Abstract
Peroxisome proliferator-activated receptor α (PPARα) and PPARγ coactivator 1α (PGC-1α) are crucial transcriptional regulators for genes involved in FA oxidation. Lipin-1 is essential for this increased capacity for β-oxidation in fasted livers, and it is also a phosphatidate phosphatase involved in triacylglycerol and phospholipid synthesis. Little is known about the regulation of these proteins in the heart during fasting, where there is increased FA esterification and oxidation. Lipin-1, lipin-2, lipin-3, carnitine palmitoyltransferase-1b (Cpt1b), and PGC-1α-b mRNA were increased by glucocorticoids and cAMP in neonatal rat cardiomyocytes. However, Cpt1b upregulation was caused by increased PPARα activation rather than expression. By contrast, the effects of PPARα in fasted livers are mediated through increased expression. During fasting, the expressions of PGC-1α-b and PGC-1α-c are increased in mouse hearts, and this is explained by increased cAMP-dependent signaling. By contrast, PGC-1α-a expression is increased in liver. Contrary to our expectations, lipin-1 expression was decreased and lipin-2 remained unchanged in hearts compared with increases in fasted livers. Our results identify novel differences in the regulation of lipins, PPARα, and PGC-1α splice variants during fasting in heart versus liver, even though the ultimate outcome in both tissues is to increase FA turnover and oxidation.
Keywords: phosphatidate phosphatase, fasting, glycerolipid, hepatic and cardiac gene regulation
Peroxisome proliferator-activated receptor α (PPARα) acts as a master transcriptional regulator to induce its target genes involved in FA uptake and oxidation (1–4). Its importance in regulating FA metabolism is underlined by the metabolic derangements in mouse models in which PPARα expression has been modified (1, 2, 5). The regulation of PPARα target genes is also dependent on PPARγ coactivator 1α (PGC-1α), which binds to PPARα in a transcriptional regulatory complex (6). In addition to FA oxidation, PGC-1α is essential in the regulation of mitochondrial biogenesis and hepatic gluconeogenesis (7–9). There are three splice variants of PGC-1α in mice, which are classified as PGC-1α-a, PGC-1α-b and PGC-1α-c (10). The PGC-1α-b and PGC-1α-c proteins are shorter by four and thirteen amino acids at the N-terminus, respectively, but are functionally similar to full-length PGC-1α-a (10). PGC-1α-a is important in liver, where its expression is induced by fasting (10). By contrast, PGC-1α-b and PGC-1α-c are more important for metabolic regulation during exercise in muscle (10).
The lipins are cytosolic, multifunctional proteins involved in the regulation of glycerolipid synthesis, FA oxidation, signal transduction, and transcriptional regulation of adipogenic, lipogenic, and inflammatory processes (11–13). There are three members in the lipin family, namely lipin-1, lipin-2, and lipin-3 (11). Lipin-1 is a crucial transcriptional coactivator, which acts in concert with PPARα and PGC-1α to induce FA oxidation genes in the liver. Lipin-2 can also coactivate PPARα (14), and lipin-3 presumably acts in a similar manner owing to the conserved transcriptional coactivator motif (LxxIL). All the lipin isoforms are phosphatidate phosphatases (PAPs), responsible for converting phosphatidate (PA) to diacylglycerol (DG) in the Kennedy pathway of glycerolipid synthesis (15–19). This depends on a conserved catalytic DxDxT motif and a characteristic secondary structure sequence (13, 14, 20, 21). To participate in glycerolipid synthesis, the lipins need to translocate from the cytosol onto endoplasmic reticulum membranes where the other enzymes in the pathway are mainly located (22–25). This translocation of lipins to membranes can also regulate signaling processes involved in cell survival and insulin signaling by controlling the conversion of PA to DG, which are both important bioactive molecules (11, 13).
Because the lipins, PGC-1α and PPARα are important in regulating lipid metabolism, it is not surprising that numerous studies have demonstrated the dynamic regulation of their expressions in the liver (6, 9, 17, 26). Previous work by our group established that lipin-1 and PAP activity are synergistically induced in hepatocytes by the combination of glucocorticoid and cAMP signaling, whereas insulin antagonizes this upregulation (26, 29). The gene expressions of PPARα and PGC-1α in liver are also increased by glucocorticoid and cAMP signaling (26). Finally, the mRNA levels of the genes encoding lipin-1, PPARα. and PGC-1α are all increased in the liver after fasting, which demonstrates the physiological roles of cAMP and glucocorticoids (6, 8, 26). Therefore, it could be expected that similar mechanisms of regulation could operate in the heart, inasmuch as FAs are the major substrates used to fuel cardiac energy requirements (30). Moreover, the turnover of cardiac triacylglycerol (TG) stores contributes FA to fuel ATP production as well as providing ligands that activate PPARα, which promotes the upregulation of genes involved in FA oxidation (30–33). During fasting, the heart receives an increased FA supply through increased adipose tissue lipolysis and hepatic VLDL production. This is coupled with decreased glucose availability, resulting in increased FA oxidation, which provides up to 70% of ATP production in the heart (30).
We hypothesized that the dynamic control of fuel utilization in fasted hearts would be facilitated by increased cardiac expressions of the lipin isoforms, PGC-1α splice variants, and PPARα through increased glucocorticoid and cAMP action, similar to the regulation in the liver. Indeed, we found that the mRNA levels of the PGC-1α splice variants PGC-1α-b and PGC-1α-c are increased by fasting in mouse hearts, which can be explained by cAMP signaling. However, these splice variants are virtually undetectable in the liver. By contrast, PGC-1α-a is the splice variant induced in fasted livers. We also show that PPARα activation in the heart rather than its expression is needed for the increased expression of carnitine palmitoyltransferase-1b (Cpt1b) mRNA during fasting. This is significantly different from the regulation of PPARα and its target genes in liver. Finally, Lpin1 mRNA was decreased and Lpin2 mRNA was unchanged in fasted mouse hearts. The results contrasted with the increases in Lpin1 and Lpin2 gene expressions in fasted livers from our own experiments and previous studies (17, 26). We attribute these differences to the relative absence of glucocorticoid receptors in adult heart compared with liver (34, 35). The present studies increase our understanding of how these important regulators of FA metabolism are differentially controlled in heart versus liver during fasting, even though increased FA utilization and oxidation occur in both organs.
MATERIALS AND METHODS
Reagents
DNase I, trypsin, and collagenase were obtained from Worthington Biochemical Corporation (Lakewood, NJ). Dexamethasone, insulin, CPT-cAMP [8-(4-chlorophenylthio)-2′-O-methyladenosine 3′,5′-cyclic monophosphate], fenofibric acid, pertussis toxin, and oleic acid were purchased from Sigma-Aldrich (St. Louis, MO). Clenbuterol was obtained from Toronto Research Chemicals (Toronto, ON). GW 6471 and Torin1 came from Tocris Bioscience (Bristol, England). Anti- GAPDH and anti-calnexin antibodies were from Sigma-Aldrich and Enzo Life Sciences (Farmingdale, NY), respectively. [1,3-3H]glycerol was acquired from Amersham (Buckinghamshire, England) and [1-14C]oleate was purchased from Perkin-Elmer (Waltham, MA).
Preparation of primary neonatal rat ventricular myocytes
Isolation of neonatal rat ventricular myocytes (NRVMs) was performed as previously described (36). Briefly, ventricles from 2 day-old Sprague-Dawley rat pups were minced and digested for 20 min at 37°C on a rotary shaker with 25,000 U DNase I, 2,250 U trypsin and 5,000 U collagenase in 19.5 ml PBS. After digestion, 20 ml Dulbecco's Modified Eagle's/Nutrient Mixture Ham's F-12 (DME/F12) medium (Sigma-Aldrich) containing 20% FBS, 1% penicillin/streptomycin, and 50 µg/ml gentamicin (denoted as DF20 medium) was added and the solution was centrifuged at 130 × g for 2 min. The supernatant was discarded and the pellet was further digested at 37°C for 20 min. DF20 medium was again added and the mixture was centrifuged as before. The pellet was digested one last time using the previous conditions. Following a final centrifugation at 650 × g for 7 min, the pellet was resuspended in plating media (DME/F12 medium containing 5% FBS, 10% horse serum, 1% penicillin/streptomycin and 50 µg/ml gentamicin). The cells were strained through a 100 µm cell strainer (BD Biosciences, Franklin Lakes, NJ) and incubated for 90 min at 37°C in a CO2 incubator. Cardiac fibroblasts and endothelial cells readily attached to the culture flask surface and the supernatant containing the myocytes was isolated and centrifuged at 300 × g for 2 min. The cells were resuspended in plating media and cells were counted after diluting with trypan blue. Cell viability was usually greater than 90% as determined by Trypan blue exclusion. Cells were plated on 35 mm Primaria-coated dishes (BD Biosciences) at a density of 1.8 million cells per dish in DME/F2 medium containing 10% FBS and 100 µM cytosine β-D-arabinofuranoside (ara-C), which inhibits DNA replication and prevents fibroblast growth.
Adenoviral infection of cultured cells
Recombinant adenovirus overexpressing N-terminal HA-tagged (hemagglutinin epitope) Mus musculus lipin-1B wild-type (Adlipin1b) and lipin-2 (Adlipin2) as well as adenovirus expressing shRNA against the genes encoding β-galactosidase (which was used as the vector control) (AdshRNA LacZ) or lipin-1 (AdshRNA Lipin1) were prepared as described previously (6, 24). NRVMs were infected with adenovirus in 10% FBS-DME/F12 medium for 38 h to obtain optimum changes in the expressions of the lipins.
Quantitative real-time PCR
RNA was isolated using the RNAqueous kit (Life Technologies, Carlsbad, CA), as specified in the manufacturer's instructions. Reverse transcription to cDNA was performed using Superscript II, RNase-OUT and random primers according to supplier's instructions (Life Technolgies). mRNA concentrations were measured by quantitative RT-PCR (26). Cyclophilin A and TATA-binding protein (Tbp) were used as reference genes. Primer sequences are listed in supplementary Tables I and II.
SDS-PAGE and Western blot analysis
SDS-PAGE and Western blots were performed as described previously (26, 36). Equal amounts of protein (35 – 50 μg) were denatured in 4 × sample loading buffer (250 mM Tris-HCl pH 6.8, 4% SDS, 30% glycerol, 0.003% bromophenol blue and 10% 2-mercaptoethanol added freshly) and then boiled for 5 min. Proteins were separated by SDS-PAGE using 6% and 8% separating gels when detecting lipin-2 and lipin-1, respectively. Antibody raised against a polypeptide derived from lipin-1 (410SKTDSPSRKKDKRSRHLGADG430) was a gift from Dr. Zemin Yao (University of Ottawa, ON) (18), and it was used to measure lipin-1 levels in NRVMs and livers. Antibodies against the last 11 amino acids in the C terminus of lipin-1 and the phosphorylated serine residue 106 in lipin-1 were prepared as described previously (24), and the antibody against the C terminus of lipin-1 was used to detect lipin-1 in hearts. Antibodies against lipin-2 were raised in rabbits using the peptide sequence N’ – PKGELIQERTKGNK – C’ followed by affinity-purification (Genscript; Piscataway, NJ) (37). Alexa Fluor 680-conjugated goat anti-mouse IgG (Life Technologies), IRDye 800-conjugated goat anti-rabbit IgG (LI-COR Biosciences; Lincoln, NE) and HRP-conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology; Santa Cruz, CA) were used as the secondary antibodies. Odyssey blocking buffer and 5% milk in TBST (20 mM Tris-HCl pH 7.6, 137 mM NaCl, 0.1% Tween-20) were used as blocking buffers for optimal detection of fluorescent dye-conjugated and HRP-conjugated secondary antibodies, respectively. Infrared fluorescence and enzymatic chemiluminescence were detected using the LI-COR Odyssey Infrared System (LI-COR Biosciences) and the Immun-Star WesternC chemiluminescence kit (Bio-Rad Laboratories; Hercules, CA) followed by autoradiography, respectively.
PAP enzymatic assays
PAP assays were performed essentially as described previously (26). Each sample was assayed in a final volume of 100 µl consisting of 100 mM Tris/maleate buffer, pH 6.5, 0.6 mM DTT, 5 mM MgCl2, protease inhibitor cocktail, 30 nM microcystin-LR (Enzo Life Sciences; Farmingdale, NY), 0.6 mM [3H]phosphatidate (PA) (approximately 6 × 104 dpm per assay), 0.4 mM phosphatidylcholine, 1 mM EDTA/1 mM EGTA, 2 mg/ml BSA, and 200 µM tetrahydrolipstatin to inhibit the degradation of the DG product by lipase activity (38). Parallel measurements were performed in the presence of 8 mM N-ethylmaleimide (NEM) to determine the contribution from lipid phosphate phosphatase (LPP) activity (26).
DG formed in these assays was extracted in 2 ml chloroform-methanol (19:5 v/v) containing 0.08% olive oil as an acylglycerol carrier. Activated alumina was added to remove the unreacted PA and any liberated [3H]palmitate. The chloroform phase was dried down, and radioactivity was determined. PAP activity was calculated by subtracting the NEM-insensitive LPP activity from the total activity. Each sample was assayed at three different protein concentrations (30–200 µg) to ensure a proportional response, and the conversion of PA to DG was restricted to <20%.
Treatment of cells with radiolabeled substrates and analysis of lipid extracts
NRVMs cultured in 10% FBS-DME/F12 medium were washed and treated with DME/F12 medium containing 0.1% BSA for 4 h. Diethyl p-nitrophenyl phosphate (E600) was then added at a final concentration of 100 µM to inhibit intracellular lipases and lipid turnover (39). After 30 min, the cells were treated with DME/F12 medium containing 0.3 mM FA-poor BSA containing 1 mM [3H]glycerol (1.7 Ci/mol), 1 mM [14C]oleate (0.25 Ci/mol), 100 μM choline chloride, and 100 µM E600. In some experiments, 0.3 mM [3H]glycerol (5.3 Ci/mol), 0.5 mM [14C]oleate (0.5 Ci/mol), and 0.1 mM BSA were used instead. When treatments were complete, cells were washed twice with ice-cold HEPES-buffered saline (HBS: 25 mM HEPES, 138 mM NaCl, and 2.7 mM KCl, pH 7.4) containing 1 mg/ml BSA, followed by one wash with ice-cold HBS. Cell extracts were collected in two consecutive volumes of 500 µl methanol, and the combined volume was sequentially extracted with 1 ml chloroform followed by 0.9 ml 2 M KCl containing 0.2 mM HCl.
Samples of the chloroform phase (800 µl) containing the extracted lipids (out of a theoretical 1.1 ml) were dried down under an N2 stream and resuspended in 100 µl chloroform. Ten microliters of each sample were used to measure total radiolabel incorporation. The majority of the lipids (80 µl) was pipetted onto glass-backed, silica-coated TLC plates (EMD Chemicals). Plates were developed for 50% of their lengths using chloroform-methanol-acetic acid-acetone-water (50:10:10:20:5, v/v/v/v/v) followed by a second development for the full length of the plates with hexane-diethyl ether-acetic acid (60:40:1, v/v/v) (40). Lipids were detected with I2 vapor, and individual lipids were scraped from the plates and collected in scintillation vials containing 10% water and EcoLite™ scintillation fluor (MP Biomedicals).
Quantification of organic phosphates and triacylglycerol
Organic phosphate was assayed to measure phospholipid content using rac-glycerol 3-phosphate as a standard (41). Extracted lipids in 100 µl chloroform phase from each sample were dried down and digested with 50 µl of perchloric acid at 180°C for 30 min. The reaction mixture was cooled, and 278 µl water, 55 µl 2.5% ammonium molybdate, and 55 µl 10% ascorbic acid were added before boiling for 15 min. Absorbance readings were quantified with a multi-well plate reader at 700 nm.
Tissue TG levels were measured after extraction with chloroform-methanol-2 M KCl containing 0.2 M HCl (1:1:0.9) using the TG GPO kit (Pointe Scientific; Canton, MI).
Separation of cytosol from cell ghosts using hypotonic digitonin lysis
Cells were treated with a hypotonic solution containing 10 mM HEPES, pH 7.4, 0.5 mM DTT, protease inhibitors, 30 nM microcystin-LR to inhibit protein phosphate phosphatases, and 80 µM digitonin for 4 min on a chilled glass plate leveled on top of an ice-filled plastic container (23, 42). The cytosolic proteins released by digitonin were isolated, and the cell ghosts were washed twice and collected in 10 mM HEPES, pH 7.4, 0.5 mM DTT, protease inhibitors, 30 nM microcystin-LR, and 250 mM sucrose. Cell ghosts were sonicated twice for 4 s with 1 min intervals. Both cytosolic and cell ghost fractions were centrifuged for 2 min at 750 g to remove unbroken cells.
Lactate dehydrogenase (LDH) activities in the cytosolic and cell ghost fractions were measured to determine the extent of cytosolic leakage after treatment with digitonin (43). An average of 91.0 ± 0.6% of the total LDH activity was released by digitonin from NRVMs, indicating a high degree of cell permeabilization (23, 42). PAP activities in the cytosol and cell ghost fractions were adjusted if the minimum LDH activity in the cytosol did not reach the average cytosolic LDH value. For example, 10% of the total PAP activity was subtracted from the PAP activity in the cell ghosts when the cytosolic LDH activity in NRVMs was 81% of the total LDH activity. The proportion of cytosolic to membrane-associated PAP activity was then recalculated and expressed relative to the corrected total PAP activity.
Animal care and breeding strategy
The breeding colony of Balb/cByJ-Lpin1fld/J mice was established as previously described (37). Wild-type and heterozygous mice were both designated as control mice. Mice were kept in 12 h/12 h light/dark cycles (0600 h to 1800 h) and were fed Lab Diet 5058 containing 9% fat by weight (PMI Nutrition International; St. Louis, MO). Female mice were used in all experiments. The research was conducted in accordance with the policies of the Canadian Council on Animal Care, as approved by the University of Alberta Animal Policy and Welfare Committee.
Statistics
Results are expressed as means ± SEM. The two-tailed Student's t-test was used to test for significance when there were only two groups/treatments to be analyzed; otherwise, the Newman-Keuls post hoc test after one-way ANOVA or the Bonferroni test after two-way ANOVA was applied to test for statistical significance (P < 0.05).
RESULTS
Mechanisms for the regulation of PAP activity and the expressions of lipins, Ppargc1a splice variants, Ppara, and carnitine palmitoyltransferase (Cpt1b) in NRVMs
Lipins act as PAP enzymes in glycerolipid synthesis, and lipin-1 also functions as a transcriptional coactivator in combination with PPARα and PGC-1α to stimulate FA oxidation in the liver (6). We therefore investigated mechanisms that could coordinate the expressions of different lipins, Ppargc1a splice variants, Ppara, and Cpt1b in cardiomyocytes. To do this, we determined the effects of hormonal signals characteristic of starvation (glucocorticoids and cAMP) versus that of the fed state (insulin). Treatment of NRVMs with dexamethasone (a glucocorticoid analog of cortisol) at 10 to 1,000 nM increased PAP activity by about 50% after 8 to 12 h (Fig. 1A, B). The dexamethasone-induced increase in PAP activity was synergized significantly by CPT-cAMP, which is a cell-permeable cAMP analog resistant to phosphodiesterase activity (Fig. 1C). Conversely, insulin inhibited the dexamethasone-induced increase in PAP activity (Fig. 1C). Insulin also partially antagonized the combined action of dexamethasone and CPT-cAMP (Fig. 1C). Importantly, the changes in PAP activity elicited by dexamethasone, CPT-cAMP, and insulin coincided with changes in lipin-1 expression with one exception: the effect of insulin in completely antagonizing the synergistic effect of dexamethasone and CPT-cAMP on lipin-1 protein expression such that it reached baseline levels (Fig. 1D).
Fig. 1.
Treatment of NRVMs with different compounds and their effects on PAP activity and lipin-1 protein expression. A: NRVMs were treated with different concentrations of dexamethasone for 8 h. PAP activities of the cell lysates were measured and expressed relative to the vehicle-treated control (n = 3). B: NRVMs were treated with 10 nM dexamethasone for different times, and PAP activities were measured and expressed relative to the control at time zero (n = 3). C: NRVMs were treated with combinations of 10 nM dexamethasone, 100 µM CPT-cAMP, or 50 nM insulin for 8 h, and PAP activities were measured and expressed relative to control (n = 5–12). D: Representative Western blot (upper panel) and quantification (lower panel) of the effects of various combinations of compounds on lipin-1 expression, which was detected using the LI-COR Odyssey Infrared system (n = 3–8). Lipin-1 overexpressed in human embryonic kidney 293 cells from a recombinant adenoviral vector serves as a positive control. GAPDH was used as the loading control. * P < 0.05 compared with respective controls and baseline groups; Φ P < 0.05 when comparing the indicated treatments. Results are means ± SEM.
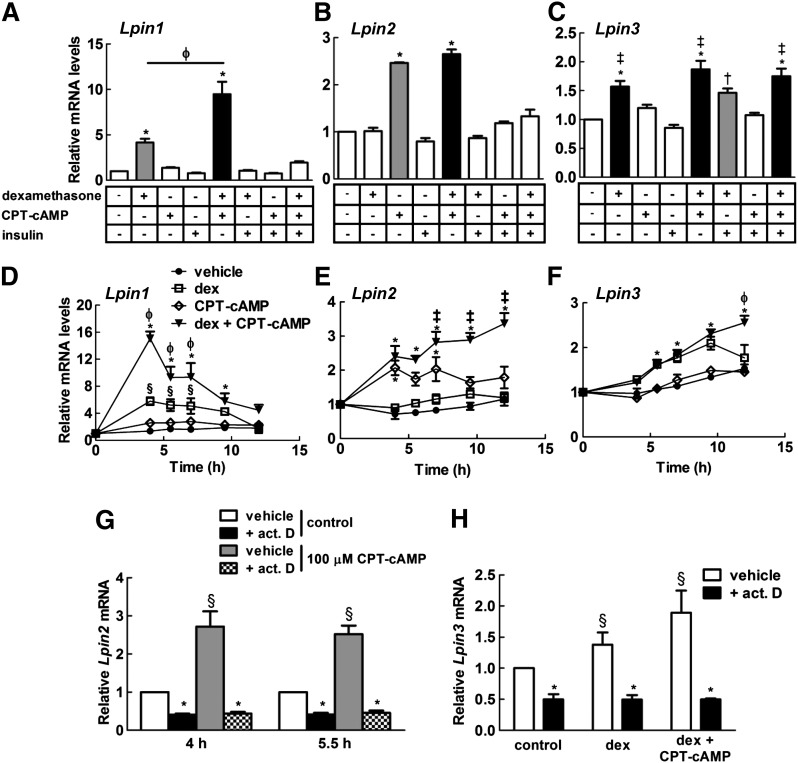
As expected, the gene expression profile of Lpin1 in NRVMs treated with the different combinations of compounds for 4 h preceded the response on PAP activity and the expression of lipin-1 protein. Dexamethasone increased the expression of mRNA for Lpin1, with CPT-cAMP acting synergistically and insulin having an antagonistic effect (Fig. 2A). Furthermore, maximal induction of Lpin1 expression with either dexamethasone alone or the combination of dexamethasone and CPT-cAMP occurred between 4 h after treatment and a decline in expression after 7 h (Fig. 2D).
Fig. 2.
The effect of various combinations of hormones and CPT-cAMP on the gene expressions of the lipins in NRVMs. The gene expressions of (A) Lpin1 and (B) Lpin2 in NRVMs were measured after treatment with combinations of 10 nM dexamethasone, 100 µM CPT-cAMP or 50 nM insulin for 4 h (n = 4–6). C: The effect of various combinations of hormones and CPT-cAMP on Lpin3 expression after 7 h in NRVMs (n = 4–8). Time course of Lpin1 (D), Lpin2 (E),and Lpin3 (F) expressions in NRVMs treated with 10 nM dexamethasone (dex) and/or 100 µM CPT-cAMP (n = 3); results are expressed relative to the controls at time zero. G: NRVMs were preincubated for 30 min with 10 µg/ml actinomycin D (act. D) or vehicle before treatment with 100 µM CPT-cAMP for 4 and 5.5 h, and mRNA levels of Lpin2 were measured (n = 3). H: Lpin3 transcript levels were also determined after 30 min preincubation with actinomycin D and treatment for 7 h with 10 nM dex and 100 µM CPT-cAMP (n = 3). * P < 0.05 compared with respective controls and baseline groups; ‡ P < 0.05 when compared with 100 µM CPT-cAMP treatment; Φ P < 0.05 when compared with 10 nM dex treatment; § P < 0.05 compared with vehicle-treated control; † P < 0.05 compared with 50 nM insulin treatment.
In contrast to hepatocytes (26), Lpin2 expression was increased by treatment with CPT-cAMP after 4 h, and dexamethasone had no effect at this time (Fig. 2B). However, CPT-cAMP and dexamethasone acted synergistically between 7 h and 12 h treatment (Fig. 2E). Similar to Lpin1 gene regulation, insulin antagonized the induction of Lpin2 expression by dexamethasone and CPT-cAMP (Fig. 2B). Furthermore, Lpin3 expression was regulated analogously to Lpin1 expression in that dexamethasone induced its expression (Fig. 2C, F). However, the induction of Lpin3 expression occurred only after 5.5 h treatment, with a significant synergism at 12 h with CPT-cAMP (Fig. 2C, F). Insulin did not antagonize the upregulation of Lpin3 expression by dexamethasone alone or in combination with CPT-cAMP after 7 h of treatment (Fig. 2C). Treatment with actinomycin D prevented the increase in Lpin2 and Lpin3 mRNA levels in NRVMs treated with dexamethasone and/or CPT-cAMP (Fig. 2G, H). This result suggests that the effect of dexamethasone and/or CPT-cAMP on Lpin2 and Lpin3 expression depends on increased mRNA synthesis. Increased Lpin1 expression by dexamethasone and CPT-cAMP also depends on transcriptional upregulation, as shown in hepatocytes (26).
There are three splice variants of PGC-1α in Mus musculus, whereas there only appear to be two PGC-1α splice variants, PGC-1α-a and PGC-1α-b in Rattus norvegicus (see supplementary Fig. I). Ppargc1a-a expression in NRVMs was increased 2- to 3-fold by 10 nM dexamethasone after 4 h, and CPT-cAMP acted antagonistically (Fig. 3A, C). Insulin can also increase Ppargc1a-a expression (Fig. 3A). More importantly, Ppargc1a-b expression was greatly induced by CPT-cAMP alone, and dexamethasone acted synergistically after 4 h of treatment, whereas insulin antagonized these actions (Fig. 3B, D). Moreover, the time course of Ppargc1a-b expression in NRVMs coincided with Lpin1 expression when stimulated with dexamethasone and CPT-cAMP (Figs. 2D, 3D).
Fig. 3.
Effects of various combinations of hormones and CPT-cAMP on the gene expressions of Ppargc1a, Ppara and Cpt1b in NRVMs. The gene expressions of (A) Ppargc1a-a and (B) Ppargc1a-b splice variants were measured after treatment of NRVMs with combinations of 10 nM dexamethasone, 100 µM CPT-cAMP, and 50 nM insulin for 4 h (n = 3). The expressions of Ppargc1a-a (C), Ppargc1a-b (D), Ppara (E), and Cpt1b (F) were measured after treating NRVMs with 10 nM dexamethasone (dex), 100 µM CPT-cAMP, or the combination of both for various time points up to 12 h. * P < 0.05 compared with respective controls and baseline groups; Φ P < 0.05 when compared with 10 nM dex treatment; Ψ P < 0.05 when compared with treatment with 10 nM dex and 100 µM CPT-cAMP; θ P < 0.05 when compared with treatment with 100 μM CPT-cAMP.
On the other hand, Ppara expression was not regulated significantly by dexamethasone and/or CPT-cAMP up to 12 h of treatment, compared with vehicle-treated cardiomyocytes (Fig. 3E). However, treatment with dexamethasone alone or in combination with CPT-cAMP, which acts synergistically, for 12 h did upregulate the gene expression of Cpt1b, which is a target of PPARα transcriptional regulation (Fig. 3E). We next determined whether this late increase in mRNA levels of Cpt1b depended on the upstream activation of PPARα. Fenofibric acid (a PPARα activator) significantly increased the gene expression of Cpt1b, but not of Lpin2 or Lpin3 (Fig. 4A–C). Not surprisingly, PPARα activation also has no significant effect on Ppargc1a-a, Ppargc1a-b, and Lpin1 mRNA levels (results not shown). Furthermore, the increase in Cpt1b levels after treatment with dexamethasone and CPT-cAMP was decreased by treatment with GW 6471 (Fig. 4D), which is a known inhibitor of PPARα (44). This novel result shows the importance of PPARα activation, which is sufficient and essential for the upregulation of Cpt1b expression (Fig. 4A, D).
Fig. 4.
Signaling downstream of PPARα activation in NRVMs. NRVMs were treated with vehicle or 100 μM fenofibric acid for various lengths of time and the gene expressions of Cpt1b (A), Lpin2 (B), and Lpin3 (C) were measured. D: NRVMs were treated with various combinations of 10 nM dexamethasone (dex), 100 μM CPT-cAMP, and 10 μM GW 6471 (PPARα inhibitor) for 12 h, and the mRNA levels of Cpt1b were measured. * P < 0.05 compared with respective controls and indicated groups.
Because β2-adrenergic receptor agonists can induce lipin-1 and PGC-1α in skeletal muscle and hearts of mice in vivo (27, 45), we examined this effect in NRVMs. Ppargc1a-a and Ppara expressions were unaffected in NRVMs treated with 100 µM clenbuterol (a β2-adrenergic receptor agonist) or with the combination of 10 nM dexamethasone and 100 µM clenbuterol (Fig. 5A, C). By contrast, Ppargc1a-b mRNA levels were significantly increased upon stimulation with clenbuterol alone (Fig. 5B). Furthermore, dexamethasone synergized the effect of clenbuterol on Ppargc1a-b expression, although the effect was not as great as that seen with dexamethasone and CPT-cAMP (Fig. 5B). β2-Adrenergic signaling activates both Gs and Gi signaling (46, 47). However, pretreatment with pertussis toxin for 24 h to inhibit Gi signaling prior to the addition of 100 µM clenbuterol showed no significant difference in Ppargc1a-b expression compared with controls (Fig. 5H). Treatment with clenbuterol alone in NRVMs has no effect on Lpin1 expression (Fig. 5D), or on the transcript levels of Lpin2, Lpin3, and Cpt1b (Fig. 5E–G). However, clenbuterol can act synergistically with dexamethasone to induce Lpin1, Lpin2, Lpin3, and Cpt1b expressions, similar to that seen with CPT-cAMP, although to a lesser extent (Fig. 5D–G).
Fig. 5.
Effect of clenbuterol on gene expression in NRVMs. The gene expressions of Ppargc1a-a (A), Ppargc1a-b (B), Ppara (C), Lpin1 (D), Lpin2 (E), Lpin3 (F), and Cpt1b (G) were measured after treatment with vehicle, 1 μM clenbuterol (clen), and 10 nM dexamethasone (dex) in combination with 100 μM CPT-cAMP, or 10 nM dex in combination with 1 μM clenbuterol for 4, 8 and 12 h. H: Ppargc1a-b mRNA levels were measured in NRVMs pretreated for 24 h with or without 200 ng/ml pertussis toxin (PTX), followed by 4 h treatment with vehicle or 1 μM clenbuterol. * P < 0.05 compared with vehicle alone; ‡ P < 0.05 compared with vehicle and clenbuterol alone; Ψ P < 0.05 compared with all other groups.
The effect of fasting on gene expression and PAP activity in hearts and livers of fld and control mice
We next determined whether the effects of dexamethasone, CPT-cAMP, and insulin on the gene expressions of the lipins, Ppargc1a, Ppara, and Cpt1b in cardiomyocytes are compatible with the changes that occur in vivo during fasting. We also determined whether the complete lack of lipin-1 in fld mice would result in compensatory changes in the regulation of the other two lipins, PGC-1α splice variants, or PPARα. First, we fasted 14 to 18 week-old female fld and control mice for 12 h, from 2200 h to 1000 h the next day. The light cycle started from 0600 h and lasted until 1800 h. We determined the heart weight-to-tibia length ratios in these mice, because this ratio is decreased in 19 to 23 week-old male fld mice while remaining unchanged in fld mice at 11 weeks of age compared with age-matched controls (37). The heart weight-to-tibia length ratios in the female fld mice compared with the controls were not significantly different, indicating that there was no detrimental loss of heart mass at this time (control, 58.8 ± 1.6 mg/cm; fld, 59.2 ± 1.2 mg/cm).
As expected, Lpin1 expression was observed in control and not in fld hearts. There was no significant change in the 12 h-fasted control animals compared with the fed mice (Fig. 6A). Similarly, cardiac lipin-2 mRNA expression was not significantly induced after fasting control mice for 12 h (Fig. 6A). However, mRNA expression for Lpin2 was higher in fld mice compared with controls (Fig. 6A). Additionally, the transcript levels for Lpin2 were also significantly increased by fasting in fld hearts (Fig. 6A). Lpin3 expression was also increased in 12 h-fasted fld hearts compared with fed fld hearts, whereas there was no difference in expression between fed and fasted control mice (Fig. 6A). Importantly, the gene expressions of the Ppargc1a-b and Ppargc1a-c splice variants were induced by fasting in both control and fld mice, with significantly greater increases in mRNA observed in fld mice (Fig. 6B). By contrast, Ppargc1a-a gene expression was not significantly different across all groups (Fig. 6B). Finally, Cpt1b and Ppara gene expressions were not significantly changed in all groups (Fig. 6C).
Fig. 6.
Effect of fasting on gene expression in lipin-1 deficient fld (fatty liver dystrophy) and control hearts. The mRNA levels of Lpin1, Lpin2, Lpin3 (A), Ppargc1a splice variants (B), and Ppara and Cpt1b (C) were measured in hearts from 14–18 week-old fld (n = 7) and control (n = 7), which were fed ad libitum or fasted for 12 h from 2200 h to 1000 h. Expressions of Lpin1, Lpin2, Lpin3 (D), Ppargc1a splice variants (E), and Ppara and Cpt1b (F) were also measured in the hearts of 14–18 week-old control mice fed ad libitum (n = 6) or fasted (n = 6) for 24 h. * P < 0.05 by unpaired t-test; Ψ P < 0.05 when comparing the indicated groups; Φ P < 0.05 compared with all other groups.
We were concerned that the absence of changes in gene expressions for the lipins, carnitine palmitoyltransferase-1 (CPT1), and PPARα after 12 h of fasting in control mice could be attributed to the relatively short length of the fast. Therefore, we studied the effects of fasting control mice for 24 h (0600 h to 0600 h the next day), but not fld mice, because they do not tolerate prolonged fasting, owing to their lack of adipose tissue. Only Cpt1b expression was significantly increased in 24 h-fasted hearts compared with controls (Fig. 6F). Cardiac Lpin1 and Ppara gene expressions were significantly decreased in the fasted compared with fed animals, and there were no significant changes in Lpin2 and Lpin3 transcript levels (Fig. 6D, F). Furthermore, the increases in Ppargc1a-b and Ppargc1a-c mRNA seen after 12 h of fasting returned to near-baseline levels after the 24 h fast (Fig. 6E).
To further validate the physiological significance of these results in the heart, we compared these with the equivalent gene expression profiles in the liver. Unlike the heart, Ppargc1a-b and Ppargc1a-c expressions are not detectable in the liver. Furthermore, Lpin2, Cpt1a and Ppara mRNA levels were significantly increased in 12 h-fasted control mice compared with fed controls (Fig. 7A, B). There were also trends toward increased Lpin1 and Ppargc1a-a transcript levels in 12 h-fasted control livers (Fig. 7A, B), which was expected (9, 17, 26). On the other hand, Cpt1a and Ppara gene expressions were not induced in 12 h-fasted fld livers compared with fed fld mice (Fig. 7B), which was demonstrated previously (6). However, Lpin2 mRNA levels in fasted fld mice were increased, as expected (Fig. 7A). Fasting-induced upregulation of Ppargc1a-a expression in 12 h-fasted fld mice was also not affected (Fig. 7B), which is not surprising, given that PGC-1α is upstream of PPARα signaling (6). Lpin3 expression was unchanged across all groups. Finally, Lpin2, Ppargc1a-a, Cpt1a, and Ppara expressions remained significantly increased in the livers of the 24 h-fasted mice (Fig. 7C, D).
Fig. 7.
Effect of fasting on gene expression in lipin-1 deficient fld (fatty liver dystrophy) and control livers. Lpin1, Lpin2, Lpin3 (A), Ppargc1a-a, Ppara and Cpt1a (B) mRNA levels were measured in livers from 14–18 week-old fld (n = 7) and control (n = 7), which were fed ad libitum or fasted for 12 h from 2200 h to 1000 h. Ppargc1a-b and Ppargc1a-c mRNA levels were undetectable. Lpin1, Lpin2, Lpin3 (C), Ppargc1a-a, Ppara and Cpt1a (D) mRNA levels were determined in the livers of the control mice, which were fed ad libitum (n = 6), or in mice fasted for 24 h (n = 6). * P < 0.05 by unpaired t-test; Ψ P < 0.05 when comparing the indicated groups.
We also validated the gene expressions of the lipins in fasted hearts and livers by measuring PAP activity, and lipin-1 and lipin-2 protein levels. Unsurprisingly, there was no significant difference in PAP activities in 12 h-fasted compared with fed control hearts (Fig. 8A), which corresponds to the mRNA expression for the lipins after 12 h of fasting (Fig. 6A). Surprisingly, there were no changes in PAP activities in the hearts of 12 h-fasted fld mice compared with fed fld mice (Fig. 8B), even though there was an increase in cardiac Lpin2 and Lpin3 gene expressions (Fig. 6A). Additionally, lipin-2 protein levels were not significantly different (0.89 ± 0.17 versus 0.83 ± 0.12, n = 7) between fed and 12 h-fasted fld hearts (Fig. 8B, inset). However, both PAP activities and lipin-1 protein levels were significantly decreased in 24 h-fasted control hearts (Fig. 8C, E), which corresponds to the decrease in cardiac Lpin1 gene expression (Fig. 6D) in the 24 h-fasted hearts compared with the fed controls. Decreased expression of lipin-1 in 24 h-fasted hearts might indicate a decrease in TG synthesis in favor of increased TG hydrolysis. However, TG levels in the hearts from 24 h-fasted control mice were increased significantly by fasting (Fig. 8G). This was not surprising, given that the complete absence of lipin-1 expression in fld hearts still does not become a rate-limiting factor and cause a decrease in phospholipid or TG synthesis (37).
Fig. 8.
Effect of fasting on PAP activity, lipin expression, and TG content in fld and control livers and hearts. Measurement of PAP activities in the livers and hearts of control (A) and fld (B) mice, which were fasted for 12 h or fed ad libitum. The inset shows a representative Western blot for the levels of lipin-2 in hearts from fed and 12 h-fasted fld mice. C: Hepatic and cardiac PAP activities for mice that were fasted for 24 h (n = 4) or fed ad libitum (n = 5). D: Representative Western blots showing the expression of lipin-1 and -2 in 24 h-fasted and fed control livers. The LI-COR Odyssey Imaging system was used to detect lipin-1, whereas lipin-2 levels were identified by enzymatic chemiluminescence and autoradiography. Calnexin was used as the loading control. E: Representative Western blots showing the expression of lipin-1 and -2 in 24 h-fasted and fed control hearts. Both lipin-1 and -2 were detected using enzymatic chemiluminescence and autoradiography. F: Lipin-1, lipin-2, and calnexin levels in 24 h-fasted (n = 4–6) or fed (n = 5) control livers and hearts were quantified using the LI-COR Odyssey Imaging software 1.2 or ImageJ. G: TG levels in 24 h-fasted and fed mouse livers and hearts were also measured. * P < 0.05 compared with mice, which were fed ad libitum; Φ P < 0.05 compared with the corresponding mouse hearts.
By contrast, PAP activities in the 12 h-fasted control livers were increased compared with those of fed control mice (Fig. 8A), as expected from previous studies (26, 48). Likewise, 12 h-fasted fld livers also had a significant increase in PAP activities compared with fed fld mice (Fig. 8B), again demonstrating the compensatory increase in lipin-2 expression in the livers of lipin-1-deficient mice (17). Similar to 12 h-fasted mice, hepatic PAP activity was increased in control mice fasted for 24 h (Fig. 8C). Furthermore, lipin-1 and lipin-2 protein levels in 24 h-fasted control livers were significantly increased compared with fed controls (Fig. 8D, F), similar to results from other studies (6, 17). Finally, TG content in the 24 h-fasted livers were also increased compared with fed controls, similar to the results found in heart (Fig. 8G). In summary, there are marked differences between the regulation of hepatic and cardiac lipin levels and PAP activities in vivo even though there are significant similarities in the regulation of lipin expression in cultured hepatocytes (26) and NRVMs. Additionally, different PGC-1α splice variants are induced in heart compared with liver during fasting. Finally, Cpt1 mRNA expression is increased in both liver and heart in the fasted state, as expected, but only hepatic PPARα transcript levels are induced by fasting. Finally, upregulation of cardiac PPARα expression during fasting is not as important as its activation state, which contrasts with the increased expression of PPARα in fasted livers.
Effect of changing lipin levels on glycerolipid synthesis in neonatal rat ventricular myocytes
Because there was no increase in lipin expression and PAP activity as a result of fasting in mouse hearts compared with the changes observed in cultured NRVMs, we determined the functional consequences of varying lipin expression in NRVMs. We investigated whether decreasing lipin-1 expression or increasing lipin-1 and -2 expressions in NRVMs using adenoviral vectors had a major effect on the rate of glycerolipid synthesis.
NRVMs either were not treated or were incubated with adenovirus encoding for shRNA against Lpin1 or lacZ (AdshRNA Lpin1 and AdshRNA LacZ) for 38 h. PAP activity and lipin-1 protein expression in NRVMs infected with AdshRNA Lpin1 were decreased by 50% compared with controls (Fig. 9A). There was no significant difference between groups in the incorporation of 1 mM [1,3-3H]glycerol or 1 mM [14C]oleate into the glycerolipids over 3 h, except for TG (Fig. 9B, C). Oleate and glycerol incorporation into TG in AdshRNA LacZ-infected cells was slightly but significantly increased compared with nontreated controls (Fig. 9B, C). Oleate incorporation into TG in AdshRNA Lpin1-infected cells was also significantly increased, compared with nontreated controls (Fig. 9C). However, this increase was significantly lower compared with NRVMs infected with AdshRNA LacZ (Fig. 9C).
Fig. 9.
Effect of knocking down lipin-1 on the rate of glycerolipid synthesis in NRVMs. A: NRVMs were infected for 38 h with recombinant adenoviral constructs expressing shRNA against lacZ (AdshRNA LacZ) or Lpin1 (AdshRNA Lipin1) at a multiplicity of infection (MOI) of 20, respectively, and the PAP activities in the cell lysates were measured and expressed relative to the no Ad control. The inset displays a representative Western blot showing the lipin-1 protein levels in NRVMs infected with AdshRNA LacZ and AdshRNA Lipin1, respectively. NRVMs were treated with 1 mM [3H]glycerol (B) and 1 mM [14C]oleate (C) in DME/F12 medium containing 0.3 mM BSA for 3 h after infection with recombinant adenovirus for 38 h (n = 3), and the rate of glycerol and oleate incorporation into glycerolipids was measured and expressed as pmol oleate or glycerol per pmol phospholipid phosphate. Similar analysis was performed for NRVMs treated with 0.3 mM [3H]glycerol (D) and 0.5 mM [14C]oleate (E) in DME/F12 medium containing 0.1 mM BSA for 3 h after infection with recombinant adenovirus for 38 h (n = 3). P < 0.05 compared with all other treatments; * P < 0.05 compared with no Ad control; † P < 0.05 compared with NRVMs infected with AdshRNA lipin1.
A previous study showed that there was a significant decrease in the rate of TG synthesis in NRVMs infected with recombinant adenovirus expressing shRNA against lipin-1. Mitra et al. (27) treated their cells with carrier-free [2-3H]glycerol (10 μCi/ml) and 0.5 mM oleate, and we decided to determine whether we could obtain similar results. When NRVMs were treated with 0.5 mM [14C]oleate and 0.3 mM [1,3-3H]glycerol for 3 h, there were no significant differences in the rate of glycerolipid synthesis in the AdshRNA Lpin1-infected cells compared with noninfected or vector controls (Fig. 9D, E). However, oleate incorporation into TG was increased slightly in AdshRNA LacZ-infected cells compared with noninfected controls (Fig. 9E). This observation was similar to the result from the previous experiment performed with 1 mM [14C]oleate and 1 mM [1,3-3H]glycerol. In summary, decreasing PAP activity by 50% in NRVMs had very little effect on the rate of glycerolipid synthesis from [1,3-3H]glycerol and [14C]oleate.
We also determined the effect of increasing lipin expression on glycerolipid synthesis. After incubation for 38 h following infection with recombinant adenoviral vectors expressing Mus musculus lipin-1B and -2 (Adlipin1b and Adlipin2), PAP activities in NRVMs were increased 14- and 5-fold, respectively (Fig. 10A). Infection with control adenovirus expressing green fluorescent protein (GFP) (AdGFP) did not affect PAP activity (Fig. 10A). Increasing PAP activity in NRVMs with either Adlipin1b or Adlipin2 did not significantly affect glycerol or oleate incorporation into phospholipids and DG when treated with 1 mM [14C]oleate and 1 mM [1,3-3H]glycerol for 3 h (Fig. 10B, C). Instead, there was a small but significant increase in oleate and glycerol incorporation into TG in NRVMs infected with Adlipin2, compared with noninfected cells (Fig. 10B, C). Moreover, a similar result was observed for oleate incorporation into TG in cells treated with Adlipin1b compared with noninfected controls (Fig. 10C). However, there were no significant differences in oleate and glycerol incorporation into TG when comparing NRVMs expressing recombinant lipin-1B and -2 to control cells infected with the control AdGFP vector (Fig. 10B, C).
Fig. 10.
Rates of glycerolipid synthesis from glycerol and oleate in NRVMs expressing recombinant lipin-1 and -2. A: NRVMs were infected for 38 h with recombinant adenoviral (Ad) constructs expressing green fluorescent protein (GFP) or lipin-1b or -2 at an MOI of 7, and the PAP activities in the cell lysates were measured and expressed relative to the no Ad control. NRVMs were treated with 1 mM [3H]glycerol (B) and 1 mM [14C]oleate (C) in DME/F12 medium containing 0.3 mM BSA for 3 h after infection with recombinant adenovirus for 38 h (n = 3), and the rate of glycerol and oleate incorporation into glycerolipids was measured and expressed as pmol oleate or glycerol per pmol phospholipid phosphate. D: NRVMs were treated with 0.3 or 1.2 mM oleate in DME/F12 medium containing 0.3 mM BSA for 1 or 4 h. NRVMs were then incubated for 4 min with a hypotonic solution containing 0.1 mg/ml digitonin on ice. The cytosolic fractions and cell ghosts were collected separately, and the PAP activity in each fraction was measured. Results were then expressed as percentage of total PAP activity on cell ghost membranes. * P < 0.05 compared with no Ad control; ‡ P < 0.05 compared with 4 h vehicle-treated control (no FA added); § P < 0.05 compared with corresponding 1 h treatments.
Because there were only minor changes in the rates of glycerol and oleate incorporation into glycerolipids when the levels of lipin-1 and/or lipin-2 were varied, we next determined the extent to which PAP activity associates with membranes when treated with FA, because this is the metabolically active form of lipin for glycerolipid synthesis. There was no significant increase in PAP activity on cell ghost membranes obtained after cell lysis with digitonin after treatment of NRVMs for 1 h with 1.2 mM oleate (Fig. 10D). However, there was a significant 2-fold increase in membrane-associated PAP activity after 4 h treatment with either 0.3 or 1.2 mM oleate (Fig. 10D). Because there were no significant changes in total PAP activities between treatments (no FA, 0.28 ± 0.02 nmol/min; 1.2 mM oleate, 0.30 ± 0.02 nmol/min; 0.3 mM oleate, 0.31 ± 0.02 nmol/min), this indicates that the increase in membrane-associated PAP activity is due to the expected translocation of cytosolic PAP activity. Importantly, only 20% of the total PAP activity was associated with membranes after 4 h of treatment with oleate. Torin1 (mTOR inhibitor) has been used previously to decrease insulin-dependent phosphorylation of lipin-1, thus decreasing its cytosolic localization (49). We hypothesized that treatment with Torin1 might decrease lipin-1 phosphorylation and increase its involvement in glycerolipid synthesis on the membranes. However, Torin1 had no effect on TG (see supplementary Fig. II) and other glycerolipids (data not shown) in NRVMs overexpressing lipin-1 or -2, even when the cells were treated with insulin, showing that the extent of lipin-1 association with membranes is not rate limiting in NRVMs. Overall, the data demonstrate that only a limited proportion of the cytosolic reservoir of lipins is required to sustain the glycerolipid biosynthetic pathway in NRVMs.
DISCUSSION
Numerous studies have shown that the relative levels of gene expression for PGC-1α, lipins, and PPARα are dynamically regulated in liver and adipose tissue with respect to altered requirements for FA esterification and oxidation (2, 6, 7, 9, 11–13). Cardiac PAP activity is acutely increased in streptozotocin-induced diabetes (50), but it is decreased in obese insulin-resistant rats (51). Moreover, mRNA levels of lipin-1 and lipin-3 in the heart are decreased in type 2 diabetic patients and in heart failure (27, 50, 52). Otherwise, little is known about the regulation of lipin expression in the heart. We, therefore, determined how the expressions of lipin isoforms are regulated during starvation with respect to the expressions of PGC-1α splice variants, PPARα, and CPT1. We also established whether these changes can be explained by increased glucocorticoid and cAMP signaling versus insulin action.
As in hepatocytes, lipin-1 expression in NRVMs was increased by dexamethasone, and CPT-cAMP and insulin acted as synergistic and antagonistic agents, respectively (Table 1). The transcript levels of Lpin2 and Lpin3 in NRVMs were also increased by the combination of dexamethasone and CPT-cAMP (Table 1). Clenbuterol also acts synergistically with dexamethasone in a cAMP-dependent manner for all three lipin isoforms (Fig. 5). The regulation of lipin gene expression by dexamethasone and CPT-cAMP in NRVMs differs quite significantly over time. Lpin1 transcript levels increased maximally at 4 h (Fig. 2D), whereas the increase in Lpin2 and Lpin3 expressions by dexamethasone and CPT-cAMP did not decline even after 12 h of treatment (Fig. 2E, F). The effect of dexamethasone and cAMP on lipin-1 is mediated through the glucocorticoid and cAMP response elements in the Lpin1 promoter (28, 53), but the regulatory sites on the Lpin2 and Lpin3 genes have yet to be identified.
TABLE 1.
Differential regulation of the mRNA expressions for PPARα, PGC-1α, CPT1, and the lipins in rat and mouse hepatocytes compared with NRVMs
| Dexamethasone | CPT-cAMP | |||
| Gene | Hepatocytes | NRVMs | Hepatocytes | NRVMs |
| Ppara | Increased (26) | No effect (Fig. 3E) | Synergista (26) | No effect (Fig. 3E) |
| Ppargc1a | Synergista (26) | Ppargc1a-a increased (Fig. 3A, C) | Increased (26) | Ppargc1a-b increased (Fig. 3A, B) |
| Synergist for Ppargc1a-b (Fig. 3B, D) | Antagonist for a (Fig. 3A,C) | |||
| Cpt1 | Not determined | Increased (Fig. 3F) | Increased (59) | SynergistaFig. 3F |
| Lpin1 | Increased (26) | Increased (Fig. 2D) | Synergista (26) | SynergistaFig. 2D |
| Lpin2 | No effect (26) | Synergista (Fig. 2E) | Increased (mouse, not rat) (26) | Increased Fig. 2E |
| Lpin3 | Increased (mouse, not rat) (26) | Increased (Fig. 2F) | Antagonist (mouse, not rat)a (26) | SynergistaFig. 2F |
The regulation of gene expression by dexamethasone alone, CPT-cAMP alone, or their synergistic or antagonistic effects in combination are shown.
The term “synergist” or “antagonist” denotes that dexamethasone or CPT-cAMP alone has no effect but can synergize or antagonize the action of the other compound to increase or decrease mRNA expression, respectively. The table shows citations or results of the present studies.
Despite increases in lipin expression observed with dexamethasone and CPT-cAMP in NRVMs, there was no increase in Lpin1 and Lpin2 mRNA in the heart after 12 h fasting in wild-type mice (Fig. 6), and there was no increase in PAP activity (Fig. 8). However, Lpin2 and Lpin3 transcript levels were increased in the 12 h-fasted versus fed fld mice, which demonstrates a compensatory transcriptional response in the absence of lipin-1, which we have reported previously (37). However, there were no significant differences in PAP activity and cardiac lipin-2 protein levels between control and fld hearts, which suggests that lipin-2 is also regulated posttranscriptionally, as was demonstrated previously (17). Finally, PAP activity was decreased in the hearts of control mice that had been fasted for 24 h (Fig. 8C) and this was explained by decreased lipin-1 expression (Figs. 6D, 8F).
By contrast, PAP activities, and lipin-1 and -2 protein levels in the livers of the same fasted control mice, were significantly increased compared with fed mice (Fig. 8AndashD), as expected from previous studies (6, 17, 26). It is important to note that although lipin-2 is the major lipin isoform in liver, lipin-1 also plays an essential role as a transcriptional coactivator and can compensate for lipin-2 deficiency (14, 17, 54). This is shown by the minor increase in PAP activity in fasted fld livers (Fig. 8D). In summary, the expressions of the lipins in the heart are not as sensitive to fasting as in the liver. We attribute this difference to the relative absence of glucocorticoid receptors in adult heart compared with liver (34, 35). Moreover, the glucocorticoid-mediated induction of lipin isoforms in NRVMs, in contrast to the absence of changes in fasted hearts, can be explained by the high numbers of glucocorticoid receptors expressed in the heart during the end stages of gestation, which then decline to become essentially absent in adult cardiac tissue (34, 35).
The three splice variants of PGC-1α, i.e., PGC-1α-a, PGC-1α-b, and PGC-1α-c, are regulated differently in liver compared with skeletal and cardiac muscle (10). Exercise increases mRNA levels for PGC-1α-b and PGC-1α-c in skeletal muscle specifically, with no changes in PGC-1α-a; similar results are produced by clenbuterol, the β2 adrenergic receptor agonist (10). We found that CPT-cAMP or cAMP-mediated signaling through clenbuterol significantly increased Ppargc1a-b expression, whereas dexamethasone and insulin were synergistic and antagonistic, respectively (Figs. 3, 5). Ppargc1a expression in hepatocytes is increased by CPT-cAMP, and this increase is synergized by dexamethasone, but insulin has no antagonistic effect (9, 26). We also showed for the first time that Ppargc1a-b and Ppargc1a-c mRNA levels are induced by fasting in mouse heart, which is compatible with the observed cAMP-mediated effect, whereas there were no changes in Ppargc1a-a (Fig. 6B). Interestingly, Lpin1 expression in NRVMs is not increased by CPT-AMP or clenbuterol alone, even though it is induced by β2-adrenergic signaling in the heart and skeletal muscle in vivo (27, 45). This result indicates that the effects of β2-adrenergic agonists on Lpin1 expression in vivo are not directly related to signaling downstream of β2-adrenergic receptors alone. In summary, the increase in mRNA of the two PGC-1α splice variants, Ppargc1a-b and Ppargc1a-c, during fasting is an important mechanism for the heart to sustain energy production by promoting mitochondrial biogenesis and increasing the capacity for FA oxidation. Conversely, PGC-1α-a is the main isoform in the liver, and this is the only splice variant increased by fasting (Fig. 7) (10).
On the other hand, Ppara expression was not affected by dexamethasone or CPT-cAMP in NRVMs, whereas Ppara transcription is induced in hepatocytes by dexamethasone, and CPT-cAMP acts synergistically (Table 1) (26). The present work and other studies (55, 56) also show that there is no increase in cardiac Ppara expression after fasting. In fact, there was a decrease in Ppara mRNA levels in 24 h-fasted hearts (Table 2). CPT-1 is one of the downstream targets of PPARα regulation involved in FA oxidation (57, 58). Because CPT-1 controls β-oxidation through regulating acyl-CoA entry into mitochondria, we determined whether it is regulated independently of PPARα expression levels. Cpt1b expression was increased by dexamethasone and CPT-cAMP treatments in NRVMs (Table 1). More importantly, PPARα activation is sufficient and essential for the increased Cpt1b expression in cultured cells in the absence of significant changes in Ppara expression (Figs. 3, 4) as well as during fasting (Fig. 6F). This result differs from the livers, where upregulation of hepatic Cpt1a expression depends on increased PPARα expression (Table 2) (6). We, therefore, conclude that the differential regulation of Ppara in livers compared with hearts during fasting is explained by this divergence in the mechanism of action in these two organs.
TABLE 2.
Differential regulation of PGC-1α splice variants, PPARα, CPT1, and the lipins in the liver and heart in fed and fasted conditions
| Gene | Fasted liver | Fasted heart | Notes |
| Ppara | Increased (3, 6, 55, 56), Fig. 7D) | Decreased (56, Fig. 6F | |
| Ppargc1a-a | Increased (10, Fig. 7D) | No change (Fig. 6B, E) | |
| Ppargc1a-b | Not detectable | Increased (Fig. 6B) | Increased by exercise and β2 adrenergic agonists in muscle (10) |
| Ppargc1a-c | Not detectable | Increased (Fig. 6B) | Increased by exercise and β2 adrenergic agonists in muscle (10) |
| Cpt1 | Increased (6, Fig. 7D) | Increased (Fig. 6F) | |
| Lpin1 | Increased (6, 17, 26) | Decreased (Fig. 6D) | Increased in muscle indirectly by β2 adrenergic agonist (27, 45) |
| Lpin2 | Increased (17, 26, Fig. 7A, C | No change (Fig. 6D) | Increase in mRNA but not protein in fld hearts (Figs. 6A, 8B) |
| Lpin3 | No change (26, Fig. 7C) | No change (Fig. 6D) | Increase in mRNA in fasted fld hearts (Fig. 6A) |
The table shows citations or results of the present studies.
Additionally, Cpt1b levels were induced by fasting, even though lipin-1 expression was decreased by approximately 50% (Fig. 6). We also decreased lipin-1 expression in NRVMs using shRNA and found that this did not affect the induction of Cpt1b mRNA by glucocorticoids and CPT-cAMP (results not shown). This is completely different from the situation in the liver, where increased lipin-1 expression during fasting is required for PPARα target gene induction, including Cpt1a expression (Fig. 7) (6). This does not rule out the transcriptional coactivator function of lipin-1 in the heart, because it is likely that only a minor proportion of the lipins is required to fulfill this function in hearts during fasting.
Finally, we studied whether changes in lipin levels affect the rate of FA incorporation in TG and phospholipids in cardiomyocytes. Overall, there were relatively minor changes in glycerol or oleate incorporation into glycerolipids when lipin levels were increased or decreased. We found no decreases in glycerol or oleate incorporation into glycerolipids when lipin-1 expression, which provides the major PAP activity in heart (24, 37), was decreased. Thus, results with the cultured NRVMs confirm the lack of detrimental effects on the synthesis of major phospholipids and TG in perfused fld hearts (37). These fld hearts did have increased accumulation of [14C]oleate or [1,3-3H]glycerol in PA and phosphatidylinositol, which we did not observe in the AdshRNA Lpin1-treated NRVMs, probably because lipin-1 expression was only decreased and was not completely absent. Furthermore, the demand for FA metabolism to perform contractile work in cultured cells is considerably less compared with perfused working hearts. Increased expression of either lipin-1 or lipin-2 only caused minor changes in the rate of oleate and glycerol accumulation in phospholipids or TG. Lipins participate in glycerolipid synthesis when they bind to the endoplasmic reticulum where PA is produced. Only 20% of the total cellular PAP activity associated with membranes after treatment with 0.3 or 1.2 mM oleate for 4 h, which explains why there were no major changes in the rate of glycerolipid synthesis when lipin levels were modified in NRVMs. These results further reinforce the hypothesis that cytosolic lipins act as a reservoir of PAP activity, and that only a relatively small proportion of PAP activity is required to fulfill its physiological role in glycerolipid synthesis and in preventing excessive FA accumulation when FA flux increases. However, we do not rule out the possibility that regulating the relative expressions of the different lipins could play important physiological roles in glycerolipid synthesis, transcriptional coactivation, and cell signaling (11, 13).
In conclusion, our findings demonstrate a difference in the regulation of PGC-1α splice variants between liver and heart during fasting. Ppargc1a-b mRNA in cardiomyocytes is increased by cAMP, and this explains the increases that occur in the heart when mice are fasted. By contrast, Ppargc1a-a mRNA is increased in fasted liver, with no expression of the other isoforms. We also show that fasting-induced increases in lipin-1 and lipin-2 in the liver are not mirrored by changes in lipin expression in the heart. In fact, lipin-1 expression is decreased and lipin-2 levels are unchanged in fasted hearts. This difference can be partly explained by the abundance of glucocorticoid receptors in liver compared with their relative absence in adult heart (34, 35). Finally, our work also demonstrates important mechanistic differences in PPARα regulation between liver and heart. PPARα activation in the heart can upregulate its target genes in fasting conditions independently of changes in its expression, whereas increased PPARα expression in the liver is required for its regulatory function. The present work provides a novel demonstration that the regulation of the lipins, PGC-1α splice variants, and PPARα in fasting is significantly different in the heart compared with the liver, even though the shared physiological response in both organs to fasting is to increase the capacity for FA esterification and oxidation.
Supplementary Material
Acknowledgments
The authors thank Suzanne Kovacic and Carrie-Lynn Soltys for help with the neonatal rat ventricular myocyte preparations and Jay Dewald for technical assistance.
Footnotes
Abbreviations:
- CPT1
- carnitine palmitoyltransferase-1
- CPT-cAMP
- 8-(4-chlorophenylthio)-2′-O-methyladenosine 3′5′-cyclic mono phosphate
- DG
- diacylglycerol
- GFP
- green fluorescent protein
- PA
- phosphatidate
- HBS
- HEPES-buffered saline
- LDH
- lactate dehydrogenase
- NEM
- N-ethylmaleimide
- LPP
- lipid phosphate phosphatase
- NRVM
- neonatal rat ventricular myocyte
- PAP
- phosphatidate phosphatase
- PGC-1α
- peroxisome proliferator-activated receptor gamma coactivator 1α
- PPARα
- peroxisome proliferator-activated receptor α
- TG
- triacylglycerol
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and the Heart and Stroke Foundation of Alberta and the Northwest Territories to D.N.B., a CIHR grant to J.R.B.D. and an American Diabetes Association Junior Investigator Award (7-11-JF-21) to T.E.H., and a 75th Anniversary Award from the University of Alberta to B.P.C.K.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures, and two tables.
REFERENCES
- 1.Leone T. C., Weinheimer C. J., Kelly D. P. 1999. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. USA. 96: 7473–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finck B. N., Lehman J. J., Leone T. C., Welch M. J., Bennett M. J., Kovacs A., Han X., Gross R. W., Kozak R., Lopaschuk G. D., et al. 2002. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J. Clin. Invest. 109: 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kersten S., Seydoux J., Peters J. M., Gonzalez F. J., Desvergne B., Wahli W. 1999. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Invest. 103: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilde A. J., van der Lee K. A., Willemsen P. H., Chinetti G., van der Leij F. R., van der Vusse G. J., Staels B., van Bilsen M. 2003. Peroxisome proliferator-activated receptor (PPAR) alpha and PPARbeta/delta, but not PPARgamma, modulate the expression of genes involved in cardiac lipid metabolism. Circ. Res. 92: 518–524 [DOI] [PubMed] [Google Scholar]
- 5.Campbell F. M., Kozak R., Wagner A., Altarejos J. Y., Dyck J. R., Belke D. D., Severson D. L., Kelly D. P., Lopaschuk G. D. 2002. A role for peroxisome proliferator-activated receptor alpha (PPARalpha) in the control of cardiac malonyl-CoA levels: reduced fatty acid oxidation rates and increased glucose oxidation rates in the hearts of mice lacking PPARalpha are associated with higher concentrations of malonyl-CoA and reduced expression of malonyl-CoA decarboxylase. J. Biol. Chem. 277: 4098–4103 [DOI] [PubMed] [Google Scholar]
- 6.Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., Jr, Kelly D. P. 2006. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 4: 199–210 [DOI] [PubMed] [Google Scholar]
- 7.Finck B. N., Kelly D. P. 2006. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 116: 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herzig S., Long F., Jhala U. S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., et al. 2001. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 413: 179–183 [DOI] [PubMed] [Google Scholar]
- 9.Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., et al. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 413: 131–138 [DOI] [PubMed] [Google Scholar]
- 10.Miura S., Kai Y., Kamei Y., Ezaki O. 2008. Isoform-specific increases in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to beta2-adrenergic receptor activation and exercise. Endocrinology. 149: 4527–4533 [DOI] [PubMed] [Google Scholar]
- 11.Reue K., Brindley D. N. 2008. Thematic Review Series: Glycerolipids. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 49: 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris T. E., Finck B. N. 2011. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol. Metab. 22: 226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kok B. P., Venkatraman G., Capatos D., Brindley D. N. 2012. Unlike two peas in a pod: lipid phosphate phosphatases and phosphatidate phosphatases. Chem. Rev. 112: 5121–5146 [DOI] [PubMed] [Google Scholar]
- 14.Donkor J., Zhang P., Wong S., O'Loughlin L., Dewald J., Kok B. P., Brindley D. N., Reue K. 2009. A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J. Biol. Chem. 284: 29968–29978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282: 3450–3457 [DOI] [PubMed] [Google Scholar]
- 16.Han G. S., Carman G. M. 2010. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 285: 14628–14638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gropler M. C., Harris T. E., Hall A. M., Wolins N. E., Gross R. W., Han X., Chen Z., Finck B. N. 2009. Lipin 2 is a liver-enriched phosphatidate phosphohydrolase enzyme that is dynamically regulated by fasting and obesity in mice. J. Biol. Chem. 284: 6763–6772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bou Khalil M., Sundaram M., Zhang H. Y., Links P. H., Raven J. F., Manmontri B., Sariahmetoglu M., Tran K., Reue K., Brindley D. N., et al. 2009. The level and compartmentalization of phosphatidate phosphatase-1 (lipin-1) control the assembly and secretion of hepatic VLDL. J. Lipid Res. 50: 47–58 [DOI] [PubMed] [Google Scholar]
- 19.Han G. S., Wu W. I., Carman G. M. 2006. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281: 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burroughs A. M., Allen K. N., Dunaway-Mariano D., Aravind L. 2006. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 361: 1003–1034 [DOI] [PubMed] [Google Scholar]
- 21.Carman G. M., Han G. S. 2006. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem. Sci. 31: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopewell R., Martin-Sanz P., Martin A., Saxton J., Brindley D. N. 1985. Regulation of the translocation of phosphatidate phosphohydrolase between the cytosol and the endoplasmic reticulum of rat liver. Effects of unsaturated fatty acids, spermine, nucleotides, albumin and chlorpromazine. Biochem. J. 232: 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cascales C., Mangiapane E. H., Brindley D. N. 1984. Oleic acid promotes the activation and translocation of phosphatidate phosphohydrolase from the cytosol to particulate fractions of isolated rat hepatocytes. Biochem. J. 219: 911–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., Lawrence J. C., Jr 2007. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282: 277–286 [DOI] [PubMed] [Google Scholar]
- 25.Karanasios E., Han G. S., Xu Z., Carman G. M., Siniossoglou S. 2010. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. USA. 107: 17539–17544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manmontri B., Sariahmetoglu M., Donkor J., Khalil M. B., Sundaram M., Yao Z., Reue K., Lehner R., Brindley D. N. 2008. Glucocorticoids and cyclic AMP selectively increase hepatic lipin-1 expression, and insulin acts antagonistically. J. Lipid Res. 49: 1056–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitra M. S., Schilling J. D., Wang X., Jay P. Y., Huss J. M., Su X., Finck B. N. 2011. Cardiac lipin 1 expression is regulated by the peroxisome proliferator activated receptor gamma coactivator 1alpha/estrogen related receptor axis. J. Mol. Cell. Cardiol. 51: 120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang P., O'Loughlin L., Brindley D. N., Reue K. 2008. Regulation of lipin-1 gene expression by glucocorticoids during adipogenesis. J. Lipid Res. 49: 1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pittner R. A., Fears R., Brindley D. N. 1985. Effects of cyclic AMP, glucocorticoids and insulin on the activities of phosphatidate phosphohydrolase, tyrosine aminotransferase and glycerol kinase in isolated rat hepatocytes in relation to the control of triacylglycerol synthesis and gluconeogenesis. Biochem. J. 225: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopaschuk G. D., Ussher J. R., Folmes C. D., Jaswal J. S., Stanley W. C. 2010. Myocardial fatty acid metabolism in health and disease. Physiol. Rev. 90: 207–258 [DOI] [PubMed] [Google Scholar]
- 31.Saddik M., Lopaschuk G. D. 1991. Myocardial triglyceride turnover and contribution to energy substrate utilization in isolated working rat hearts. J. Biol. Chem. 266: 8162–8170 [PubMed] [Google Scholar]
- 32.Haemmerle G., Moustafa T., Woelkart G., Buttner S., Schmidt A., van de Weijer T., Hesselink M., Jaeger D., Kienesberger P. C., Zierler K., et al. 2011. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat. Med. 17: 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kienesberger P. C., Pulinilkunnil T., Nagendran J., Dyck J. R. 2013. Myocardial triacylglycerol metabolism. J. Mol. Cell. Cardiol. 55: 101–110 [DOI] [PubMed] [Google Scholar]
- 34.Giannopoulos G., Hassan Z., Solomon S. 1974. Glucocorticoid receptors in fetal and adult rabbit tissues. J. Biol. Chem. 249: 2424–2427 [PubMed] [Google Scholar]
- 35.Pujols L., Mullol J., Roca-Ferrer J., Torrego A., Xaubet A., Cidlowski J. A., Picado C. 2002. Expression of glucocorticoid receptor alpha- and beta-isoforms in human cells and tissues. Am. J. Physiol. Cell Physiol. 283: C1324–C1331 [DOI] [PubMed] [Google Scholar]
- 36.Kovacic S., Soltys C. L., Barr A. J., Shiojima I., Walsh K., Dyck J. R. 2003. Akt activity negatively regulates phosphorylation of AMP-activated protein kinase in the heart. J. Biol. Chem. 278: 39422–39427 [DOI] [PubMed] [Google Scholar]
- 37.Kok B. P., Kienesberger P. C., Dyck J. R., Brindley D. N. 2012. Relationship of glucose and oleate metabolism to cardiac function in lipin-1 deficient (fld) mice. J. Lipid Res. 53: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin A., Gómez-Muñoz A., Jamal Z., Brindley D. N. 1991. Characterization and assay of phosphatidate phosphatase. Methods Enzymol. 197: 553–563 [DOI] [PubMed] [Google Scholar]
- 39.Ko K. W., Erickson B., Lehner R. 2009. Es-x/Ces1 prevents triacylglycerol accumulation in McArdle-RH7777 hepatocytes. Biochim. Biophys. Acta. 1791: 1133–1143 [DOI] [PubMed] [Google Scholar]
- 40.Mangiapane E. H., Brindley D. N. 1986. Effects of dexamethasone and insulin on the synthesis of triacylglycerols and phosphatidylcholine and the secretion of very-low-density lipoproteins and lysophosphatidylcholine by monolayer cultures of rat hepatocytes. Biochem. J. 233: 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin A., Duffy P. A., Liossis C., Gómez-Muñoz A., O'Brien L., Stone J. C., Brindley D. N. 1997. Increased concentrations of phosphatidate, diacylglycerol and ceramide in ras- and tyrosine kinase (fps)-transformed fibroblasts. Oncogene. 14: 1571–1580 [DOI] [PubMed] [Google Scholar]
- 42.Mackall J., Meredith M., Lane M. D. 1979. A mild procedure for the rapid release of cytoplasmic enzymes from cultured animal cells. Anal. Biochem. 95: 270–274 [DOI] [PubMed] [Google Scholar]
- 43.Saggerson E. D., Greenbaum A. L. 1969. The effect of dietary and hormonal conditions on the activities of glycolytic enzymes in rat epididymal adipose tissue. Biochem. J. 115: 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu H. E., Stanley T. B., Montana V. G., Lambert M. H., Shearer B. G., Cobb J. E., McKee D. D., Galardi C. M., Plunket K. D., Nolte R. T., et al. 2002. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 415: 813–817 [DOI] [PubMed] [Google Scholar]
- 45.Pearen M. A., Myers S. A., Raichur S., Ryall J. G., Lynch G. S., Muscat G. E. 2008. The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology. 149: 2853–2865 [DOI] [PubMed] [Google Scholar]
- 46.Xiao R. P. 2001. Beta-adrenergic signaling in the heart: dual coupling of the beta2-adrenergic receptor to G(s) and G(i) proteins. Sci. STKE. 104: re15. [DOI] [PubMed] [Google Scholar]
- 47.Xiao R. P., Zhu W., Zheng M., Cao C., Zhang Y., Lakatta E. G., Han Q. 2006. Subtype-specific alpha1- and beta-adrenoceptor signaling in the heart. Trends Pharmacol. Sci. 27: 330–337 [DOI] [PubMed] [Google Scholar]
- 48.Vavrecˇka M., Mitchell M. P., Hübscher G. 1969. The effect of starvation on the incorporation of palmitate into glycerides and phospholipids of rat liver homogenates. Biochem. J. 115: 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson T. R., Sengupta S. S., Harris T. E., Carmack A. E., Kang S. A., Balderas E., Guertin D. A., Madden K. L., Carpenter A. E., Finck B. N., et al. 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 146: 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoonderwoerd K., Broekhoven-Schokker S., Hülsmann W. C., Stam H. 1990. Properties of phosphatidate phosphohydrolase and diacylglycerol acyltransferase activities in the isolated rat heart. Effect of glucagon, ischaemia and diabetes. Biochem. J. 268: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jamal Z., Martin A., Gómez-Muñoz A., Hales P., Chang E., Russell J. C., Brindley D. N. 1992. Phosphatidate phosphohydrolases in liver, heart and adipose tissue of the JCR:LA corpulent rat and the lean genotypes: implications for glycerolipid synthesis and signal transduction. Int. J. Obes. Relat. Metab. Disord. 16: 789–799 [PubMed] [Google Scholar]
- 52.Burgdorf C., Hansel L., Heidbreder M., Johren O., Schutte F., Schunkert H., Kurz T. 2009. Suppression of cardiac phosphatidate phosphohydrolase 1 activity and lipin mRNA expression in Zucker diabetic fatty rats and humans with type 2 diabetes mellitus. Biochem. Biophys. Res. Commun. 390: 165–170 [DOI] [PubMed] [Google Scholar]
- 53.Ryu D., Oh K. J., Jo H. Y., Hedrick S., Kim Y. N., Hwang Y. J., Park T. S., Han J. S., Choi C. S., Montminy M., et al. 2009. TORC2 regulates hepatic insulin signaling via a mammalian phosphatidic acid phosphatase, LIPIN1. Cell Metab. 9: 240–251 [DOI] [PubMed] [Google Scholar]
- 54.Dwyer J. R., Donkor J., Zhang P., Csaki L. S., Vergnes L., Lee J. M., Dewald J., Brindley D. N., Atti E., Tetradis S., et al. 2012. Mouse lipin-1 and lipin-2 cooperate to maintain glycerolipid homeostasis in liver and aging cerebellum. Proc. Natl. Acad. Sci. USA. 109: E2486–E2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Escher P., Braissant O., Basu-Modak S., Michalik L., Wahli W., Desvergne B. 2001. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 142: 4195–4202 [DOI] [PubMed] [Google Scholar]
- 56.Van der Lee K. A., Willemsen P. H., Samec S., Seydoux J., Dulloo A. G., Pelsers M. M., Glatz J. F., Van der Vusse G. J., Van Bilsen M. 2001. Fasting-induced changes in the expression of genes controlling substrate metabolism in the rat heart. J. Lipid Res. 42: 1752–1758 [PubMed] [Google Scholar]
- 57.Mascaro C., Acosta E., Ortiz J. A., Marrero P. F., Hegardt F. G., Haro D. 1998. Control of human muscle-type carnitine palmitoyltransferase I gene transcription by peroxisome proliferator-activated receptor. J. Biol. Chem. 273: 8560–8563 [DOI] [PubMed] [Google Scholar]
- 58.Brandt J. M., Djouadi F., Kelly D. P. 1998. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 273: 23786–23792 [DOI] [PubMed] [Google Scholar]
- 59.Brady P. S., Brady L. J. 1989. Regulation of carnitine palmitoyltransferase in vivo by glucagon and insulin. Biochem. J. 258: 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.