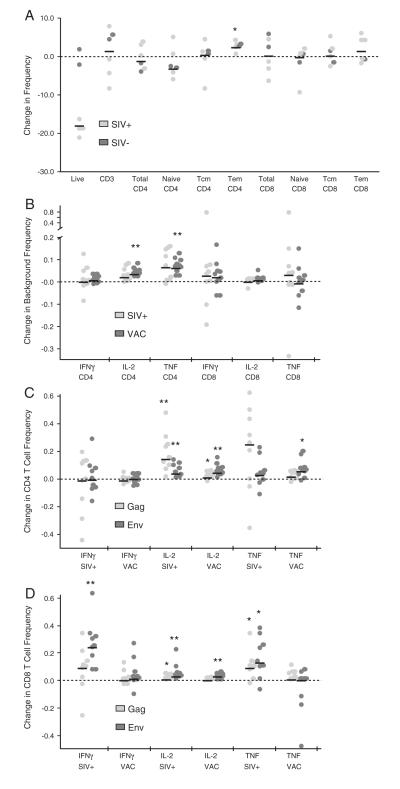
Fig. 1.
Effect of overnight rest on memory populations, background, and cytokine production. Frozen cells were thawed and subjected to either surface staining (A) or ICS (B, C, and D) immediately or following overnight rest. For C and D, data was background subtracted and the frequency of cytokine positive cells from unrested cells was subtracted from rested cells. P value * 0.01 to 0.05, ** 0.001 to 0.01, ***<0.001, Wilcoxon matched-pairs signed rank test. Bars represent the median.
Fig. 2.
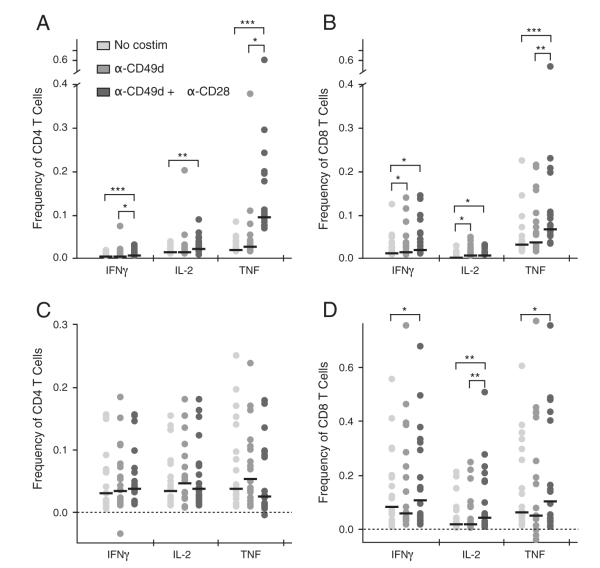
Effect of co-stimulation on cytokine production. PBMC from 18 SIV-vaccinated animals were stimulated with Gag peptides with and without costimulation then subjected to ICS. A and B are background values (no peptide stimulation) and C and D are peptide-stimulated, background subtracted values. * indicates P<0.05, ** P<0.001 to 0.01, Wilcoxon matched-pairs signed rank test. Bars represent the median.
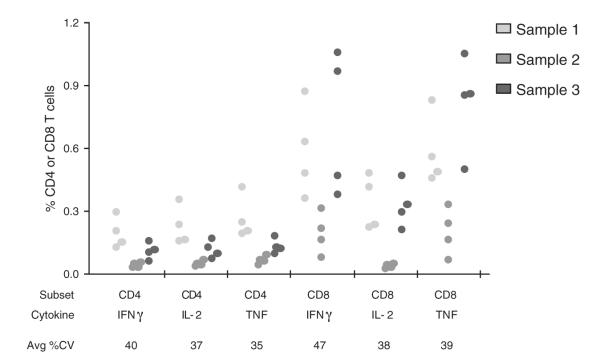
Fig. 3.
Multi-laboratory comparison of the 8-color ICS panel and assay. Four laboratories performed ICS using the same protocol and panel on aliquots of the same 3 samples. Each dot represents the response from an individual laboratory for a given sample, T cell subset, and cytokine. The average CV% for each marginal response is given.
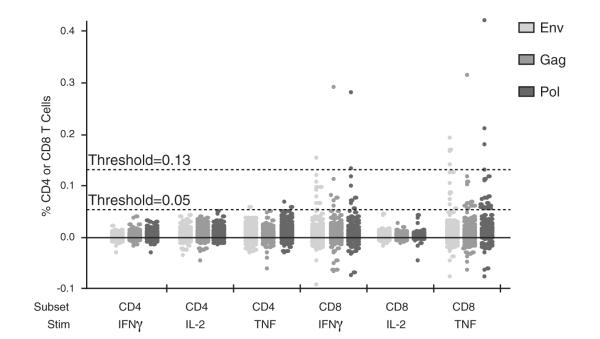
Fig. 4.
Qualification of the 8-color ICS panel and assay for specificity. 100 seronegative animals were assayed per protocol. Positivity thresholds were set at 0.05% for all cytokines for CD4 T cells and for IL-2 for CD8 T cells and 0.13% for IFNγ and TNF for CD8 T cells. Pass if: ≤6% of SIV− NHP have positive responses for each of the T cell subsets.
Table 2.
Pre-qualification specificity results. Values are the 97th percentile of background-subtracted responses from 30 SIV naïve rhesus macaques.
| Cytokine | CD4 (%) Env/Gag/Pol |
CD8 (%) Env/Gag/Pol |
|---|---|---|
| IFNγ | 0.01/0.01/0.01 | 0.09/0.09/0.13 |
| IL-2 | 0.03/0.03/0.03 | 0.02/0.02/0.01 |
| TNF | 0.03/0.03/0.02 | 0.17/0.12/0.13 |
Table 3.
Qualification positivity criteria. Shown are minimum values for response positivity based on background-subtracted cytokine measurements from the pre-qualification study. The values are approximately at or higher than the 97th percentile of 90 separate stimulations on SIV naïve samples (30 animals×3 peptide pools).
| Cytokine | % of CD4 | % of CD8 |
|---|---|---|
| IFNγ | 0.05 | 0.13 |
| IL-2 | 0.05 | 0.05 |
| TNF | 0.05 | 0.13 |
Table 4.
Pre-qualification precision results. Coefficients of variation (CVs) were computed from linear mixed effects models using the natural log of the response. One model was fitted per animal (3 animals) for each cell type/stimulation/cytokine combination. Values are shown for animals with an overall mean≥0.1%. N/A means no animals with mean≥0.1% for that measurement.
| Measurement | Mean response (% of CD4 or CD8 T cells) |
Avg CV% day |
Avg CV% vial |
Avg CV% operator |
Avg CV% residual error |
Avg CV% total |
|---|---|---|---|---|---|---|
| CD4 IFNγ | 0.17 | 7.8 | 0 | 29.6 | 19.2 | 38.3 |
| CD4 IL-2 | 0.20 | 5.5 | 0 | 26.1 | 16.6 | 32.3 |
| CD4 TNF | 0.22 | 7.2 | 0 | 32.3 | 19.2 | 40.0 |
| CD8 IFNγ | 0.10 | 16.5 | 8.8 | 14.8 | 18.9 | 31.0 |
| CD8 IL-2 | N/A | N/A | N/A | N/A | N/A | N/A |
| CD8 TNF | 0.15 | 15.8 | 0 | 10.8 | 17.9 | 26.5 |
Table 6.
Qualification specificity results. Values in parentheses indicate the percent of SIV seronegative samples with responses greater than threshold. Pass if: the total false positive rate for each of the 3 cytokine responses (all 3 peptide pools) is ≤6% for each T cell subset. In some cases, the percent positives for each peptide pool or cytokine don’t add up to the total because the same animal is positive for more than one pool or cytokine.
| Stimulation | CD4 IFNγ | CD4 IL-2 | CD4 TNF | CD8 IFNγ | CD8 IL-2 | CD8 TNF |
|---|---|---|---|---|---|---|
| Env | 0/102 (0%) | 0/102 (0%) | 2/102 (2%) | 1/102 (1%) | 0/102 (0%) | 4/102 (4%) |
| Gag | 0/103 (0%) | 0/103 (0%) | 0/103 (0%) | 1/103 (1%) | 0/103 (0%) | 1/103 (1%) |
| Pol | 0/102 (0%) | 0/102 (0%) | 2/102 (2%) | 2/102 (2%) | 0/102 (0%) | 3/102 (3%) |
| Total % Pos | 0% | 0% | 4/103 (4%) | 2/103 (2%) | 0% | 5/103 (5%) |
| Total % Pos for CD4 T cells=3/103 (3%) | ||||||
| Total % Pos for CD8 T cells=5/103 (5%) | ||||||
Table 7.
Qualification precision results. Assay precision was assessed via the coefficient of variation (CV) for samples with net mean responses ≥0.1%. Values are averaged over animal and peptide pool. All 4 CVs were evaluated, but only day and vial were used for qualification pass/fail. Pass if: for samples with mean response %<0.2, CV must be≤70%; for all other samples, CV≤35%.
| Measurement | Mean response (% of CD4 or CD8 T cells) |
Avg CV% day |
Avg CV% vial |
Avg CV% operator |
Avg CV% residual error |
Avg CV% total |
|---|---|---|---|---|---|---|
| CD4 IFNγ | 0.27 | 7.7 | 0 | 6.4 | 21.7 | 26.1 |
| CD4 IL-2 | 0.29 | 10.3 | 0 | 7.8 | 19.5 | 25.6 |
| CD4 TNF | 0.60 | 7.0 | 0.5 | 5.2 | 16.3 | 20.0 |
| CD8 IFNγ | 0.71 | 13.2 | 2.0 | 14.3 | 28.2 | 39.7 |
| CD8 IL-2 | 0.26 | 17.4 | 0 | 21.7 | 24.9 | 44.1 |
| CD8 TNF | 0.58 | 11.2 | 0.9 | 15.6 | 18.3 | 30.1 |
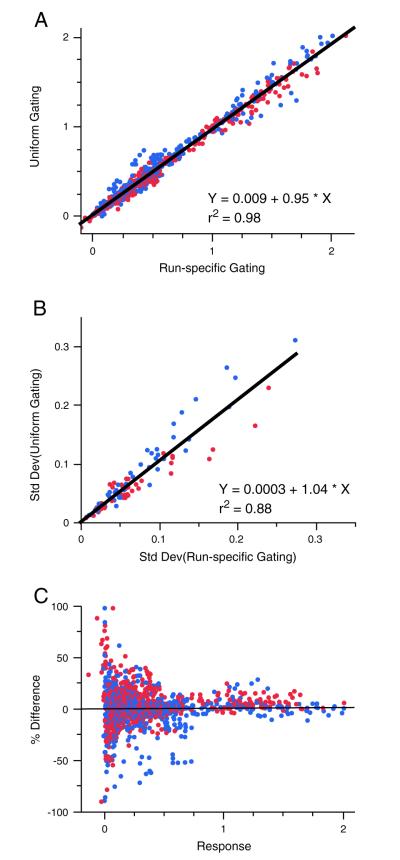
Fig. 6.
Comparison of gating by run or instrument. Background subtracted values for the six marginal cytokines obtained by the two different gating methods were plotted. Red and blue depict the two different operators. For A and B, linear regression was used to calculate the slope of the line. A) All days, vials, and animals are plotted for both operators. B) The standard deviations of the average of all of the measurements made (days and vials) for each animal and operator are plotted. C) The percent difference (defined as run-specific gating – uniform gating/average of the 2 values) plotted versus the response (measured as % cytokine positive CD4 or CD8 T cells).
2.4.1. Evaluation of assay specificity
The proportion of positive responses of the 6 marginal cytokine combinations among the tested ~100 SIV seronegative samples was examined for each of the three peptide pools individually and combined. A positive response occurred if the percent positive cells for a peptide minus the percent positive cells for the negative control (background subtracted percent) exceeded the cut-off criterion defined in Table 3. A sample was declared positive if there was a positive response for any cytokine to any peptide pool. To pass the qualification, ≤6% of samples could be positive for CD4 or CD8 T cells (Table 6).
2.4.2. Evaluation of assay precision
Linear mixed-effects models were used to estimate the components of the total variation in the data; the components include day, vial, and operator, in addition to the underlying assay variance. A model was fit for each of the 4 individual NHP samples for each of the 6 background subtracted outcomes. A natural log transformation was employed on the background-subtracted responses to stabilize the variance because the variability of the observed response often increases as the mean increases. The factors of day, vial, and operator were fit as random effects in the model. The residual error (assay error) is the remaining variance after the random effects are considered. The estimated grand mean on the original scale was used to define three different levels of response categories (PT=positivity threshold from Table 3): <PT, PT – 0.2%, >0.2%. Coefficients of variation were computed for day, vial, and operator and residual error via the formula: CV for Factor X=sqrt(exp(VEX) - 1), where VEX is the variance estimate for the random effect X. This CV is an exact estimate for data which follow a log normal distribution on the original scale. For outcomes which include background subtracted values less than zero, the percentage of such values were reported, and the models were fit without including these data points (Table 7).
3. Results
3.1. Effect of overnight rest on cytokine production from T cells
A previous report demonstrated that resting thawed human cells in R10 at 37C/5% CO2 overnight prior to stimulation increases T cell cytokine responses (Horton et al., 2007). We tested the effect of overnight rest on thawed rhesus macaque cells from either SIV-infected or SIV-vaccinated animals. Cryovials were thawed and cells were rested as described in Methods; the next morning, additional aliquots from the same blood draws were thawed and processed in parallel (i.e., without a rest). Samples were then either surface stained to determine changes in cell populations, or stimulated with SIV Gag and/or Env peptide pools and subjected to intracellular cytokine staining to characterize any changes in cytokine production.
Following overnight rest, there were ~15% fewer live cells as determined by Aqua blue live/dead amine staining, with less death in PBMC from SIV− than SIV+ NHP (Fig. 1A). There was no change in the frequency within lymphocytes of CD3+ cells or of total CD4 and CD8 T cells. However, there was a small (~2%) but statistically significant increase in effector memory CD4 T cells, as defined by CD28 and CD95 staining, with a corresponding decrease in naïve CD4 T cells that itself was not statistically significant. The same trend was apparent for CD8 T cells. Thus, overall, resting thawed cells overnight does not have a dramatic impact on the representation of major T cell populations.
Next, we examined the effect of overnight rest on cytokine production, both in our mock controls (background) as well as for peptide-stimulated samples following background subtraction. Overnight rest increased the frequency of background IL-2 and TNF production from SIV-vaccinated CD4 T cells (Fig. 1B). However, the increase in cytokine production from stimulated cells was far greater, such that background-subtracted cytokine responses were significantly increased for IL-2 and TNF from CD4 T cells (Fig. 1C) and IFNγ, IL-2, and TNF for CD8 T cells (Fig. 1D). Based on the lack of changes in phenotype and an increased sensitivity for peptide-specific responses, we included an overnight rest in our procedure.
3.2. Effect of co-stimulation on cytokine production from T cells
Since it has been previously shown that adding co-stimulation significantly enhances responses from protein-stimulated human PBMC (Waldrop et al., 1998), we examined the impact of adding the co-stimulatory antibodies anti-CD49d and anti-CD28 during the stimulation on cytokine detection from peptide-stimulated rhesus macaque T cells. PBMC from 18 vaccinated animals were stimulated with an SIV Gag peptide pool in the absence of co-stimulatory antibodies, with anti-CD49d only, or with anti-CD49d and anti-CD28 combined. As shown in Fig. 2A, adding co-stimulation increased background values for all three cytokines for both T cell subsets however IL-2 and TNF from CD4 T cells increased the most, by more than 10-fold for a number of samples. For some samples, this large increase in background resulted in an inability to detect low-level cytokine responses from peptide-stimulated samples compared to parallel assays that did not use co-stimulation. In contrast, co-stimulation did not lead to as much of an increase in background responses for CD8 T cells (Fig. 2B). Overall, the addition of co-stimulatory antibodies did not significantly increase background-subtracted peptide-stimulated cytokine responses from CD4 T cells (Fig. 2C). For CD8 T cells, co-stimulation led to a statistically significant increase in all three cytokine responses (Fig. 2D). However, since the mean increase with co-stimulation was very low for CD8 T cells, averaging 0.03% for IFNγ, 0.05% for IL-2, and 0.05% for TNF, we decided not to include co-stimulatory antibodies because of the increase in background responses and the risk of loss of detection of cytokine responses from CD4 T cells.
3.3. Prequalification study
Once the method for ICS for NHP samples was finalized, it was tested in two rounds of multi-lab comparisons for an initial assessment of reproducibility. According to ICH guidelines, reproducibility describes the precision between laboratories. Four different laboratories performed ICS on aliquots of the same three samples using the same reagents. A fifth laboratory also participated but used different cells due to international shipping limitations on nonhuman primate cells. Overall, the results were fairly consistent, more so for CD4 than CD8 T cells (Fig. 3). The average coefficient of variation (CV) for the six outcome variables for the 3 samples from the 4 labs was 39%. Based on these results, the panel and assay procedure was approved by all participating laboratories.
In preparation for qualifying the assay, it was necessary to establish parameters for evaluating the success of the qualification, therefore a formal prequalification study was performed. Data from the pre-qualification study was used to assess the performance characteristics of the assay, including precision, and to define the baseline parameters and final assay format for the qualification study as well as to explore potential statistical analysis procedures for use in the qualification study.
The pre-qualification study consisted of two experiments, one to assess the specificity and one to assess precision (repeatability and intermediate precision) of the assay. The 8-color assay format was used for both experiments and the same analysis was performed on six outcome variables, which comprised the three cytokine responses (IFNγ, IL-2, or TNF) for each of CD4 or CD8 T cell subsets. All six outcome variables were treated independently. The expression of multiple cytokines can be evaluated several ways. For three cytokines, as with this study, a total of 7 different cytokine subsets can be measured based on expression of one or multiple cytokines. For this qualification, analyses only examined the marginal distributions for these three cytokines; for example, the marginal response for IFNγ includes all cells making IFNγ, alone or in combination with IL-2 and TNF.
The purpose of the pre-qualification specificity experiment was to determine the range of peptide-stimulated responses in non-immune animals and in combination with data from the precision experiment, to determine positivity criteria (the threshold a response must be above to be considered positive) for the qualification study. The aim was to minimize the false-positive rate while not reducing the ability to detect true positive responses. Samples from 30 healthy, SIV-naïve NHP were assayed per protocol for responses to each of the SIV peptide pools (Gag, Env, Pol). A mock stimulation well was also included for each sample. The 97th percentile of the background-subtracted responses was determined for each of the six cytokine responses and three peptide pools (Table 2). Since the desired false positive rate for the qualification study was determined to be ≤6% for each T cell subset (3 cytokines and 3 peptide pools combined), the thresholds for positivity for each of the 6 cytokine responses were set approximately at or above the 97th percentile to allow for ~2% error per cytokine; with 3 cytokines per T cell subset, this would minimize the chance that false positives would exceed 6%. The cut-off values that were selected for the qualification study are listed in Table 3.
The precision experiment was designed to assess the variability of the assay across aliquots, operators, and experimental days. The data from this study was used to establish acceptance criteria and appropriate design of the qualification protocol. Parallel frozen aliquots from three healthy, SIV-vaccinated NHP were assayed per protocol for responses to Gag and Env peptide pools. A mock stimulation well was also included for each aliquot on every run day. For each of the three samples, three different vials were assayed each day. The same three samples were assayed on three different days and two different operators conducted the experiments (thus, a total of 18 aliquots from a single cell preparation from each animal were used). This allowed us to compare the assay inter-vial (within day), inter-day (within operator), and inter-operator variability. For each of the comparisons, the coefficient of variation was determined from linear mixed effects models using the natural log of the response (Table 4). Based on these values and past experience, minimal assay performance target criteria were set for the qualification study (Table 5). These criteria were used for the intra-assay and inter-day comparisons but since the CV for operator was based only on 2 operators, and therefore was less reliable, the allowable cut-offs for operator were higher than for vial and day in the qualification study. In order for the assay to become qualified, these targets needed to be met in >90% of samples with peptide-stimulated responses above the pre-defined positivity cut-off.
Table 5.
Qualification precision criteria. Minimal assay performance target criteria allowed for inter-day and inter-vial comparisons during qualification.
| Overall Mean % Cytokine Positive |
CV |
|---|---|
| <cut-off | N/A |
| Cut-off – 0.2 | ≤70% |
| >0.2 | ≤35% |
3.4. Qualification study
The qualification study was composed of two experiments, similar to the pre-qualification study but larger in scale. In addition, success of the qualification study was predicated on meeting the pre-defined criteria for assay performance. The first experiment was designed to evaluate specificity of the ICS assay. Immune responses (six outcome variables) to SIV Env, Gag, Pol and a mock stimulation were measured in samples from ~100 healthy, SIV-negative NHP. Positivity was assessed according to threshold criteria defined in Table 3. In this study, samples for which the background-subtracted responses exceed this cut-off are false positives. The false positive rate for the ~100 samples was calculated along with a 95% confidence interval. The results showed that for each of the T cell subsets, there were fewer than 6% false positive responses (Fig. 4 and Table 6). The two samples that were positive for CD8 IFNγ were also positive for CD8 TNF and were not counted twice in the total percent positive for CD8 T cells.
The second experiment was designed to qualify precision. Parallel aliquots from two healthy, SIV-vaccinated and two SIV-infected NHP were assayed per protocol for responses to Gag and Env peptide pools. The same four samples were assayed in triplicate on five different days by two operators (thus, a total of 30 vials from the same cell preparation for each animal were used). Precision was assessed using the coefficient of variation. Standard mixed effects models were used to derive the precision estimates for inter-day, inter-vial, and inter-operator variability. The inter-day and inter-vial variance components were assessed for each operator separately and combined. The range of reliable responses was defined as the range over which the precision estimates are within the pre-specified criteria listed in Table 5. Due to limitations in number of operators, only day and vial were used for qualification pass/fail. As shown in Table 7, all values for day, vial, and operator were within the target criteria. These results demonstrate that the variability due to the assay being run on different days and by different operators was low, and practically negligible for different vials of the same cryopreserved blood draw. Residual error has the highest CV% values indicating that most of the variability in the study comes from components outside day, vial and operator, i.e. the biological nature of the assay. While there are no specified acceptable CVs for method qualification since each method is different, the target CVs for cell-based assays like ICS and ELISpot are usually≤~30%. CVs for bio-assays like ELISA, are usually≤ ~15%. Because this qualification study met all pre-defined criteria for specificity and precision, the NHP ICS assay was formally qualified as defined by ICH guidelines Q2 (R1).
3.4.1. Exploration of sensitivity of results to exact gate placement
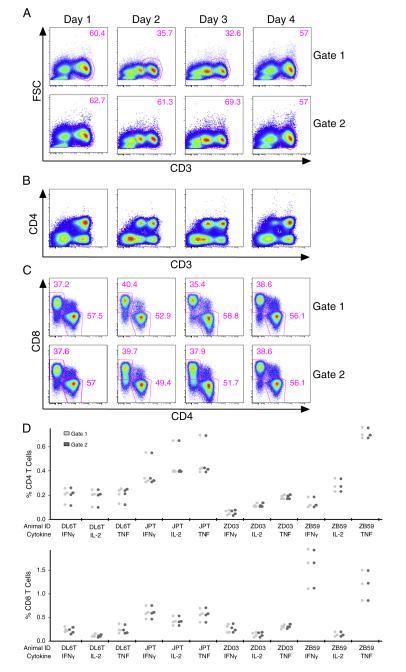
Because of the large number of samples included in our testing, we were able to investigate additional variables that may affect ICS quantitation. Analysis of flow cytometry data usually involves subjective gate placement to identify cell subsets. It is a good practice to try to use the same gates for all samples, where possible, to minimize analyst bias. This often means that gates may not be “ideally” placed for individual samples. Moreover, when samples are run on multiple days, using the same gates for all samples can be problematic since the staining and instrumentation may vary from day to day. Using the precision data set, we were able to compare two different analysis approaches for two different scenarios. In the first scenario, we observed that for the same sample run on multiple days, the CD3 population appeared as either a single or double population (Fig. 5A). On the days, where the CD3 population appeared as a doublet, the CD3 low cells fell outside the gate used for the qualification analysis. These cells were determined to be CD3 positive because including them increased the CD3+ frequency to approximately same level as when the population appeared as a singlet (Fig. 5A) and these CD3 low cells expressed both CD4 (Fig. 5B) and CD8 (data not shown). We examined the effect of adjusting the CD3 gate on these samples to include the CD3 low cells and found that it did not significantly change the frequency of CD4+, CD8+, or cytokine+ cells (Fig. 5C and D). Thus, the imperfect CD3 gate for the samples with the double population had little impact on the results. However, the number of events increased for these samples, which might improve sensitivity and precision.
Fig. 5.
Comparison of results with different CD3 gate placements. Plots are from a representative replicate from each day for one animal. A) The top row depicts the gate used for the qualification analysis. The bottom row shows how the gates were adjusted to include the CD3 low cells. B) The CD3 low cells express CD4. C) Frequencies of CD4 and CD8 T cells by both gating methods. D) Frequencies of cytokine+ CD4 and CD8 T cells by animal.
In the second scenario, the 336 qualification precision study samples that were stained and acquired on multiple days and instruments were analyzed by two different methods. With the first method, samples were grouped and analyzed by run (~36 samples per run) so that uniform gates were used for each staining and acquisition episode, but could vary across runs. With the second method, samples were grouped and analyzed by instrument (132 samples acquired on one cytometer, 36 on another, and 168 on a third) so that uniform gates were selected for samples over multiple runs (but were unique to each instrument). For both analysis methods, samples were compensated with controls acquired during the same run as the samples. Overall, the results obtained with the two different gating methods were very similar. Responses obtained by the second method using uniform gates (minimizing subjectivity) were only a fraction (~5%) less on average, as determined by a slope of 0.95, than those obtained by grouping analyses by individual run (Fig. 6A). Also, the standard deviations from the samples with uniform rather than run-specific gates were ~4% higher, indicating only slightly more variability with the uniform gates (Fig. 6B). Finally, the percent differences of the values obtained by the two gating methods were equally distributed around zero over the range of responses (Fig. 6C), suggesting that any differences observed between the two methods didn’t correlate with the magnitude of the response. Overall, these results demonstrate that there was very little difference (less than 5%) between results obtained when gating was done on single or multiple runs. The analysis for the qualification study reported in Section 3.4 was performed with the second method, the less stringent of the two methods, yet still met the qualification criteria. These results further demonstrate how robust the assay is.
3.4.2. Background responses by animal origin
Animals with high responses to the negative control may be more likely to be false positive for the SIV peptide pools and are often excluded from studies during prescreening. For the qualification study, the decision was made to exclude any samples that have a mock response over 0.2% for two or more of the six outcome variables. As a result, 136 samples were screened to ensure at least 100 samples would meet the study criterion for inclusion. When we were initially exploring testing different values for exclusion, we noticed that when the animals were grouped by origin, more animals from Primate Center D had background (no specific peptides included) responses for IFNγ, IL-2, or TNF of 0.15% or above, than those from other centers (only 110 animals were included in this analysis because animal origin could not be reliably traced for several animals). When we considered whether the animals would fail based on the exclusion criterion of mock responses over 0.15% for two or more of the six outcome variables, animals from centers B and D had considerably higher failure rates than the other centers, 50 and 30.23% respectively (Supplementary Table 1). Animals from center D had a statistically significant higher failure rate than the other centers using a Fisher’s exact test, while center B wasn’t significant since there were only 4 animals from this center. Of note, only Center D had animals that failed due to high CD4 background, the animals that failed from other centers all failed due to high CD8 background. This may be biologically relevant for SIV challenge studies if the presence of more activated CD4 T cells could make animals more susceptible to infection. While these results were not statistically significant with the exclusion criterion of 0.2% for two or more of the six outcome variables as with the qualification study, the trend is still apparent. These results highlight the need to prescreen animals prior to study initiation and include such data during randomization of animals into different study groups.
4. Discussion
Because SIV infection in the Indian rhesus macaque is the preferred model for understanding HIV infection in humans, it is important for the study of HIV vaccine research. Current methods such as ELISpot provide very limited, one-dimensional information. In order to more thoroughly characterized vaccine responses, it is necessary to take a more comprehensive, multi-dimensional approach. In this paper, we describe the qualification of such an assay for measuring SIV vaccine-induced Ag-specific T cell responses in the rhesus macaque. Through flow cytometry and intracellular cytokine staining, we are able to screen both qualitatively and quantitatively vaccine-induced T cell immune responses.
Before qualifying this assay, we investigated several methodological variables including overnight rest and co-stimulation with antibodies to CD28 and/or CD49d. Our results from overnight rest experiments indicate that there is a slight loss in CD4 naïve cells and a slight increase in the CD4 effector memory cells, but overall no changes that are expected to have an impact on this assay occurred. In addition, our data indicate that while overnight rest increases background IL-2 and TNF responses slightly, it dramatically increases net cytokine responses for IL-2 and TNF from CD4 T cells along with IFNγ, IL-2, and TNF from CD8 T cells. We concluded that overnight rest increases sensitivity to peptide stimulation without any detrimental impact on subset frequencies.
In addition, we assessed anti-CD28 and anti-CD49d co-stimulation on rhesus T cell populations during peptide stimulation. Previous data suggest that co-stimulation increases responses to whole protein stimulation in human T cells (Waldrop et al., 1998). We confirmed that co-stimulation increases T cell responses during peptide stimulation for CD8 T cells, albeit only slightly; however, increases were not statistically significant for CD4 responses. In addition, our data indicate an increase in background responses with co-stimulation that results in decreased sensitivity, particularly in detecting low-level peptide responses. For this reason, we omitted antibodies to CD28 and CD49d co-stimulation from the assay. Similar results were found for experiments with samples from cynomologous macaques (data not shown). It should be noted that other forms of stimulation, particularly using proteins as antigens, can show dramatically improved responses when co-stimulation is included (data not shown).
Since we acquired the same samples multiple times for the qualification study, we were able to evaluate how different gate placement strategies can affect the final results. We discovered that drawing best-fit gates over multiple runs combined rather than on each individual run had a minimal impact on the results, only decreasing our responses by 5% and increasing variability by 4%. In terms of a cost-benefit analysis, this 5% loss of response from the less stringent analysis is offset by the greater subjectivity, additional time, and need for a highly experienced analyst with the more precise analysis.
To date, a qualification study has yet to be done on multi-functional measurements from either human or NHP cells. Flow cytometric measurement of three different cytokines allows for the measurement of multifunctional responses using Boolean gating. For example, cells can be IFNγ+IL-2−TNF−, IFNγ+IL-2+TNF−, or IFNγ+IL-2+TNF+. In fact, all possible combinations from 3 cytokines generates 7 different populations. The power of multiparametric flow cytometry is that coexpression of multiple proteins can be delineated. However, including all combinations of cytokine expression from two T cell populations in a qualification study would produce 14 outcome measurements. Qualifiying all responses would require screening many hundreds of specimens in order to find samples with sufficient responses in each of the 14 measurements. In addition, since a large number of cells (~500 million) from one time point are required, animals would need to undergo a daylong apheresis procedure (or be euthanized), which is not practical on hundreds of animals. Therefore, such a resource demand makes qualifying multi-functional responses highly impractical for human immunology and nearly impossible for NHP experiments. For this reason, we chose to qualify only the 6 marginal cytokine responses.
In conclusion, we optimized and qualified an eight-color ICS assay for detecting vaccine induced antigen-specific T cell responses in the Indian rhesus macaque model. We show that our assay is both highly repeatable with low inter-day, inter-vial, and inter-operator variability and highly sensitive for detecting low level responses; therefore, it is ideal for both quantitative and qualitative measure of T cell responses in pre-clinical trials.
Supplementary Material
Acknowledgments
We thank Drs. Joanne Yu and Pratip Chattopadhyay for custom antibody conjugation, and members of the ImmunoTechnology Section, Flow Cytometry Core, and Immunology Core at the Vaccine Research Center for advice and assistance. We thank Gail Levine and Anna Sambor of the Foundation for the NIH for management support and logistical support. This work was supported by the Intramural Research Program of the NIAID, NIH, and by the Collaboration for AIDSVaccine Discovery (CAVD), grant #OPP1032325, from the Bill & Melinda Gates Foundation.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jim.2012.08.011.
References
- Betts MR, Nason MC, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay PK, Yu J, et al. Application of quantum dots to multicolor flow cytometry. Methods Mol. Biol. 2007;374:175–184. doi: 10.1385/1-59745-369-2:175. [DOI] [PubMed] [Google Scholar]
- Darrah PA, Patel DT, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 2007;13(7):843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- De Rosa SC, Lu FX, et al. Vaccination in humans generates broad T cell cytokine responses. J. Immunol. 2004;173(9):5372–5380. doi: 10.4049/jimmunol.173.9.5372. [DOI] [PubMed] [Google Scholar]
- Foulds KE, Donaldson M, et al. OMIP-005: Quality and phenotype of antigen-responsive rhesus macaque T cells. Cytometry A. 2012;81(5):360–361. doi: 10.1002/cyto.a.22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MB. SIV infected rhesus macaques: an AIDS model for immunoprevention and immunotherapy. Adv. Exp. Med. Biol. 1989;251:279–293. doi: 10.1007/978-1-4757-2046-4_26. [DOI] [PubMed] [Google Scholar]
- Horton H, Thomas EP, et al. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J. Immunol. Methods. 2007;323(1):39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH), Q2 (R1): Validation of Analytical Procedures: Text and Methodology; 2005. http://www.ich.org/products/guidelines/quality/article/quality-guidelines.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin NL, Mascola JR, et al. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312(5779):1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil JJ, Douek DC, et al. Vaccination preserves CD4 memory T cells during acute simian immunodeficiency virus challenge. J. Exp. Med. 2006;203(6):1533–1541. doi: 10.1084/jem.20060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure HM, Anderson DC, et al. Spectrum of disease in macaque monkeys chronically infected with SIV/SMM. Vet. Immunol. Immunopathol. 1989;21(1):13–24. doi: 10.1016/0165-2427(89)90126-8. [DOI] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Ritter N, Advant SJ, et al. What Is Test Method Qualification? Proceedings of the WCBP CMC Strategy Forum, 24 July 2003. BioProcess International. 2004:2–11. [Google Scholar]
- Russell ND, Hudgens MG, et al. Moving to human immunodeficiency virus type 1 vaccine efficacy trials: defining T cell responses as potential correlates of immunity. J. Infect. Dis. 2003;187(2):226–242. doi: 10.1086/367702. [DOI] [PubMed] [Google Scholar]
- Sun Y, Santra S, et al. Magnitude and quality of vaccine-elicited T-cell responses in the control of immunodeficiency virus replication in rhesus monkeys. J. Virol. 2008;82(17):8812–8819. doi: 10.1128/JVI.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrop SL, Davis KA, et al. Normal human CD4+ memory T cells display broad heterogeneity in their activation threshold for cytokine synthesis. J. Immunol. 1998;161(10):5284–5295. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.