Abstract
The past 5 years have witnessed extraordinary advances in the field of DNA sequencing technology. What once took years to accomplish with Sanger sequencing can now be accomplished in a matter of days with next-generation sequencing (NGS) technology. This has allowed researchers to sequence individual genomes and match combinations of mutations with specific diseases. As cancer is inherently a disease of the genome, it is not surprising to see NGS technology already being applied to cancer research with promises of greater understanding of carcinogenesis. While the task of deciphering the cancer genomic code remains ongoing, we are already beginning to see the application of genetic-based testing in the area of colorectal cancer. In this article we will provide an overview of current colorectal cancer genetic-based biomarkers, namely mutations and other genetic alterations in cancer genome DNA, discuss recent advances in NGS technology and speculate on future directions for the application of NGS technology to colorectal cancer diagnosis and treatment.
Keywords: adenocarcinoma, antineoplastic agents, biological analysis, biomarkers, cetuximab, colorectal neoplasms, EGFR inhibition, etiology genetics, genomics, humans, panitumumab, sequencing, targeted therapies, tumor markers
Colorectal cancer is the third leading cause of cancer mortality worldwide with an estimated 639,000 deaths per year [101]. In the USA, it is the third most common cause of cancer deaths for both men and women with an estimated 140,000 new cases diagnosed and 50,000 deaths in 2010 [102]. Prognosis for patients with colorectal cancer is directly related to the timing of diagnosis. When detected early, it is often cured with surgery alone. For more advanced or metastatic disease, chemotherapy is added to surgical treatment [1,2]. Given the global burden of disease from colorectal cancer, there is a great need for more accurate diagnostic tests and better-targeted treatments for patients.
Much of our understanding of the fundamental genetic and molecular biological processes driving colorectal cancer pathogenesis has come from studies of somatic mutations in sporadic, noninherited colorectal cancers and germline mutations in familial and inherited colorectal cancers. The current genetic-based paradigm of colorectal tumorigenesis as described by Vogelstein et al. is partially based on molecular analysis of sporadic primary colorectal cancer in which compounding genetic mutations lead to the transformation of a benign polyp into invasive colorectal cancer [3]. Studies of germline mutations found in inherited forms of colorectal cancer, such as familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer (HNPCC), also helped to shed light on additional genes involved in malignant transformation. One by one, tumor suppressor genes, DNA mismatch repair genes and proto-oncogenes have been identified and their roles elucidated in the progression from normal colonic epithelium to carcinoma.
With the advent of next-generation sequencing (NGS) technology and its associated massive output in DNA sequence, the newfound ability to resequence individual cancer genomes opens a new chapter in biomedical cancer research with the potential for future tailoring of medical care. In this article, we will first discuss the current methods for prognosticating and predicting response to chemotherapy in colorectal cancer with a focus on current genetic-based biomarkers. We will then examine the advances in NGS technology and discuss their application to future colorectal cancer research and clinical practice.
Clinical management of colorectal cancer & the need for improved diagnostics
Current clinical methods for prognostication in colorectal cancer are based on the American Joint Committee on Cancer (AJCC) tumor, node and metastases (TNM) staging classification. Stage is determined by a combination of TNM characteristics. T represents the tumor depth of invasion through the colonic wall; N represents metastases to regional lymph nodes; and M represents the presence of distant metastases. The TNM stage was designed such that increasing stages would theoretically correlate with worse prognoses: the larger the size of the tumor, the more likely it is to have regional metastases to the lymph nodes; the more regional lymph nodes involved with the disease, the more likely the cancer is to metastasize to distant organs. Large tumors would be expected to have a worse prognosis than small, locally contained tumors. In addition, cancers with distant metastases to the lung or liver would be considered far worse in terms of prognosis.
In reality, the relationship between TNM stage and prognosis is much more complex. For example, stage II cancers are a heterogeneous group containing various sized tumors without regional metastases. It is the presence of these regional metastases (i.e., lymph node involvement) that distinguish stage II from stage III colon cancer. However, the relative 5-year survival rates for stage IIB and IIC cancers are worse than those for some subsets of stage IIIA and IIIB cancers [4]. Surgical resection is the primary step for treatment in stages I—III with chemotherapy recommended for more aggressive tumors, including all stage III tumors and some stage II tumors. However, reliable identification of the aggressive stage II tumors has been elusive due to the aforementioned heterogeneity of this group. While there are certain clinicopathologic characteristics that coincide with more aggressive cancers (e.g., lymphovascular invasion or high grade on histology or intestinal perforation at clinical presentation), these characteristics are neither sensitive nor specific and are somewhat subjective. Thus, the dilemma for the clinician and the patient with stage II colorectal cancer is treatment choice. Should patients with stage II colorectal cancer undergo chemotherapy with the associated side effects and cost in exchange for a potential increase in survival?
The reality is that the majority (~75%) of stage II colon cancer is cured by surgery alone [5]. Moreover, only a small proportion of the remaining 25% actually derive any benefit from adjuvant chemotherapy. According to the Quick and Simple and Reliable (QUASAR) clinical trial in which stage II colorectal cancer patients were randomized to receive fluorouracil (5-FU)-based therapy or observation, patients who received 5-FU-based therapy demonstrated an 18% risk reduction for death and 22% risk reduction for recurrence (relative risk for death = 0.82, p = 0.008; relative risk for recurrence = 0.78, p = 0.001) [6]. Based on the study numbers, 36 patients would need to be treated with chemotherapy in order for one person to benefit from that therapy. In other words, 35 patients would not benefit from the therapy or worse yet, might suffer adverse effects.
Identifying the one patient in 36 who would benefit from chemotherapy provides a major motivation to find clinically relevant biomarkers for colorectal cancer. In this instance, clinically relevant biomarkers can be broken down into two distinct groups – predictive biomarkers and prognostic biomarkers. Predictive markers refer to those markers that provide information about the likelihood of a positive response to a treatment. The HER-2/neu overexpression is an example of such a predictive biomarker in that it predicts the response to trastuzumab (Herceptin®, Genentech, San Francisco, CA, USA) in breast cancer [7]. On the other hand, prognostic biomarkers help to identify patients at risk of aggressive disease while they are in an earlier stage. Specifically, prognostic markers refer to those markers that provide information about the natural course of the disease independent of treatment effects.
Like all clinical tests, the utility of genetic biomarkers depends on the sensitivity and specificity of the test. The higher the sensitivity or specificity of a given test, the lower the chance of a false-negative or false-positive test result, respectively. For example, the use of MRI for general population screening of breast cancer is not recommended despite its extremely high sensitivity. This is because MRI’s specificity for detecting breast cancer is low compared with mammography, measuring approximately 90%. This leads to a high false-positive rate and increases the number of biopsies needed to rule out breast cancer. By contrast, the HIV test (which is actually a series of tests including the antibody-detecting ELISA test and confirmatory western blot) has both high sensitivity and high specificity, virtually eliminating the false-positive and false-negative results.
Clinically relevant biomarkers come in various forms and can be measured in a variety of ways – from mutations in the actual coding DNA found by PCR amplification to dysfunctional proteins found by specific activity assays. There are advantages and disadvantages to all methods of detection. Ultimately, the adoption of any one particular test into clinical practice will depend on the sensitivity and specificity of the test, as well as cost and inconvenience to the patients.
Current colorectal cancer genetic biomarkers
Despite significant progress in colon cancer research, the translation of genetic discoveries into diagnostic tests for colon cancer patients has been difficult. As an example, one only has to refer to the recent 2006 guideline recommendations for gastrointestinal malignancies from the American Society of Clinical Oncology (ASCO) [8]. Among the hundreds of cancer gene mutations implicated in colorectal cancer development and phenotype, only one has had adequate validation to warrant clinical use in diagnosis, staging, surveillance or treatment [8]. In this section, we will review those genetic biomarkers – somatic and germline – currently under investigated for colorectal cancer.
Microsatellite instability
Within the human genome, the mismatch repair (MMR) genes comprise one component of a complex proofreading system dedicated to ensuring the replicative fidelity of the genome by preventing the accumulation of DNA mutations. Some MMR genes that have been identified thus far include MLH1, MSH2, MSH3, PMS1, PMS2 and MSH6. Loss of function or expression in these genes leads to a higher than normal frequency of frameshift mutations and base-pair substitutions in regions of short tandem repeated nucleotide sequences. These regions, also known as microsatellites, are found ubiquitously throughout the genome [9]. Mutations in the MMR genes were first identified by studying colon cancer in HNPCC families. Affected members of this cancer syndrome exhibited germline mutations in MMR genes. However, scientists soon found that MMR gene mutations are not exclusive to HNPCC families. Sporadic colon cancers also display somatic mutations in the MMR genes.
In 1997, a National Cancer Institute consortium was convened to resolve the heterogeneity in the scientific literature around the diagnosis of microsatellite instability (MSI). As a result, the Bethesda Criteria emerged and established guidelines for testing and diagnosing MSI. Per the criteria, a diagnosis of MSI involves the examination of a reference panel of five micro-satellite loci – D5S346, D2S123, D17S250, BAT25 and BAT26. Prior studies had shown these five microsatellite loci to be highly sensitive and specific for MMR gene perturbation. The detection of MSI in two or more of the above loci is considered high instability (MSI-H). Microsatellite-stable tumors show no evidence of instability in any of the five loci. Low-MSI (MSI-L) tumors exhibit changes in only one of the loci [10]. In current clinical practice, PCR amplification with commercially available kits followed by fluorescent capillary electrophoresis is used to assess the lengths of the mononucleotide or dinucleotide repeat elements in these five loci from tumor and normal tissue.
Approximately 15% of colorectal cancers show MSI. Tumors with MSI-H tend to be more proximal, poorly differentiated, mucinous and show marked lymphocytic infiltration. Recent clinical studies suggest that MSI is associated with improved prognosis [11–13] but decreased response to 5-FU-based chemotherapy [14–17]. Ribic and colleagues examined tumor samples from 570 patients with stage II or stage III colon cancer who had previously been enrolled in Phase III clinical trials of 5-FU-based adjuvant chemotherapy versus surgical resection alone. They found that the benefit of chemotherapy differed according to MSI status with low-MSI or microsatellite-stable tumors showing a statistically significant increased survival rate (hazard ratio = 0.72; p = 0.04) with chemotherapy compared with surgery alone. By contrast, patients with MSI-H tumors showed no benefit to 5-FU therapy compared with surgery alone and actually had worse survival rates when given 5-FU [14].
TP53
Cancer-specific, somatic point mutations in the tumor suppressor gene TP53 play a critical role in colorectal cancer development and phenotype. There are some data to suggest that the mutations affect prognosis; however, the variability in the assessment of p53 status and the disparities in reporting results make it difficult to validate its prognostic significance. In a recent systematic review of clinical studies investigating the effect of TP53 mutations on prognosis and therapy outcome in colorectal cancer, results of the reported studies were often found to be conflicting and heterogeneous [18]. This was a direct result of the assorted methodologies lacking adequate sensitivity to assess TP53 mutations, lack of concordance among studies and the limited examination of both alleles of the gene.
Chromosome 18q deletion
Somatic deletion mutations and large-scale genomic deletions in the chromosome arm 18q may also have prognostic significance in determining risk of developing metastatic cancer. For stage II and III colorectal cancer patients, a number of retrospective studies have shown strong correlations between genomic deletion events on chromosome arm 18q and reduced survival for patients with colorectal carcinoma [19–22]. The deleterious effects for such a large chromosomal deletion are understandable given that cancer genes DCC, SMAD2 and SMAD4 are located in the region of deletion [23]. However, isolated genomic deletions of SMAD2 or SMAD4 are not sufficient to account for the prognostic significance of 18q deletion [24]. This suggests that other candidate colorectal cancer genes may exist in the 18q region [24]. Complicating the possible clinical utility of 18q, a number of other studies have failed to correlate the 18q deletion with poor prognosis [25,26].
Thymidylate synthase & methylene tetrahydrofolate reductase
Theoretically, responsiveness to chemotherapies may be impacted by a patient’s germline variations that affect drug metabolism. For example, almost all chemotherapy regimens for colorectal cancer use 5-FU. One mechanism by which 5-FU exerts its anticancer effect is through the inhibition of thymidylate synthase (TS), which is encoded by the TYMS gene. TS catalyzes the reductive methylation of deoxyuridine monophosphate to deoxythymidine monophosphate using 5,10-methylenetetrahydrofolate as the methyl donor. Methylenetetrahydrofolate reductase (MTHFR; MTHFR gene) regulates the amount of 5,10-methylenetetrahydrofolate by irreversibly converting it to 5-methyl-hydrofolate. The above process provides the sole de novo source of thymidylate, which is necessary for DNA replication and repair [27]. Thus, one would expect that mutations that alter TS or MTHFR activity would affect chemotherapeutic outcomes.
Unfortunately, despite being grounded in theoretical logic, the effects of TYMS [28–31] and MTHFR [29,32,33] polymorphisms have yielded conflicting results in clinical studies. As in the case of TP53, these studies have suffered from a lack of consistency in experimental design. Heterogeneity of study populations, methodologies in detecting the polymorphisms and measurements of outcomes has made a direct comparison across studies difficult.
EGF receptor signaling pathway & KRAS
In contrast to the abovementioned mutations, KRAS mutations are one of the strongest negative predictive markers for EGF receptor (EGFR) inhibitor chemotherapy in the setting of metastatic colorectal cancer. Mutations leading to EGFR activation or overexpression have been associated with a number of cancers. This led to the development of EGFR inhibitors as targeted anticancer therapy. Cetuximab (Erbitux®, ImClone Systems Inc., New York, NY, USA) and panitumumab (Vectibix®, Amgen, Thousand Oaks, CA, USA) are two monoclonal antibodies targeting EGFR. They are approved for use in combination with 5-FU, leucovorin and oxaliplatin (FOLFOX) or 5-FU, leucovorin and irinotecan (FOLFIRI) for stage IV metastatic colorectal cancer [2]. Unfortunately, efficacy of these regimens remains modest with 8–25% objective response rates [34].
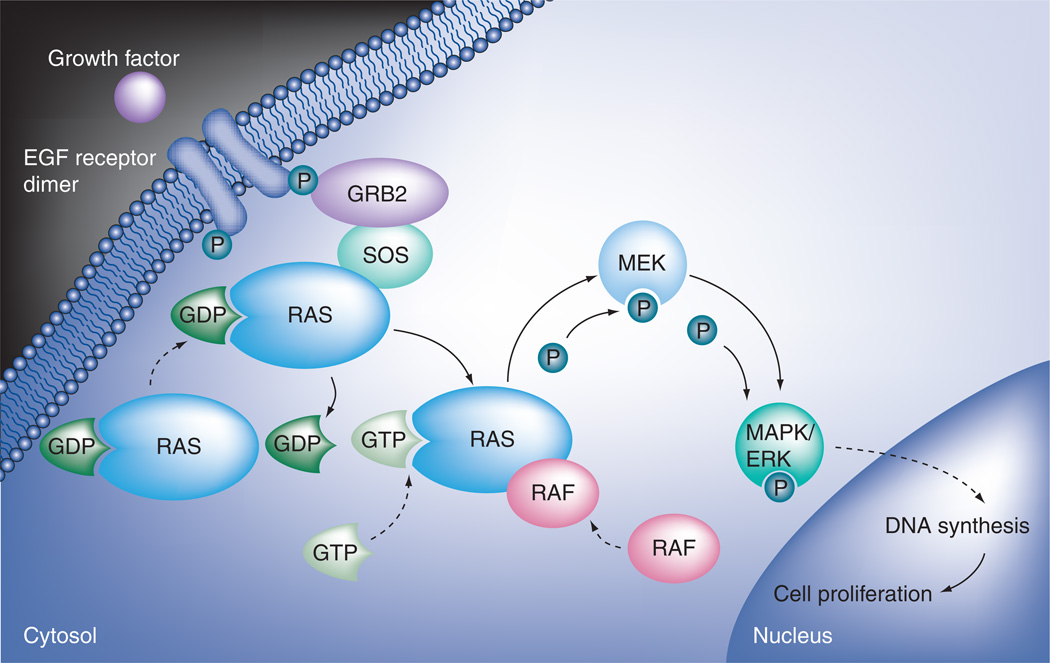
In order to understand the mechanism of the KRAS mutation in the resistance to EGFR inhibitors, it is necessary to understand the EGFR signaling pathway. EGFR is a member of the ErbB family of receptor tyrosine kinases. It is a survival and proliferation factor for a variety of tumor types. EGFR signals through the RAS/RAF/MEK/ERK pathway. Upon activation by extracellular ligands, EGFR dimerizes and autophosphorylates the tyrosine residues in the C-terminal domain. This leads to RAS GTPase complexing with RAF to phosphorylate and thereby activate two MAPK kinases which in turn phosphorylate ERK. Phosphorylation of multiple nuclear transcription factors by activated ERK ultimately leads to DNA synthesis and cell proliferation (Figure 1). Tumors with mutations that constitutively activate RAS/RAF/MEK/ERK-pathway downstream EGFR can bypass the block by EGFR inhibitors, leading to drug resistance.
Figure 1. EGF receptor signaling pathway.
Upon activation by extracellular ligands, EGF receptor dimerizes and autophosphorylates. This leads to activation of RAS, which then complexes with RAF. The RAS/RAF complex then phosphorylates and thereby activates MEK. MEK in turn phosphorylates MAPK/ERK. Phosphorylation of multiple nuclear transcription factors by activated MAPK/ERK ultimately leads to DNA synthesis and cell proliferation.
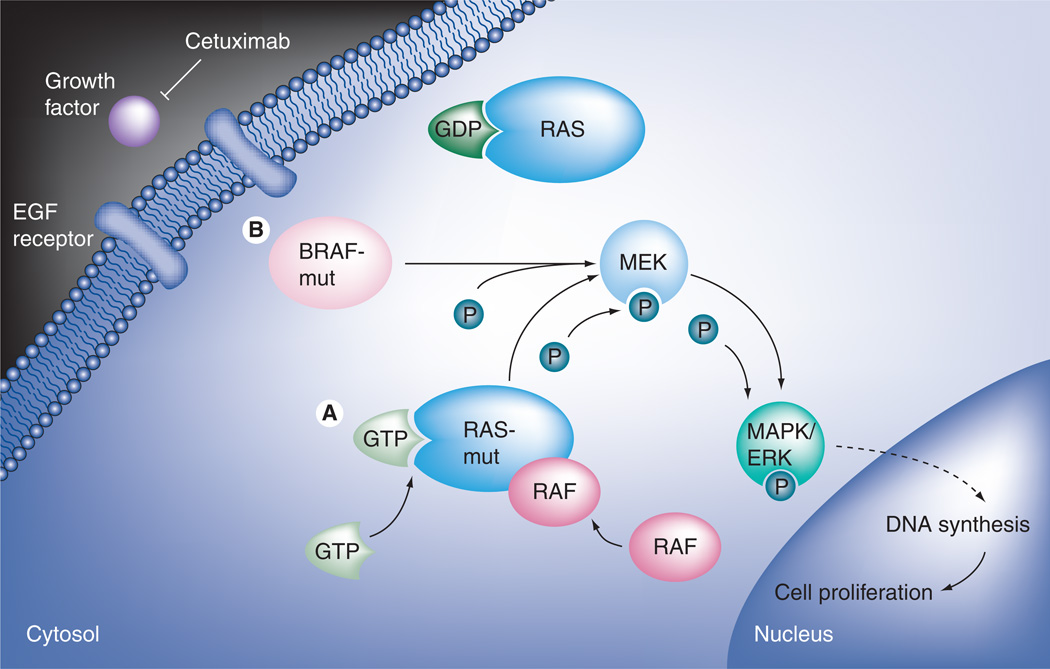
KRAS is a proto-oncogene encoding a GTPase whose mutations can result in the constitutive activation of the RAS/RAF pathway (Figure 2A). Such activating mutations are found in 35–42% of colorectal cancers. Seven different missense base substitutions within codons 12 and 13 of exon 2 constitute more than 97% of the observed genetic events within the KRAS gene in colorectal cancer [35]. The KRAS mutation testing traditionally relies on allele-specific PCR amplification using commercially available kits that detect the seven most common mutations in codons 12 and 13.
Figure 2. Constitutive activation of RAS/RAF pathway.
(A) KRAS is a proto-oncogene encoding a GTPase whose mutations can result in the constitutive activation of the RAS/RAF pathway. (B) The BRAF gene encodes a serine/threonine protein kinase that operates downstream of KRAS in the RAS/RAF pathway. Mutations in BRAF lead to constitutive activation of the BRAF kinase activity regardless of RAS activity.
Early studies suggested that patients harboring a KRAS mutation had worse prognosis with increased risk of recurrence (p = 0.007) and death (p = 0.004) compared with patients with wild-type KRAS [36]. But this was later shown to be applicable only to stage III colorectal cancers [37]. More recent clinical trials have consistently shown that patients with KRAS mutant cancers receive little to no benefit from EGFR inhibitors [38–40]. In fact, Bokemeyer and colleagues found that patients with KRAS mutations actually fared worse if treated with FOLFOX plus cetuximab (hazard ratio = 1.83; p = 0.02) [41]. Subsequent clinical trials confirmed the negative predictive value of the KRAS mutation for responsiveness to EGFR inhibitor therapy with cetuximab or panitumumab alone or in combination with FOLFOX or FOLFIRI, administered to patients with metastatic colorectal cancer as initial or as salvage therapy [42–49]. Taken together, these studies point to a KRAS mutation as a strong predictive biomarker for EGFR inhibitor therapy in both chemo-resistant and chemo-naive cancers.
The number of studies is so overwhelming that KRAS mutation tests are the only ones that have been integrated into clinical practice in patients with metastatic colorectal cancer. The European Medicines Agency and the US FDA now recommend the determination of KRAS status before initiating treatment with an EGFR inhibitor and restrict these treatments to wild-type KRAS patients [34].
BRAF
Cetuximab or panitumumab are ineffective in a large proportion (~70–90%) of metastatic colorectal cancer patients. Patients with KRAS mutations, however, account for only 30–40% of the nonresponders. It is likely that other downstream effectors of the RAS/RAF pathway, such as BRAF, may be involved in resistance to EGFR inhibitors.
The BRAF gene encodes a serine/threonine protein kinase that also belongs to the RAS/RAF kinase pathway. BRAF operates downstream of KRAS and has been shown to be involved in colorectal carcinogenesis. A DNA missense mutation leading to a valine to glutamic acid amino acid change (V600E; formerly known as V599E) is the most frequent BRAF mutation observed [35]. This mutation leads to constitutive activation of the BRAF kinase activity (Figure 2B). Interestingly, KRAS and BRAF mutations have been found to be mutually exclusive [35,50].
Several studies of wild-type KRAS tumors have examined BRAF mutation status and suggest its contribution to EGFR inhibitor resistance [35,39,51]. Di Nicolantonio and colleagues retrospectively analyzed tumor responses, time to progression, overall survival and the mutational status of KRAS and BRAF in 113 tumors from cetuximab- or panitumumab-treated metastatic colorectal cancer patients. The BRAF V600E mutation was detected in 11 of 79 patients who had wild-type KRAS. None of the BRAF-mutated patients responded to the anti-EGFR treatment, whereas none of the responders carried BRAF mutations (p = 0.029). Moreover, patients with the BRAF mutation had significantly shorter progression-free survival (p = 0.011) and overall survival (p < 0.0001) than patients with wild-type BRAF [52].
Other downstream effectors of the EGFR pathway
Given the clinical significance of KRAS and BRAF mutations in predicting the response to anti-EGFR chemotherapy, additional downstream effectors of the EGFR pathway are under investigation and have yielded promising results with implications for multigenic testing.
De Roock and colleagues examined 773 tumors from chemotherapy-refractory metastatic colorectal cancer patients treated with cetuximab and found a 24.4% objective response rate in the study population. This rate nearly doubled to 41.2% when the population was narrowed to include only KRAS, BRAF, NRAS and PIK3CA wild-type tumors. Practically speaking, the authors showed that dramatic improvements in the response rate to anti-EGFR therapy could be gained by tailoring therapy to patients with genetic susceptibility to the medication [39].
In a second retrospective analysis of tumor response, progression-free survival and overall survival, 132 colorectal tumors from patients with cetuximab- or panitumumab-treated metastatic disease were analyzed for mutational status of KRAS, BRAF and PIK3CA and expression of PTEN. The authors found that up to 70% of patients with metastatic disease who are unlikely to respond to anti-EGFR therapy can be identified by examining mutations in KRAS, BRAF and PIK3CA and expression of PTEN. The probability of anti-EGFR response was 51% among patients with no genetic alterations in these genes. This probability of response dropped to 4% among patients with one alteration and 0% for patients with more than two alterations (p < 0.0001). Similarly, progression-free survival and overall survival were increasingly worse for patients with tumors harboring 0, 1 or 2 or more alterations (p < 0.001) [53].
These two studies suggest that chemotherapy with EGFR inhibitors can be tailored to those patients who have an intact EGFR pathway. At this time, given that the cost of PCR testing of these three genes plus immunohistochemistry of a fourth protein product (PTEN) would run into the thousands of US dollars, NGS would be competitive in terms of cost. As the monetary and time cost of sequencing continue to decrease, NGS is likely to become the clinical test of choice given the vast amount of data that can be generated regarding all the genes in the EGFR pathway, as well as those involved with all other pathways implicated in cancer progression and chemotherapeutic response.
Advances in NGS technology
Significant advances have been made in the development of genetic-based tests for colorectal cancer over the past 5 years. With the use of NGS technology, even greater strides are to be made in the next few years as NGS technology brings greater understanding of the mechanism of the disease, leading to rational drug design. In the next section, we will give a brief review the recent advances in sequencing technology.
Brief history of sequencing technology
The mid-1970s gave birth to a new era in biochemical research with the development of chain-terminator sequencing or Sanger sequencing. The following two decades saw the expansion of the understanding of genetics and DNA, culminating in the concerted international scientific effort to sequence the human haploid genome through the Human Genome Project [103]. Beginning in 1990 under the direction of James Watson at the NIH with contributions from universities and research centers around the world, the Human Genome Project made use of a hierarchical shotgun approach to sequence approximately 3 billion bp or 20,000–25,000 genes. Sanger-based shotgun genomic sequencing involved the use of bacterial plasmid vectors for subcloning fragments of human genomic DNA. Following replication, these large segments were randomly fragmented into smaller 100–1000-bp segments for sequencing. With the assistance of complex computational alignment programs, the sequences of the 150,000-bp segments were generated based on the overlapping reads from the shorter fragments. The first draft of the human genome was published in 2000. A final version was released 3 years later [54].
Even with technological advances to the Sanger sequencing method through automation, the process of sequencing was time consuming and costly. The growing interest in the sequencing of personal genomes fueled the development of new and improved technologies. Over the past 5 years, NGS technologies have matured and thus lowered cost and dramatically increased throughput by eliminating the time-consuming and labor-intensive step to generate single clones via bacterial cloning or PCR, and using parallel processing to simultaneously sequence a large number of DNA templates. Thus, instead of generating hundreds of longer reads (800–1000 bp), NGS technology produces millions of shorter reads (40–400 bp) ranging on the order of giga-bases (Gb) per run [55]. Consequently, the major work of sequencing has shifted from the bench-top to the desktop. Analysis of the NGS data presents new challenges mainly due to the short read lengths and requires significant investment in informatics (including hardware, software and technical support) to handle the massive amount of generated data. But, as a result of this trade-off, what once took years to sequence using Sanger sequencing can now be accomplished in a matter of weeks.
NGS overview
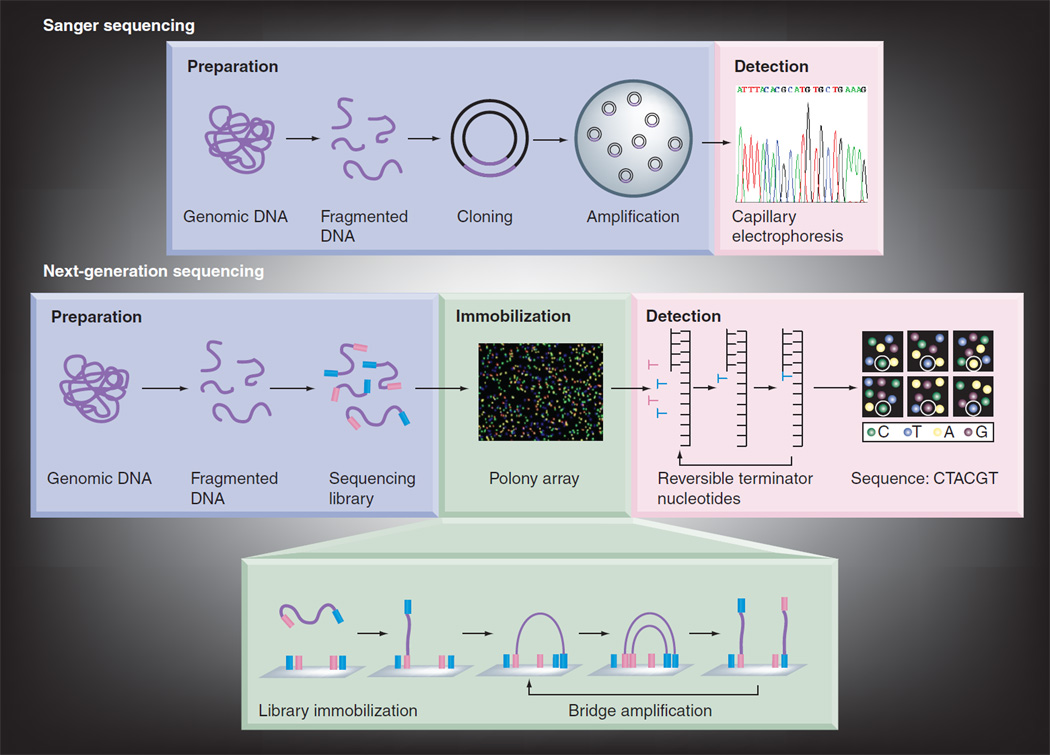
In-depth reviews of the biochemistry [56,57], commercially available sequencing platforms [56,57], computational challenges [58,59] and experimental approach caveats [60] for NGS technology have been previously published. Briefly, NGS technology can be roughly divided into three phases: template preparation, sequencing and detection and sequence analysis. For template preparation, the production of a representative, nonbiased pool of fragmented DNA is necessary. To enable the parallel processing of large numbers of sequencing reactions, these fragments are immobilized or restricted to spatially separated sites on a solid support or in a nanosized chamber. The spatially separated templates are then either clonally amplified or remain as single molecules. Commercially available platforms employ a variety of principles for the actual sequencing reaction. These include cyclic reversible termination, single-nucleotide addition, real-time sequencing and sequencing by ligation. Once NGS reads have been generated they are aligned to either a known reference sequence or assembled de novo [56–58]. Because of the short read-length in general, the accuracy of the alignment or assembly processes depends on the depth of coverage or oversampling. To detect nucleotide alterations with high sensitivity, at least 30-fold coverage is needed [60]. Figure 3 gives a comparison of Sanger sequencing to NGS.
Figure 3. Comparison of Sanger sequencing with next-generation sequencing.
In a typical Sanger sequencing, genomic DNA is fragmented and cloned as a plasmid vector in transfected bacteria. A single bacterial colony is picked for each sequencing reaction. Each sequencing reaction makes use of ddNTP-terminated, fluorescently labeled products which are subjected to high-resolution electrophoresis separation. As labeled fragments of discrete sizes pass a detector, a four-channel emission spectrum is used to generated a sequencing trace. In next-generation sequencing, genomic DNA is fragmented. Common adaptors are ligated to the fragments. The resulting sequencing library is then immobilized onto an array of millions of spatially separated PCR colonies, or Polonies. Millions of sequencing reactions occur in parallel on the Polony array. As each nucleotide is added, image-based detection of fluorescent labels can be used to acquire sequencing data. Successive iterations of enzymatic interrogation and imaging are used to build up a continuous sequencing read.
There are two main types of experimental approaches for genomic DNA sequencing – whole-genome and targeted sequencing. Whole-genome sequencing, as its name implies, provides a comprehensive characterization of the entire genome. This allows for a greater ability to identify genomic alterations in both coding and noncoding regions. Shotgun sequencing is a lower-cost approach to whole-genome sequencing which relies on the sequencing of randomly derived fragments. Researchers often opt for shotgun sequencing due to the simpler sequencing library preparation, lower cost and decreased amount of required input DNA as compared with the traditional whole-genome sequencing using ordered, overlapping clones or fragments. While the shotgun approach is sufficient to identify somatic rearrangements and copy number alterations, it is often not accurate enough to allow for identification of nucleotide substitutions due to the limited depth of coverage which can be achieved reasonably in terms of DNA input and cost. This is particularly relevant in diseases such as cancer which exhibit a high degree of heterogeneity [60].
An alternative strategy is targeted sequencing. This approach has the advantage of providing increased depth of coverage while generating information quickly and cheaply. In this approach, any subregion of the genome can be targeted or enriched by one of several methods. Hybrid selection-based technologies use oligonucleotides complementary to a region of interest. The oligonucleotides are tagged to allow for isolation, and the captured DNA is then amplified and sequenced. A major limitation to this technique is that it requires prior knowledge of a specific region of interest and will miss alterations in other parts of the genome. In addition, uniformity of the capture method can have a significant impact on the sequencing results, both in terms of coverage and the quality of the genotype calls.
Approximately 99.9% of the human genome is identical between two different individuals. Despite this high level of concordance, there are still millions of variations that lead to the phenotypic differences we see everyday among and within different groups of individuals. The most common variations are SNPs. In using NGS to decode the human genome, scientists are attempting to understand which SNPs and other structural variants produce observable phenotypes. These phenotypes can range from physical characteristics as simple as eye or hair color to diseases as complex as obesity or cancer.
Technical limitations
To date, Sanger sequencing has been regarded as the ‘gold standard’ metric by which other methods are compared because of its high accuracy and the fact that it was the method used to sequence the first human genome. Compared with Sanger sequencing, NGS has several disadvantages, such as a greater reliance on bioinformatics and computational science to generate alignment protocols. There is also a real concern regarding the amount of disk space required to store and analyze the massive amounts of data generated from every sequencing run. In addition, data safety poses a significant obstacle, as genetic data are considered protected health information.
From a technical standpoint, NGS is limited in its ability for de novo sequencing of large repeated segments of the genome. For example, Huntington’s disease is a neurodegenerative genetic disorder characterized by trinucleotide repeat expansions. Patients with the disease generally have more than 40 repeated glutamine residues (or 120 bases of ‘CAG’ repeats) in their Huntingtin protein. Unaffected people typically have fewer than 36 glutamine residues (or 108 bases of ‘CAG’ repeats). NGS would have difficulty resolving the actual number of repeated glutamine residues coded by the Huntingtin gene because the read lengths are shorter than the repeated stretch of coding DNA.
Advantages
Next-generation sequencing has considerable advantages over Sanger sequencing in terms of improved accuracy of nonrepeated elements, lower cost and ability to detect low-frequency alleles. Using the Illumina platform, Bentley and colleagues sequenced 162,752 bp of a MHC complex that had been previously sequenced with traditional Sanger sequencing. Approximately 90% of the raw 35-bp reads matched perfectly to the reference. Using consensus data (based on 30-fold average depth coverage), they were able to raise this accuracy to 99.96% coverage of the reference genome [61]. Ahn and colleagues sequenced the first Korean genome and attained 99.9% coverage of the National Center for Biotechnology Information (NCBI) reference genome. Moreover, the authors were able to identify 3.4 million SNPs of which 0.4 million (12.2%) were novel [62]. Similarly, when the genome of James D Watson was sequenced using the Roche/454 NGS platform, the results showed 99.4% agreement with the reference allele when the allele in question was homozygous for the variant. An additional 0.61 million novel SNPs were identified [63]. There is ongoing debate as to whether these novel SNPs are true polymorphisms or errors in the NGS technology. Given that no direct head-to-head comparison has been performed to compare the various sequencing platforms, we will only know if these newly discovered SNPs are true polymorphisms as personal genomes continue to be sequenced and resequenced.
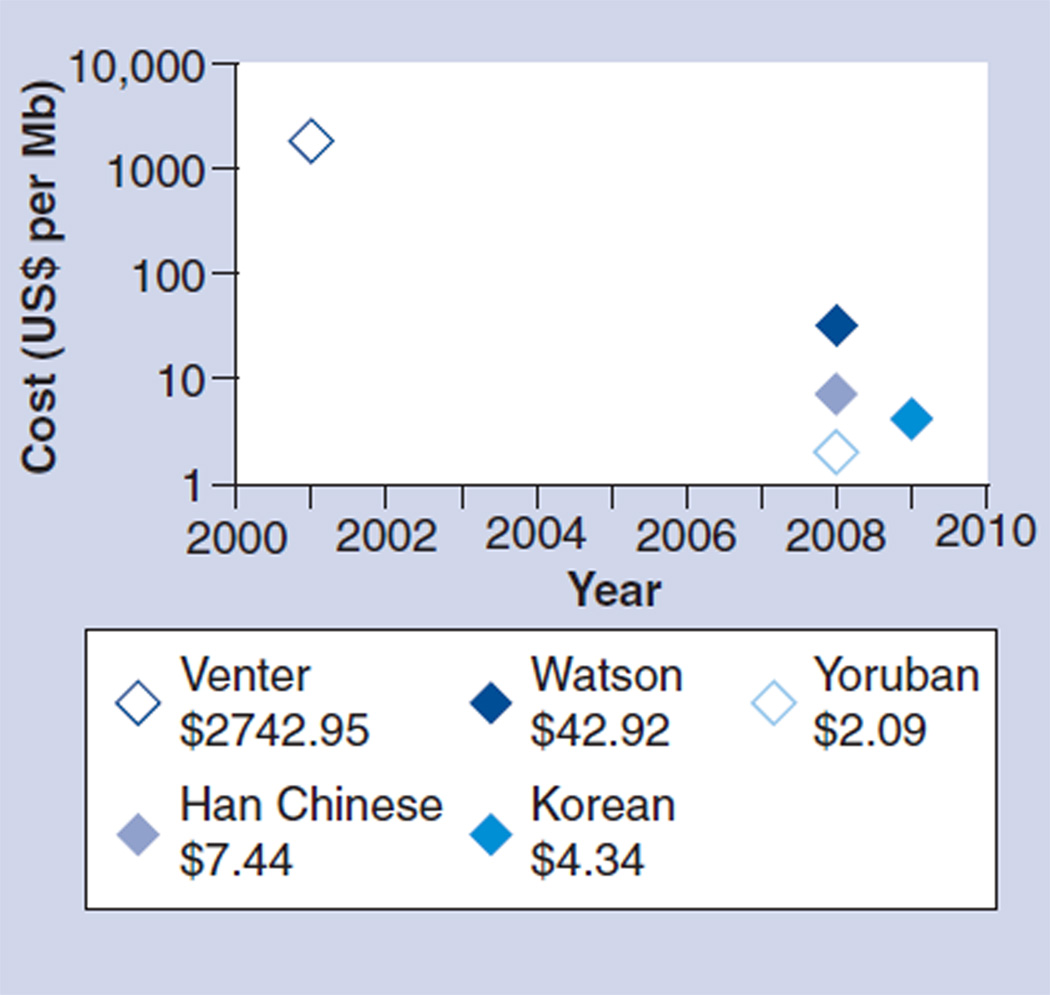
One thing is certain, the financial and time cost of sequencing using NGS is dramatically less compared with Sanger sequencing. This is mainly due to the Sanger sequencing requirement for clonal templates that are individually amplified by PCR or in Escherichia coli. As a result, the reagent cost is relatively high. As can be seen in Figure 4, the cost of sequencing whole genomes has decreased dramatically over the past 10 years [57]. Costs are predicted to continue dropping with the promise of a US$1000 genome in the next few years. Indeed, after the initial investment in the sequencing platform, NGS whole-genome tests are expected to be far less expensive than the current PCR-based tests. For example, the current PCR test for RAS mutations costs several thousand dollars. Using NGS technology the RAS genes can be sequenced for approximately one-third of the cost. Even more relevant for future application, the remaining genes in the same pathway or multiple samples can be simultaneously sequenced in a single sequencing run without incurring additional operating costs.
Figure 4. Cost of whole-genome sequencing over time.
Reagent costs only for genomes sequenced after 2008.
Data taken from [57].
This has ramifications from the health systems perspective. Genetic testing is publicly funded in most European countries and in Canada and Australia. In the USA, testing is offered mainly through private facilities. For people with publicly funded health insurance, coverage for genetic testing is decided on a case-by-case basis [64]. There is no doubt that genetic testing is expensive; however, the cost of a one-time test and its associated counseling may be cheaper in the long-term than the recurrent annual or biannual cost of screening for patients with a germline cancer predisposition. Using HNPCC as an example, a one-time blood test for a defect in MMR genes is far more cost effective and less invasive than annual or biannual colonoscopy screening for an entire family. In the case of therapeutic biomarker tests, the cost of the genetic tests themselves should be more than offset by the savings gained by avoiding costly and ineffective treatment.
In one analysis of 96 unaffected family members who met the Amsterdam criteria for HNPCC, 48 patients were offered predictive genetic testing for HNPCC with 39 (81%) undergoing the test. Of these, seven (18%) were positive for the mutation. As a result of the test, the total number of colonoscopies recommended for the tested group and all their offspring was half the number they had undertaken in the previous 5 years even though many had not been compliant with recommendations prior to the test [65].
In addition to highlighting the potential cost savings, the above study exposed a real clinical concern in that many family members did not follow the recommended screening guidelines due to the invasiveness, frequency or time and cost component of screening tests (in this case, colonoscopy). For germline genetic testing, a blood sample is usually sufficient for genomic DNA isolation.
Lastly, NGS has the added advantage over Sanger sequencing of having higher sensitivity towards low-frequency mutations. This is because Sanger sequencing relies on relative peak heights at a given position when determining a nucleotide base call. A minor allele will likely have a low signal-to-noise ratio that is indistinguishable from the background. This will be particularly problematic for cancer genomic tests since cancer cells are heterogeneous in nature and many clinically relevant changes or mutations may only be present in a fraction of the cells. NGS, on the other hand, makes use of high base coverage which provides a higher sensitivity toward minor alleles or mutations. For example, using the Roche/454 sequencing platform, Thomas and colleagues were able to identify novel EGFR kinase domain mutations in 22 patients with lung cancer. All mutations that had previously been detected using Sanger sequencing could reliably be identified with the Roche/454 sequencing platform [66]. In reviewing their previous Sanger output, the authors found low-level peaks that had been missed by the Sanger analysis software. Combined with the very low sequencing cost per base, the increased sensitivity for low-frequency mutations makes NGS a very powerful tool for cancer genomic studies and diagnosis.
NGS & colorectal cancer
The power of NGS is its ability to detect multiple types of genomic alterations, including nucleotide substitutions, small insertions and deletions, copy number variations and chromosomal rearrangements. For cancer, NGS technology has the potential to elucidate the mechanism of pathogenesis and improve diagnostic and therapeutic testing. At the genomic level, cancer manifests as compounding mutations and genomic aberrations. Already, studies have shown the ability of NGS to identify new somatic mutations associated with acute myeloid leukemia [67,68], melanoma [69], mesothelioma [70,71], small-cell lung cancer [71] and prostate cancer [72]. While these studies by individual researchers have increased our understanding of cancer development in a wide array of cancer types, the future of understanding cancer genomes is likely to come from concerted efforts through projects such as The Cancer Genome Atlas (TCGA). TCGA aims to systematically explore the entire spectrum of genomic changes involved in over 20 types of cancers using NGS technology through the collaboration of the National Cancer Institute and the National Human Genome Research Institute [104].
Two important developments in the field of colorectal cancer research so far include the description of the colorectal genetic landscape by Wood and colleagues and the establishment of the aforementioned Cancer Genome Atlas. Wood and colleagues performed one of the first large-scale unbiased colorectal cancer genomic surveys. They examined 18,191 genes in 11 fully progressed metastatic colorectal cancers. Using pairs of tumor tissue from liver metastases and normal tissues from peripheral blood, they were able to identify those genes mutated in tumor progression [73]. Genes of interest were identified using the Reference Sequence (RefSeq) database. RefSeq is a comprehensive, nonredundant collection of annotated gene sequences consolidated from major gene databases. Statistical analyses of the tumor/normal pairs suggested that most of the mutations in an individual tumor were nonsignificant. The authors postulated that fewer than 15 were likely to be driving the initiation, progression or maintenance of the tumor. A total of 280 candidate cancer genes were identified accounting for 38 colorectal cancer pathways that include PIK3CA, IRS2, IRS4, PTEN, as well as cell adhesion, cytoskeleton and the extracellular matrix pathways.
While Wood and colleagues have revealed some general trends in the genomic landscape of colorectal cancer from a limited number of cases, other genetic events are still yet to be uncovered. Concerted efforts, such as TCGA, will catalogue a large number of these mutations and genomic aberrations. For colorectal cancer, TCGA aims to characterize up to 400 samples of both primary tumor and matched normal tissues to determine mutations and other genomic aberrations using targeted exome sequencing and other genomic analysis approaches. Ultimately, TCGA will be a source catalogue for SNPs and structural variants and their associated phenotypes of cancer genomes.
Future challenges: protecting patients from genetic discrimination based on heritable genetic variation & establishing clinical utility
Even though the Human Genome Project was completed in 2003, the reality is that genetic testing has not become widespread in the clinical arena, particularly in the realm of germline inherited genetic variation. One should note that cancer-specific mutations are not heritable, but represent unique genetic errors specific to the tumor alone.
For heritable genetic testing, there are a number of reasons for the less than enthusiastic uptake into the clinical arena. Concerns for discrimination and questions of clinical utility are the two most common. Discrimination based on genetic testing remains a particular concern, especially when such testing revolves around cancer predisposition. At issue is the right of patients to access genetic tests that will aid in the screening, diagnosis and treatment of life-threatening cancer while safeguarding against discrimination in the workplace and the insurance market. In 1996 ASCO published a statement on genetic testing for cancer susceptibility with the primary goal of expanding access to and promoting scientific advances in medical care to all patients and families affected by hereditary cancer syndromes. An updated statement was published in 2003 with specific recommendations for genetic testing to be offered to patients who present with a personal or family history suggestive of a genetic cancer susceptibility condition. The caveat to this recommendation was that the genetic tests should be adequately interpretable and the results should be clinically meaningful (i.e., aid in diagnosis or influence the medical or surgical management of the disease). In addition, ASCO recommended that genetic testing be carried out in the setting of pre- and post-test counseling [74].
While there are few documented cases of genetic-based insurance discrimination, societal fear has remained high. Medical societies, such as ASCO, have worked with political leaders to create legislation to protect patients’ rights. In the USA, 47 states and the District of Columbia have already passed laws restricting the use of genetic information by health insurers [75]. In addition, the Health Insurance Portability and Accountability Act (HIPAA) defined genetic information as a component of the ‘health status’. As a result, genetic information cannot be used by employers or insurers to exclude employees from group coverage or to charge them higher rates [76]. In 2008, former President George W Bush signed into law the Genetic Information Nondiscrimination Act, which outlawed genetic discrimination in health insurance and employment. Genetic Information Nondiscrimination Act defines genetic information as information about an individual’s genetic tests, the genetic tests of family members and the manifestation of a disease or disorder in a family. A genetic test is considered any analysis of human DNA, RNA, chromosome, protein or metabolite that detects genotypes, mutations or chromosomal changes. The law prohibits group health plans from adjusting premiums or contributions on the basis of genetic information [75]. While these laws are not comprehensive, they are the first step in breaking down societal obstacles to genetic testing.
Concern over clinical utility also poses an obstacle to widespread use of genome-based testing. This is partially due to the recent advances in the technology itself. NGS has only recently become accessible to the individual researcher. As a result, genomic sequencing has not been able to establish itself as a meaningful clinical test. We are just now attempting to match cancer phenotype to cancer genotype through the analysis of clinical outcomes and genomic SNPs. Not unexpectedly, the handful of published studies on genome-wide associations is less than conclusive [77–79]. A recent review of colon cancer genetic association studies showed that of the ten identified susceptibility loci, no individual SNPs or panels of SNPs enhanced the predictive or prognostic models that are currently based on clinical factors such as age, and personal and family history of cancer [77].
In the near future, it is likely that clinically useful tests will come from multiple modalities, as genotypic variations do not always correlate with phenotypic variations. There are several layers of DNA, RNA and protein standing between genotypic mutations and phenotypic expression. One can imagine that changes in gene copy number may not necessarily translate into similar changes in mRNA transcripts. In addition, mRNA abundance has been shown over and over to poorly correlate with protein abundance. A comprehensive understanding of cancer phenotypes and the potential discovery of the cancer stem cell are to come from an integration of genomics research with the emerging fields of transcriptomics, proteomics and interactomics using a systems biology approach [80–83].
Conclusion & future perspective
In summary, the past 5 years have witnessed significant advances in the understanding of colorectal cancer genetics and pathogenesis. With the identification of genetic-based biomarkers, clinicians will be able to better characterize individual colorectal cancers and tailor chemotherapy treatment. Already, the genetic testing of colorectal cancer tumors for KRAS mutation is being applied clinically to determine which patients should undergo treatment with monoclonal antibodies against EFGR. Additionally, tests for MSI, while not required, are in the oncologists’ armamentarium and influence recommendation for starting chemotherapy.
In parallel with the advances in understanding colorectal cancer, our capacity to sequence DNA has increased exponentially. We are only now beginning to see NGS technology being applied to cancer research. TCGA will hopefully assist with the application of genomic knowledge. By examining multiple tumors and identifying common genetic mutations or molecular pathway perturbations, scientists will be able to better understand cancer development. This will allow for the development of more accurate screening and diagnostic tests and the identification of new drug targets for treatment. At the same time, as NGS technology becomes more accessible to private institutions, patients’ cancer genomes will soon be routinely resequenced allowing for the characterization of individual cancers. This will allow for the precise characterization of a single patient’s tumor, providing information on individual prognosis and guiding treatment decisions.
In the next decade, new sequencing technologies promise to decode colorectal cancer genomes rapidly and cheaply, yielding massive amounts of data. Sifting through these data to extract clinically meaningful tests will require the multidisciplinary efforts of biologists, computer scientists and bioinformatics specialists. Yet, despite its limitations, NGS technology will be a valuable tool to compliment Sanger sequencing in exploring the cancer genome. Its true utility will likely come in the form of improved clinical diagnostics and drug-target discovery due to its speed, accuracy and low cost.
Executive Summary.
Clinical management of colorectal cancer & the need for improved diagnostics
-
▪
Given the current limitations to clinicopathologic staging of colorectal cancer based on American Joint Committee on Cancer Tumor, Node and Metastases Classification, there is an unmet need for the development of more accurate clinical tests to steer diagnosis and treatment. As colorectal cancer is inherently a genetic disease driven by point mutations and other chromosomal aberrations, more accurate tests for prognostication and resistance to chemotherapy are likely to come from genetic-based tumor biomarkers. We explicitly focus on mutations and other genetic aberrations given their rapid adoption. Other biomarkers derived from protein analysis or gene expression exist, but the focus of this article is on genetics and DNA-based analysis.
Genetic mutations as colorectal cancer biomarkers
-
▪
A variety of mutations have proven to be clinically informative in colorectal cancer. Microsatellite instability (MSI) is a well-known biomarker for improved survival in colorectal cancer and decreased response to chemotherapy. KRAS and BRAF mutations are predictive of nonresponse to EGF receptor (EGFR) inhibitors, such as cetuximab and panitumumab, in patients with metastatic colorectal cancer. KRAS mutation-positive tumors display a profound resistance to EFGR inhibitor treatment. Testing for its mutation has become a standard molecular diagnostic in clinical practice. Likewise, BRAF and other downstream effectors of the EGFR pathway are being assessed for their clinical utility as predictive biomarkers.
Advances in next-generation sequencing technology
-
▪
Advances in next-generation sequencing (NGS) technology have allowed for faster and more accessible sequencing of individual human genomes, putting the ability to sequence into the hands of independent researchers and clinicians. This has opened a new chapter in oncology research with the potential to decipher the genetic code for cancer. In turn, decoding individual cancer genomes will allow for the personalization of oncologic treatment.
NGS & colorectal cancer
-
▪
There are substantial advances in the field of colorectal cancer genome research which are taking advantage of NGS technology. For example, The Cancer Genome Atlas is a concerted effort to catalogue the genomic changes and their associated phenotypes involved in 20 different cancer genomes, including colorectal cancer. As part of this study, a significant proportion of the colorectal cancer genome will be sequenced.
Conclusion
-
▪
With the potential for whole cancer genome sequencing, researchers will have the ability to sequence many more individual genomes. This will assist in the discovery of novel mutations correlated to clinical outcomes, raising the possibility of developing more accurate diagnostic tests and more effective targeted therapies. Additionally, clinicians will have improved tools to assess prognosis and predict the response to various chemotherapeutic regimens, allowing for the individual tailoring of treatment.
Acknowledgments
Hanleeji is supported by the following grants from the NTH: 5K08CA96879–6, DK56339, and 2P01HG000205. In addition, Hanlee Ji received support from the Doris Duke Clinical Foundation, Reddere Foundation, the Liu Bie Ju Cha and Family Fellowship in Cancer, the Wang Family Foundation and the Howard Hughes Medical Foundation. Samuel Myllykangas received support from the Sigrid Jusélius Foundation and the Academy of Finland.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
- 1.Fry R, Mahmoud N, Maron D, Ross H, Rombeau J. Colon and rectum. In: Townsend C, Beauchamp R, Evers B, Mattox K, editors. Sabiston Textbook of Surgery. 8th Edition. Saunders, Elsevier, PA: USA; 2008. [Google Scholar]
- 2.Engstrom P, Arnoletti J, Benson AB, 3rd, et al. NCCN guidelines: Colon Cancer. 2010 Version 1.2011. [Google Scholar]
- 3.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988;319(9):525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 4.Gunderson LL, Jessup JM, Sargent DJ, Greene FL, Stewart AK. Revised TN categorization for colon cancer based on national survival outcomes data. J. Clin. Oncol. 2010;28(2):264–271. doi: 10.1200/JCO.2009.24.0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall JL. Risk assessment in stage II colorectal cancer. Oncology (Williston Park) 2010;24(1 Suppl. 1):9–13. [PubMed] [Google Scholar]
- 6.Gray R, Barnwell J, McConkey C, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370(9604):2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 7.Becquemont L. Pharmacogenomics of adverse drug reactions: practical applications and perspectives. Pharmacogenomics. 2009;10(6):961–969. doi: 10.2217/pgs.09.37. [DOI] [PubMed] [Google Scholar]
- 8.Locker GY, Hamilton S, Harris J, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin. Oncol. 2006;24(33):5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 9.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin. Oncol. 2003;21(6):1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 10.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248–5257. [PubMed] [Google Scholar]
- 11.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin. Oncol. 2005;23(3):609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 12.Benatti P, Gafa R, Barana D, et al. Microsatellite instability and colorectal cancer prognosis. Clin. Cancer Res. 2005;11(23):8332–8340. doi: 10.1158/1078-0432.CCR-05-1030. [DOI] [PubMed] [Google Scholar]
- 13.Kim GP, Colangelo LH, Wieand HS, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute–National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin. Oncol. 2007;25(7):767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 14.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2003;349(3):247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertagnolli MM, Niedzwiecki D, Compton CC, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin. Oncol. 2009;27(11):1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Des Guetz G, Schischmanoff O, Nicolas P, et al. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur. J. Cancer. 2009;45(10):1890–1896. doi: 10.1016/j.ejca.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin. Oncol. 2010;28(20):3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munro AJ, Lain S, Lane DP. P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br. J. Cancer. 2005;92(3):434–444. doi: 10.1038/sj.bjc.6602358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Font A, Abad A, Monzo M, et al. Prognostic value of K-ras mutations and allelic imbalance on chromosome 18q in patients with resected colorectal cancer. Dis. Colon Rectum. 2001;44(4):549–557. doi: 10.1007/BF02234328. [DOI] [PubMed] [Google Scholar]
- 20.Jernvall P, Makinen MJ, Karttunen TJ, Makela J, Vihko P. Loss of heterozygosity at 18q21 is indicative of recurrence and therefore poor prognosis in a subset of colorectal cancers. Br. J. Cancer. 1999;79:5–6. 903–908. doi: 10.1038/sj.bjc.6690144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanza G, Matteuzzi M, Gafa R, et al. Chromosome 18q allelic loss and prognosis in stage II and III colon cancer. Int. J. Cancer. 1998;79(4):390–395. doi: 10.1002/(sici)1097-0215(19980821)79:4<390::aid-ijc14>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe T, Wu TT, Catalano PJ, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2001;344(16):1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fearon ER. Molecular genetics of colorectal cancer. Ann. NY Acad. Sci. 1995;768:101–110. doi: 10.1111/j.1749-6632.1995.tb12114.x. [DOI] [PubMed] [Google Scholar]
- 24.Kirley SD, DApuzzo M, Lauwers GY, et al. The cables gene on chromosome 18Q regulates colon cancer progression in vivo. Cancer Biol. Ther. 2005;4(8):861–863. doi: 10.4161/cbt.4.8.1894. [DOI] [PubMed] [Google Scholar]
- 25.Carethers JM, Hawn MT, Greenson JK, Hitchcock CL, Boland CR. Prognostic significance of allelic lost at chromosome 18q21 for stage II colorectal cancer. Gastroenterology. 1998;114(6):1188–1195. doi: 10.1016/s0016-5085(98)70424-x. [DOI] [PubMed] [Google Scholar]
- 26.Cohn KH, Ornstein DL, Wang F, et al. The significance of allelic deletions and aneuploidy in colorectal carcinoma. Results of a 5-year follow-up study. Cancer. 1997;79(2):233–244. [PubMed] [Google Scholar]
- 27.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003;3(5):330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 28.Popat S, Matakidou A, Houlston RS. Thymidylate synthase expression and prognosis in colorectal cancer: a systematic review and meta-analysis. J. Clin. Oncol. 2004;22(3):529–536. doi: 10.1200/JCO.2004.05.064. [DOI] [PubMed] [Google Scholar]
- 29.Boige V, Mendiboure J, Pignon JP, et al. Pharmacogenetic assessment of toxicity and outcome in patients with metastatic colorectal cancer treated with LV5FU2, FOLFOX, and FOLFIRI: FFCD 2000–2005. J Clin. Oncol. 2010;28(15):2556–2564. doi: 10.1200/JCO.2009.25.2106. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Balibrea E, Abad A, Martinez-Cardus A, et al. UGT1A and TYMS genetic variants predict toxicity and response of colorectal cancer patients treated with first-line irinotecan and fluorouracil combination therapy. Br. J. Cancer. 2010;103(4):581–589. doi: 10.1038/sj.bjc.6605776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakobsen A, Nielsen JN, Gyldenkerne N, Lindeberg J. Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphism in normal tissue as predictors of fluorouracil sensitivity. J. Clin. Oncol. 2005;23(7):1365–1369. doi: 10.1200/JCO.2005.06.219. [DOI] [PubMed] [Google Scholar]
- 32.Etienne-Grimaldi MC, Milano G, Maindrault-Goebel F, et al. Methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms and FOLFOX response in colorectal cancer patients. Br. J. Clin. Pharmacol. 2010;69(1):58–66. doi: 10.1111/j.1365-2125.2009.03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zintzaras E, Ziogas DC, Kitsios GD, et al. MTHFR gene polymorphisms and response to chemotherapy in colorectal cancer: a meta-analysis. Pharmacogenomics. 2009;10(8):1285–1294. doi: 10.2217/pgs.09.59. [DOI] [PubMed] [Google Scholar]
- 34.Lievre A, Blons H, Laurent-Puig P. Oncogenic mutations as predictive factors in colorectal cancer. Oncogene. 2010;29(21):3033–3043. doi: 10.1038/onc.2010.89. [DOI] [PubMed] [Google Scholar]
- 35.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60–00 trial. J Clin. Oncol. 2010;28(3):466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 36.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90(9):675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 37.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten Ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br. J. Cancer. 2001;85(5):692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peeters M, Price TJ, Cervantes A, et al. Randomized Phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin. Oncol. 2010;28(31):4706–4713. doi: 10.1200/JCO.2009.27.6055. [DOI] [PubMed] [Google Scholar]
- 39.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 40.Qiu LX, Mao C, Zhang J, et al. Predictive and prognostic value of KRAS mutations in metastatic colorectal cancer patients treated with cetuximab: a meta-analysis of 22 studies. Eur. J. Cancer. 2010;46(15):2781–2787. doi: 10.1016/j.ejca.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Bokemeyer C, Bondarenko I, Makhson A, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin. Oncol. 2009;27(5):663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 42.Ocvirk J, Brodowicz T, Wrba F, et al. Cetuximab plus FOLFOX6 or FOLFIRI in metastatic colorectal cancer: CECOG trial. World J. Gastroenterol. 2010;16(25):3133–3143. doi: 10.3748/wjg.v16.i25.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tol J, Koopman M, Cats A, et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N. Engl. J. Med. 2009;360(6):563–572. doi: 10.1056/NEJMoa0808268. [DOI] [PubMed] [Google Scholar]
- 44.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin. Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 45.Lievre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin. Oncol. 2008;26(3):374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 46.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 2008;359(17):1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 47.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67(6):2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 48.Loupakis F, Pollina L, Stasi I, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin. Oncol. 2009;27(16):2622–2629. doi: 10.1200/JCO.2008.20.2796. [DOI] [PubMed] [Google Scholar]
- 49.Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N. Engl. J. Med. 2009;360(14):1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 50.Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418(6901):934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 51.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin. Oncol. 2009;27(35):5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 52.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin. Oncol. 2008;26(35):5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 53.Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, et al. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS ONE. 2009;4(10):e7287. doi: 10.1371/journal.pone.0007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers J. The finished genome sequence of Homo sapiens. Cold Spring Harh. Symp. Quant. Biol. 2003;68:1–11. doi: 10.1101/sqb.2003.68.1. [DOI] [PubMed] [Google Scholar]
- 55.Ansorge WJ. Next-generation DNA sequencing techniques. Nat. Biotechnol. 2009;25(4):195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 56.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 57. Metzker ML. Sequencing technologies – the next generation. Nat. Rev. Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. ▪ Comprehensive overview of next-generation sequencing technology. This review includes details on the biochemistry of sequencing and a summary of commercially available sequencing platforms.
- 58.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat. Methods. 2009;6(Suppl. 11):S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 59.McPherson JD. Next-generation gap. Nat. Methods. 2009;6(Suppl. 11):S2–S5. doi: 10.1038/nmeth.f.268. [DOI] [PubMed] [Google Scholar]
- 60. Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010;11(10):685–696. doi: 10.1038/nrg2841. ▪ Recent and thorough review of next-generation sequencing technology as it applies to cancer research. This review highlights the technical considerations of next-generation sequencing that are specific to oncologic research.
- 61.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahn SM, Kim TH, Lee S, et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19(9):1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 64.Harris M, Winship I, Spriggs M. Controversies and ethical issues in cancer – genetics clinics. Lancet Oncol. 2005;6(5):301–310. doi: 10.1016/S1470-2045(05)70166-2. [DOI] [PubMed] [Google Scholar]
- 65.Stanley AJ, Gaff CL, Aittomaki AK, et al. Value of predictive genetic testing in management of hereditary non-polyposis colorectal cancer (HNPCC) Med. J. Aust. 2000;172(7):313–316. doi: 10.5694/j.1326-5377.2000.tb123976.x. [DOI] [PubMed] [Google Scholar]
- 66.Thomas RK, Greulich H, Yuza Y, et al. Detection of oncogenic mutations in the EGFR gene in lung adenocarcinoma with differential sensitivity to EGFR tyrosine kinase inhibitors. Cold Spring Harb. Symp. Quant. Biol. 2005;70:73–81. doi: 10.1101/sqb.2005.70.056. [DOI] [PubMed] [Google Scholar]
- 67.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl. J. Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berger MF, Levin JZ, Vijayendran K, et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20(4):413–427. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bueno R, De Rienzo A, Dong L, et al. Second generation sequencing of the mesothelioma tumor genome. PLoS ONE. 2010;5(5):el06l2. doi: 10.1371/journal.pone.0010612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pleasance ED, Stephens PJ, O’Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463(7278):184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu J, Mani RS, Cao Q, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2–ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17(5):443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. ▪ One of the first large-scale unbiased genomic surveys of colorectal cancer. It established the genomic landscape, identifying 280 genetic mutations involved in 38 pathways in metastatic colorectal cancer.
- 74.American Society of Clinical Oncology policy statement update: genetic testing for cancer susceptibility. J. Clin. Oncol. 2003;21(12):2397–2406. doi: 10.1200/JCO.2003.03.189. [DOI] [PubMed] [Google Scholar]
- 75.Health law – genetics – Congress restricts use of genetic information by insurers and employers. Genetic Information Nondiscrimination Act of 2008, Pub. L. No. 110–233, 122 Stat. 881 (to be codified in scattered sections of 26, 29, and 42 U.S.C.) Harv. Law Rev. 2009;122(3):1038–1045. [PubMed] [Google Scholar]
- 76.Offit K, Thorn P. Ethical and legal aspects of cancer genetic testing. Semin. Oncol. 2007;34(5):435–443. doi: 10.1053/j.seminoncol.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 77.Stadler ZK, Thorn P, Robson ME, et al. Genome-wide association studies of cancer. J. Clin. Oncol. 2010;28(27):4255–4267. doi: 10.1200/JCO.2009.25.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J. Natl Cancer Inst. 2010;102(7):464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garman KS, Acharya CR, Edelman E, et al. A genomic approach to colon cancer risk stratification yields biologic insights into therapeutic opportunities. Proc. Natl Acad. Sci. USA. 2008;105(49):19432–19437. doi: 10.1073/pnas.0806674105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Ali MA, Sjoblom T. Molecular pathways in tumor progression: from discovery to functional understanding. Mol. Biosyst. 2009;5(9):902–908. doi: 10.1039/b903502h. [DOI] [PubMed] [Google Scholar]
- 81.Vilar E, Gruber SB. Microsatellite instability in colorectal cancer – the stable evidence. Nat. Rev. Clin. Oncol. 2010;7(3):153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 2009;10(9):617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 83.Kandpal R, Saviola B, Felton J. The era of ‘omics unlimited. Biotechniques. 2009;46(5):351–352. 4–5. doi: 10.2144/000113137. [DOI] [PubMed] [Google Scholar]
Websites
- 101.WHO. Cancer. 2009 www.who.int/mediacentre/factsheets/fs297/en/index.html.
- 102.Altekruse SF, Krapcho M, Neyman N, et al., editors. SEER Cancer Statistics Review 1975–2007. 2010 http://seer.cancer.gov/csr/1975_2007/
- 103.Programs UDoEG. Human Genome Project Information. http://genomics.energy.gov.
- 104. NIH. The Cancer Genome Atlas – Mission and Goals. http://cancergenome.nih.gov/about/mission.asp. ▪ Provides up-to-date information on The Cancer Genome Atlas project and access to the data portal. The data portal contains all data pertaining to clinical information, genomic characterization and sequencing analysis of tumor genomes in a searchable format.