Abstract
The PE/PPE family of proteins which are in high abundance in pathogenic species such as Mycobacterium tuberculosis and M. marinum, play the critical role in generating antigenic variation and evasion of host immune responses. However, little is known about their functional roles in mycobacterial pathogenesis. Previously, we found that PPE38 is associated with the virulence of mycobacteria, presumably by modulating the host immune response. To clarify the link between PPE38 and host response, we employed a subcellular, amino acid-coded mass tagging (AACT)/SILAC-based quantitative proteomic approach to determine the proteome changes during host response to M. marinum PPE38. As a result, 291 or 290 proteins were found respectively to be up- or down-regulated in the nucleus. Meanwhile, 576 upregulated and 272 downregulated proteins were respectively detected in the cytosol. The data of quantitative proteomic changes and concurrent biological validations revealed that M. marinum PPE38 could trigger extensive inflammatory responses in macrophages, probably through interacting with toll-like receptor 2 (TLR2). We also found that PPE38 may arrest MHC-1 processing and presentation in infected macrophages. Using bioinformatics tools to analyze global changes in the host proteome, we obtained a PPE38-respondor network involved in various transcriptional factors (TFs) and TF-associated proteins. The results of our systems investigation now indicate that there is cross-talk involving a broad range of diverse biological pathways/processes that coordinate the host response to M. marinum PPE38.
Keywords: PPE38, M. marinum, Amino acid-coded mass tagging/stable isotope labeling with amino acid in cell culture, LC−MS/MS, toll-like receptor 2, major histocompatibility complex 1, transcription factors
Introduction
Tuberculosis, caused by Mycobacterium tuberculosis, is still a serious threat to public health. At present, approximately one-third of the world’s population is infected by M. tuberculosis (WHO, http://www.who.int/mediacentre/factsheets/fs104/en/). As the major host cells, macrophages are the primary niche for infection by mycobacteria. Many macrophage receptors are involved in phagocytosis of mycobacteria, such as complement receptor, mannose receptor, and CD14.1,2 To promote their survival under the pressure of host response, M. tuberculosis bacteria counteract certain cell biological and immune processes involved in the host response, including antigen presentation, pro-inflammatory cytokine secretion and phagosome maturation, so it can survive inside host cells.3
In the M. tuberculosis genome, two distinctive protein families have been known as the proline–glutamic acid (PE) and the proline–proline–glutamic acid (PPE) families, which represent about the 10% of the coding genes of the genome.4 The PE/PPE families contain a large number of repeat units and have been implicated as restructuring and mutation hotspots.5,6 As such, researchers have speculated that M. tuberculosis may undergo antigenic variation within these regions, thereby escaping the immune response of the host cells.4,6 Among the Mycobacterium genus, Mycobacterium marinum has the most PE/PPE family members,7 and is closely related to M. tuberculosis,8 which therefore is an ideal model system to study M. tuberculosis pathogenesis.9,10 PPE24 and PPE53 are major virulence factors of M. marinum, playing regulatory roles in cell invasion and survival inside macrophages.11 Mutation of M. marinum ESX-5, which functions in the secretion of many PE and PPE family members, is capable of triggering a macrophage-mediated immune response.12 Exogenous expression of the PPE37 protein in M. smegmatic-infected macrophages results in lowered expression levels of tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6), contributing to the suppression of the proinflammatory cytokine-mediated response as well as inhibited clearance of Mycobacterium.13 Recently, emerging attention has been paid to possible functions of PPE proteins in host response although preliminary studies described PPE proteins as potential antigens.14−16
The PPE38 gene (Rv2352c) is located in the region of difference 7 (RD7) of M. tuberculosis. The RD regions generally exist in the genome of virulent strains of M. tuberculosis, but they are absent in the bacillus Calmette–Guérin(BCG) genome. Due to gene recombination, it has been suggested that the ppe38 region may have little impact on the pathogenicity of Mycobacterium, as indicated by the high degree of polymorphisms in this region.5 However, PPE38 was found highly expressed 90 days after guinea pigs were infected with M. tuberculosis, implicating the function of PPE38 in the pathogenicity of M. tuberculosis.17
Our previous studies identified the mutant strains of ppe38 (05B1) by screening a MycoMarT7 mariner transposon mutagenesis library.18 Further study found that PPE38 was localized in the cell wall, and disruption of PPE38 resulted in reduced secretion of TNF-α and IL-6 and a decreased ability to invade macrophages. Adult zebrafish infected with the PPE38 mutant survived longer and exhibited reduced pathology. On the basis of these observations, we reason that PPE38 may play a direct role in the virulence of Mycobacteria, presumably by modulating the host immune response.18
In the present study, we employed our amino acid-coded mass tagging (AACT)- or SILAC-assisted quantitative proteomics19−21 to comparatively determine the host proteome changes when the host macrophages were infected respectively by the wild-type (WT) vs a mutant M. marinum strain, 05B1. Following AACT metabolic labeling, the infection, the subcellular fractionation, and equally mixing of WT-infected and 05B1-infected to minimize possible experimental variations,20,22 LC–MS/MS was performed to identify and quantify those host proteins showing changes during the macrophage exposure to the WT vs mutant strain. We found that the PPE38 protein of M. marinum not only could promote TLR2-mediated secretion of TNF-α and IL-6 but could also participate in antigen processing and presentation. Furthermore, those transcription factors (TFs) and TF-associated proteins which related to the function of PPE38 were studied using bioinformatics methods, and the links between TFs and the corresponding biological processes were determined. Regarding the systems view, our results facilitate understanding how PPE modulates the host response through particular regulatory pathways and provide insight into the prevention and treatment of tuberculosis.
Materials and Methods
Chemicals
The components of common and labeling cell culture media were from Gibco and Sigma, respectively. Fetal bovine serum (FBS) was purchased from Gibco, and dialyzed FBS was obtained from Invitrogen (26400-044). Deuterium-labeled leucine (leucine-d3) was purchased from Cambridge Isotope (Andover, MA). Trypsin was purchased from Promega (Madison, WI).
TLR2 antibody was purchased from Millipore (NG1897884); CD14 antibody was obtained from BD Biosciences (553738); PCNA antibody was purchased from Bioworld Consulting Laboratories (BS1289); antibodies against CHEK1 (10362-1-AP), GAPDH (60004-1-Ig), IRF5 (10547-1-AP), and BAK (14673-1-AP) were obtained from Proteintech Group; ERK1/2 and P38 antibodies were purchased from ANBO (CO185, BO798); antibody for PARP1 was obtained from Cell Signaling Technology (#9542); anti-human TLR2 Ab was purchased from Biolegend (309709); isotype control for IgG2a,κ was obtained from Ebioscience (14-4724-81).
Bacterial Strains, Media, and Growth Conditions
The M. marinum M strain (ATCC BAA-535) was obtained from L. Ramakrishnan (University of Washington, Seattle). Strain 05B1 (MMAR_3661::φMycoMar) was generated by transposon mutagenesis of the M strain of M. marinum.15M. marinum cells were grown at 32 °C in Middlebrook 7H9 broth (BD, 3349030) supplemented with 0.2% glycerol and 10% oleic acid-albumin-dextrose-catalase (OADC) (Difco) or on Middlebrook 7H10 agar (BD, 1097997) supplemented with 0.5% glycerol and 10% OADC. When necessary, the growth medium was supplemented with kanamycin at 50 μg/mL.
Cell Culture and AACT/SILAC Labeling
The murine macrophage cell line RAW264.7 (ATCC TIB71) was maintained at 37 °C in 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% fetal bovine serum (FBS), and 10 mM HEPES. Similar to the procedures of AACT/SILAC-labeling previously reported,23 we labeled the cells through seven passages. Leu-d3-containing peptides derived from β-actin were extracted from the labeled cells, the efficiency/extent of the label was examined by using MALDI-TOF 4700 (Applied Biosystems) (Figure 1 in the Supporting Information [SI]), the results were the same as reported previously.23
Infection of RAW264.7 Macrophages by M. marinum
The single-cell suspension of bacteria was prepared by shaking with glass beads, followed by low-speed centrifugation, and passing through a 5-μm syringe filter to remove bacterial aggregates. Prior to infection, RAW264.7 cells were seeded into a 10 cm dish at a density of 5 × 105 cells per well and maintained at 37 °C in 5% CO2. After overnight growth, the RAW264.7 cells were infected. Spent media was replaced with 20 mL of fresh DMEM media containing 10% FBS and sufficient mycobacteria to achieve a multiplicity of infection (MOI) of 10 (i.e., ten bacteria for one macrophage). The infection was allowed to proceed for 4 h at 32 °C in 5% CO2. The number of intracellular mycobacteria was enumerated by plating appropriate dilutions on Middlebrook 7H10 agar plates.
The Neutralization of TLR2 Receptor and Cytokine Assay
The THP-1 cells were maintained at 37 °C in 5% CO2 in RPMI-1640 media (Gibco) supplemented with 10% FBS. Prior to experiment, THP-1 cells were differentiated into macrophages in 96-well plates (0.5 × 106 cells/well) with 200 nM PMA (Sigma) for 24 h and then incubated with fresh media without PMA for 24 h. THP-1 cells were treated with anti-human TLR2 Ab or isotype (IgG2a,κ) control Ab for 120 min at 37 °C and then infected with mycobacteria at MOI of 10 for 4 h at 32 °C. The extracellular bacteria were removed by washing and then treated with 200 μg/mL gentamicin for 2 h. THP-1 cells were then maintained in RPMI-1640 (10% FBS) supplemented with 20 μg/mL of gentamicin. At 24 h post infection, cell-free supernatants were collected and cytokine productions determined by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s instructions (BD Biosciences).
Subcellular Fractionation
Cytosolic and nuclear fractions were prepared using a nuclear protein extraction kit (Beyotime Biotechnology). The protein concentrations were determined using the bicinchoninic acid assay kit (Beyotime Biotechnology).
Protein Separation, Band Excision, In-Gel Trypsin Digestion, and Peptides Extraction
The cytoplasmic or nuclear fractions derived from WT-stimulated and 05B1-stimulated cells were mixed 1:1 on the basis of the total protein mass, respectively. Samples were separated by 12% SDS-PAGE and stained with Coomassie Brilliant Blue (CBB).
Following the protocol described previously,24 the SDS-PAGE gel of cytoplasmic fractions were cut into 19 slices, and those of nuclear factions were cut into 13 slices. The gel slices were washed with Milli-Q water, and CBB dye was removed with 50% ACN/50 mM ammonium bicarbonate for 15 min. Gel slices were dehydrated twice in 100% ACN for 30 min and reconstituted overnight at 37 °C with an in-gel digestion reagent containing 10 ng/μL sequencing grade trypsin. The tryptic peptides were extracted from the gel pieces with 50% ACN/0.1% TFA and lyophilized for 4 h.
LC–MS/MS
LC–MS/MS experiments were performed on a hybrid linear quadrupole ion trap/Orbitrap (LTQ Orbitrap) mass spectrometer (Thermo Finnigan, Bremen, Germany) coupled to a Shimadzu LC-20AD LC system (Shimadzu, Japan) and SIL-20AC autosampler (Shimadzu, Japan). Tryptic peptides were redissolved in 30 μL of 0.1% FA solution and chromatographically separated on a C18 column (0.1 mm × 150 mm 5μ 200A; Michrom). Each sample was loaded in solvent A (95% H2O/5% ACN/0.1% FA) and followed by gradient elution of 5–45% solvent B (5% H2O/95% ACN/0.1% FA) over 90 min with a flow rate of 500 nL/min. The entire eluant was sprayed into the mass spectrometer via a dynamic nanospray probe (Thermo Fisher Scientific) and analyzed in positive mode. The 10 most abundant precursor ions detected in the full MS survey scan (m/z range of 400–2000, R = 60,000) were isolated for further MS/MS analyzing. Spectra were acquired under automatic gain control (AGC) in one microscan for survey spectra (AGC: 106) and in three microscans for MS/MS spectra (AGC: 104).
Database Search, Protein Identification, and Quantification
Protein identification and quantification were performed with MaxQuant version 1.2.0.18. Data were searched using the Andromeda search engine against the IPI mouse database (3.68; 56,729 entries). The mass tolerances for precursor and fragment ions were initially set to 20 ppm and 0.5 Da, and then recalibration was done in silico. Variation modifications included methione oxidation (15.9994 Da) and protein N-terminus acetylation (42.0106 Da) and leucine-d3 (3.0188 Da). Peptides with lengths of a minimum of six amino acids were considered with both the peptide and protein FDR set to 1%. The light- (L) and heavy-isotope (H) peptide elution profiles were isolated automatically. The area of each peptide peak was determined, and the abundance ratios (H/L) based on these areas were calculated to determine the threshold of the quantitative baseline that could be used to distinguish these different protein expressions. The nuclear and cytoplasmic proteins were normalized by nucleolin and tubulin, respectively. With these controls of subcellular markers, the ratios of all other proteins were calibrated accordingly.
Immunoblotting
Protein samples were separated by SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with TBST containing 5% nonfat milk and were incubated with the specified primary antibodies overnight followed by incubation with secondary antibody conjugated with horseradish peroxidase. ECL substrate (Millipore, WBKLS0500) was added on the membranes and exposed using ECL systems (Las 3000, Fujifilm, Japan)
Functional Clustering and Network Analysis
The quantified proteins were submitted to DAVID (http://david.abcc.ncifcrf.gov/) to obtain their known biological processes and molecular functions. Proteins involved in signaling pathways were categorized by PANTHER (http://www.pantherdb.org/). According to their categorized function, a network was constructed by STRING (http://string-db.org/). The links in the network were edited by the software Medusa (http://www.foofus.net/∼jmk/medusa/medusa.html) or Cytoscape (http://www.cytoscape.org/).
Results and Discussion
Identification and Quantification of Proteins Showing PPE38-Dependent Changes in Their Expressions
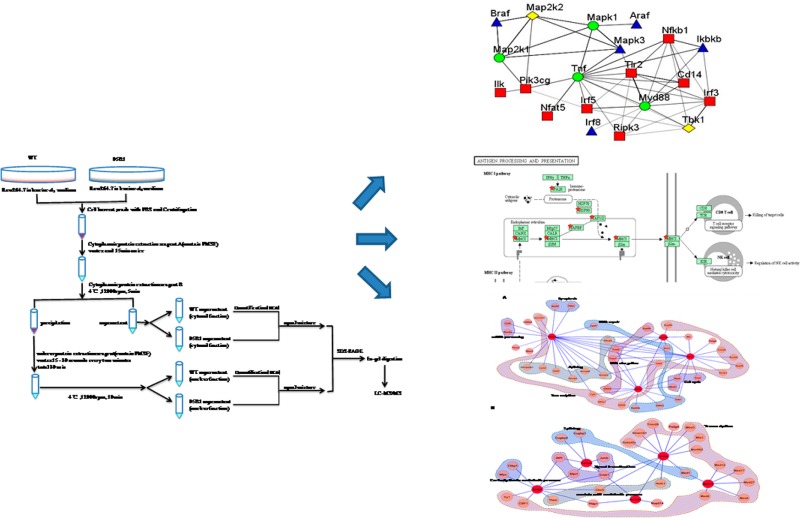
To identify/profile the proteins associated with PPE38 function, we designed an AACT-based quantitative proteomic approach. As shown by Figure 1, the macrophage cells (RAW264.7) grown in either “light” (leucine-d0) (L) or “heavy” medium (leucine-d3) (H) were respectively infected by either 05B1 mutant or wild-type (WT) M. marinum for 4 h. Either the cytoplasmic or the nuclear fractions derived from each cell pool were mixed at 1:1 on the basis of the total protein mass, respectively, followed by SDS-PAGE separation, in-gel trypsin digestion, peptide extraction, and LC–MS/MS analysis. The effectiveness of subcellular fractionation was also examined by both cytoplasmic and nuclear markers (Figure 2 in SI), indicating the purity of the separation.
Figure 1.
Schematic describing the AACT/SILAC quantitative proteomic approach for protein extraction.
Using the threshold for protein identification as previously described,25 a total of 3875 proteins were identified and quantified. Among them, 2394 are in the cytoplasmic fraction, and 1481 are in the nuclear fraction (see Tables 1 and 2 in SI). The H/L (wild type/mutant) ratio of either nucleolin or tubulin, which are high-abundant proteins known to locate in the nucleus or the cytosol, respectively, were found at close to 1:1. We then used these ratios to normalize other proteins identified in nuclear or cytoplasmic fractions accordingly. Referring to the criteria previously described,23,26,27 we considered the proteins showing 20% or more changes in their abundances as the differentially expressed proteins. Meeting this threshold, 291 upregulated and 290 downregulated proteins were found in the nucleus along with 576 upregulated and 272 downregulated proteins in the cytosol. Meanwhile, only 103 differentially expressed proteins were identified in both the nucleus and cytosol (Figure 2B in the SI), indicating the high purity of our subcellular fractionation.
Functional Categorization of the Differentially Expressed Proteins in the Macrophages Infected by Wild-Type vs Its 05B1 Mutant
To clarify those biological processes of the differentially expressed proteins, we utilized PANTHER (http://www.pantherdb.org/) to classify these proteins on the basis of their known functions. As shown in Figure 2, most of the identified cytosolic proteins were involved in protein metabolism, signal transduction, immunology, cell cycle, or apoptosis. The nuclear proteins were primarily involved in nucleic acid metabolism, signal transduction, and cell cycle. The proteins that were differentially expressed in cytosol and nucleus following PPE38 stimulation were involved in each large functional category including signal transduction, immunity and defense, apoptosis and response to stress (Figure 2). It is inferred that PPE38 could elicit a series of intracellular signaling cascades such as TLR signaling, NF-κB-regulated signaling, and PI3K signaling. Here our data further indicated that PPE38 played a coordinated role in promoting the cross-talk among these pathways involved in the host immune response and defense.
Figure 2.
Analyses of biological processes and distribution of differentially expressed proteins in PPE38-stimulated macrophages identified by MS. The proteins whose H/L ratios were greater than 1.2 or less than 0.8, were submitted to PANTHER (http://www.pantherdb.org/) to obtain information about their associated biological processes.
We further analyzed the relevance of the functions of all of the differentially expressed proteins. The primary functional categories included nucleic acid binding, oxidoreductase, protein binding, hydrolase, and cytoskeletal proteins (Figure 3 in the SI). Mitochondrial damage is known to regulate the outcome of macrophage infection with M. tuberculosis,28,29 and mitochondrial intermediates are also involved in the apoptosis induced by PE-PGRS33.30,31 In our results, many differentially expressed proteins were functionally classified as oxidation hydrolases, indicating that mitochondrial intermediates were involved in the response of macrophages to PPE38 stimulation.
PPE38 Activates Multiple Pathways Downstream of TLR2, Involving the Activation of Proinflammatory Programming
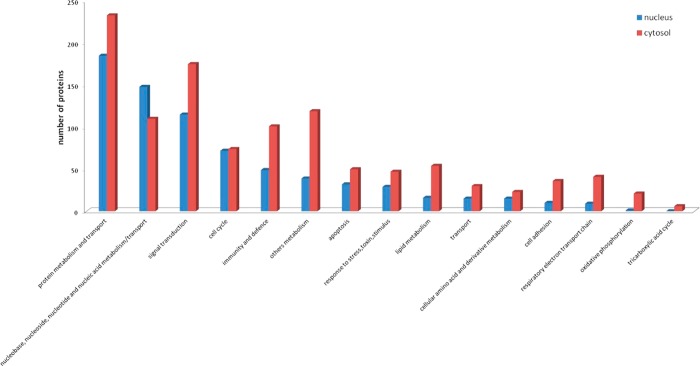
To clarify the mechanism by which PPE38 interacts with macrophages, we used PANTHER (http://www.pantherdb.org/) to determine the proteins engaged in the TLR, NF-κB, MAPK, and IRF signaling pathways (Table 1). Subsequently, we compared this list with those differently expressed proteins identified in the present study to reconstruct an interaction network (Figure 3) using STRING (http://string-db.org/).
Table 1. Quantified Proteins Involved in Toll-Like Receptor, NF-κB, MAPK, and IRF Signal Cascades.
| Uniprot_ID | locationa | gene symbol | protein descriptions | Pepb(pepc) | coverage [%] | H/L ratiod | variabilitye [%] |
|---|---|---|---|---|---|---|---|
| Q63932 | cytosol | Map2k2 | dual specificity mitogen activated protein kinase kinase 2 | 6(2) | 7.50 | 1.167 | 20.18 |
| P31938 | cytosol | Map2k1 | dual specificity mitogen activated protein kinase kinase 1 | 11(7) | 21.40 | 0.746 | 123.62 |
| P63085 | cytosol | Mapk1 | mitogen-activated protein kinase 1; ERK2 | 11(8) | 29.30 | 0.917 | 27.47 |
| P63085 | nuclear | Mapk1 | mitogen-activated protein kinase 1 | 5(5) | 14.00 | 0.758 | 62.99 |
| Q9JHG7 | cytosol | Pi3kcg | phosphatidylinositol 3-kinase gamma isoform | 4(4) | 3.80 | 1.311 | 79.70 |
| Q9JHG7 | nuclear | Pi3kcg | phosphatidylinositol 3-kinase gamma isoform | 2(2) | 3.20 | 0.025 | 483.36 |
| O55222 | cytosol | Ilk | integrin-linked protein kinase | 6(6) | 13.70 | 1.206 | 9.54 |
| O55222 | nuclear | Ilk | integrin-linked protein kinase | 9(9) | 21.50 | 1.244 | 43.81 |
| Q8VHJ6 | cytosol | Nfat5 | nuclear factor of activated T-cells 5 isoform b | 11(11) | 10.30 | 1.966 | 82.61 |
| Q8VHJ6 | nuclear | Nfat5 | nuclear factor of activated T-cells 5 isoform b | 7(7) | 5.30 | 0.947 | 36.16 |
| P56477 | cytosol | Irf5 | interferon regulatory factor 5 | 7(7) | 14.90 | 1.398 | 23.37 |
| P56477 | nuclear | Irf5 | interferon regulatory factor 5 | 6(6) | 13.30 | 0.971 | 11.02 |
| Q9QUN7 | cytosol | Tlr2 | toll-like receptor 2 | 11(11) | 17.10 | 1.879 | 33.01 |
| P25799-5 | cytosol | Nfkb1 | nuclear factor kappa-B, subunit 1 | 2(2) | 2.50 | 0.959 | 34.05 |
| P25799-5 | nuclear | Nfkb1 | nuclear factor kappa-B, subunit 1 | 1(1) | 1.60 | 2.063 | 61.06 |
| P06804 | cytosol | Tnfa | tumor necrosis factor alpha | 1(1) | 5.10 | 0.485 | 112.41 |
| P10810 | cytosol | Cd14 | monocyte differentiation antigen CD14 | 2(2) | 6.60 | 3.094 | 24.80 |
| P22366 | cytosol | Myd88 | myeloid differentiation primary response protein MyD88 | 4(4) | 15.20 | 0.754 | 39.31 |
| P70671 | cytosol | Irf3 | interferon regulatory factor 3 | 2(2) | 5.00 | 1.320 | 7.94 |
| Q9WUN2 | cytosol | Tbk1 | serine/threonine-protein kinase TBK1 | 5(5) | 8.60 | 0.846 | 25.02 |
| Q9QZL0 | cytosol | Ripk3 | receptor-interacting serine/threonine protein kinase 3 | 8(8) | 21.00 | 1.220 | 25.58 |
Protein identified in cytosol or nuclear fraction.
Peptide number matched to the protein.
Peptide number used to quantify the protein.
Protein expression changes of WT-stimulated (H) vs 05B1-stimulated (L) macrophages.
Variability was calculated by the isotope intensity ratio from multiple leucine-containing peptides.
Figure 3.
Reconstructed network involving NF-κB, MAPK, IRF, and TLR signaling pathways in the early response to PPE38 stimulation. We used PANTHER (http://www.pantherdb.org/) to acquire the key components (Table 1 in the SI), such as NF-κB, MAPK, IRF, and TLR, and submitted them to STRING (http://string.embl.de/) for network construction. The network was modified using Medusa software.
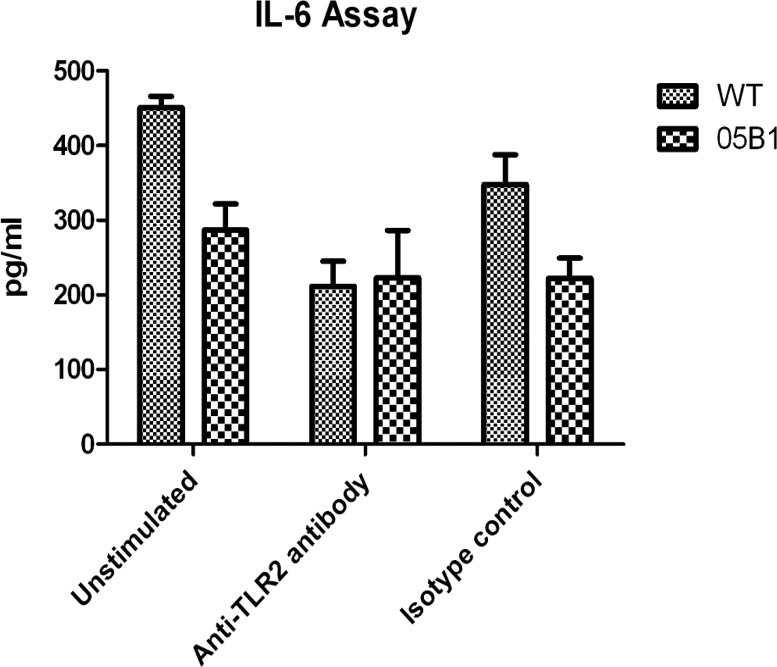
Our MS analysis indicated that TLR2 was increased by 1.2-fold in WT-infected macrophages compared to TLR2 in 05B1-infected macrophages while other TLR family members were not detected. Previous studies have found that the PE/PPE proteins could induce macrophages to secrete TNF-α, IL-10, IL-12, and other cytokines, resulting in apoptosis or dendritic cell (DC) maturation and activation, and these processes are mostly TLR2-dependent.31−33 To validate the interaction between TLR2 and PPE38, we incubated PMA-differentiated THP-1 macrophages respectively with WT or 05B1 (MOI = 10) strain in the presence of anti-TLR2 neutralization antibody (Ab) or isotype control Ab, and found that anti-TLR2 Ab, and not isotype control Ab, prevented PPE38-mediated IL-6 production by 53% (Figure 4), all indicating that TLR2 was the possible interacting partner of PPE38 during IL-6 activation.
Figure 4.
PPE38 induced IL-6 production by TLR2. IL-6 production in PMA-differentiated THP-1 macrophages was stimulated with WT and 05B1 (MOI = 10) in the presence of anti-TLR2 or isotype (IgG2a,κ) control Ab (10 μg/mL). Results are mean ± SD of three different experiments.
Myeloid differentiation primary response protein 88 (Myd88) is the common adapter protein of most of the TLR molecules except for TLR3 to activate downstream signaling pathways.34 Our results showed that Myd88 was reduced by 25% in the cytosol. Meanwhile, TLR signaling mainly results in activation of three major families of proteins downstream that are critical for activating inflammatory gene expression, including NF-κB/Rel, interferon regulatory factors (IRFs), and MAPKs (i.e., ERK1/2, JNK, and p38 mitogen-activated protein kinases).
NF-κB/Rel is a regulatory complex involved in transcription and immune response to infection.35 Our results indicated that NF-κB (p105/p50) was upregulated by 2.06-fold in the nucleus and slightly downregulated in the cytosol. Given that the nuclear translocation of NF-κB indicates the activation of its regulated cytokine production, we concluded that PPE38 triggered the activation of NF-κB-regulated inflammatory response of the infected macrophages, which subsequently induced the secretion of TNF-α and IL-6.
Interferon regulatory factor 5 (IRF5) and interferon regulatory factor 3 (IRF3) were both upregulated by 1.4- and 1.3-fold respectively in the cytosol. IRF3 is a key transcriptional factor (TF) responsible for IFN-γ production.36,37 A recent report demonstrated that IRF-3 could bind to the TNF promoter, resulting in TNF dysregulation induced by chronic ethanol.38 Unlike IRF3, IRF5 plays a key role in the induction of pro-inflammatory cytokines, including TNF, IL-6, and IL-12.39 IRF5 activation is not well understood, but it has been shown that TLR signaling induces the formation of Myd88–IRF5–TRAF6 complexes,40 and this is probably followed by phosphorylation at specific sites within the IRF5 C-terminal autoinhibitory domain.41 Nuclear factor of activated T cells 5 (Nfat5) was upregulated by 2.0-fold in the cytosol. This protein regulates gene expression induced by osmotic stress in mammalian cells and plays a central role in inducible gene transcription during the immune response. NFAT5 binds to the TNF promoter and activates TNF transcription under hypertonic conditions.42 In other reports, NFAT5 has been shown to regulate the expression of the TNF-α and lymphotoxin-b genes in osmotically stressed T cells.43 On the basis of these observations, we speculated that IRF and NFAT5 might be involved in the expression of IL-6 and TNF-α.
The MAPK cascades are evolutionary conserved and responsible for transducing diverse extracellular signals that regulate multiple processes, including cell growth, proliferation, differentiation, stress responses, apoptosis, and cytokine secretion.44,45 ERK, JNK, and p38 are the major effectors in the MAPK pathways and play key roles in activating downstream signaling.46 Mitogen-activated protein kinase 1 (MAPK1, also named ERK2) is localized to the cytoplasm. Upon activation by dual phosphorylation, MAPK1 is translocated into the nucleus and phosphorylates its nuclear targets.46 In our results, MAPK1 was decreased by 24% in the nucleus .Mitogen-activated protein kinase kinase 1 (MAP2K1), which can phosphorylate ERK1 and ERK2 to active them, was also decreased by 25% in the cytosol. Hence, we speculated that the ERK pathway might not participate in the function of PPE38. MAP kinase kinase kinase kinase 5 (MAP4K5) acts as a link to the JNK pathway and47 was upregulated by 1.7- fold in the cytosol. MAP kinase kinase 3 (MAP2K3), an upstream kinase related to p38 activation,48 was upregulated by 1.6-fold in the cytosol. MAP kinase-activated protein kinase 2 (MAPKAPK2), which is regulated through direct phosphorylation by p38 MAP kinase,49 was decreased by 40% in the nucleus. Although the expression of JNK or p38 remained little changed in the cytosol, their phosphorylated forms may play the functional roles. Results above suggested that PPE38 might active the JNK and p38 pathways rather than that of ERK.
Phosphoinositide-3-kinase (PI3K) is known with its established roles in regulation of cellular growth and inhibition of apoptosis.50 Previous reports also suggested that the role of PI3K in mycobacterial phagocytosis and in TNF-α expression is dependent on the PI3K/Akt pathway.51 On the basis of the results of our proteomic analysis, we found that phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma (PIK3CG), an enzyme that phosphorylates phosphoinositides and belongs to the PI3-kinase family, was upregulated by 1.3-fold in the cytosol. However, in the cytosol, the expression of Akt was unchanged, indicating that the expression of TNF-α could be PI3K dependent. Stimulation of integrin-linked kinase (ILK) activity is inhibited by inhibitors of PI3K, and expression of constitutively active PI3K also results in constitutively increased ILK activity, suggesting that ILK is a PI3K-dependent kinase.52 Consistent with previous reports, ILK was upregulated by 1.2-fold in the cytosol in our experiments.
Previous studies have shown that cluster of differentiation 14 (CD14) is closely related to phagocytosis.2,53 Our previous study found that the phagocytic ability of 05B1-stimulated macrophages was significantly decreased compared to WT-stimulated macrophages.18 The present results further showed that CD14 was upregulated by approximately 3.0-fold in the cytosol, suggesting that PPE38 played a role in phagocytosis via CD14.
In our data set, TNF-α was decreased by 52% in the cytosol, whereas IL-6 was not detected. However, WT-stimulated macrophages could promote higher expressions of TNF-α and IL-6 compared to 05B1-stimulated macrophages, as shown by our previous work.18 We therefore reasoned that undetectable IL-6 could be caused by its secretion.
Our results indicated that a possible ‘cross-talk’ connection involving key components of the MAPK, NF-κB, IRF, and PI3K pathways could coordinate modulation of inflammatory factors for the PPE38-induced TLR2-mediated early response. Together, these results suggested that the PPE38 protein of M. marinum could combine with the cell surface receptor TLR2, activate signaling pathways of PI3K and MAPKs, influence IRF, NF-κB, and NFAT5 expression, and effect altered expression of TNF-α and IL-6.
PPE38 Participates in Antigen Processing and Presentation
In the cytosol, we found that H2-L and H2-K were downregulated by 59% and 50%, respectively, and that many proteins involved in antigen processing and presentation were also downregulated (Table 2), suggesting that the PPE38 protein of M. marinum may participate in antigen processing and presentation.
Table 2. Quantified Proteins Involved in Antigen Processing and Presentation.
| Uniprot _ID | location | gene symbol | protein descriptions | pepb (pepc) | coverage [%] | H/L ratio | variability [%] |
|---|---|---|---|---|---|---|---|
| Q99LN3 | cytosol | Pa28b1 | proteasome activator subunit 2 isoform 2 | 10(10) | 35.70 | 0.781 | 42.65 |
| P11499 | cytosol | Hsp90b | heat shock protein HSP 90-beta | 38(1) | 1.40 | 0.053 | 128.80 |
| Q3TCU5 | cytosol | Tapbp | TAP binding protein isoform 1 | 3(3) | 5.40 | 0.338 | 11.59 |
| P21958 | cytosol | Tap1 | antigen peptide transporter 1 | 4(4) | 5.00 | 0.391 | 8.08 |
| P01897 | cytosol | H2-L | H-2 class I histocompatibility antigen, L-D alpha chain | 8(1) | 5.00 | 0.409 | 54.96 |
| P36371 | cytosol | Tap2 | antigen peptide transporter 2 | 2(2) | 2.70 | 0.355 | 30.71 |
| O35641 | cytosol | H2-K | H-2 class I histocompatibility antigen, K-D alpha chain | 11(1) | 2.10 | 0.503 | 20.22 |
M. marinum and M. tuberculosis are intracellular bacteria and are able to subvert the host immune response. Furthermore, the host immune response is largely available to control infection. In this process, CD4+ T cells play a vital role, and CD8+ T cells are also essential due to their ability to recognize intracellular infection. Antigen presentation by major histocompatibility complex (MHC) class I molecules enable CD8+ T cells to recognize intracellular infection.
MHC class I molecules consist of two polypeptide chains, α- and β2-microglobulin, whose assembly depends on multiprotein peptide loading complex (PLC), which contains antigen peptide transporter (TAP), TAP binding protein isoform 1 (TAPBP), ERp57, calreticulin, and calnexin.54 In our study, TAP1, TAP2, and TAPBP were shown to be decreased by 61%, 64%, and 66% in the cytosol, respectively. TAP is an ATP-binding transporter involved in translocation of peptides from the cytosol into the endoplasmic reticulum and is involved in the final stage of MHC class I folding.55 TAPBP facilitates loading of high-affinity peptides onto MHC class I molecules. Subsequently, MHC class I molecules are separated from the PLC and transported to the cell surface for antigen presentation. MHC class I ligands are mainly modified by proteasomes. The proteasome activator, PA28, which can be induced by IFN-γ,56 has been implicated in the regulation of MHC class I antigen processing.57,58 In our work, PA28 was downregulated by 22%. The chaperone heat shock protein 90 (Hsp90) is one of the most abundant proteins in eukaryotic cells. Hsp90 can bind proteins through N- and COOH-terminal domains to refold and play an important role in maintenance of functional integrity of some fragile proteins.58 Some evidence suggests that Hsp90 could bind to MHC class I ligands or their precursors, and accelerate the processing of C-terminal flanking regions of MHC class I ligands.58,59 Our results indicated that Hsp90b was decreased by 95% in the cytosol, but HSP90a was not changed.
On the basis of the above observations, we hypothesized that the PPE38 protein of M. marinum might arrest MHC class I processing and presentation in macrophages, and then would allow the bacteria to escape the immune response mediated by CD8+ T cells (Figure 4 in the SI).
PPE38 Induces a Complex Regulatory Cross-Talk Network Involving Multiple Transcriptional Factors (TFs)
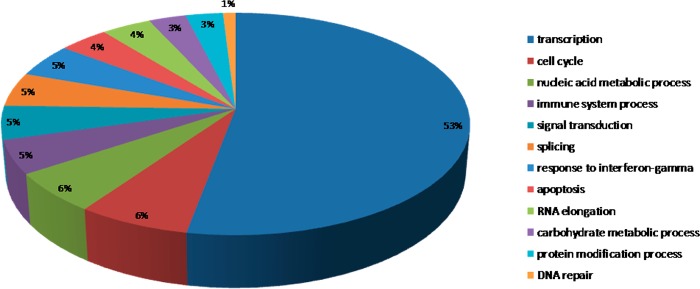
With the use of PANTHER, 102 TFs and 117 TF-associated proteins were identified and quantified in the nuclear fraction. These proteins were then clustered on the basis of their biological processes according to the Gene Ontology database (http://www.geneontology.org). The identified TFs were mainly involved in transcription; cell cycle; nucleic acid metabolic processes; immune system processes; signal transduction; splicing; response to interferon-γ; apoptosis; RNA elongation; carbohydrate metabolic processes; protein modification processes; and DNA repair (Figure 5). The TF-associated proteins mainly participated in nucleic acid metabolic processes; transcription; splicing; cell proliferation and differentiation; rRNA, DNA, and tRNA metabolic processes; DNA repair; transport; mRNA processing; cell cycle; immune system processes; signal transduction; lipid metabolic processes; and protein targeting (Figure 5 in the SI). On the basis of our quantitative proteomic data for network analysis using STRING, we generated a global functional map among these TFs and their associated proteins, which illustrates how the TF-regulatory network operates in conjunction with upstream signal cascades to generate the response following PPE38 induction (Figure 6 in the SI). On the basis of the global functional map, we found that some subnetworks connected to a variety of biological processes (Figure 6).
Figure 5.
Functional analysis of transcriptional factors (TFs). The TFs were submitted to PANTHER (http://www.pantherdb.org/) to identify related functional processes and corresponding percentages.
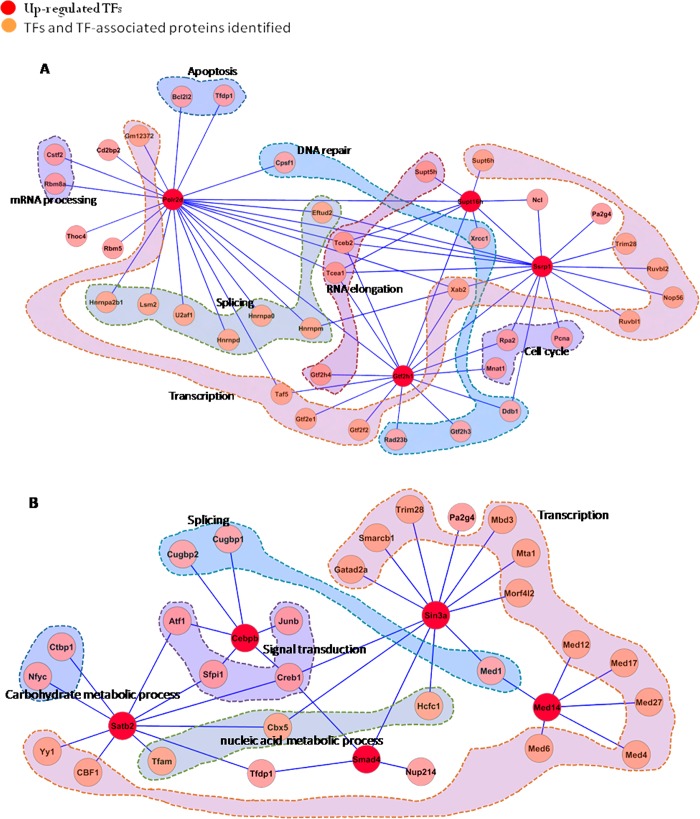
Figure 6.
Subnetworks identified by data-dependent network analysis and involved in different biological processes. The previously known functions of the proteins in the network were based on PANTHER (http://www.pantherdb.org/). The “zoom-in” maps of these subnetworks extracted from the global regulatory network (Figure 3 in the SI) are given as follows: (A) the subnetwork involving Polr2d, Supt16h, Gtf2h1, and Ssrp1; and (B) the subnetwork involving Satb2, C/EBP-β, Sin3a, Smad4, and MED14.
FACT (facilitates chromatin transcription) complex subunit SPT16 (SUPT16H), which is a general chromatin factor that functions to reorganize nucleosomes, is a component of the FACT complex. This protein was upregulated by 1.29-fold in the nucleus. The FACT complex is involved in multiple processes, such as mRNA elongation, DNA replication, and DNA repair. Nucleosome binding activity of the FACT complex is regulated by poly(ADP-ribosyl)ation. The level of SUPT16H poly(ADP-ribosyl)ation has been found to coincide with the activation of poly(ADP-ribose)polymerase 1 (PARP1) and to play a role in chromatin remodeling.60 In the nucleus, we also found that PARP1 was upregulated by 1.27-fold, which was consistent with previously published results.60 Structure specific recognition protein 1 (SSRP1) is also a component of the FACT complex and can form a heterodimer with SUPT16H. SSRP1 has SPT16-dependent and -independent roles in the regulation of gene transcription.61 SUPT16H, SSRP1, and CK2 (casein kinase 2) can form a complex that is likely involved in phosphorylation of p53 at Ser-392.62 Our results indicated that SSRP1 was increased by 1.29-fold, suggesting that our MS results were reliable. PPE38 stimulation led to a 1.37-fold and 1.21-fold increase in the abundance of polymerase (RNA) II (DNA directed) polypeptide D (Polr2d) and general transcription factor IIH subunit 1 (GTF2H1), respectively. Polr2d is a component of RNA polymerase II, which plays a role in mRNA processing and production of noncoding RNAs. GTF2H1 is important to the architecture and function of TFIIH,63 which plays an important role in the nucleotide excision repair pathway.
SIN3 transcription regulator homologue A (Sin3a), a transcriptional repressor, was upregulated by 1.21-fold. Sin3a has been found to repress STAT3 activity by modifying its acetylation status.64 In the cytosol, STAT3 was downregulated by 51%. Mothers against decapentaplegic homologue 4 (SMAD4) was upregulated by 1.43-fold. This protein is a member of the SMAD family of signal transduction factors, recognizes an 8-bp palindromic sequence (GTCTAGAC), and is involved in TGF-β signaling. It is able to heterodimerize with phosphorylated Smad2 or Smad3, subsequently translocate to the nucleus, and regulate transcription. CCAAT/enhancer-binding protein beta (C/EBP-β), a bZIP TF, was increased by 1.55-fold. This protein is important for the regulation of inflammatory responses genes and can regulate the expression of IL-665 and TNF-α.66 Mediator of RNA polymerase II transcription subunit 14 (MED14) was upregulated by 1.23-fold. This protein is a coactivator involved in the regulated transcription of RNA polymerase II-dependent genes and required for activity of the enhancer-binding protein Sp1. Special AT-rich sequence-binding protein 2 (SATB2) was increased by 3.09-fold. This protein is a nuclear matrix attachment region-binding protein and functions as a TF controlling nuclear gene expression. SATB2 was found to play an important role in resisting oxidative stress-induced apoptosis.67
Transcriptional regulation is coordinately controlled by multiple TFs and TF-associated proteins and plays a central role in the inflammatory response. The global functional map generated from our quantitative proteomic data-dependent bioinformatics analysis provided direct evidence about how these TFs regulate biological processes related to PPE38 challenge.
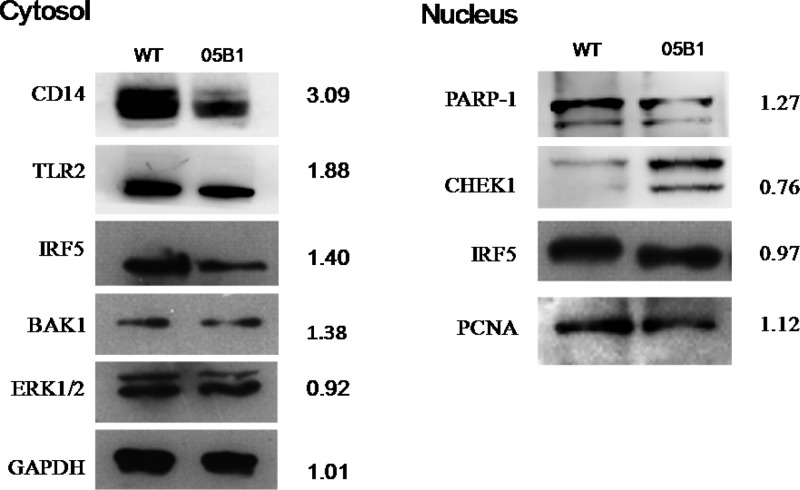
Immunoblotting Validation of Quantitative Proteomics Data
To verify the reliability of our MS results, we used Western blot analysis to confirm the observed changes in some key proteins (Figure 7). In the cytosol, we selected CD14, TLR2, IRF5, ERK1/2, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). As shown in Figure 6, we found that the expression of ERK1/2 was not changed and that expression levels of CD14, TLR2, and IRF5 were the same as those from the MS results.
Figure 7.
Validated proteins identified in our MS analysis via Western blot analysis. Following a 4-h infection, the nuclear and cytosolic fractions were separated by SDS-PAGE and submitted to immunoblotting. GAPDH and PCNA were used as the reference for the proteins in the cytoplasmic and nuclear fractions, respectively. The H/L ratio refers to the protein expression changes (heavy/light) between WT-stimulated and 05B1-stimulated macrophages, respectively.
In the nucleus, we selected PARP1, serine/threonine-protein kinase 1 (CHEK1), IRF5, and proliferating cell nuclear antigen (PCNA) for further validation. PARP1 is a nuclear enzyme activated by DNA strand breaks and plays a key role in repairing DNA damage.68 Overactivation of PARP1 potentially leads to NAD+ reduction, ATP depletion, cellular energy failure, and release of apoptosis-inducing factor (AIF) from the mitochondria, all of which result in massive DNA damage and necrotic cell death.69 CHEK1 is a kinase that phosphorylates cdc25 and plays an important role in cell-cycle control. Studies have reported that inhibition of CHEK1 enhances the cytotoxicity of DNA-damaging agents.70,71 The results of our Western blot analyses were consistent with those obtained via MS.
Conclusion
In previous work, PPE38 has been implicated in the virulence and immune response of M. marinum, but the function of PPE38 was not clear. We used AACT/SILAC to study the function of PPE38 of M. marinum. On the basis of our quantitative data, we hypothesized that the PPE38 protein of M. marinum could combine with the cell surface receptor, TLR2, and induce downstream signaling pathways related to inflammatory factors. Furthermore, PPE38 of M. marinum might arrest MHC class I processing and presentation. Finally, we generated an interaction network of TFs and TF-associated proteins, generating system-wide insights into various associated biological processes.
Acknowledgments
We thank E. J. Rubin, Harvard University, for providing the MycoMarT7 mariner transposon phage; Jun Ren for helpful discussions; Chen Niu and Lu Meng for comments on the manuscript. This work was supported by the National Natural Science Foundation of China Grants 91231115 and 81271790 (to Q.G.) and U.S. NIH R01AI064806 and 863 High Technology Funding of China (Grant 2006AA02A310) (to X.C.).
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Ocampo M.; Rodriguez D. M.; Curtidor H.; Vanegas M.; Patarroyo M. A.; Patarroyo M. E. Peptides derived from Mycobacterium tuberculosis Rv2301 protein are involved in invasion to human epithelial cells and macrophages. Amino Acids 2012, 4262067–2077. [DOI] [PubMed] [Google Scholar]
- Devitt A.; Moffatt O. D.; Raykundalia C.; Capra J. D.; Simmons D. L.; Gregory C. D. Human CD14 mediates recognition and phagocytosis of apoptotic cells. Nature 1998, 3926675505–509. [DOI] [PubMed] [Google Scholar]
- Schnappinger D.; Ehrt S.; Voskuil M. I.; Liu Y.; Mangan J. A.; Monahan I. M.; Dolganov G.; Efron B.; Butcher P. D.; Nathan C.; Schoolnik G. K. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 2003, 1985693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T.; Brosch R.; Parkhill J.; Garnier T.; Churcher C.; Harris D.; Gordon S. V.; Eiglmeier K.; Gas S.; Barry C. R.; Tekaia F.; Badcock K.; Basham D.; Brown D.; Chillingworth T.; Connor R.; Davies R.; Devlin K.; Feltwell T.; Gentles S.; Hamlin N.; Holroyd S.; Hornsby T.; Jagels K.; Krogh A.; McLean J.; Moule S.; Murphy L.; Oliver K.; Osborne J.; Quail M. A.; Rajandream M. A.; Rogers J.; Rutter S.; Seeger K.; Skelton J.; Squares R.; Squares S.; Sulston J. E.; Taylor K.; Whitehead S.; Barrell B. G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 3936685537–544. [DOI] [PubMed] [Google Scholar]
- McEvoy C. R.; van Helden P. D.; Warren R. M.; Gey V. P. N. Evidence for a rapid rate of molecular evolution at the hypervariable and immunogenic Mycobacterium tuberculosis PPE38 gene region. BMC Evol. Biol. 2009, 9, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karboul A.; Mazza A.; Gey V. P. N.; Ho J. L.; Brousseau R.; Mardassi H. Frequent homologous recombination events in Mycobacterium tuberculosis PE/PPE multigene families: Potential role in antigenic variability. J. Bacteriol. 2008, 190237838–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottai D.; Brosch R. Mycobacterial PE, PPE and ESX clusters: novel insights into the secretion of these most unusual protein families. Mol. Microbiol. 2009, 733325–328. [DOI] [PubMed] [Google Scholar]
- Stinear T. P.; Seemann T.; Harrison P. F.; Jenkin G. A.; Davies J. K.; Johnson P. D.; Abdellah Z.; Arrowsmith C.; Chillingworth T.; Churcher C.; Clarke K.; Cronin A.; Davis P.; Goodhead I.; Holroyd N.; Jagels K.; Lord A.; Moule S.; Mungall K.; Norbertczak H.; Quail M. A.; Rabbinowitsch E.; Walker D.; White B.; Whitehead S.; Small P. L.; Brosch R.; Ramakrishnan L.; Fischbach M. A.; Parkhill J.; Cole S. T. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008, 185729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin D. M.; Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 2008, 1051027–1039. [DOI] [PubMed] [Google Scholar]
- Stamm L. M.; Brown E. J. Mycobacterium marinum: The generalization and specialization of a pathogenic mycobacterium. Microbes Infect. 2004, 6151418–1428. [DOI] [PubMed] [Google Scholar]
- Mehta P. K.; Pandey A. K.; Subbian S.; El-Etr S. H.; Cirillo S. L.; Samrakandi M. M.; Cirillo J. D. Identification of Mycobacterium marinum macrophage infection mutants. Microb. Pathog. 2006, 404139–151. [DOI] [PubMed] [Google Scholar]
- Abdallah A. M.; Savage N. D.; van Zon M.; Wilson L.; Vandenbroucke-Grauls C. M.; van der Wel N. N.; Ottenhoff T. H.; Bitter W. The ESX-5 secretion system of Mycobacterium marinum modulates the macrophage response. J. Immunol. 2008, 181107166–7175. [DOI] [PubMed] [Google Scholar]
- Daim S.; Kawamura I.; Tsuchiya K.; Hara H.; Kurenuma T.; Shen Y.; Dewamitta S. R.; Sakai S.; Nomura T.; Qu H.; Mitsuyama M. Expression of the Mycobacterium tuberculosis PPE37 protein in Mycobacterium smegmatis induces low tumour necrosis factor alpha and interleukin 6 production in murine macrophages. J. Med. Microbiol. 2011, 60Pt 5582–591. [DOI] [PubMed] [Google Scholar]
- Bertholet S.; Ireton G. C.; Kahn M.; Guderian J.; Mohamath R.; Stride N.; Laughlin E. M.; Baldwin S. L.; Vedvick T. S.; Coler R. N.; Reed S. G. Identification of human T cell antigens for the development of vaccines against Mycobacterium tuberculosis. J. Immunol. 2008, 181117948–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M.; Rindi L.; Korf H.; Bonanni D.; Adnet P. Y.; Jurion F.; Garzelli C.; Huygen K. Immunogenicity and protective efficacy of tuberculosis subunit vaccines expressing PPE44 (Rv2770c). Vaccine 2008, 26486053–6063. [DOI] [PubMed] [Google Scholar]
- Wang J.; Qie Y.; Zhu B.; Zhang H.; Xu Y.; Wang Q.; Chen J.; Liu W.; Wang H. Evaluation of a recombinant BCG expressing antigen Ag85B and PPE protein Rv3425 from DNA segment RD11 of Mycobacterium tuberculosis in C57BL/6 mice. Med. Microbiol. Immunol. 2009, 19815–11. [DOI] [PubMed] [Google Scholar]
- Kruh N. A.; Troudt J.; Izzo A.; Prenni J.; Dobos K. M. Portrait of a pathogen: The Mycobacterium tuberculosis proteome in vivo. PLoS One 2010, 511e13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D.; Wang D.; Li M.; Wang H.; Yu J.; Wang C.; Liu J.; Gao Q. PPE38 modulates the innate immune response and is required for Mycobacterium marinum virulence. Infect. Immun. 2012, 80143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S. E.; Blagoev B.; Kratchmarova I.; Kristensen D. B.; Steen H.; Pandey A.; Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 2002, 15376–386. [DOI] [PubMed] [Google Scholar]
- Zhu H.; Pan S.; Gu S.; Bradbury E. M.; Chen X. Amino acid residue specific stable isotope labeling for quantitative proteomics. Rapid Commun. Mass Spectrom. 2002, 16222115–2123. [DOI] [PubMed] [Google Scholar]
- Chen X.; Smith L. M.; Bradbury E. M. Site-specific mass tagging with stable isotopes in proteins for accurate and efficient protein identification. Anal. Chem. 2000, 7261134–1143. [DOI] [PubMed] [Google Scholar]
- Ibarrola N.; Kalume D. E.; Gronborg M.; Iwahori A.; Pandey A. A proteomic approach for quantitation of phosphorylation using stable isotope labeling in cell culture. Anal. Chem. 2003, 75226043–6049. [DOI] [PubMed] [Google Scholar]
- Du R.; Long J.; Yao J.; Dong Y.; Yang X.; Tang S.; Zuo S.; He Y.; Chen X. Subcellular quantitative proteomics reveals multiple pathway cross-talk that coordinates specific signaling and transcriptional regulation for the early host response to LPS. J. Proteome Res. 2010, 941805–1821. [DOI] [PubMed] [Google Scholar]
- Shevchenko A.; Tomas H.; Havlis J.; Olsen J. V.; Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 162856–2860. [DOI] [PubMed] [Google Scholar]
- Pattin K. A.; Moore J. H. Exploiting the proteome to improve the genome-wide genetic analysis of epistasis in common human diseases. Hum. Genet. 2008, 124119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooncer E.; Brouard N.; Nilsson S. K.; Williams B.; Liu M. C.; Unwin R. D.; Blinco D.; Jaworska E.; Simmons P. J.; Whetton A. D. Developmental fate determination and marker discovery in hematopoietic stem cell biology using proteomic fingerprinting. Mol. Cell. Proteomics 2008, 73573–581. [DOI] [PubMed] [Google Scholar]
- Xue Y.; Yun D.; Esmon A.; Zou P.; Zuo S.; Yu Y.; He F.; Yang P.; Chen X. Proteomic dissection of agonist-specific TLR-mediated inflammatory responses on macrophages at subcellular resolution. J. Proteome Res. 2008, 783180–3193. [DOI] [PubMed] [Google Scholar]
- Gan H.; He X.; Duan L.; Mirabile-Levens E.; Kornfeld H.; Remold H. G. , Enhancement of antimycobacterial activity of macrophages by stabilization of inner mitochondrial membrane potential. J. Infect. Dis. 2005, 19181292–1300. [DOI] [PubMed] [Google Scholar]
- Abarca-Rojano E.; Rosas-Medina P.; Zamudio-Cortez P.; Mondragon-Flores R.; Sanchez-Garcia F. J. Mycobacterium tuberculosis virulence correlates with mitochondrial cytochrome c release in infected macrophages. Scand. J. Immunol. 2003, 584419–427. [DOI] [PubMed] [Google Scholar]
- Balaji K. N.; Goyal G.; Narayana Y.; Srinivas M.; Chaturvedi R.; Mohammad S. Apoptosis triggered by Rv1818c, a PE family gene from Mycobacterium tuberculosis is regulated by mitochondrial intermediates in T cells. Microbes Infect. 2007, 93271–281. [DOI] [PubMed] [Google Scholar]
- Basu S.; Pathak S. K.; Banerjee A.; Pathak S.; Bhattacharyya A.; Yang Z.; Talarico S.; Kundu M.; Basu J. Execution of macrophage apoptosis by PE_PGRS33 of Mycobacterium tuberculosis is mediated by Toll-like receptor 2-dependent release of tumor necrosis factor-alpha. J. Biol. Chem. 2007, 28221039–1050. [DOI] [PubMed] [Google Scholar]
- Nair S.; Ramaswamy P. A.; Ghosh S.; Joshi D. C.; Pathak N.; Siddiqui I.; Sharma P.; Hasnain S. E.; Mande S. C.; Mukhopadhyay S. The PPE18 of Mycobacterium tuberculosis interacts with TLR2 and activates IL-10 induction in macrophage. J. Immunol. 2009, 183106269–6281. [DOI] [PubMed] [Google Scholar]
- Bansal K.; Elluru S. R.; Narayana Y.; Chaturvedi R.; Patil S. A.; Kaveri S. V.; Bayry J.; Balaji K. N. PE_PGRS antigens of Mycobacterium tuberculosis induce maturation and activation of human dendritic cells. J Immunol 2010, 18473495–504. [DOI] [PubMed] [Google Scholar]
- Kawai T.; Akira S. TLR signaling. Cell Death Differ. 2006, 135816–25. [DOI] [PubMed] [Google Scholar]
- Perkins N. D. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol. 2007, 8149–62. [DOI] [PubMed] [Google Scholar]
- Hiscott J.; Pitha P.; Genin P.; Nguyen H.; Heylbroeck C.; Mamane Y.; Algarte M.; Lin R. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 1999, 1911–13. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S.; Negishi H.; Asagiri M.; Nakajima C.; Mizutani T.; Takaoka A.; Honda K.; Taniguchi T. Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem. Biophys. Res. Commun. 2003, 3064860–866. [DOI] [PubMed] [Google Scholar]
- Zhao X. J.; Dong Q.; Bindas J.; Piganelli J. D.; Magill A.; Reiser J.; Kolls J. K. TRIF and IRF-3 binding to the TNF promoter results in macrophage TNF dysregulation and steatosis induced by chronic ethanol. J. Immunol. 2008, 18153049–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K.; Taniguchi T. Toll-like receptor signaling and IRF transcription factors. IUBMB Life 2006, 585–6290–295. [DOI] [PubMed] [Google Scholar]
- Takaoka A.; Yanai H.; Kondo S.; Duncan G.; Negishi H.; Mizutani T.; Kano S.; Honda K.; Ohba Y.; Mak T. W.; Taniguchi T. Integral role of IRF-5 in the gene induction programme activated by toll-like receptors. Nature 2005, 4347030243–249. [DOI] [PubMed] [Google Scholar]
- Chen W.; Lam S. S.; Srinath H.; Jiang Z.; Correia J. J.; Schiffer C. A.; Fitzgerald K. A.; Lin K.; Royer W. J. Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat. Struct. Mol. Biol. 2008, 15111213–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esensten J. H.; Tsytsykova A. V.; Lopez-Rodriguez C.; Ligeiro F. A.; Rao A.; Goldfeld A. E. NFAT5 binds to the TNF promoter distinctly from NFATp, c, 3 and 4, and activates TNF transcription during hypertonic stress alone. Nucleic Acids Res. 2005, 33123845–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rodriguez C.; Aramburu J.; Jin L.; Rakeman A. S.; Michino M.; Rao A. Bridging the NFAT and NF-kappaB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity 2001, 15147–58. [DOI] [PubMed] [Google Scholar]
- Raman M.; Chen W.; Cobb M. H. Differential regulation and properties of MAPKs. Oncogene 2007, 26223100–3112. [DOI] [PubMed] [Google Scholar]
- Shaul Y. D.; Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim. Biophys. Acta 2007, 177381213–1226. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Liu H. T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 1219–18. [DOI] [PubMed] [Google Scholar]
- Shi C. S.; Huang N. N.; Harrison K.; Han S. B.; Kehrl J. H. The mitogen-activated protein kinase kinase kinase kinase GCKR positively regulates canonical and noncanonical Wnt signaling in B lymphocytes. Mol. Cell. Biol. 2006, 26176511–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett T. D.; Brautigan D. L. Cytokine activation of p38 mitogen-activated protein kinase and apoptosis is opposed by alpha-4 targeting of protein phosphatase 2A for site-specific dephosphorylation of MEK3. Mol. Cell. Biol. 2007, 27124217–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abnous K.; Dieni C. A.; Storey K. B. Suppression of MAPKAPK2 during mammalian hibernation. Cryobiology 2012, 653235–241. [DOI] [PubMed] [Google Scholar]
- Fruman D. A.; Meyers R. E.; Cantley L. C. Phosphoinositide kinases. Annu. Rev. Biochem. 1998, 67, 481–507. [DOI] [PubMed] [Google Scholar]
- Yang C. S.; Lee J. S.; Jung S. B.; Oh J. H.; Song C. H.; Kim H. J.; Park J. K.; Paik T. H.; Jo E. K. Differential regulation of interleukin-12 and tumour necrosis factor-alpha by phosphatidylinositol 3-kinase and ERK 1/2 pathways during Mycobacterium tuberculosis infection. Clin. Exp. Immunol. 2006, 1431150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcommenne M.; Tan C.; Gray V.; Rue L.; Woodgett J.; Dedhar S. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc. Natl. Acad. Sci. U.S.A. 1998, 951911211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff D. E.; Kline L.; Soldau K.; Lee J. D.; Pugin J.; Tobias P. S.; Ulevitch R. J. Phagocytosis of Gram-negative bacteria by a unique CD14-dependent mechanism. J. Leukocyte Biol. 1997, 626786–794. [DOI] [PubMed] [Google Scholar]
- Wearsch P. A.; Cresswell P. The quality control of MHC class I peptide loading. Curr. Opin. Cell Biol. 2008, 206624–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abele R.; Tampe R. The TAP translocation machinery in adaptive immunity and viral escape mechanisms. Essays Biochem. 2011, 501249–264. [DOI] [PubMed] [Google Scholar]
- Tanaka K.; Kasahara M. The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-gamma-inducible proteasome activator PA28. Immunol. Rev. 1998, 163, 161–176. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M.; Realini C.; Ustrell V. The proteasome activator 11 S REG (PA28) and class I antigen presentation. Biochem. J. 2000, 345Pt 11–15. [PMC free article] [PubMed] [Google Scholar]
- Yamano T.; Murata S.; Shimbara N.; Tanaka N.; Chiba T.; Tanaka K.; Yui K.; Udono H. Two distinct pathways mediated by PA28 and Hsp90 in major histocompatibility complex class I antigen processing. J. Exp. Med. 2002, 1962185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T.; Udono H.; Yamano T.; Ohta H.; Uenaka A.; Ono T.; Hizuta A.; Tanaka N.; Srivastava P. K.; Nakayama E. Isolation of MHC class I-restricted tumor antigen peptide and its precursors associated with heat shock proteins hsp70, hsp90, and gp96. J. Immunol. 1999, 16231303–1309. [PubMed] [Google Scholar]
- Huang J. Y.; Chen W. H.; Chang Y. L.; Wang H. T.; Chuang W. T.; Lee S. C. Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res. 2006, 3482398–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Zeng S. X.; Landais I.; Lu H. Human SSRP1 has Spt16-dependent and -independent roles in gene transcription. J. Biol. Chem. 2007, 282106936–6945. [DOI] [PubMed] [Google Scholar]
- Keller D. M.; Zeng X.; Wang Y.; Zhang Q. H.; Kapoor M.; Shu H.; Goodman R.; Lozano G.; Zhao Y.; Lu H. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 2001, 72283–292. [DOI] [PubMed] [Google Scholar]
- Gervais V.; Lamour V.; Jawhari A.; Frindel F.; Wasielewski E.; Dubaele S.; Egly J. M.; Thierry J. C.; Kieffer B.; Poterszman A. TFIIH contains a PH domain involved in DNA nucleotide excision repair. Nat. Struct. Mol. Biol. 2004, 117616–622. [DOI] [PubMed] [Google Scholar]
- Icardi L.; Mori R.; Gesellchen V.; Eyckerman S.; De Cauwer L.; Verhelst J.; Vercauteren K.; Saelens X.; Meuleman P.; Leroux-Roels G.; De Bosscher K.; Boutros M.; Tavernier J. The Sin3a repressor complex is a master regulator of STAT transcriptional activity. Proc. Natl. Acad. Sci. U.S.A. 2012, 1093012058–12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsuka S.; Akira S.; Nishio Y.; Hashimoto S.; Sugita T.; Isshiki H.; Kishimoto T. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood 1992, 792460–466. [PubMed] [Google Scholar]
- Liu H.; Sidiropoulos P.; Song G.; Pagliari L. J.; Birrer M. J.; Stein B.; Anrather J.; Pope R. M. TNF-alpha gene expression in macrophages: regulation by NF-kappa B is independent of c-Jun or C/EBP beta. J. Immunol. 2000, 16484277–4285. [DOI] [PubMed] [Google Scholar]
- Wei J. D.; Lin Y. L.; Tsai C. H.; Shieh H. S.; Lin P. I.; Ho W. P.; Chen R. M. SATB2 participates in regulation of menadione-induced apoptotic insults to osteoblasts. J. Orthop. Res. 2012, 3071058–1066. [DOI] [PubMed] [Google Scholar]
- Godon C.; Cordelieres F. P.; Biard D.; Giocanti N.; Megnin-Chanet F.; Hall J.; Favaudon V. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008, 36134454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G.; Xing J.; Xiao X.; Liou A. K.; Gao Y.; Yin X. M.; Clark R. S.; Graham S. H.; Chen J. Critical role of calpain I in mitochondrial release of apoptosis-inducing factor in ischemic neuronal injury. J. Neurosci. 2007, 27359278–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z.; Chen Z.; Gunasekera A. H.; Sowin T. J.; Rosenberg S. H.; Fesik S.; Zhang H. Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J. Biol. Chem. 2003, 2782421767–21773. [DOI] [PubMed] [Google Scholar]
- Maude S. L.; Enders G. H. Cdk inhibition in human cells compromises chk1 function and activates a DNA damage response. Cancer Res. 2005, 653780–786. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.