Absence of the transcriptional regulator Zfp521 results in decreased bone formation by osteoblasts and increased osteoclast differentiation, largely via Zfp521’s regulation of the transcription factor Ebf1.
Abstract
Bone homeostasis is maintained by the coupled actions of hematopoietic bone-resorbing osteoclasts (OCs) and mesenchymal bone-forming osteoblasts (OBs). Here we identify early B cell factor 1 (Ebf1) and the transcriptional coregulator Zfp521 as components of the machinery that regulates bone homeostasis through coordinated effects in both lineages. Deletion of Zfp521 in OBs led to impaired bone formation and increased OB-dependent osteoclastogenesis (OC-genesis), and deletion in hematopoietic cells revealed a strong cell-autonomous role for Zfp521 in OC progenitors. In adult mice, the effects of Zfp521 were largely caused by repression of Ebf1, and the bone phenotype of Zfp521+/− mice was rescued in Zfp521+/−:Ebf1+/− mice. Zfp521 interacted with Ebf1 and repressed its transcriptional activity. Accordingly, deletion of Zfp521 led to increased Ebf1 activity in OBs and OCs. In vivo, Ebf1 overexpression in OBs resulted in suppressed bone formation, similar to the phenotype seen after OB-targeted deletion of Zfp521. Conversely, Ebf1 deletion led to cell-autonomous defects in both OB-dependent and cell-intrinsic OC-genesis, a phenotype opposite to that of the Zfp521 knockout. Thus, we have identified the interplay between Zfp521 and Ebf1 as a novel rheostat for bone homeostasis.
The mammalian skeleton is continuously remodeled. This process needs to be tightly regulated to maintain skeletal homeostasis while ensuring structural integrity and support of metabolic functions. The number and activity of bone-resorbing osteoclasts (OCs) and bone-forming osteoblasts (OBs) are balanced to ensure homeostasis of the postdevelopmental skeleton. A complex network of endocrine and paracrine signals orchestrates bone remodeling by controlling mesenchymal and hematopoietic progenitor cell differentiation and/or the activity of the mature cells. These signals converge to control the expression and activity of specific transcription factors that modulate cellular functions by regulating the expression of their target genes (Karsenty et al., 2009). The activity of these transcription factors is controlled by association with different activator or repressor complexes, some of which are cell lineage specific, whereas others are relevant in multiple cell types but specifically regulated (MacDonald et al., 2009).
The regulation of bone homeostasis during remodeling involves three essential components: (1) OB differentiation and bone matrix production, (2) OB-dependent regulation of osteoclastogenesis (OC-genesis) through the secretion of receptor activator of NF-κB ligand (RANKL) and osteoprotegerin (OPG; Boyle et al., 2003), and (3) cell-autonomous regulation of OC differentiation and bone resorption within hematopoietic OC precursors (Negishi-Koga and Takayanagi, 2009). Although much is known about the transcriptional program regulating OB and OC differentiation and function, our understanding of the coordinated regulation of these two lineages and bone remodeling as a whole is still only partial. For a single transcription factor to achieve such coordination, it would have to affect all the three components of bone remodeling to regulate bone formation and bone resorption in an opposite manner, i.e., increasing bone formation and reducing bone resorption to control bone mass. However, many transcription factors affect bone formation and resorption in parallel, not in an opposite manner. For instance, several AP1 transcription factors and Nfatc1 are positive regulators of OB function (Yang and Karsenty, 2004; Yang et al., 2004; Koga et al., 2005; Bozec et al., 2010) but enhance OC-genesis either indirectly via OBs (ATF4) or in OC precursors (Nfatc1). Thus, these factors act more as rheostats for bone turnover, increasing both bone formation and resorption, rather than a rheostat for bone mass, a critical consideration in the clinic. FoxO family transcription factors, PPARγ, and β-catenin all function both in mesenchymal and hematopoietic progenitors to regulate bone formation and resorption in an opposite manner, but even these key regulators do not affect all three components of bone remodeling (Glass et al., 2005; MacDonald et al., 2009; Wan, 2010; Almeida, 2011; Kousteni, 2011; Wei et al., 2011; Otero et al., 2012).
The transcriptional regulators Zfp521 and early B cell factor 1 (Ebf1), both first identified in the hematopoietic system (Warming et al., 2003; Lukin et al., 2008), have recently emerged as important players in bone biology. Zfp521 interacts with and suppresses Runx2 activity to regulate early skeletal development, whereas overexpression of Zfp521 in mature OBs promotes bone formation (Wu et al., 2009; Hesse et al., 2010). Conversely, deletion of Ebf1 in mice results in increased bone formation and increased BM adiposity (Hesslein et al., 2009).
We show here that the interplay of Zfp521 and Ebf1 can coordinately regulate bone mass. Through its activity in both OBs and OCs, Zfp521 affects all three components of bone remodeling, positively affecting bone homeostasis. Furthermore, we show that the negative effects of Ebf1 on bone mass are endogenously repressed by the transcriptional modulator Zfp521 in both OBs and OCs, such that the latter exerts a positive and coordinated influence on bone homeostasis. We have therefore identified the interaction between Zfp521 and Ebf1 as a novel regulator of bone homeostasis that functions in both mesenchymal and hematopoietic cells to modulate bone formation and bone resorption in a coordinated manner, acting as a rheostat for bone homeostasis.
RESULTS
Germline deletion of Zfp521 decreases bone formation and increases bone resorption, leading to osteopenia
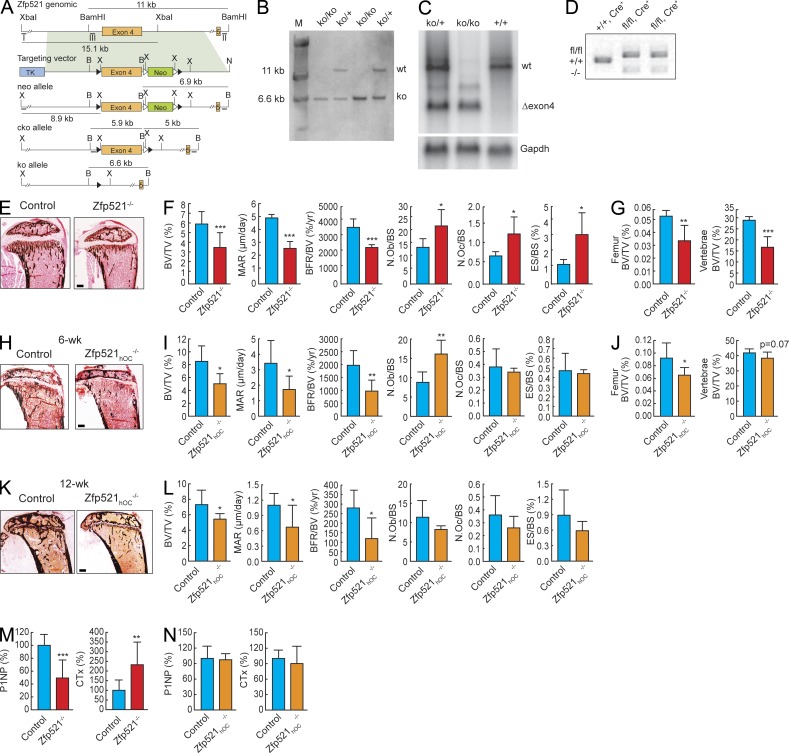
OB-targeted overexpression of Zfp521 leads to a high postdevelopmental bone mass with a high bone formation rate (BFR; Wu et al., 2009). Here we sought to determine whether Zfp521 was required for normal skeletal homeostasis by generating a germline deletion in mice (Fig. 1, A–D). The skeleton of Zfp521−/− mice developed normally, but these mice were runted and survived only 4–5 wk. We therefore analyzed their bones at 3 wk of age. Histomorphometric analysis showed that Zfp521−/− mice were osteopenic, with decreased mineral apposition rate (MAR; the activity of individual OBs) and BFR (BFR/bone volume [BV]; overall bone formation activity). Interestingly, this occurred despite increased OB numbers, suggesting that Zfp521 affects mostly OB function (Fig. 1, E and F; and Table S1). Low bone mass was confirmed by microcomputed tomography (μCT) analysis, and lower P1NP levels in Zfp521−/− mice verified the decrease in bone formation (Fig. 1, G and M). These mice also showed an increased number of OCs and eroded surfaces (ESs; ES/bone surface [BS]), as well as elevated levels of serum CTX (C-terminal telopeptide of type I collagen), showing that germline deletion of Zfp521 not only decreases bone formation but also increases bone resorption (Fig. 1, F and M).
Figure 1.
Zfp521 favors bone formation in mature OBs. (A) Generation of null and conditional Zfp521 alleles. Zfp521 genomic region encoding exon 4 is shown. Restriction fragment sizes are indicated as well as the positions of the internal and the two flanking probes used for genotyping analysis (Roman numerals). The shaded area indicates the part of the genomic region included in the targeting vector, and the three different alleles are shown. “neo” is the result of the gene-targeting event. “cko” is the conditional knockout allele derived from the neo allele after Flpe-mediated excision of the PGK-neo cassette. One Frt site and two loxP sites remain in the locus. “ko” is the null allele derived from the “neo” or from the “cko” allele by Cre-mediated recombination between the two loxP sites. Only a single loxP site remains in the modified locus. Splicing of exon 3 to exon 5 generates a frameshift. loxP and Frt sites are indicated as closed and open triangles, respectively. Neo, PGK-em7-neomycin dual selection cassette for bacteria and embryonic stem cells; TK, thymidine kinase cassette for counter-selection in embryonic stem cells; X, XbaI; B, BamHI; N, NotI. The genomic region is not drawn to scale. (B) Results of a Southern blot analysis of BamHI-digested tail DNA, probed with the internal probe (III). (C) Northern blot analysis of whole-brain RNA from 3-wk-old mice using a full-length Zfp521 cDNA probe. The blot was rehybridized with a Gapdh probe as a control for RNA quality. wt, wild type; Δexon4, position of the residual mRNA after removal of exon 4. (D) Genotyping PCR showing deletion of Zfp521 allele in genomic DNA extracted from Zfp521hOC−/− long bones cleaned of soft tissues and BM. (E) Von Kossa staining of tibia sections in 3-wk-old global Zfp521−/− mice and Zfp521+/+ littermate controls. (F) Histomorphometric analysis of samples in E (n = 5). (G) Trabecular BV (BV/tissue volume [TV]) at distal femoral metaphysis and in second lumbar vertebra in 3-wk-old Zfp521−/− and control mice measured by μCT (n = 5). (H) Von Kossa staining of tibia sections in 6-wk-old Zfp521hOC−/− mice and littermate controls. (I) Histomorphometric analysis of samples in H (n = 6). (J) Trabecular BV (BV/TV) at distal femoral metaphysis and in second lumbar vertebra in 12-wk-old Zfp521hOC−/− and control mice measured by μCT (n = 5–6). (K) Von Kossa staining of tibia sections in 12-wk-old Zfp521hOC−/− mice and littermate controls. (L) Histomorphometric analysis of samples in K (n = 6). (M) Serum PINP and CTX levels in 3-wk-old global Zfp521−/− mice and Zfp521+/+ littermate controls (n = 6–9). (N) Serum PINP and CTX levels in 6-wk-old Zfp521hOC−/− and control mice (n = 5–6). N.Ob, number of OBs; N.Oc, number of OCs. All data are means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars: (E) 400 μm; (H and K) 600 μm. See also Tables S1–S3.
Conditional deletion of Zfp521 in mature OBs decreases bone formation but does not affect bone resorption
To determine whether the effects of Zfp521 on bone homeostasis were OB dependent, and to circumvent the early lethality of the full deletion, we generated conditional Zfp521 knockouts targeted to mature OBs using the human osteocalcin (hOC)–Cre mouse line (Zhang et al., 2002). Unlike Zfp521−/− mice, Zfp521hOC−/− mice grew normally, allowing us to perform analysis in more mature skeletons, at 6 and 12 wk. Similar to the global knockout, histomorphometric analysis revealed that deletion of Zfp521 in OBs resulted in decreased BV (Fig. 1, H, I, K, and L; and Tables S2 and S3). The osteopenic phenotype was also confirmed by μCT (Fig. 1 J). Bone formation was impaired, with both MAR and BFR/BV significantly decreased, although serum PINP (N-terminal propeptide of type I procollagen) was not significantly decreased (Fig. 1, I, L, and N). Interestingly, and again similar to the germline deletion, the number of OBs was increased in 6-wk-old Zfp521hOC−/− mice, despite the decrease in BFR/BV (Fig. 1 I). This discrepancy was maintained at 12 wk, with the BFR continuing to be low and the number of OBs remaining as high as in controls (Fig. 1 L and Tables S2 and S3). Thus, the deletion of Zfp521 in mature OBs leads to a bone formation phenotype that is reversed compared with what we observed in mice overexpressing Zfp521, also under the control of the osteocalcin (Ocn) promoter (Wu et al., 2009). These findings demonstrate that Zfp521 has a cell-autonomous, nonredundant function in mature OBs in vivo to promote bone formation. However, and in contrast with germline deletion, there was no difference in OC number, ES, or serum CTX in Zfp521hOC−/− mice (Fig. 1, I, L, and N).
Zfp521 is required for OB maturation and function
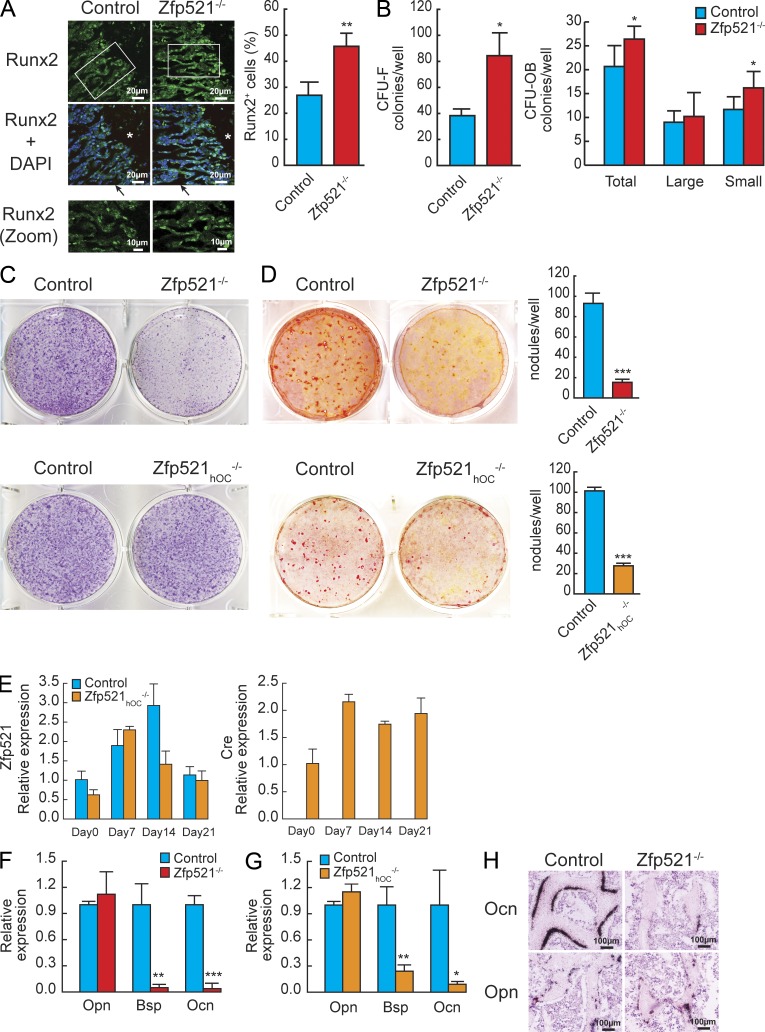
The association of low MAR and BFR with an increase in OB numbers in Zfp521-deficient mice suggested that deletion of Zfp521 induces the accumulation of poorly functional OBs along trabecular surfaces and therefore that OB maturation could be altered in the absence of Zfp521. Consistent with this hypothesis, we detected more OBs at early stages of differentiation along Zfp521−/− trabeculae, using Runx2 as an osteoprogenitor marker (Fig. 2 A), and an increased number of alkaline phosphatase (ALP)–positive fibroblast CFUs (CFU-Fs) in Zfp521−/− BM (Fig. 2 B). Interestingly, the number of Alizarin red–positive colonies (CFU-OB) was only modestly increased, with only the number of small nodules being significantly increased, which suggests that the Zfp521−/− progenitors are increased in numbers but are impaired in their OB function, i.e., their capacity to form large bone nodules.
Figure 2.
Zfp521 is required for OB maturation. (A) Cryosections of 3-wk-old Zfp521−/− and control mice showing the distal femoral metaphysis were immunostained for Runx2 (green). Nuclear DAPI staining (dark blue) and colocalization with Runx2 (light blue) are shown. Higher magnification images of the marked areas show trabecular surfaces. Asterisks indicate the growth plate, and arrows indicate the trabecular bone. Bar graph shows the ratio of Runx2+/total cell number quantified in the primary spongiosa from confocal images (n = 3 mice/genotype). (B) An equal number of BM cells flushed from Zfp521−/− and control mice was plated on 6-well plates and cultured in osteogenic medium (OM) for 10 and 21 d. The cells were stained for ALP activity on day 10 to count the number of CFU-Fs and with Alizarin red for osteoblastic colonies (CFU-OB) on day 21. (C) Calvarial cells from Zfp521−/−, Zfp521hOC−/−, and respective control newborn mice were harvested and cultured on 6-well plates for 7 d in OM and stained for ALP activity. (D) Zfp521−/−, Zfp521hOC−/−, and control calvarial cells were cultured for 21 d in OM and stained with Alizarin red to detect mineralized bone nodules. Alizarin red–positive nodules were quantified (bar graphs). (E) Time course of Zfp521 and hOC-Cre expression in Zfp521hOC−/− and control calvarial cells during OB differentiation was measured using quantitative RT-PCR (qRT-PCR) in RNA extracted from cultures in C and D. (F) Expression of late OB marker genes Bsp and Ocn was measured by qRT-PCR in Zfp521−/− and control calvarial cells cultured for 21 d in OM. (G) Expression of late OB marker genes Bsp and Ocn was measured by qRT-PCR in Zfp521hOC−/− and control calvarial cells cultured for 21 d in OM. (H) Expression of early (Opn) and late (Ocn) marker genes by in situ hybridization in Zfp521−/− and control bones. All data are means ± SD. Similar results were obtained in at least three separate experiments performed in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In vitro, Zfp521−/− calvarial cells exhibited decreased ALP staining (Fig. 2 C) and mRNA expression (not depicted) at day 7. As expected, there was no detectable change in ALP in Zfp521hOC−/− cultures because hOC-Cre deleted Zfp521 in more mature cells after they became ALP positive (Fig. 2 C). The formation of mineralized bone nodules was comparably impaired in both Zfp521−/− and Zfp521hOC−/− cultures (Fig. 2 D). In vitro, hOC-Cre deleted Zfp521 only in the fraction of cells that became hOC-Cre positive, reducing the apparent expression of Zfp521 in the total cell pool at day 14 when Zfp521 expression is normally the highest (Fig. 2 E). The Zfp521−/− and Zfp521hOC−/− cultures shared nearly identical gene expression fingerprints, where the expression of early OB marker genes such as Osteopontin (Opn) was unchanged but the expression of the mature OB marker genes Bone sialoprotein (Bsp) and Ocn was markedly decreased, further demonstrating impaired OB maturation in the absence of Zfp521 (Fig. 2, F and G). This finding was confirmed in vivo by in situ hybridization for Opn and Ocn (Fig. 2 H).
Thus, global and hOC-Cre–targeted Zfp521 deletions resulted in similar impairment of OB maturation and function, but germline deletion increased bone resorption, whereas OB-targeted deletion with the Ocn promoter did not. This discrepancy suggested that the regulation of OCs by Zfp521 may occur in cells other than OBs and/or at earlier stages of differentiation within the OB lineage, before efficient expression of Ocn. To address this question, we used two different but complementary approaches.
Zfp521 affects OB-dependent OC-genesis when deleted early
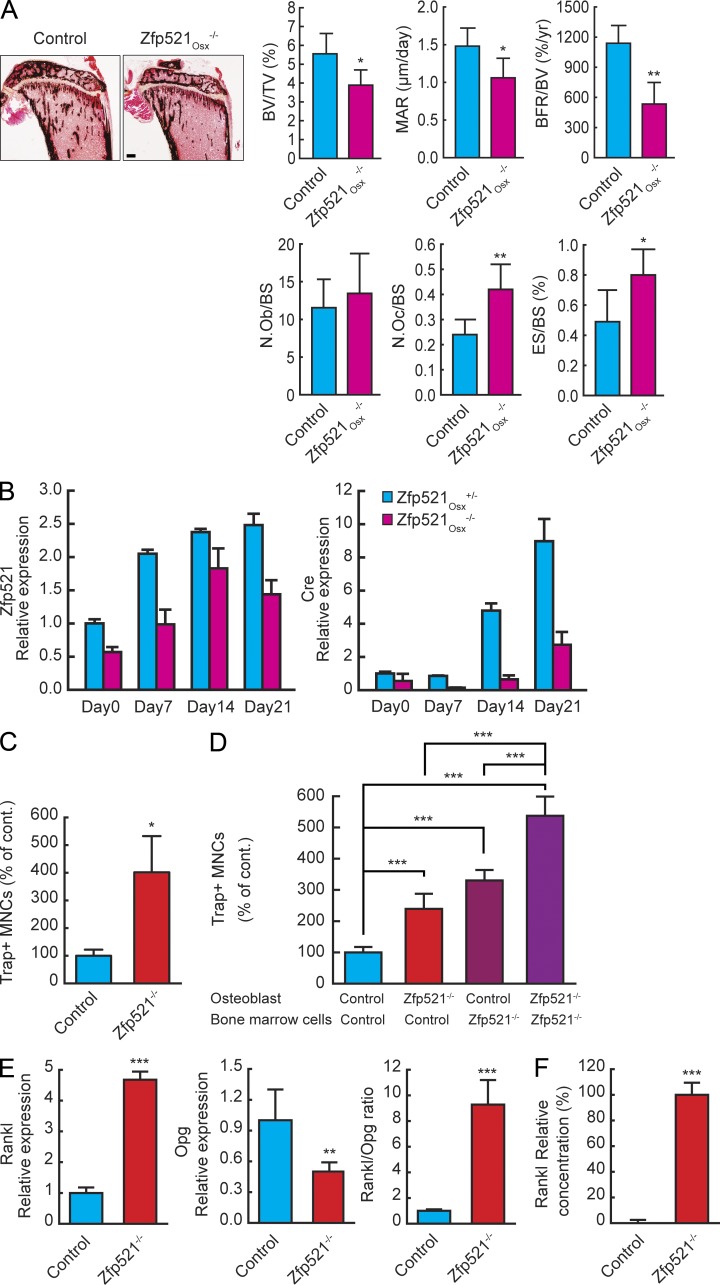
In vivo, we used osterix (Osx)-Cre mice to target Zfp521 deletion to early OBs (Rodda and McMahon, 2006). Similar to Zfp521−/− and Zfp521hOC−/− mice, Zfp521Osx−/− mice were osteopenic and exhibited impaired bone formation when compared with Cre-expressing Zfp521Osx+/+ control mice (Fig. 3 A and Table S4). However, like the Zfp521−/− mice, Zfp521Osx−/− mice exhibited increased number of OCs and ES, a change not observed in Zfp521hOC−/−. This result shows that Zfp521 controls OB-dependent OC-genesis in vivo in early cells within the OB lineage that express (or have expressed) Osx-Cre but in which the hOC promoter is not yet active (Fig. 3 B).
Figure 3.
Zfp521 controls OB-dependent OC-genesis. (A) Von Kossa staining and histomorphometric analysis of tibia sections in 6-wk-old Zfp521Osx−/− mice and littermate controls (n = 6–8). N.Ob, number of OBs; N.Oc, number of OCs; TV, tissue volume. Bar, 520 μm. (B) Calvarial cells from Zfp521Osx−/− and control newborn mice were harvested and cultured on 6-well plates in OM. RNA was extracted at the indicated times, and Zfp521 and Osx-Cre expression were measured by qRT-PCR. (C) Total BM cells from Zfp521−/− and control mice were plated on 48-well plates, stimulated with 10 nM hPTH(1–34), and stained for TRAP. Number of TRAP+ multinucleated cells (MNCs) per well is shown. (D) Calvarial and nonadherent BM cells from control and Zfp521−/− mice were mixed as indicated in 24-well plates, stimulated with vitD3 and PGE2, and stained for TRAP. Bar graphs indicate the number of TRAP+ multinucleated cells per well. (E) RNA was extracted from the co-culture experiment in D, and the mRNA expression of Rankl and Opg and the Rankl/Opg ratio were quantified using qRT-PCR. (F) Control and Zfp521−/− calvarial cells were cultured in 24-well plates and stimulated with vitD3 and PGE2 as in co-culture experiments. RANKL concentration in the controls was below detection limit (n = 3). All data are means ± SD. Similar cell number and mRNA data were obtained from at least three experiments with three to six replicates per condition. *, P < 0.05; **, P < 0.01; ***, P < 0.001. See also Table S4.
The main mechanism by which OBs regulate OC-genesis involves the secretion of RANKL and OPG. We therefore measured the production of these two cytokines in cells derived from Zfp521−/− mice. In vitro, OC production was strikingly increased in Zfp521 total BM cultures when stimulated with parathyroid hormone (PTH), a known inducer of endogenous Rankl expression in OBs (Fig. 3 C). Comparison of classical co-cultures of calvarial cells from control or Zfp521−/− mice mixed with nonadherent BM from wild-type or Zfp521−/− animals showed that Zfp521−/− calvarial cells were more efficient in supporting OC-genesis than control cells (Fig. 3 D), implicating Zfp521 in OB-dependent OC-genesis. In contrast, calvarial cells from control and Zfp521hOC−/−, in which OC numbers were not altered in vivo, did not differ in their capacity to support OC-genesis (not depicted). This confirmed that Zfp521 acts at a defined stage within the OB lineage to control OC-genesis. Zfp521−/− calvarial cells expressed significantly more Rankl and less Opg mRNAs than control cells, resulting in a pronounced increase in the Rankl/Opg ratio (Fig. 3 E), and the culture medium from Zfp521−/− OBs contained significantly more Rankl protein than control medium (503.3 ± 38.6 pg/ml vs. undetectable; Fig. 3 F). Thus, deletion of Zfp521 at early stages within the OB lineage increases the Rankl/Opg ratio, enhancing OC-genesis.
However, these mix-and-match experiments also showed increased OC-genesis in Zfp521−/− BM even when these cells were cultured with control calvarial cells, i.e., in the presence of a normal RANKL/OPG ratio. This suggests that Zfp521 could also be involved in the regulation of OC-genesis through its activity within hematopoietic cells (Fig. 3 D).
Zfp521 deletion in hematopoietic cells also increases OC-genesis
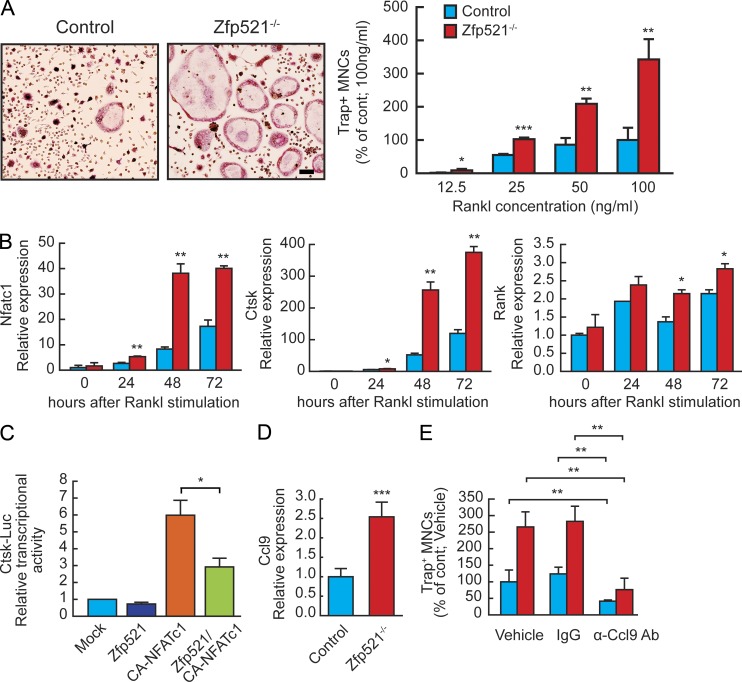
Confirming this hypothesis, RANKL-induced differentiation of BM and spleen-derived Zfp521−/− OC progenitors was both more rapid and markedly increased (Fig. 4 A and not depicted, respectively). This was consistent across a range of RANKL concentrations (Fig. 4 A). To elucidate the mechanisms by which Zfp521 could function within the OC lineage, we compared transcriptional profiling of known regulators of OC-genesis downstream of RANKL in control and Zfp521−/− OC progenitors. The expression of Nfatc1 and of its target genes, Ctsk and Rank, was increased after RANKL stimulation in Zfp521−/− OCs derived from BM macrophages (BMMs; Fig. 4 B). Moreover, Zfp521 repressed Nfatc1 transcriptional activity in an in vitro Cathepsin K promoter-luciferase assay in Raw-267 cells (Fig. 4 C). In addition, the expression of Ccl9, an Ebf1 target gene which has been reported to enhance RANKL-stimulated OC-genesis (Okamatsu et al., 2004), was up-regulated at steady-state (not depicted) and was markedly increased 4 h after RANKL stimulation in Zfp521−/− cultures (Fig. 4 D). Blocking Ccl9 activity by neutralizing anti-Ccl9 antibody normalized OC-genesis in Zfp521−/− OC progenitor cells (Fig. 4 E). These data suggest that Zfp521 controls OC-genesis by two complementary mechanisms. First, Zfp521 regulates the autocrine expression of Ccl9 in OC progenitors, enhancing RANKL-stimulated OC-genesis. Second, Zfp521 controls the activity of RANK downstream signaling by inhibiting Nfatc1 transcriptional activity. Together, these two mechanisms contribute to the increased OC-genesis observed in vitro and in vivo when Zfp521 is absent within the hematopoietic lineage.
Figure 4.
Zfp521 acts in OC progenitor cells to control OC-genesis. (A) Control and Zfp521−/− spleen cells were stimulated with 20 ng/ml M-CSF and then with increasing doses of RANKL for 3 d, stained for TRAP activity, and quantified. Bar, 64 μm. (B) BMM-derived OCs were cultured with 20 ng/ml M-CSF and then M-CSF + 100 ng/ml RANKL for the indicated times, and expression of Nfatc1, Ctsk, and Rank was measured by qRT-PCR. (C) Ctsk-Luciferase reporter construct was transfected to RAW264.7 cells together with Zfp521, constitutively active NFATc1, or both. Luciferase activity was normalized to cotransfected Renilla activity. (D) Nonadherent BM cells were cultured for 2 d with 20 ng/ml M-CSF and then stimulated for 4 h with 100 ng/ml RANKL and 20 ng/ml M-CSF, and expression of Ccl9 was measured by qRT-PCR. (E) Control and Zfp521−/− spleen cells were cultured with 20 ng/ml M-CSF and 50 ng/ml RANKL for 5 d in the presence of blocking anti-Ccl9 antibody, IgG, or vehicle. Cells were fixed and stained for TRAP activity and quantified. All data are means ± SD. Similar cell number and mRNA data were obtained from three independent experiments with three to six replicates per condition. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Thus, Zfp521 appears to regulate in a coordinated manner the three main aspects of bone homeostasis, i.e., bone formation, OB-dependent OC-genesis, and OC lineage–dependent OC-genesis, the net result being a net increase in bone mass. We then turned to identifying the mechanism by which Zfp521 could mediate such a positive effect on the postdevelopmental skeleton.
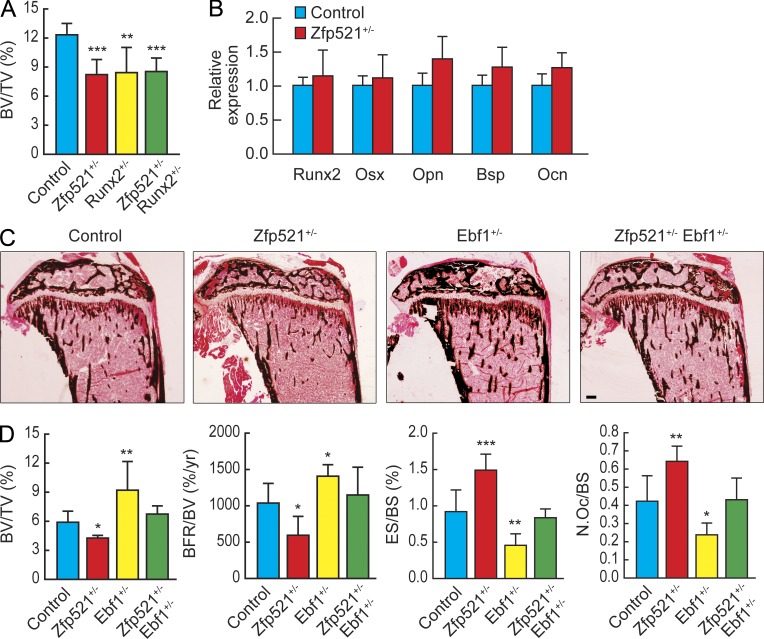
Runx2 haploinsufficiency fails to rescue bone homeostasis in the Zfp521+/− postdevelopmental skeleton
We have previously shown that Zfp521 interacts with and represses Runx2 activity such that haploinsufficiency of Zfp521 rescues some aspects of the developmental bone phenotype of Runx2+/− mice (Hesse et al., 2010). We have also shown that this repressive activity of Zfp521 on Runx2 can prevent the negative influence of artificially increased Runx2 expression in OBs on adult skeleton (Hesse et al., 2010). Because in this model both proteins were artificially overexpressed, we sought to determine whether the interaction between Zfp521 and Runx2 was involved in the regulation of postdevelopmental bone homeostasis under more physiological conditions. For this purpose, we crossed Zfp521+/− with Runx2+/− mice and analyzed their bone density by μCT at the postdevelopmental stage of 6 wk. As shown in Fig. 5 A, deletion of one allele of Runx2, which induced a mild osteopenia by itself, failed to restore the osteopenic phenotype induced by Zfp521 haploinsufficiency, demonstrating that increased Runx2 activity is not influencing the Zfp521 postdevelopmental phenotype. Furthermore, we measured several Runx2 target genes in undifferentiated Zfp521-deficient calvarial cells (day 0) and in fully differentiated OBs (day 21) but found no significant changes in the expression of any of the Runx2 target genes examined (Osx, Opn, Bsp, and Ocn) at early stages (Fig. 5 B). In contrast Bsp and Ocn were not only unaltered but even markedly down-regulated in fully differentiated OBs (Fig. 2 F). This data suggested that, although Zfp521 regulation of Runx2 plays a critical role at early developmental stages, this interaction does not contribute to the physiological regulation of postdevelopmental bone homeostasis. Because OB-targeted overexpression of Zfp521 increases bone formation and bone mass (Wu et al., 2009) and OB-targeted Zfp521 deletion leads to osteopenia, we then asked which transcription factor other than Runx2 could be both regulated by Zfp521 and important for adult bone homeostasis.
Figure 5.
Ebf1 haploinsufficiency rescues the bone phenotype of Zfp521+/− mice. (A) μCT analysis of tibias from control, Zfp521+/−, Runx2+/−, and Zfp521+/−:Runx2+/− mice at 6 wk (n = 4–6). (B) mRNA expression of Runx2 target genes in Zfp521−/− and control calvarial cells at day 0 of the culture. (C) Von Kossa staining of tibia sections of 6-wk-old control, Zfp521+/−, Ebf1+/−, and Zfp521+/−:Ebf1+/− mice. Bar, 450 μm. (D) Histomorphometric analysis of samples in C (n = 6). All data are means ± SD. Similar mRNA data were obtained from three independent experiments with three replicates per condition. N.Oc, number of OCs; TV, tissue volume. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Ebf1 haploinsufficiency rescues bone homeostasis in the postdevelopmental Zfp521+/− skeleton
Several lines of evidence called our attention to the transcription factor Ebf1 as a possible candidate. First, Zfp521 has been previously shown to interact with and repress Ebf1 in B cells in vitro (Mega et al., 2011). Second, deletion of Ebf1 has been reported to increase bone formation and bone mass in vivo (Hesslein et al., 2009), and third, Ccl9, which we found markedly increased in BM cells in the absence of Zfp521, is a well-known target gene of Ebf1.
To test the hypothesis that some of the effects we observed in bone in the absence of Zfp521 could be related to dysregulation of Ebf1 transcriptional activity, we crossed Zfp521+/− mice with Ebf1+/− heterozygous mice. Unlike haploinsufficiency of Runx2, deletion of one allele of Ebf1 was able to rescue the osteopenic phenotype of Zfp521+/− mice (Fig. 5, C and D). Furthermore, full histomorphometric analysis (Fig. 5, C and D; and Table S5) of these mice at 6 wk of age showed that both bone formation and bone resorption parameters were altered, and in an opposite manner: BFR was decreased and OC numbers (N.Oc/BS) and activity (ES/BS) were increased in Zfp521+/− mice, whereas the opposite was true in Ebf1+/− mice. Each and all of these parameters were normalized in the compound heterozygote mutants, strongly suggesting that the dysregulation of Ebf1 transcriptional activity was a critical contributor to the postdevelopmental alterations of bone homeostasis after deletion of Zfp521, affecting both bone formation and bone resorption.
Ebf1 target genes are markedly up-regulated in OBs in the absence of Zfp521
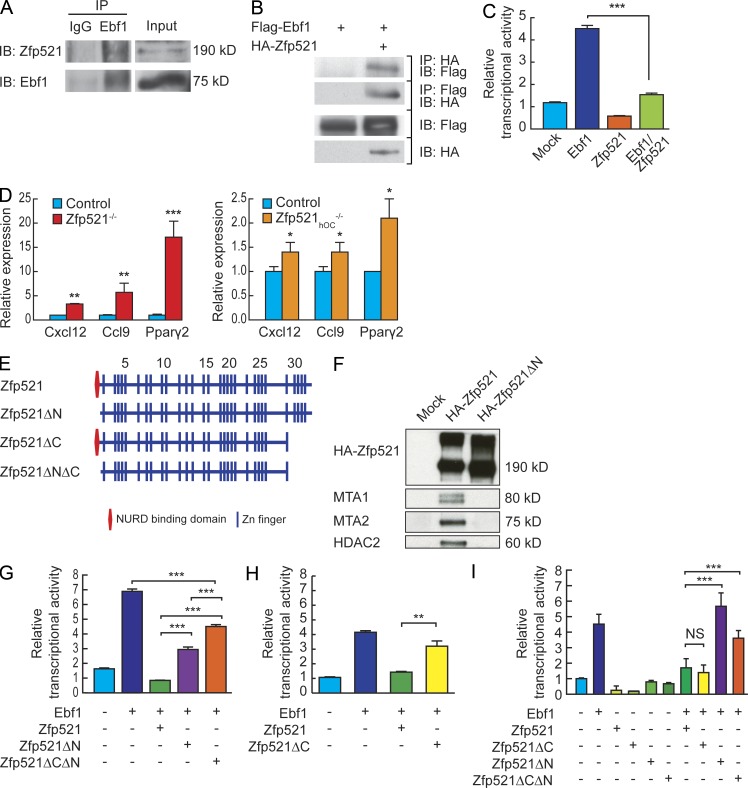
These findings suggested that deletion of Zfp521 enhanced Ebf1 activity. As reported, we found that both endogenous and overexpressed Zfp521 and Ebf1 coimmunoprecipitate (Fig. 6, A and B) and Zfp521 suppresses Ebf1 transcriptional activity (Fig. 6 C). Consequently, and in contrast with Runx2 target genes, we found a strong and consistent increase in the expression of several Ebf target genes (Ccl9, Cxcl12, and PPARγ; Jimenez et al., 2007; Lagergren et al., 2007) in Zfp521−/− and in Zfp521hOC−/− cells, showing that Ebf transcriptional activity is indeed markedly increased in the absence of Zfp521 (Fig. 6 D).
Figure 6.
Zfp521 antagonizes Ebf1 activity. (A) Ebf1 was immunoprecipitated with α-Ebf1 antibody. Mouse IgG was used as control. The immune complexes (left) and 10% input were blotted with α-Ebf1 and α-Zfp521 as indicated. (B) HA-Zfp521 and Flag-Ebf1 proteins were overexpressed in 293T cells. Proteins were immunoprecipitated with α-HA, and the immune complexes (two top panels) and 5% of the original cell lysates (two bottom panels) were blotted with α-HA and α-Flag as indicated. (C) Ebf1-responsive B29-luc plasmid was transfected into 293T cells together with Ebf1, Zfp521, or both. Luciferase activity was normalized to cotransfected Renilla activity. (D) The expression of Ebf1 target genes Cxcl12, Ccl9, and Pparγ was measured by qRT-PCR in Zfp521−/−, Zfp521hOC−/−, and respective control calvarial cells cultured for 7 d in OM. (E) Schematic presentation of Zfp521 domain structure and mutants. (F) HA-Zfp521 and HA-Zfp521ΔN were overexpressed in 293T cells and immunoprecipitated with anti-HA antibody. The Western blot was performed with antibodies against endogenous NuRD complex proteins (MTA1, MTA2, and HDAC2) and anti-HA antibody. (G) B29-luc plasmid was transfected into 293T cells together with Ebf1 and Zfp521 or Zfp521-deletion mutants as indicated. Luciferase activity was normalized to cotransfected Renilla activity. (H) B29-luc plasmid was transfected into 293T cells together with Ebf1 and Zfp521 or Zfp521ΔC as indicated. Luciferase activity was normalized to cotransfected Renilla activity. (I) Ccl9-luc plasmid was transfected into 293T cells together with Ebf1 and Zfp521 or Zfp521-deletion mutants as indicated. Luciferase activity was normalized to cotransfected Renilla activity. All data are means ± SD. Similar results were obtained in at least three independent experiments, and the assays were performed in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Zfp521 contains a nucleosome remodeling and deacetylase (NuRD) complex–binding domain at its N terminus (Fig. 6 E; Matsubara et al., 2009), and deletion of the first 13 aa (Zfp521ΔN) abolished its interaction with several NuRD components (Fig. 6 F). In contrast, Ebf1 interacts with the last four zinc fingers of Zfp521 (ZFs1-26), and deletion of these zinc fingers (Zfp521ΔC) reduced the binding of Zfp521 to Ebf1 (not depicted; Mega et al., 2011). On B cell–specific B29 promoter, the Zfp521ΔN mutant was significantly less efficient in repressing Ebf1, although Ebf1 did not lose all of its transcriptional activity (Fig. 6 G). Deletion of the last four zinc fingers also resulted in partial loss of suppression of Ebf1 activity (Fig. 6 H). Consequently, a double mutant that lacked both the N-terminal NuRD and C-terminal Ebf1-binding sites (Zfp521ΔCΔN; Fig. 6 E) had a markedly reduced ability to repress Ebf1 (Fig. 6 G). More relevant to bone, Ebf1 efficiently activated the Ccl9-Luc reporter, which was suppressed by Zfp521 (Fig. 6 I). In contrast to the B29 promoter, Zfp521ΔC retained full suppressive capability, suggesting the presence of another binding site or an indirect interaction with Ebf1. Most importantly, the Zfp521ΔN and Zfp521ΔCΔN mutants lost all ability to repress Ebf1 activity on the Ccl9 promoter (Fig. 6 I). Based on our data, Zfp521-dependent recruitment of the NuRD complex to Ebf1 contributes significantly to the suppression of Ebf1 activity on the Ccl9 promoter, whereas it is only partially or not required for the repression of Ebf1 activity on several other Ebf1 target promoters (Mega et al., 2011; Kang et al., 2012). Thus, it appears that the mechanisms by which Zfp521 interacts with and/or represses Ebf1 activity are promoter and possibly cellular context dependent.
OB-targeted overexpression of Ebf1 mimics the effects of conditional deletion of Zfp521 on bone formation
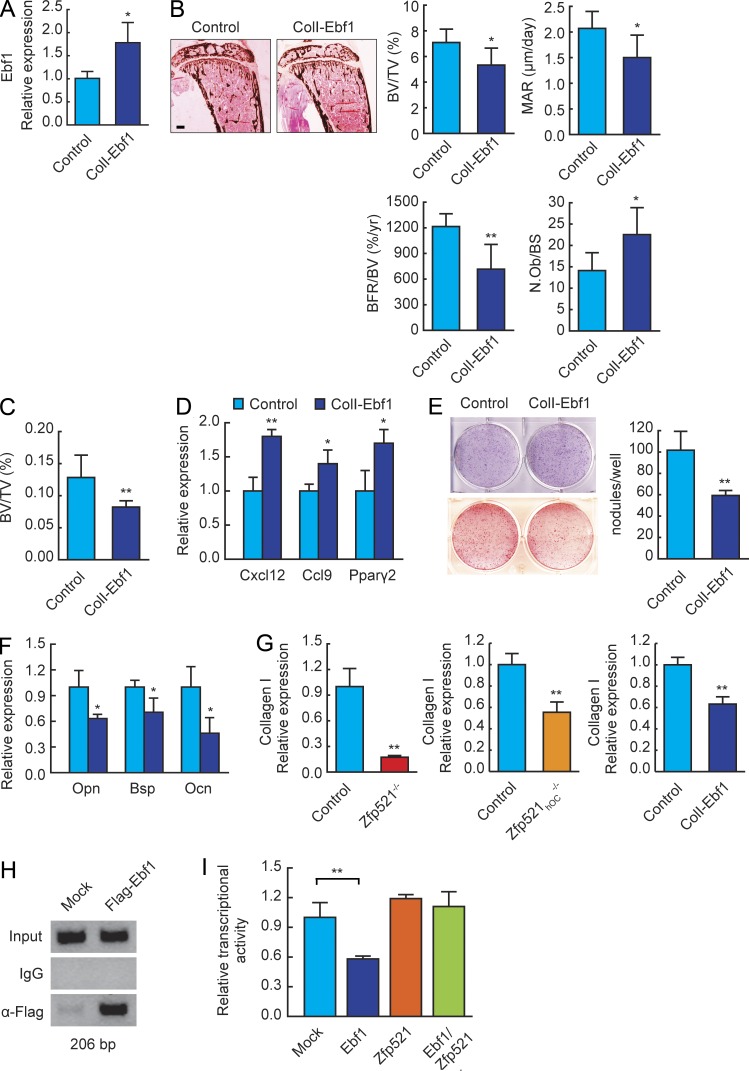
To further establish that increased Ebf1 activity contributed to the phenotype of Zfp521-deficient mice, we targeted overexpression of Ebf1 in mature OBs, hypothesizing that this could result in a bone phenotype similar to that of Zfp521hOC−/− mice. To test this, we generated transgenic mice that overexpress Ebf1 in OBs using the rat collagen I 2.3-kb promoter (Woitge et al., 2001; Wu et al., 2009), leading to a twofold overexpression of Ebf1 in long bones (Fig. 7 A). The bone phenotype of the ColI-Ebf1 mice was a phenocopy of Zfp521hOC−/− mice, with low BV and low MAR and mineralizing surface/BS leading to decreased BFR (Fig. 7, B and C; and Table S6). The ColI-Ebf1 phenotype was also the opposite of the Ebf1−/− mice (Hesslein et al., 2009), thereby confirming the negative role of Ebf1 on bone formation and that it is cell autonomous to OBs. As with Zfp521 deletion, the decrease in BFR occurred despite increased OB numbers, indicating that the function of individual OBs was impaired. Importantly, overexpression of Ebf1 targeted to mature OBs led to increased expression of the same Ebf1 target genes as in Zfp521-deleted OBs (Ccl9, Cxcl12, and PPARγ; Figs. 7 D and 6 D). Both Zfp521hOC−/− and ColI-Ebf1 calvarial cells formed less mineralized bone nodules in vitro, whereas early OB differentiation and ALP activity were not affected (Fig. 7 E). Accordingly, ColI-Ebf1 cells expressed lower levels of mature OB markers (Bsp and Ocn), indicating impaired maturation and function (Fig. 7 F).
Figure 7.
Ebf1 suppresses bone formation in vivo and in vitro. (A) Ebf1 expression in bone RNA from control and ColI2.3-Ebf1 transgenic mice measured by qRT-PCR (n = 6). (B) Von Kossa staining and histomorphometric analysis of tibia sections in 6-wk-old ColI2.3-Ebf1 transgenic and control mice (n = 6). N.Ob, number of OBs. Bar, 600 μm. (C) Metaphyseal trabecular BV (BV/tissue volume [TV]) in distal femurs of 6-wk-old ColI2.3-Ebf1 mice measured by μCT (n = 6). (D) ColI2.3-Ebf1 and control calvarial cells were harvested from newborn mice, plated on 6-well plates, and cultured in OM for 21 d. Expression of Ebf1 target genes was measured by qRT-PCR. (E) ALP (blue) and Alizarin red (red) staining of ColI2.3-Ebf1 and control calvarial cells cultured in OM for 7 d or 21 d, respectively. Bar graph shows quantification of bone nodules in the Alizarin red–stained plates. (F) Expression of OB marker genes Opn, Bsp, and Ocn in ColI2.3-Ebf1 and control calvarial cells after 21 d in OM was measured by qRT-PCR. (G) Col1a1 expression in Zfp521−/−, Zfp521hOC−/−, and ColI2.3-Ebf1 and respective control calvarial cells after 21 d in OM was measured by qRT-PCR. (H) Ebf1-containing chromatin complexes were immunoprecipitated from control and Flag-Ebf1–overexpressing MC3T3-E1 cells using anti-Flag antibody. IgG was used as negative control. The region of the ColI2.3kb promoter containing the putative Ebf1-binding site was amplified by PCR. (I) ColI2.3kb-luc reporter was transfected into C3H10T1/2 cells together with Ebf1, Zfp521, or both. Luciferase activity was normalized to cotransfected Renilla activity. All data are mean ± SD. Similar mRNA and reporter assay data were obtained from at least three experiments performed in triplicate. *, P < 0.05; **, P < 0.01.
Interestingly, gene expression profiling of calvarial cells isolated from Zfp521-deleted and Ebf1-overexpressing mice revealed that in all cases, expression of the Col1a1 gene was decreased (Fig. 7 G). In silico analysis of the Col1a1 promoter identified a putative Ebf1-binding site, and a chromatin immunoprecipitation (ChIP) assay showed that Ebf1 did in fact occupy this promoter region (Fig. 7 H). Furthermore, although Zfp521 had no effect by itself, Ebf1 suppressed the activity of ColI 2.3-kb promoter-luciferase construct, and this suppression was relieved by coexpressing Zfp521 (Fig. 7 I). These data show that increased Ebf1 activity inhibits bone formation in vivo and in vitro, at least partly by directly suppressing type I collagen expression, a negative effect which is reversed by Zfp521. Thus, OB-targeted overexpression of Ebf1 recapitulated all the hallmarks of the phenotype of Zfp521hOC−/− mice in vivo and in vitro, suggesting that the mechanism by which Zfp521 deletion impairs OB maturation and bone formation involves the up-regulation of Ebf1 transcriptional activity.
Ebf1 regulates OB-dependent and hematopoietic lineage–dependent OC-genesis
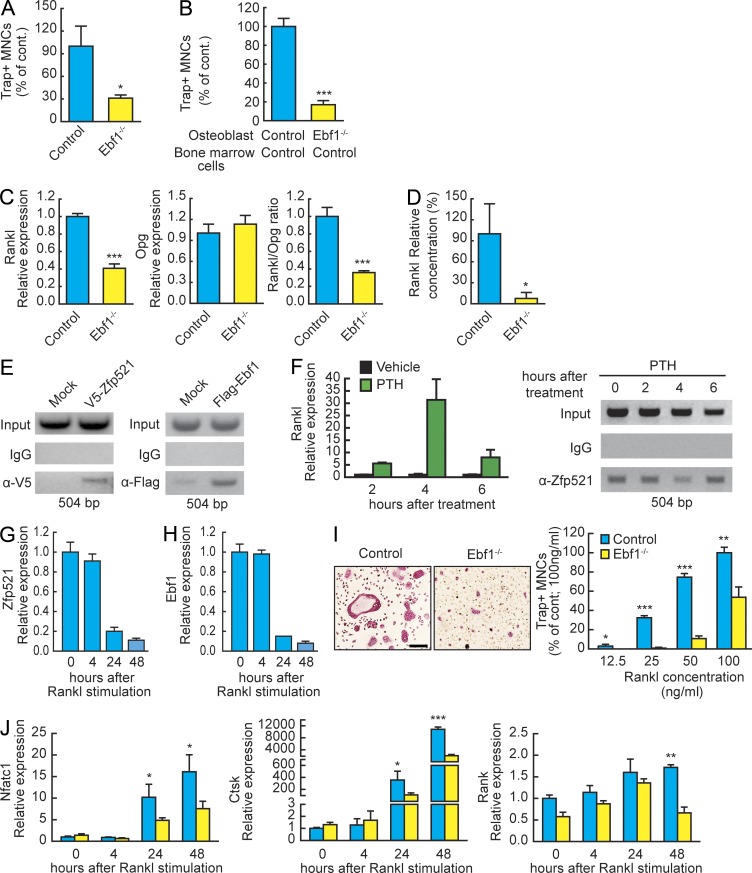
To test whether Ebf1 also regulates OB-dependent OC-genesis, we performed BM cultures and mix-and-match experiments, now using Ebf1−/− cells. Stimulation of total BM from Ebf1−/− mice with PTH resulted in significantly decreased OC-genesis compared with controls (Fig. 8 A). Similarly, Ebf1−/− calvarial cells exhibited impaired support of OC-genesis from wild-type BM, demonstrating a role for Ebf1 in the regulation of OB-dependent OC-genesis (Fig. 8 B). Deletion of Ebf1 resulted in a threefold decrease in Rankl/Opg ratio, opposite to the Zfp521−/− phenotype (Fig. 8 C). In agreement with the mRNA data, Rankl protein levels were significantly decreased in the Ebf1−/− cultures (Fig. 8 D).
Figure 8.
Ebf1 and Zfp521 regulate OB-dependent and hematopoietic lineage–dependent OC-genesis. (A) Total BM cells from Ebf1−/− and control mice were plated on 48-well plates, stimulated with 10 nM hPTH(1–34), and stained for TRAP. Number of TRAP+ multinucleated cells (MNCs) per well is shown. (B) Calvarial cells from control and Ebf1−/− mice were co-cultured with nonadherent BM cells as indicated in 24-well plates, stimulated with vitD3 and PGE2, and stained for TRAP. Bar graphs indicate the number of TRAP+ multinucleated cells per well. (C) Rankl and Opg mRNA expression and Rankl/Opg ratio measured by qRT-PCR in the co-culture experiment in B. (D) RANKL protein levels were measured with RANKL ELISA in medium samples from control and Ebf1−/− calvarial cells cultured in 12-well plates and stimulated with vitD3 and PGE2 as in co-culture experiments (n = 3). (E) ChIP with anti-V5 antibody for V5-Zfp521 and anti-Flag antibody for Flag-Ebf1 in MC3T3-E1 cells overexpressing the tagged proteins. IgG was used as control. The PCR-amplified promoter area contained a putative Ebf1 consensus site in the active distal promoter region. (F) The time courses of PTH-induced Rankl mRNA expression and displacement and rebinding of endogenous Zfp521 from the Rankl distal promoter region after stimulation by 10 nM PTH were compared. Zfp521 binding to the promoter was analyzed in a ChIP assay using the anti-Zfp521 antibody. IgG was used as control. (G) Expression of Zfp521 mRNA by qRT-PCR in OC progenitors cultured with 20 ng/ml M-CSF for 2 d and then stimulated with 100 ng/ml RANKL for the indicated times. (H) Expression of Ebf1 mRNA by qRT-PCR in OC progenitors cultured as in G. (I) Control and Ebf1−/− spleen cells were stimulated with 20 ng/ml M-CSF and then with increasing doses of RANKL for 5 d, stained for TRAP activity, and quantified. Bar, 100 μm. (J) Expression of Nfatc1, Ctsk, and Rank mRNAs by qRT-PCR in Ebf1−/− spleen cell–derived OCs cultured with 20 ng/ml M-CSF and then M-CSF + 100 ng/ml RANKL for the indicated times. All data are mean ± SD. Similar cell number and mRNA data were obtained from three independent experiments with four to six replicates per condition. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
As Zfp521 and Ebf1 had divergent effects on the expression of Rankl, we analyzed the effects of the two proteins on the regulation of this promoter. A search of the proximal promoter and distal enhancer regions revealed an Ebf1 consensus sequence in the distal enhancer region, previously shown to be required for PTH-induced activation (Fig. 8 E; Fu et al., 2002, 2006; Kitazawa and Kitazawa, 2002). ChIP analysis showed that both Zfp521 and Ebf1 could bind the distal enhancer but not the proximal promoter (Fig. 8 E and not depicted). Endogenous Zfp521 occupied the distal enhancer at steady-state but was rapidly displaced by treatment of the cells with PTH, which led to a robust (30-fold) increase in Rankl expression (Fig. 8 F). With time, Zfp521 returned to the enhancer region, followed by a decrease in Rankl expression. Collectively, our data demonstrates that Zfp521 and Ebf1 regulate Rankl expression in an opposite manner in cells of the OB lineage by binding to its distal enhancer region.
Zfp521 and Ebf1 were both expressed in OC progenitors, and their expression decreased identically upon induction of differentiation with RANKL (Fig. 8, G and H). We therefore tested whether the interaction between Zfp521 and Ebf1 also had cell-autonomous effects in OC progenitors. In contrast with the accelerated OC-genesis of BM and spleen-derived Zfp521−/− OC progenitors, spleen cells and BMMs derived from Ebf1−/− mice required longer time and higher concentration of RANKL to form OCs, confirmed by decreased expression of OC marker genes in a time course experiment (Fig. 8, I and J; and not depicted, respectively). These in vitro findings were therefore consistent with histomorphometric analysis of bone resorption in both Zfp521+/− and Ebf1+/− mice (Fig. 5 D). Thus, in addition to their role within OBs in regulating bone formation and the RANKL/OPG ratio, the interplay of these molecules in OC precursors is also an important cell-autonomous determinant of OC differentiation, most likely through regulation of Ccl9 expression, an Ebf1 target gene reported to enhance RANKL-stimulated OC-genesis (Okamatsu et al., 2004).
DISCUSSION
Bone mass is under complex regulation by endocrine and paracrine signals, which modulate bone formation via OB differentiation and function, and bone resorption through both OB-dependent and hematopoietic lineage–autonomous mechanisms. We found that alterations in the balance between Ebf1 and the 30–zinc finger transcriptional coregulator Zfp521 affects in a coordinated manner the regulation of all three aspects of bone homeostasis, i.e., bone formation and both OB-dependent and OC precursor–intrinsic OC-genesis. Thus, the interaction between Ebf1 and Zfp521 in both the mesenchymal and the hematopoietic lineages acts as a rheostat for bone homeostasis that coordinates the activities of OBs and OCs to regulate bone mass.
Zfp521 represses Ebf1 to maintain bone homeostasis
Zfp521 interacts with Ebf1 and suppresses its transcriptional activity, such that deletion of Zfp521 enhanced Ebf1 target gene expression in cells of both the OB and the OC lineages. In OBs, high Ebf1 activity results in impaired bone formation. Targeted overexpression of Ebf1 in mature OBs in vivo recapitulated the low bone formation phenotype of Zfp521hOC−/− mice. Further demonstrating this relationship, the impaired bone formation and low bone mass of Zfp521+/− mice was rescued in Zfp521+/−:Ebf1+/− double heterozygous mice. Importantly, although Zfp521 can repress Runx2 and some of the early skeletal defects in Runx+/− mice are rescued in Zfp521+/−:Runx2+/− pups (Wu et al., 2009; Hesse et al., 2010), Runx2 haploinsufficiency did not rescue the Zfp521+/− low bone mass phenotype. The lack of rescue of osteopenic phenotype in Zfp521+/−:Runx2+/− may be at least in part caused by the low endogenous Runx2 levels in mature OBs and in the postdevelopmental skeleton (Maruyama et al., 2007). Thus, in addition to its interaction with Runx2 to regulate early OB lineage differentiation at developmental stages (Wu et al., 2009; Hesse et al., 2010), Zfp521 also interacts with and controls the activity of Ebf1 to regulate OB maturation and bone formation, the latter being predominant in the mature skeleton.
Derepression of Runx2 activity may nevertheless also contribute to the phenotype observed after deletion of Zfp521. We have previously shown that supraphysiological levels of Zfp521 suppress early OB differentiation in vitro but also promote bone formation in vivo (Wu et al., 2009). Our findings may now help explain this apparent contradiction. Deletion of Zfp521 increased the number of Runx2-positive cells as well as the number of CFU-Fs and CFU-OBs, confirming that Zfp521 tends to repress early OB differentiation, and this most likely through repression of Runx2. Yet these accumulating early OBs exhibit impaired capacity to progress to full maturity and to form bone in the absence of Zfp521, suggesting that Runx2-positive cells cannot differentiate further without Zfp521. Our results suggest that this late stage effect on OBs is caused by derepression of Ebf1. We therefore propose that in the regulation of the OB lineage, Runx2 is predominant at early stages and Ebf1 at later stages. By repressing both, Zfp521 would decrease early differentiation but favor late maturation of OBs and bone formation. As a matter of fact, Zfp521 expression kinetics with increased expression at later stages during OB differentiation would indeed favor the latter, consistent with its recently proposed positive role during neuronal differentiation (Wu et al., 2009; Kamiya et al., 2011).
Zfp521 and Ebf1 regulate bone resorption in both hematopoietic and mesenchymal cells
Bone resorption is a critical component of bone remodeling and of the regulation of bone homeostasis. Deletion of Zfp521 led to an increase in OC numbers and serum CTX as the result of effects in both OBs and OCs. In OBs, Ebf1 stimulated the expression of Rankl and Zfp521 had opposite effects. ChIP analysis showed that both Ebf1 and Zfp521 associate directly with the Rankl promoter to reciprocally regulate its transcription. In contrast, Ebf2 has been reported to regulate OC-genesis through OPG and not RANKL expression in OBs (Kieslinger et al., 2005). Recent studies have suggested that the osteocytes may be the major source of RANKL in adult bone, whereas OBs and hypertrophic chondrocytes would be an important source only during growth (Nakashima et al., 2011; Xiong et al., 2011). Interestingly, we found that although deletion of Zfp521 in early OBs (Osx-Cre) affects OC-genesis, deletion of Zfp521 (hOC-Cre) or targeted overexpression of Ebf1 (ColI2.3-Ebf1) in more mature cells does not. We cannot however exclude the possibility that some of these regulatory events could also take place in matrix-embedded cells.
Most importantly, our study shows for the first time that besides their opposing function in OBs, Ebf1 and Zfp521 also act cell-autonomously within the monocyte-macrophage lineage to regulate OC-genesis. Zfp521 acts within OC progenitors to inhibit their differentiation, whereas Ebf1 is required for normal differentiation. These data are consistent with the hypothesis that increased Zfp521 expression is involved in the maintenance of undifferentiated hematopoietic cell phenotypes (Warming et al., 2003; Bond et al., 2004; Hentges et al., 2005). Again confirming the opposition between Zfp521 and Ebf1, but now in hematopoietic cells, OC numbers were decreased after deletion of one Ebf1 allele, increased after deletion of one allele of Zfp521, and normalized in the double heterozygous mutants. Notably, the osteoclastogenic potential of Ebf1−/− progenitors in vitro was also clearly decreased, establishing firmly that Ebf1 promotes OC-genesis within hematopoietic cells.
What is the mechanism by which the interaction between Zfp521 and Ebf1 regulates OC-genesis? Besides the opposing roles of Zfp521 and Ebf1 in the regulation of the Rankl promoter in OBs, the Ebf1–Zfp521 interaction also affected OCs directly. We show that the up-regulation of OC-genesis in Zfp521−/− mice occurred by at least two separate but synergistic mechanisms. First, deletion of Zfp521 resulted in increased expression of the Ebf1 target gene Ccl9 (Lagergren et al., 2007). Ccl9 and its receptor Ccr1 have been reported to be the main chemokine ligand receptor pair expressed in OCs, enhancing RANKL-induced OC-genesis (Lean et al., 2002; Okamatsu et al., 2004). Consistent with Ccl9 playing a critical role in our model, blocking Ccl9 with a neutralizing antibody was sufficient to normalize OC-genesis in Zfp521−/− cultures. However, in this experiment, Zfp521−/− OCs still showed enhanced OC-genesis compared with controls, suggesting that an additional mechanism was involved. Indeed, we found that Zfp521 inhibits Nfatc1 transcriptional activity in OCs such that the expression of several known Nfatc1 target genes was increased after Zfp521 deletion. Thus, Zfp521 represses OC-genesis not only by altering the RANKL/OPG ratio in cells of the OB lineage but also by suppressing RANK signaling at several levels. It inhibits an Ebf1-dependent Ccl9 autocrine loop in OC precursors, decreasing their sensitivity to RANKL, and suppresses Nfatc1 activity, a critical downstream signaling effector of RANK.
Zfp521 as a transcriptional modulator
What is the molecular mechanism by which Zfp521 represses Ebf1? Zfp521 is a component of transcriptional complexes, but it lacks any classical transactivation domains. Instead it contains clusters of C2H2 Krüppel-like zinc fingers, suggesting that it could act as a platform to assemble transcriptional complexes. Indeed, Zfp521 interacts with its N terminus with components of the NuRD complex and binds to Ebf1 via its C-terminal zinc fingers, but whether NuRD was required for transcriptional repression by Zfp521 remained unresolved. We and others have previously found that the Zfp521–NuRD interaction is dispensable for the repression of Runx2 (Correa et al., 2010; Hesse et al., 2010) or of some Ebf1-responsive promoters (Mega et al., 2011; Kang et al., 2012). However, the NuRD-binding domain is required for the efficient repression of GATA-1 activity by Zfp521 during erythroid differentiation (Matsubara et al., 2009). Thus, Zfp521 might exert its repressive action on transcription through several mechanisms, one of which requires the recruitment of NuRD.
Indeed, we show here that on a B cell–specific B29 promoter, deletion of the N-terminal NuRD-binding domain partially impaired the ability of Zfp521ΔN to repress Ebf1, which was further reduced in the Zfp521ΔNΔC mutant, although it still retained some repressive function. In contrast, on the Ccl9 promoter, the NuRD interaction was required for efficient repression of Ebf1, whereas the C-terminal Ebf1-binding domain was dispensable. These results confirm that several mechanisms are involved in the repressive function of Zfp521 on gene transcription and suggest that (a) the requirement of NuRD interaction for Zfp521 to repress target genes of the same transcription factor, here Ebf1, or of different transcription factors, such as Runx2 or GATA1, may be promoter specific and (b) direct interaction of Zfp521 to Ebf1 (or to other target factor) may not be required for efficient repression. The latter could be mediated by an intermediate, such as the NuRD complex itself. More studies will be required to unravel the molecular mechanisms by which Zfp521 represses transcription in these different contexts.
The specific function of Zfp521 could go as far as reversing its role from a repressor to an activator of transcription, as suggested by Kamiya et al. (2011) in the regulation of neuronal differentiation or by Hentges et al. (2005) in murine lymphomas. Based on their data in lymphoma cells, Hentges et al. (2005) actually propose that on B cell–related Ebf1 target genes, Zfp521 could even enhance Ebf1 activity, whereas on other promoters Zfp521 would be a repressor. Whether this activation would be direct or indirect through Zfp521 binding to and inhibiting repressors remains to be explored. Supporting this latter hypothesis, we found here that Zfp521 enhances Col1a1 expression, a key target gene for bone formation in mature OBs, but this occurs through the relief of the Ebf1-mediated suppression of Col1a1. Collectively, these data suggest that Zfp521 regulates the transcriptional program in different cell types through interactions with a specific subset of transcription factors, recruiting specific transcriptional complexes to modulate cell differentiation and function.
In summary, by repressing Ebf1, Zfp521 exerts a cell-autonomous positive influence on OB maturation and bone formation and a negative influence on OB-dependent and on OC precursor cell–autonomous regulation of OC-genesis and bone resorption. Collectively, our results demonstrate that one single transcription factor, Ebf1, and its regulation by one single repressor, Zfp521, affect in a coordinated manner three key components of bone remodeling to regulate bone homeostasis. Thus, the interplay of these two factors acts as a rheostat to regulate bone homeostasis.
MATERIALS AND METHODS
Generation of conditional knockout construct for Zfp521.
The DNA construct for simultaneous generation of a null and a conditional allele for Zfp521 was made using recombineering, essentially as described previously (Liu et al., 2003; Warming et al., 2006). First, a retrieval vector was prepared by three-way ligation of mini-homology arms into pBlight-TK (a plasmid backbone containing the thymidine kinase from HSV) for counter-selection in embryonic stem cells using Ganciclovir (Warming et al., 2006). The homology arms were amplified from a mouse BAC containing the Zfp521 genomic region using the primers listed below. The BAC (CITB 454L20) was identified by screening a 129-based BAC library (CJ7 embryonic stem cell DNA, CITB, Research Genetics/Invitrogen), and BAC DNA was prepared using the BAC Nucleobond kit (Takara Bio Inc., BD). All primers were from Integrated DNA Technologies, and all PCR reactions were performed using Expand High Fidelity (Roche): 5′ retrieval F, 5′-AATAAAGGATCCGTGCTCCAGGCACTATAGAT-3′; 5′ retrieval R, 5′-AATAAAAAGCTTGACCTGGCCAGGTCATTTAA-3′; 3′ retrieval F, 5′-AATAAAAAGCTTGCCTTTCCAAGTTCCTGACA-3′; and 3′ retrieval R, 5′-AATAAAGCGGCCGCGTTCAAGGCCACTGTGGTTT-3′. Cloning sites were BamHI, HindIII, and NotI.
The Zfp521 BAC was transferred into DY380 cells. Next, heat-shocked and electrocompetent DY380/Zfp521 cells were electroporated with HindIII-linearized retrieval vector to subclone 13.7 kb of genomic Zfp521 DNA, to give rise to pZfp521. For further manipulation of pZfp521, two mini-targeting vectors for recombineering in Escherichia coli were prepared essentially as described previously (Liu et al., 2003). The following primers were used to amplify mini-homology arms, using Zfp521 BAC DNA as template: 5′ F1, 5′-AAATAAGTCGACCTTTTGGTCTGCAGAATTGCCT-3′; 5′ R1, 5′-AAATAAGAATTCGAGCCCCAAGCTTTTACTCTT-3′; 5′ F2, 5′-AAATAAAGATCTAGAGCGATGCTCACTCCTTCC-3′; 5′ R2, 5′-AAATAAGCGGCCGCCGGCTAAAGGACTTGTCACA-3′; 3′ F1, 5′-AAATAAGTCGACGCACAACAGGAGTTTTTCAAAGC-3′; 3′ R1, 5′-AAATAAGAATTCGGATCCCAAGGGACAAGGTTTTCCCT-3′; 3′ F2, 5′-AAATAAAGATCTCAGGTGTGGTTCCTGTAAGT-3′; and 3′ R2, 5′-AAATAAGCGGCCGCCAGGAACCACTATCCAAGCT-3′.
5′ F1 + 5′ R1 PCR product was digested with SalI + EcoRI, 5′ F2 + 5′ R2 was digested with BglII + NotI, 3′ F1 + 3′ R1 with SalI and EcoRI, and 3′ F2 + 3′ R2 with BglII + NotI. An EcoRI–BamHI loxP-PGK-em7-Neo-loxP fragment was isolated from PL452, and an EcoRI–BamHI Frt-PGK-em7-Neo-Frt-loxP fragment was isolated from PL451 (Liu et al., 2003). A SalI–NotI pBluescript backbone fragment was prepared from PL452 as well.
A 5′MTV (5′ mini-targeting vector) was made by a four-way ligation of the two 5′ PCR products, the EcoRI–BamHI loxP-Neo-loxP fragment from PL452, and the SalI–NotI backbone. Likewise, a 3′MTV was made by a four-way ligation of the two 3′ PCR products, the EcoRI–BamHI Frt-PGK-em7-Neo-Frt-loxP fragment, and the SalI–NotI backbone.
Next, using recombineering, a floxed neo cassette was inserted upstream of Zfp521 exon 4: A NotI + SalI digested fragment from the 5′MTV containing the floxed neo cassette flanked by mini-homology arms was co-electroporated with pZfp521 into heat-shocked and electrocompetent DY380 cells to give rise to pZfp521-5′neo. Then, pZfp521-5′neo was electroporated into Cre-induced EL350 cells to remove the neo cassette and leave behind a single loxP site, to give rise to pZfp521-5′loxP. Finally, a NotI + SalI–digested fragment from 3′MTV, containing the frted neo cassette with a single loxP site flanked by mini-homology arms, was co-electroporated with pZfp521-5′loxP into heat-shocked and electrocompetent DY380 cells to give rise to pZfp521-CKO. This targeting vector contains the genomic Zfp521 region with one loxP site and an engineered XbaI site for genotyping upstream of exon 4 and a loxP-frt-neo-frt cassette along with an engineered BamHI site for genotyping downstream of exon 4. The pZfp521-CKO targeting vector was linearized using NotI and electroporated into CJ7 embryonic stem cells (129 background) using standard methods. G418 and FIAU double-resistant embryonic stem cell clones were selected and analyzed using Southern blot hybridization on XbaI (5′)- and BamHI (3′)-digested genomic DNA. 7/47 clones were targeted (15%), and one of them had the single 5′ loxP site (1/47 = 2%). The correctly targeted embryonic stem cell clone was injected into C57BL/6 blastocysts according to standard methods to give rise to chimaeras.
Production of mice.
After germline transmission of the targeted neo allele, heterozygous neo mice were crossed to β-actin Flp mice to remove the neo cassette and produce mice carrying the fl/+ allele. Heterozygous fl/+ mice were then either intercrossed to produce homozygous fl/fl mice or crossed to β-actin Cre mice to produce heterozygous Zfp521+/− mice. Heterozygous Zfp521+/− mice were then intercrossed to produce homozygous Zfp521−/− mice.
Experimental animals.
Osx-Cre and hOC-Cre transgenic mice have been previously described (Zhang et al., 2002; Rodda and McMahon, 2006). Experimental mice were produced by heterozygous matings. Zfp521+/− mice were maintained on C57BL background (over 10 backcrosses). Homozygous deletion of Zfp521 on C57BL/6 background was lethal during the first 24 h. To generate Zfp521−/− animals, we backcrossed Zfp521+/− mice once to 129/Sv, and then these F1 heterozygous mice on mixed background were intercrossed to produce control and Zfp521−/− animals. These animals survived until weaning and some up to 4–5 wk of age. Runx2+/− mice have been previously described (Otto et al., 1997) and were maintained on pure C57BL background (over 10 backcrosses). Zfp521+/−:Runx2+/− mice were produced by heterozygous matings, and wild-type littermates were used as controls.
Wild-type littermates were used as controls for Zfp521−/− mice. Either Osx-Cre+; Zfp521+/+ or hOC-Cre+; Zfp521+/+ littermates served as controls in experiments examining the effects of the conditional deletions, and these animals were maintained on mixed 129/Sv-C57BL background. ColI2.3-Ebf1 transgenic mice were generated by pronuclear injection of a construct containing the 2.3-kb fragment of the rat Col1a1 promoter (Woitge et al., 2001) linked to mouse Ebf1 cDNA. Mice were generated and maintained on pure C57BL background and genotyped by transgene-specific primers. Ebf1+/− mice were provided by J. Hagman (National Jewish Health, Denver, CO) and R. Grosschedl (Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany; Lin and Grosschedl, 1995), maintained on C57BL background, and crossed with Zfp521+/− mice to produce the experimental animals. Primer sequences for genotyping are provided per request. All procedures involving animals were approved by the Harvard Medical Area Standing Committee on Animals and conform to the relevant regulatory standards.
Histology, histomorphometry, μCT, serum markers, in situ hybridization, and immunohistochemistry.
Bone histomorphometry was performed on secondary spongiosa 400 µm under the growth plate as previously described (Sabatakos et al., 2000). Five to eight animals were analyzed per group. μCT was performed with Scanco CT-35 for femurs and with Skyscan 1072 for vertebrae. 3D structural analysis was performed with software provided by the supplier. Serum samples were analyzed for PINP and CTX using Rat/Mouse PINP and RatLaps EIA assays (IDS) at Pharmatest Ltd. In situ hybridization was performed on decalcified, paraffin-embedded sections using [35S]UTP-labeled riboprobes for Opn and Ocn as described previously (Lanske et al., 1998). Three samples were analyzed per genotype. Immunohistochemistry was performed on frozen sections of decalcified tibias from 3-wk-old Zfp521−/− and control mice as previously described (Wu et al., 2009).
Cell culture, cell lines, and medium RANKL.
Calvarial cells were prepared and induced to differentiate to OBs as described previously (Sabatakos et al., 2000). OC progenitors were harvested either from the nonadherent fraction of flushed BM or from spleen. The cells were first cultured in 20 ng/ml M-CSF (R&D Systems) for 2 d and then stimulated with 20 ng/ml M-CSF and 100 ng/ml RANKL unless otherwise stated for the indicated times. To block Ccl9 activity, anti-Ccl9 blocking antibody (R&D Systems) or 5 µg/ml IgG control was added to the OC culture medium (20 ng/ml M-CSF and 50 ng/ml of RANKL) from the beginning of the culture as previously described (Okamatsu et al., 2004). For co-cultures, calvarial cells were plated at 104/well on 24-well plates. The next day, 1–2 × 106 nonadherent BM cells were plated into the same wells in medium containing 10−8 M vitamin D3 and 10−6 M prostaglandin E2. ALP and tartrate-resistant acid phosphatase (TRAP) staining and RNA extraction were performed at indicated time points as previously described (Takahashi et al., 1988; Sabatakos et al., 2000). Soluble RANKL was measured in the pooled medium samples with Quantikine Mouse RANKL assay (R&D Systems) after concentrating the samples with Microcon YM-10 columns (EMD Millipore). C3H10T1/2 and MC3T3-E1 cells were infected with either Flag-tagged Ebf1 retrovirus or V5-tagged Zfp521 lentivirus or corresponding empty viruses and selected with puromycin or blasticidin, respectively. Cells were cultured in α-MEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin.
Reporter assays and coimmunoprecipitation.
For reporter assays, 293T or C3H10T1/2 cells were transfected with appropriate plasmids including pRL promoterless Renilla plasmid using Fugene 6 reagent (Invitrogen). Reporter activity was measured with the Dual Luciferase Reporter assay (Promega) 24–48 h after transfection and normalized for Renilla activity. Coimmunoprecipitations were performed as described previously (Wu et al., 2009). The CMV-LacZ plasmid was included in each transfection condition, and all coimmunoprecipitations were normalized according to the β-galactosidase activity in the cell lysate.
Plasmids.
CMV-HA-Zfp521 was previously described (Wu et al., 2009). pCDNA3-Flag-Ebf1 and pMSCV-Flag-Ebf1 constructs (Jimenez et al., 2007) were a gift from E.D. Rosen. B29-Luc and Ccl9-Luc were gifts from M. Sigvardsson (Linköping University, Linköping, Sweden; Lagergren et al., 2007; Sigvardsson, 2000). ColI2.3kb-Luc and Ctsk-Luc were gifts from H. Takayanagi (Tokyo Medical and Dental University, Tokyo, Japan; Koga et al., 2005). HAHA-Zfp521ΔN was generated by using PCR primers removing the first 13 5′ codons and cloning this truncated Zfp521 cDNA in frame to CMV-HAHA vector according to standard methods. HA-Zfp521ΔC and HAHA-Zfp521ΔNΔC were generated from CMV-HA-Zfp521 and from HAHA-Zfp521ΔN by replacing the methionine 1181 by a premature stop codon (in bold) using Multi Site-directed Mutagenesis kit (Agilent Technologies) with primer 5′-ACGCCCCAAGTGTCACCCTAGCCCAGAATCAGTCCCTCCCAGTCCG-3′.
Antibodies.
The in-house–generated Zfp521 antibody α-Zfp521(369) was previously described (Wu et al., 2009). α-Zfp521, monoclonal anti-HA clone 12CA5 (Roche), and anti-Flag clone M2 (Sigma-Aldrich) were used for immunoprecipitation and Western blotting as indicated. Goat anti–mouse and anti–rabbit HRP were used as a secondary antibodies, and HRP activity was detected with Amersham ECL Detection reagents (GE Healthcare).
ChIP.
ChIP was performed using the Chromatin Immunoprecipitation Assay kit (EMD Millipore) according to the manufacturer’s protocol with slight modifications. In brief, calvarial cells or MC3T3-E1 cells were fixed in 1% formaldehyde for 15 min at room temperature. The cross-linking was stopped with 0.1 M glycine. After chromatin shearing, antibodies were added to samples and incubated overnight. Magnetic Dynabeads Protein A beads (Invitrogen) were used to harvest bound protein–chromatin complexes. Extracted chromatin was subjected to PCR using promoter-specific primer sequences: RANKL promoter CNS1A-F, 5′-GGTCAAGAGGGGCCTGACTT-3′; and CNS1A-R, 5′-GCAGTGTGTAAACAAAGAGA-3′; and Col1a1 promoter Col1a1-F3, 5′-TGGCTCCCCCTCTCCGAG-3′; and Col1a1-R3, 5′-TCTAGACCCTAGACATGTAG-3′.
Measurement of gene expression.
Total RNA was isolated using RNeasy mini kit (QIAGEN), cDNA was prepared with SuperScript VILO kit (Invitrogen), and quantitative real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad Laboratories). The data were normalized using GAPDH as internal control. Primer sequences are provided per request.
Statistical analysis.
Results are presented as mean ± SD. Statistical analysis was performed by two-tailed Student’s t test. P-values <0.05 were considered significant.
Online supplemental material.
Table S1 shows full histomorphometric data from 3-wk-old Zfp521−/− and control mice. Tables S2 and S3 show full histomorphometric data from 6-wk-old and 12-wk-old, respectively, Zfp521hOC−/− and control mice. Table S4 shows full histomorphometric data from 6-wk-old Zfp521Osx−/− and control mice. Table S5 shows full histomorphometric data from 6-wk-old control, Zfp521+/−, Ebf1+/−, and Zfp521+/−:Ebf1+/− mice. Table S6 shows full histomorphometric data from 6-wk-old ColI2.3-Ebf1 and control mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20121187/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Beate Lanske and Dr. Despina Sitara for help with the in situ hybridizations and Lynn Neff for expert help in immunohistochemistry. We also thank Dr. Francesca Gori for discussions and her help with the manuscript. Dr. Thomas Clemens is acknowledged for providing the hOC-Cre mice.
This work was supported in part by National Institutes of Health–National Institute of Arthritis and Musculoskeletal and Skin Diseases grants to R. Baron (AR048218 and AR57769). Additional support was provided by the Academy of Finland (Projects 10945 and 120116 to R. Kiviranta), the Gideon and Sevgi Rodan Fellowship from the International Bone and Mineral Society (to R. Kiviranta and E. Hesse), the Finnish Cultural Foundation (to R. Kiviranta), the Emil Aaltonen Foundation (to R. Kiviranta), and the Sigrid Juselius Foundation (R. Kiviranta).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ALP
- alkaline phosphatase
- BFR
- bone formation rate
- BMM
- BM macrophage
- BS
- bone surface
- Bsp
- bone sialoprotein
- BV
- bone volume
- ChIP
- chromatin immunoprecipitation
- ES
- eroded surface
- hOC
- human osteocalcin
- MAR
- mineral apposition rate
- μCT
- microcomputed tomography
- NuRD
- nucleosome remodeling and deacetylase
- OB
- osteoblast
- OC
- osteoclast
- OC-genesis
- osteoclastogenesis
- Ocn
- osteocalcin
- OM
- osteogenic medium
- Opg
- osteoprotegerin
- Opn
- osteopontin
- Osx
- osterix
- PTH
- parathyroid hormone
- qRT-PCR
- quantitative RT-PCR
- RANKL
- receptor activator of NF-κB ligand
- TRAP
- tartrate-resistant acid phosphatase
References
- Almeida M. 2011. Unraveling the role of FoxOs in bone—insights from mouse models. Bone. 49:319–327 10.1016/j.bone.2011.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond H.M., Mesuraca M., Carbone E., Bonelli P., Agosti V., Amodio N., De Rosa G., Di Nicola M., Gianni A.M., Moore M.A., et al. 2004. Early hematopoietic zinc finger protein (EHZF), the human homolog to mouse Evi3, is highly expressed in primitive human hematopoietic cells. Blood. 103:2062–2070 10.1182/blood-2003-07-2388 [DOI] [PubMed] [Google Scholar]
- Boyle W.J., Simonet W.S., Lacey D.L. 2003. Osteoclast differentiation and activation. Nature. 423:337–342 10.1038/nature01658 [DOI] [PubMed] [Google Scholar]
- Bozec A., Bakiri L., Jimenez M., Schinke T., Amling M., Wagner E.F. 2010. Fra-2/AP-1 controls bone formation by regulating osteoblast differentiation and collagen production. J. Cell Biol. 190:1093–1106 10.1083/jcb.201002111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa D., Hesse E., Seriwatanachai D., Kiviranta R., Saito H., Yamana K., Neff L., Atfi A., Coillard L., Sitara D., et al. 2010. Zfp521 is a target gene and key effector of parathyroid hormone-related peptide signaling in growth plate chondrocytes. Dev. Cell. 19:533–546 10.1016/j.devcel.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Jilka R.L., Manolagas S.C., O’Brien C.A. 2002. Parathyroid hormone stimulates receptor activator of NFkappa B ligand and inhibits osteoprotegerin expression via protein kinase A activation of cAMP-response element-binding protein. J. Biol. Chem. 277:48868–48875 10.1074/jbc.M208494200 [DOI] [PubMed] [Google Scholar]
- Fu Q., Manolagas S.C., O’Brien C.A. 2006. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Mol. Cell. Biol. 26:6453–6468 10.1128/MCB.00356-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass D.A., II, Bialek P., Ahn J.D., Starbuck M., Patel M.S., Clevers H., Taketo M.M., Long F., McMahon A.P., Lang R.A., Karsenty G. 2005. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev. Cell. 8:751–764 10.1016/j.devcel.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Hentges K.E., Weiser K.C., Schountz T., Woodward L.S., Morse H.C., Justice M.J. 2005. Evi3, a zinc-finger protein related to EBFAZ, regulates EBF activity in B-cell leukemia. Oncogene. 24:1220–1230 10.1038/sj.onc.1208243 [DOI] [PubMed] [Google Scholar]
- Hesse E., Saito H., Kiviranta R., Correa D., Yamana K., Neff L., Toben D., Duda G., Atfi A., Geoffroy V., et al. 2010. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J. Cell Biol. 191:1271–1283 10.1083/jcb.201009107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesslein D.G., Fretz J.A., Xi Y., Nelson T., Zhou S., Lorenzo J.A., Schatz D.G., Horowitz M.C. 2009. Ebf1-dependent control of the osteoblast and adipocyte lineages. Bone. 44:537–546 10.1016/j.bone.2008.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez M.A., Akerblad P., Sigvardsson M., Rosen E.D. 2007. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol. Cell. Biol. 27:743–757 10.1128/MCB.01557-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya D., Banno S., Sasai N., Ohgushi M., Inomata H., Watanabe K., Kawada M., Yakura R., Kiyonari H., Nakao K., et al. 2011. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature. 470:503–509 10.1038/nature09726 [DOI] [PubMed] [Google Scholar]
- Kang S., Akerblad P., Kiviranta R., Gupta R.K., Kajimura S., Griffin M.J., Min J., Baron R., Rosen E.D. 2012. Regulation of early adipose commitment by Zfp521. PLoS Biol. 10:e1001433 10.1371/journal.pbio.1001433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G., Kronenberg H.M., Settembre C. 2009. Genetic control of bone formation. Annu. Rev. Cell Dev. Biol. 25:629–648 10.1146/annurev.cellbio.042308.113308 [DOI] [PubMed] [Google Scholar]
- Kieslinger M., Folberth S., Dobreva G., Dorn T., Croci L., Erben R., Consalez G.G., Grosschedl R. 2005. EBF2 regulates osteoblast-dependent differentiation of osteoclasts. Dev. Cell. 9:757–767 10.1016/j.devcel.2005.10.009 [DOI] [PubMed] [Google Scholar]
- Kitazawa R., Kitazawa S. 2002. Vitamin D(3) augments osteoclastogenesis via vitamin D-responsive element of mouse RANKL gene promoter. Biochem. Biophys. Res. Commun. 290:650–655 10.1006/bbrc.2001.6251 [DOI] [PubMed] [Google Scholar]
- Koga T., Matsui Y., Asagiri M., Kodama T., de Crombrugghe B., Nakashima K., Takayanagi H. 2005. NFAT and Osterix cooperatively regulate bone formation. Nat. Med. 11:880–885 10.1038/nm1270 [DOI] [PubMed] [Google Scholar]
- Kousteni S. 2011. FoxO1: a molecule for all seasons. J. Bone Miner. Res. 26:912–917 10.1002/jbmr.306 [DOI] [PubMed] [Google Scholar]
- Lagergren A., Månsson R., Zetterblad J., Smith E., Basta B., Bryder D., Akerblad P., Sigvardsson M. 2007. The Cxcl12, periostin, and Ccl9 genes are direct targets for early B-cell factor in OP-9 stroma cells. J. Biol. Chem. 282:14454–14462 10.1074/jbc.M610263200 [DOI] [PubMed] [Google Scholar]
- Lanske B., Divieti P., Kovacs C.S., Pirro A., Landis W.J., Krane S.M., Bringhurst F.R., Kronenberg H.M. 1998. The parathyroid hormone (PTH)/PTH-related peptide receptor mediates actions of both ligands in murine bone. Endocrinology. 139:5194–5204 10.1210/en.139.12.5194 [DOI] [PubMed] [Google Scholar]
- Lean J.M., Murphy C., Fuller K., Chambers T.J. 2002. CCL9/MIP-1gamma and its receptor CCR1 are the major chemokine ligand/receptor species expressed by osteoclasts. J. Cell. Biochem. 87:386–393 10.1002/jcb.10319 [DOI] [PubMed] [Google Scholar]
- Lin H., Grosschedl R. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 376:263–267 10.1038/376263a0 [DOI] [PubMed] [Google Scholar]
- Liu P., Jenkins N.A., Copeland N.G. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13:476–484 10.1101/gr.749203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukin K., Fields S., Hartley J., Hagman J. 2008. Early B cell factor: Regulator of B lineage specification and commitment. Semin. Immunol. 20:221–227 10.1016/j.smim.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B.T., Tamai K., He X. 2009. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 17:9–26 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Z., Yoshida C.A., Furuichi T., Amizuka N., Ito M., Fukuyama R., Miyazaki T., Kitaura H., Nakamura K., Fujita T., et al. 2007. Runx2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev. Dyn. 236:1876–1890 10.1002/dvdy.21187 [DOI] [PubMed] [Google Scholar]
- Matsubara E., Sakai I., Yamanouchi J., Fujiwara H., Yakushijin Y., Hato T., Shigemoto K., Yasukawa M. 2009. The role of zinc finger protein 521/early hematopoietic zinc finger protein in erythroid cell differentiation. J. Biol. Chem. 284:3480–3487 10.1074/jbc.M805874200 [DOI] [PubMed] [Google Scholar]
- Mega T., Lupia M., Amodio N., Horton S.J., Mesuraca M., Pelaggi D., Agosti V., Grieco M., Chiarella E., Spina R., et al. 2011. Zinc finger protein 521 antagonizes early B-cell factor 1 and modulates the B-lymphoid differentiation of primary hematopoietic progenitors. Cell Cycle. 10:2129–2139 10.4161/cc.10.13.16045 [DOI] [PubMed] [Google Scholar]
- Nakashima T., Hayashi M., Fukunaga T., Kurata K., Oh-Hora M., Feng J.Q., Bonewald L.F., Kodama T., Wutz A., Wagner E.F., et al. 2011. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med. 17:1231–1234 10.1038/nm.2452 [DOI] [PubMed] [Google Scholar]
- Negishi-Koga T., Takayanagi H. 2009. Ca2+-NFATc1 signaling is an essential axis of osteoclast differentiation. Immunol. Rev. 231:241–256 10.1111/j.1600-065X.2009.00821.x [DOI] [PubMed] [Google Scholar]
- Okamatsu Y., Kim D., Battaglino R., Sasaki H., Späte U., Stashenko P. 2004. MIP-1 gamma promotes receptor-activator-of-NF-kappa-B-ligand-induced osteoclast formation and survival. J. Immunol. 173:2084–2090 [DOI] [PubMed] [Google Scholar]
- Otero K., Shinohara M., Zhao H., Cella M., Gilfillan S., Colucci A., Faccio R., Ross F.P., Teitelbaum S.L., Takayanagi H., Colonna M. 2012. TREM2 and β-catenin regulate bone homeostasis by controlling the rate of osteoclastogenesis. J. Immunol. 188:2612–2621 10.4049/jimmunol.1102836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W., Beddington R.S., Mundlos S., Olsen B.R., et al. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 89:765–771 10.1016/S0092-8674(00)80259-7 [DOI] [PubMed] [Google Scholar]
- Rodda S.J., McMahon A.P. 2006. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 133:3231–3244 10.1242/dev.02480 [DOI] [PubMed] [Google Scholar]
- Sabatakos G., Sims N.A., Chen J., Aoki K., Kelz M.B., Amling M., Bouali Y., Mukhopadhyay K., Ford K., Nestler E.J., Baron R. 2000. Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat. Med. 6:985–990 10.1038/79683 [DOI] [PubMed] [Google Scholar]
- Sigvardsson M. 2000. Overlapping expression of early B-cell factor and basic helix-loop-helix proteins as a mechanism to dictate B-lineage-specific activity of the lambda5 promoter. Mol. Cell. Biol. 20:3640–3654 10.1128/MCB.20.10.3640-3654.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Yamana H., Yoshiki S., Roodman G.D., Mundy G.R., Jones S.J., Boyde A., Suda T. 1988. Osteoclast-like cell formation and its regulation by osteotropic hormones in mouse bone marrow cultures. Endocrinology. 122:1373–1382 10.1210/endo-122-4-1373 [DOI] [PubMed] [Google Scholar]
- Wan Y. 2010. PPARγ in bone homeostasis. Trends Endocrinol. Metab. 21:722–728 10.1016/j.tem.2010.08.006 [DOI] [PubMed] [Google Scholar]
- Warming S., Liu P., Suzuki T., Akagi K., Lindtner S., Pavlakis G.N., Jenkins N.A., Copeland N.G. 2003. Evi3, a common retroviral integration site in murine B-cell lymphoma, encodes an EBFAZ-related Krüppel-like zinc finger protein. Blood. 101:1934–1940 10.1182/blood-2002-08-2652 [DOI] [PubMed] [Google Scholar]
- Warming S., Rachel R.A., Jenkins N.A., Copeland N.G. 2006. Zfp423 is required for normal cerebellar development. Mol. Cell. Biol. 26:6913–6922 10.1128/MCB.02255-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Zeve D., Suh J.M., Wang X., Du Y., Zerwekh J.E., Dechow P.C., Graff J.M., Wan Y. 2011. Biphasic and dosage-dependent regulation of osteoclastogenesis by β-catenin. Mol. Cell. Biol. 31:4706–4719 10.1128/MCB.05980-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woitge H., Harrison J., Ivkosic A., Krozowski Z., Kream B. 2001. Cloning and in vitro characterization of alpha 1(I)-collagen 11 beta-hydroxysteroid dehydrogenase type 2 transgenes as models for osteoblast-selective inactivation of natural glucocorticoids. Endocrinology. 142:1341–1348 10.1210/en.142.3.1341 [DOI] [PubMed] [Google Scholar]
- Wu M., Hesse E., Morvan F., Zhang J.P., Correa D., Rowe G.C., Kiviranta R., Neff L., Philbrick W.M., Horne W.C., Baron R. 2009. Zfp521 antagonizes Runx2, delays osteoblast differentiation in vitro, and promotes bone formation in vivo. Bone. 44:528–536 10.1016/j.bone.2008.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Onal M., Jilka R.L., Weinstein R.S., Manolagas S.C., O’Brien C.A. 2011. Matrix-embedded cells control osteoclast formation. Nat. Med. 17:1235–1241 10.1038/nm.2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Karsenty G. 2004. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J. Biol. Chem. 279:47109–47114 10.1074/jbc.M410010200 [DOI] [PubMed] [Google Scholar]
- Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H.C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T.M., et al. 2004. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 117:387–398 10.1016/S0092-8674(04)00344-7 [DOI] [PubMed] [Google Scholar]
- Zhang M., Xuan S., Bouxsein M.L., von Stechow D., Akeno N., Faugere M.C., Malluche H., Zhao G., Rosen C.J., Efstratiadis A., Clemens T.L. 2002. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J. Biol. Chem. 277:44005–44012 10.1074/jbc.M208265200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.