Neutralization of IL-22 production from colonic innate lymphoid cells reduces dysplasia in bacterial-induced colon cancer by reducing proliferation of epithelial cells via reduced activation of Stat3.
Abstract
Patients with inflammatory bowel disease (IBD) have an increased risk of colon cancer. However, the immune cells and cytokines that mediate the transition from intestinal inflammation to cancer are poorly understood. We show that bacteria-induced colon cancer is accompanied by differential accumulation of IL-17+IL-22+ colonic innate lymphoid cells (cILCs), which are phenotypically distinct from LTi and NK-22 cells, and that their depletion in mice with dysplastic inflammation blocks the development of invasive colon cancer. Analysis of the functional role of distinct Type 17 cytokines shows that although blockade of IL-17 inhibits some parameters of intestinal inflammation, reduction in dysplasia and colorectal cancer (CRC) requires neutralization of IL-22 indicating a unique role for IL-22 in the maintenance of cancer in this model. Mechanistic analyses showed that IL-22 selectively acts on epithelial cells to induce Stat3 phosphorylation and proliferation. Importantly, we could detect IL-22+CD3+ and IL-22+CD3− cells in human CRC. Our results describe a new activity of IL-22 in the colon as a nonredundant mediator of the inflammatory cascade required for perpetuation of CRC, highlighting the IL-22 axis as a novel therapeutic target in colon cancer.
Microbe-induced inflammatory pathways are important drivers of carcinogenesis in malignancies like Helicobacter pylori–induced gastric cancer and colitis-associated cancer (CAC; Trinchieri, 2012). In inflammatory bowel disease (IBD) patients, deranged inflammatory responses toward the intestinal microbiota increase the risk of colon cancer twofold (Eaden et al., 2001). However, the immune cells and cytokines that mediate the transition from colitis to colon cancer are poorly understood.
Cancer and chronic inflammation share common features and often use shared molecular pathways. The IL-23 axis is a key pathway for intestinal inflammation in many mouse models (Hue et al., 2006; Izcue et al., 2008), and its components IL-23R and Stat3 are not only associated with IBD susceptibility (Duerr et al., 2006; Barrett et al., 2008) but also involved in inflammation-associated cancer in the skin and colon (Langowski et al., 2006; Bollrath et al., 2009; Kortylewski et al., 2009; Grivennikov et al., 2012).
IL-23 was found to be increased in human colon adenocarcinoma and through induction of proinflammatory responses such as up-regulation of MMP9 and evasion of tumor immunosurveillance by exclusion of cytotoxic T cells promotes tumor growth and incidence (Langowski et al., 2006). One of the key functions of IL-23 is the ability to promote Th17-type responses characterized by the induction of the cytokines IL-17 and IL-22 (Liang et al., 2006). Although there is a good understanding of the role of other T cell subsets like Th1 cells and cytotoxic T cells in cancer and their contribution to tumor immunosurveillance, the role of Th17 responses seems to be more contextual, depending on tumor type, models, and experimental setup (Murugaiyan and Saha, 2009). Th17 responses and the action of IL-17 itself were initially thought to be proangiogenic, proinvasive, and tumor growth promoting (Numasaki et al., 2003). In contrast, other studies described a host-protective role in tumor surveillance, inhibition of proliferation, and inhibition of metastasis (Muranski et al., 2008; Kryczek et al., 2009; Martin-Orozco et al., 2009). However, in patients with colorectal cancer (CRC), Th17 cells and increased IL-17 seem to be associated with a poor prognosis (Liu et al., 2011; Tosolini et al., 2011). Th17 cell infiltration was also found in animal models of colon cancer, such as the ApcMin model driven by the enterotoxigenic bacterium Bacteroides fragilis, where blockade of IL-17 decreased tumor incidence (Wu et al., 2009).
It is now appreciated that a Type 17 cytokine expression signature is not restricted to Th-17 cells, and that cytokines such as IL-17 and IL-22 can be produced by unconventional T cell populations and more recently described RORγt-expressing innate lymphoid populations (Pearson et al., 2012). Indeed we recently showed that Helicobacter hepaticus (Hh)–driven colitis in genetically susceptible 129SvEv.RAG−/− mice involved the actions of a novel innate lymphoid population with the phenotype ROR-γt+, CD45+, lineage−, IL-7Rα+, Thy1hi, Sca1hi, Nkp46−, and CD4−. These ILCs accumulated in the inflamed colon and contributed to colitis through IL-23–driven IL-17 and IFN-γ production (Buonocore et al., 2010). Similar IL-17–producing ILCs accumulate in the intestine of Crohn’s disease patients (Geremia et al., 2011). Colonic ILCs, together with CD4+ LTi cells and Nkp46+ NK-22 cells, are members of a growing family of ROR-γt+ ILCs, which all express IL-7Rα and Thy1 and are negative for the lineage markers CD11b, B220, and CD3 and contribute to host defense and inflammation (Pearson et al., 2012).
Infection of most strains of immune-deficient mice with the mouse pathogen Hh leads to a mild inflammatory response. In contrast, 129SvEv.RAG−/− mice develop a chronic innate colitis, which in the presence of the DNA damaging agent 2-azoxymethane (AOM) progresses to invasive adenocarcinoma. We have recently mapped a genetic locus on chromosome 3, Hiccs (Hh-induced colitis and associated cancer susceptibility), that controls the bacteria-driven innate response and progression to CRC (Boulard et al., 2012). This mouse model of colon cancer therefore bears hallmarks of human CAC being driven by the interaction between genetic susceptibility and environmental factors. Although IL-23–driven ILCs promote colitis in this model, the role of ILCs and their downstream cytokines in CAC is not known.
IL-22 is an important component of Type 17 responses and functions as a double-edged sword in the intestinal inflammatory response. On the one hand, it has well described protective functions through induction of epithelial regeneration and the production of antimicrobial peptides, mucins, and chemokines via epithelial Stat3 activation (Sugimoto et al., 2008; Zheng et al., 2008; Witte et al., 2010). On the other hand, IL-22 has been shown to promote colitis in some settings (Kamanaka et al., 2011) and IBD patients express increased levels of IL-22 in the colon (Brand et al., 2006; Geremia et al., 2011). The heterodimeric receptor for IL-22, IL-22R, and IL-10R2 is exclusively expressed on nonhematopoietic cells like intestinal epithelial cells (IECs) and signals through Stat3, a molecule involved in several models of CAC (Bollrath et al., 2009; Grivennikov et al., 2009). A procarcinogenic role for IL-22 via Stat3 activation was previously shown in several extracolonic cancers like hepatocellular carcinoma and nonsmall cell lung cancer (Zhang et al., 2008; Jiang et al., 2011). Surprisingly little is known about the role of IL-22 in CRC although polymorphisms in IL-22 were shown to be associated with a 1.46-fold increased risk for development of CRC (Thompson et al., 2010). Furthermore, IL-22 in conjunction with IFN-γ can induce iNOS production and procarcinogenic nitric oxygen species in human colon carcinoma cell lines (Ziesché et al., 2007).
In this paper, we have assessed the role of ILC and Type 17 cytokines in the development and maintenance of bacteria-driven CAC. Our results reveal a hitherto unappreciated role for ILC in the progression from colitis to colon cancer and, furthermore, show a specific role for IL-22 but not IL-6 or IL-17 in this process.
RESULTS
Colonic innate lymphoid cells (cILCs) accumulate during cancer development
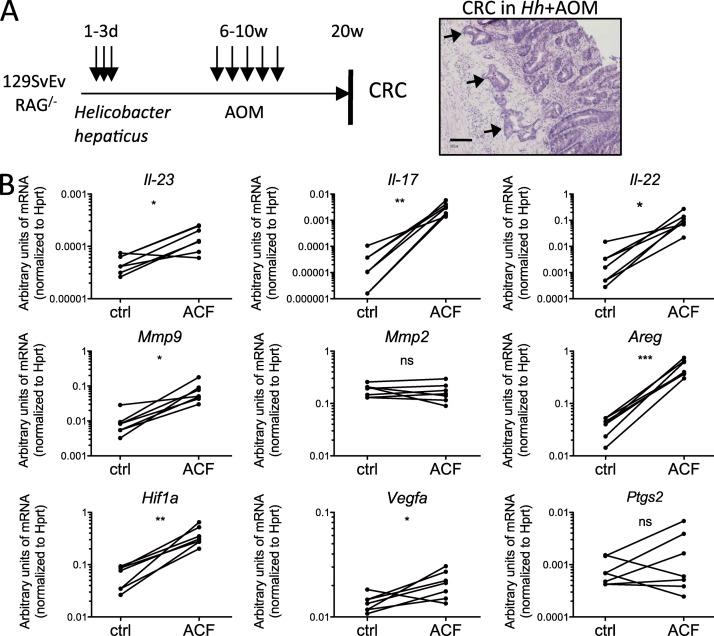
To investigate whether ILCs are involved in bacteria-induced CAC, we used a new innate model in which genetically susceptible 129SvEv.RAG−/− mice are infected with Hh and treated with the carcinogen AOM. In this setting, mice progress from having chronic inflammation of the cecum and colon to invasive CRC within 3–5 mo (model based on Nagamine et al., 2008; Boulard et al., 2012; Fig. 1 A, left). CRCs are mainly located at the sites of highest inflammation like the cecum and distal colon (Boulard et al., 2012). Approximately 75% of mice develop between 1 and 4 (average per mouse, 1.2) colorectal carcinomas. In contrast to other CRC models, for example, DSS+AOM, the tumor number is low. However, the pathological features closely resemble human colitis-associated carcinoma and contained atypical glands breaching the muscularis mucosa accompanied by the induction of a desmoplastic stromal response (Fig. 1 A, right).
Figure 1.
IL-23 signature genes are increased in aberrant crypt lesions of Hh+AOM-treated mice. (A) Treatment scheme (left) and H&E-stained invasive colorectal-carcinoma in Hh+AOM-treated 129SvEv.Rag−/− mice (right; bar, 100 µm). Arrows represent atypical glands breaching the muscularis mucosa. (B) Quantitative PCR analysis showing levels of mRNA of the cytokines Il-23, Il-17, and Il-22, the matrix metalloproteinases Mmp9 and Mmp2, the EGF receptor ligand amphiregulin (Areg), and Hif1a, Vegfa, and Ptgs2 (Cox-2) in tumors versus surrounding normal tissue (n = 5–7). Graphs show a representative experiment out of two independently performed. *, P < 0.05; **, P < 0.01; ***, P < 0.001, paired Student’s t test.
In Hh+AOM-treated mice, we found increased levels of IL-23, IL-17, and IL-22 mRNA in precarcinogenic aberrant crypt foci (ACF) compared with adjacent control tissue (Fig. 1 B). This was accompanied by increased levels of molecules involved in intestinal epithelial neoplasia, such as the EGFR ligand amphiregulin (Areg), a mediator of proliferation, and the matrix metalloproteinase Mmp9 (Langowski et al., 2006), involved in invasion and angiogenesis, and Hif1a and Vegfa, but not Ptgs2 (Cox2) and Mmp2 (Fig. 1 B). To characterize the populations of ILC present, we stained colonic lamina propria leukocytes (cLPs) for different ILC markers. The majority of CD45+, IL-7Rα+, lineage− cells expressed Thy1 in uninfected as well as in Hh+AOM-treated mice (Fig. 2 A). The composition of ILC changed upon inflammation and cancer development as the percentage of Nkp46+ cells decreased from ∼20 to 2% and the percentage of CD4− Nkp46− Sca1hi cILC increased from 70 to 95%, whereas CD4+ LTi cells remained stable (Fig. 2, A and B). Although the overall frequency of all three populations in the colonic lamina propria decreased upon Hh+AOM treatment as a result of the recruitment of other inflammatory cells, cILC numbers increased two- to threefold, whereas Nkp46+ ILC decreased and LTi cells were unchanged (Fig. 2 C).
Figure 2.
Nkp46−CD4− cILCs accumulate during H. hepaticus–driven cancer. (A–C) Flow cytometry on cLP cells of uninfected and Hh+AOM mice. Composition (B), frequency, and numbers of cILC (n = 5; C). (D) Surface marker expression on cILC from uninfected or Hh+AOM mice. (E) Representative immunofluorescence of an Hh+AOM CRC. Arrows indicate invasive crypts. Data are shown as means and SEM. Results are representative of at least two independent experiments. *, P < 0.05; **, P < 0.01, Mann-Whitney nonparametric test.
Further characterization of cILC surface markers showed that they expressed CD44, the IL-2 receptor α chain CD25, ICOS, and CCR6 (Fig. 2 D). They were negative for c-Kit and the IL-33R subunit ST2, markers which characterize Th2-type nuocytes and natural helper cells. cILCs, therefore, are phenotypically similar to ILCs previously identified by us during chronic Hh-induced inflammation. ILCs (IL-7Rα+ ROR-γ+) were localized in clusters (Fig. 2 E), here shown close to an E-cadherin+ invasive crypt, but also distributed within the lamina propria (not depicted).
ILCs promote bacteria-driven colon cancer
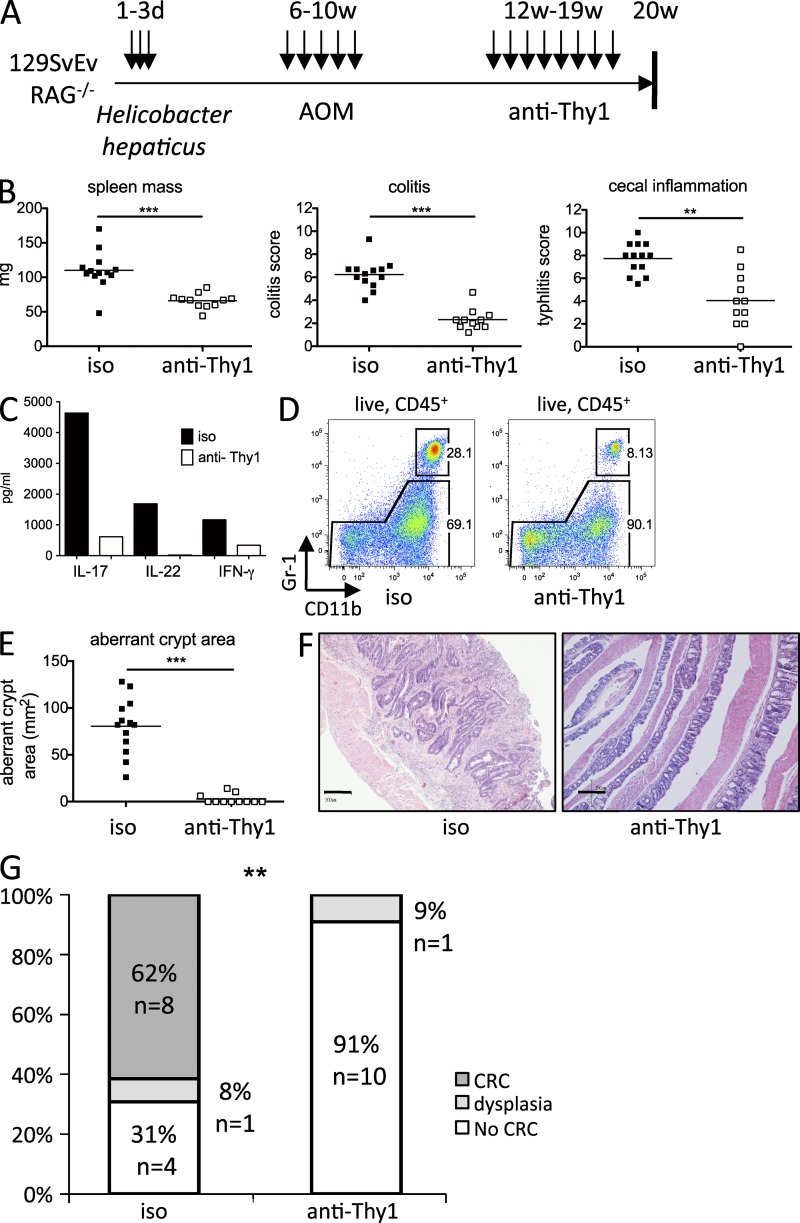
It was previously shown that depletion of ILC with a Thy1 antibody could block the development of colitis and also impair protective immunity to Citrobacter rodentium infection (Buonocore et al., 2010; Sonnenberg et al., 2011). To assess if ILCs were players in the transition from established chronic inflammation to colon cancer, we injected ILC-depleting anti-Thy1 into mice subjected 3 mo earlier to Hh infection and AOM treatment (Fig. 3 A). This therapeutic intervention led to a significant reduction of spleen mass, as well as cecal and colonic inflammation (Fig. 3 B), accompanied by reduced levels of IL-17, IL-22, and IFN-γ, cytokines previously shown to be produced by cILC (Buonocore et al., 2010; Fig. 3 C). We also detected reduced granulocyte recruitment to the colon (Fig. 3 D). To determine the impact of the treatment on carcinogenesis, we first analyzed methylene blue–stained colons for ACF. The area of ACF was strongly reduced upon treatment with anti-Thy1 (Fig. 3 E). Histopathological analysis of sequential H&E-stained sections of the colon revealed no invasive carcinomas but only dysplasia (9%) in the ILC depleted group, whereas in the isotype-treated group 62% of mice progressed to invasive adenocarcinoma (Fig. 3 G). Invasive adenocarcinomas were characterized by architecturally abnormal crypts penetrating the muscularis mucosa (Fig. 3 F, left). In contrast, in the colon of ILC-depleted mice, epithelial hyperplasia and inflammation was abrogated (Fig. 3 F, right).
Figure 3.
Colitis to cancer transition is dependent on ILC. (A) Treatment scheme. (B) Spleen mass (left), colitis (middle), and typhlitis (right) in isotype (n = 13) or anti-Thy1 (n = 11)–treated Hh+AOM mice. (C) Cytokines in supernatants of pooled cLP cells cultured overnight from one representative experiment of three independently performed experiments. (D) Flow cytometry of colonic CD11bhi Gr1hi granulocytes. (E) Aberrant crypt area on methylene blue–stained colons. (F) Representative photomicrographs (bars, 200 µm) of invasive CRC in an isotype treated mouse (left) and normal colon in an anti-Thy1–treated mouse (right). (G) Highest tumor grade per mouse. Data are shown as means. Results show a pool of two independent experiments (B, E, and G). **, P < 0.01; ***, P < 0.001, Mann-Whitney non parametric t test (B and E), Fisher’s square test (G).
IL-22 in tumor maintenance
Selective accumulation of cILC in the colon was accompanied by increased levels of IL-17, IL-22, and proinflammatory factors like TNF-α and IL-1β, but not IL-6, produced by cLPs (Fig. 4 A). We investigated which of those cytokines were produced by the ILC populations predominantly present in uninfected and Hh+AOM-treated mice. Whereas in uninfected mice the two dominant populations, cILC and Nkp46+ cells, expressed none of these cytokines in detectable amounts when cultivated ex vivo in the presence of monensin, cILC from Hh+AOM-treated mice expressed IL-17 and IL-22 (Fig. 4 B). We could also detect low levels of intracellular IL-6 and TNF in cILC.
Figure 4.
Nkp46− CD4− lin− Thy1hi ILCs are the major source of IL-22 in Hh+AOM CRC. (A) Secretion of cytokines into the supernatant of overnight cultured cLP cells from uninfected or Hh+AOM-treated mice. (B) Intracellular cytokine stain gated on different ILC populations from uninfected or Hh+AOM-treated mice after 3 h culture of cLPs in the presence of monensin. (C–E) Characterization of IL-22 producers in cLPs of Hh+AOM-treated mice stimulated overnight in the presence of IL-23 and for 3 h with PMA, Ionomycin, and monensin (n = 5). (C) Intracellular stain with isotype (left) or anti–IL-22 (right). (D) Anti–IL-22 stains mostly Thy1hi (left) Nkp46− CD4− (right) cells. (E) IL-17 and IL-22 expression in Thy1hi cells. Data are shown as means and SEM. Results are representative of at least two independent experiments. *, P < 0.05, Mann-Whitney nonparametric test.
CD4+ LTi and Nkp46+ NK-22, as well as CD11c+ cells, were previously shown to be major producers of IL-22 in response to IL-23 in different models of colitis, especially in a tissue-protective response to C. rodentium infection and DSS colitis (Cella et al., 2009; Pickert et al., 2009; Sonnenberg et al., 2011). To further investigate which cells were the main producers of IL-22 in Hh+AOM cancer, we stained for intracellular IL-22 in cLPs stimulated overnight in the presence of IL-23. The ability to produce IL-22 was restricted to lineage-negative CD45+ cells (Fig. 4 C). Within the lineage-negative population, almost exclusively Thy1hi cells produced IL-22, of which 94.9 ± 1% (n = 5) were CD4−Nkp46− cILCs (Fig. 4 D). Of all Thy1hi ILCs, 37.4 ± 2.5% were producers of both IL-17 and IL-22, 11.8 ± 1% were IL-22 single positive, and 26 ± 4.9% IL-17 single positive (Fig. 4 E).
Next, we investigated if IL-22 or any of the other known cILC-associated cytokines could sustain bacteria-induced cancer. Hh+AOM-treated mice with established epithelial dysplasia and CRC were administered neutralizing antibodies against IL-17, IL-22, or IFN-γ and analyzed for intestinal inflammation and CRC 8 wk later (Fig. 5 A). Treatment with these cytokine-blocking antibodies did not influence the number or frequency of cILCs present (unpublished data). Analysis of a group of Hh+AOM-treated mice at the time of antibody treatment showed a high tumor incidence (100%; Fig. 5 E). Blockade of IL-17 or IL-22 significantly reduced systemic inflammation, as measured by spleen mass as well as cecal inflammation (Fig. 5 B). In both anti–IL-17 and anti–IL-22–treated groups there was a trend toward reductions in the number of infiltrating colonic granulocytes (Gr1hi, CD11bhi, Ly6Cint, F4-80− cells) but this did not reach significance (Fig. 5 D). Similarly, accumulation of Ly6Chi, F4-80+ inflammatory monocytes was not reduced (unpublished data). Strikingly, IL-17 and IL-22 appear to have distinct functional roles in inflammation and tumor development, as only IL-22 blockade ameliorated established colitis and reduced tumor burden significantly (Fig. 5, B, C, and E). The differential effects of anti-cytokine treatment cannot be explained by effects on Hh infection, as we could not detect differences in cecal Hh colonization levels upon antibody treatment (Fig. 5 F).
Figure 5.
IL-22 drives cancer perpetuation. (A) Treatment scheme. (B) Spleen mass (left), colitis (middle), and typhlitis (right). Means (n = 9–14). Results are a pool of two independent experiments. (C) Representative photomicrographs of colons (bars, 200 µm). (D) Granulocyte frequencies (left) and numbers (right) in cLPs as analyzed by flow cytometry for Gr-1hi, CD11bhi, and F4-80−, Ly6Cint cells (n = 3–5). Data shown represent one out of two independent experiments. (E) CRC in antibody-treated Hh+AOM mice. Highest tumor grade per mouse. (F) Colonization with Hh. Results are a pool of two independent experiments (B, E, and F). Bars represent the mean value (B, D, and F). Significance refers to isotype control (B and E). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, one-way ANOVA with Bonferroni’s post test (B) or Fisher’s square test (E).
Remarkably, in both the anti–IL-22 and anti-Thy1 group the numbers of mice that showed high-grade dysplasia and invasive CRC were significantly lower (30%) than in the isotype-treated group (78%). As all mice sacrificed before therapeutic treatment with anti–IL-22 had developed CRC, these results suggest that IL-22 is a pivotal player not only in the transition from inflammation to CRC but also in the perpetuation of the cancerous state.
IL-22 regulates epithelial functions via Stat3
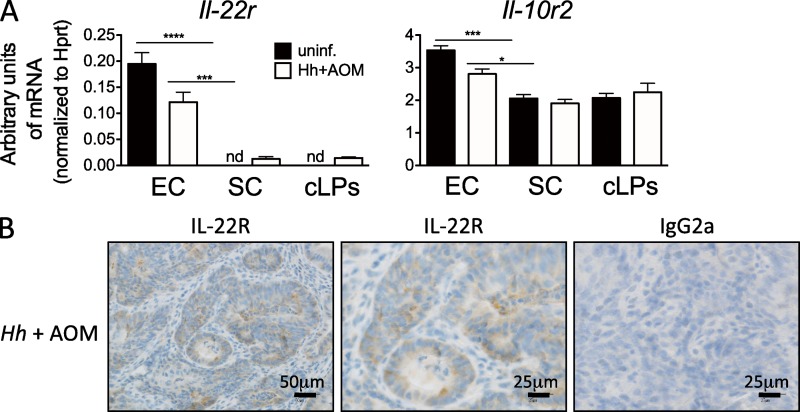
To investigate the mechanisms of how IL-22 sustains inflammation and cancer, we hypothesized that IL-22 acting on epithelial cells could lead to an inflammatory response and tissue repair program (Brand et al., 2006; Cella et al., 2009; Pickert et al., 2009) which becomes deregulated and promotes CRC. The heterodimeric receptor for IL-22 consisting of IL-22R and IL-10R2 is supposedly only expressed on nonhematopoietic cells (Witte et al., 2010). Indeed, detectable IL-22R expression was found only in colonic epithelial cells but not in the stromal cell fraction containing intestinal fibroblasts, myofibroblasts, and endothelial cells or cLPs (Fig. 6 A). Expression of IL-22R was also found in dysplastic epithelial cells in the colon of Hh+AOM-treated mice (Fig. 6 B). We further dissected the therapeutic effect of blocking IL-22 on epithelial cells in our cancer model by treating mice in vivo for a short time period with anti–IL-22.
Figure 6.
IL-22R expression in colonic epithelial cells in the Hh+AOM model. (A) Expression of IL-22R is restricted to colonic epithelial cells. Quantitative PCR analysis of mRNA levels of Il-22r and Il-10r2, the two subunits of IL-22 receptor, in colonic epithelial cells (EC), stromal cells (SC), and cLPs. Mean and SEM of one representative experiment (n = 3) of two is shown. (B) Expression of IL-22R in dysplastic epithelial cells of 5-mo Hh+AOM-treated mice as assessed by immunohistochemistry. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001, one-way ANOVA with Bonferroni’s post test (A).
We found that phosphorylated Stat3 accumulates in the nuclei of colonic epithelial cells in isotype-treated Hh+AOM mice (Fig. 7 A, top left). However, upon anti–IL-22 treatment for 1 wk the phosphorylation was almost undetectable in the epithelium (Fig. 7, A [top right] and B) but was still found at reduced levels in cells of the lamina propria (Fig. 7, A and B). Interestingly short-term treatment with anti–IL-6R or anti–IL-17 either failed or only minimally reduced Stat3 phosphorylation, indicating that epithelial Stat3 activation in this model is not regulated by these pathways (Fig. 7, A [bottom left and right] and B). We could detect an overall reduction of proliferating epithelial cells in colons of Hh+AOM mice treated with anti–IL-22 using flow cytometry for Ki-67 (Fig. 7 C). Furthermore, Cyclin D1 levels were reduced in dysplastic areas of anti–IL-22–treated mice (Fig. 7 D, top), as well as Cyclin D1 message levels in colonic epithelial cell extracts (Fig. 7 D, bottom).
Figure 7.
IL-22 regulates epithelial functions through Stat3 phosphorylation. (A–E) Hh+AOM-treated mice were injected with isotype control, anti–IL-22, anti-IL6R, anti–IL-17, or anti-Thy1 (B and E) for 1 wk. (A) Colonic phospho-Stat3-Y705 expression (bars, 25 µm). Arrows indicate pStat3+ cells in the cLP. (B) Immunoblot for phospho-Stat3 and total Stat3 in colonic epithelial and lamina propria cells. (C and D) Blocking of IL-22 reduces epithelial proliferation. (C) Ki-67 expression in freshly isolated EC from Hh+AOM mice (n = 4) and anti–IL-22 treated mice (n = 4). Gated on live, Epcam+, CD45− cells. (D) Cyclin D1 expression in dysplastic epithelial cells of Hh+AOM mice after 1 wk isotype or anti–IL-22 treatment (top). Cyclin D1 mRNA expression in isolated EC (bottom). (E) mRNA expression in EC. Data are shown as means and SEM of n = 2–5 per group. Results are representative of at least two independent experiments. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001, one-way ANOVA with Bonferroni’s post test (E) or unpaired Student’s t test (C and D).
Blockade of IL-22 also affected other target genes of the IL-22–Stat3 axis. We found that RNA levels of the antimicrobial peptides RegIIIg, RegIIIb, and S100A8, the suppressor of cytokine signaling 3 (Socs3), and the neutrophil-attracting chemokine Cxcl1, which were up-regulated in Hh+AOM-treated animals, were strongly decreased upon IL-22 blockade and, to a lower extent, by anti-Thy1 treatment but not upon IL-6R or IL-17 blockade (Fig. 7 E). The effect of anti–IL-22 treatment on Cxcl2 expression was less pronounced, indicating the possible involvement of other factors in granulocyte recruitment. The expression of the antiapoptotic molecules Bcl2 and Bclxl was also not inhibited by IL-22 blockade. Together, these results indicate a strong and almost exclusive role for IL-22 in the epithelial–Stat3 axis in this model, regulating epithelial proliferation, myeloid cell recruitment, and antimicrobial defense.
IL-22 in human CRC
To investigate the presence of IL-22 in human CRC, we obtained fresh and paraffin-embedded CRC tissue from patients who underwent surgery. Tumors were from different colonic locations and patients did not have IBD. As IBD patients undergo a rigorous screening procedure to avoid cancer development, IBD-associated CRC samples are rare and sporadic CRC samples provided the best available alternative for our study.
In tissue sections of human CRC specimens, cancerous areas showed an increased and aberrant expression of Ki-67 (Fig. 8 A, left). We could identify IL-22–positive cells close to dysplastic areas (Fig. 8 A, right) but also in normal tissue areas of the same patients. Immunofluorescence revealed that IL-22 was expressed by CD3+ and CD3− cells within tumors (Fig. 8 B). Although the majority of IL-22–positive cells within tumors are T cells, a lower percentage of lineage− cells produce IL-22 (Fig. 8 C). We also identified IL-17 and IL-22 double-producing T cells to be present in human tumor tissue (Fig. 8 D).
Figure 8.
Presence of IL-22+ T cells and IL-22+ lineage-negative cells in human CRC. (A) huCRCs show an aberrant pattern of Ki-67 expression (left). IL-22+ cells are located in proximity to dysplastic areas (right). (B) Representative immunofluorescence image of a tumor and adjacent normal tissue (n = 4). CD3 (green), IL-22 (red), and DAPI (blue) are shown. Yellow arrows indicate CD3+ IL-22+ double-positive cells. White arrows indicate CD3− IL-22+ cells. (C and D) Flow cytometry of cLPs isolated from a huCRC and treated overnight with IL-23 followed by 3 h in the presence of PMA, ionomycin, and monensin (n = 3). (C) Lineage+ and lineage− CD45+ cells express intracellular IL-22. (n = 3). (D) Presence of IL-22 and IL-17 double-producing cells in human tumors (n = 3). (E) Cytokine expression in huCRC. mRNA expression of IL22, IL17, IL10, and IFNg in the colonic mucosa of matched tumors and adjacent normal tissue (n = 12) relative to bACT. Graph shows the percentage of matched tumor (T)-normal (N) tissue specimens, expressing the same levels of cytokines or more than twofold increased levels in either tumor or normal tissue.
We analyzed cytokine expression levels in 12 matched pairs of human CRC and adjacent tissue by quantitative real-time PCR (Fig. 8 E). In 58% of pairs, IL-22 mRNA expression was more than twofold higher in tumor versus normal tissue. Similarly, IL-17 was overexpressed in 75% of tumors and IFN-γ was more associated with CRC. In contrast, IL-10 was predominantly expressed in normal tissue. The presence of IL-22–producing cells within tumors could point toward a possible therapeutic application of IL-22 blockade in some CRC patients.
DISCUSSION
It is increasingly appreciated that inflammatory responses can be both protective and deleterious in cancer. In human CRC, Type 17 responses are associated with a poor prognosis, and in animal models of CRC, IL-23 and Th17 responses can provide an inflammatory environment that supports tumorigenesis (Wu et al., 2009; Liu et al., 2011; Tosolini et al., 2011). However, to date it has not been possible to separate pathways that are required for the inflammatory response from those that promote and sustain cancer. In this paper, using a bacteria-driven CAC model we identify Type 17 ILC and IL-22 in particular as pivotal for the development and maintenance of invasive CAC.
IL-22–producing ILCs are present in the intestine and contribute to the early innate response to enteric pathogens such as C. rodentium through induction of antimicrobial peptides and mucus production by IEC (Sugimoto et al., 2008; Zheng et al., 2008). IL-22–producing host-protective responses are contained within CD4+ LTi cells (Sonnenberg et al., 2011) and Nkp46+ NK-22 populations (Satoh-Takayama et al., 2008). A subpopulation of IL-12–responsive Nkp46+ LTi cells was also shown to be involved in antitumor immunity (Eisenring et al., 2010). In contrast with early protective responses, we observed accumulation of pathogenic double-negative Nkp46− CD4− ILC in chronic bacteria-induced inflammation (Buonocore et al., 2010). This population also differentially expands in bacteria-driven CAC at the expense of Nkp46+ and CD4+ ILC populations and produces IL-17 and IL-22. Thus, microbial insult in a genetically susceptible host leads to a shift from tissue-protective and potentially anti-tumorigenic ILC populations to chronic inflammatory pathogenic cILC. Although we describe ILC as the major source of IL-22 in this model, in lymphocyte-replete models of CRC, functionally similar IL-17– and IL-22–producing T cells may contribute to pathogenesis. Indeed, in human CRC we observed both T cell and non–T cell sources of IL-22. Further studies are required to determine the contribution of IL-22–producing T cells and innate cells to the natural history of human cancer development using normal and premalignant tissue samples.
IL-23 is a key driver of ILC-mediated colitis in the Hh model (Hue et al., 2006). IL-23 also drives colon cancer in an APC model, where it is induced in tumor-associated myeloid cells by microbial products, which are thought to specifically penetrate tumors through local down-regulation of mucins and tight junction proteins (Grivennikov et al., 2012). However, in that model and in the enterotoxigenic bacterium Bacteroides fragilis–driven colonic ApcMin model, IL-17 is considered to be the key downstream mediator (Wu et al., 2009). Although the exact mechanism of IL-17 function in tumorigenesis is unclear, it was suggested to induce epithelial hyperproliferation and production of reactive oxygen and nitrogen species from granulocytes. However, it should be noted that both of these studies analyzed cancer development and not signals required for maintenance of the cancerous state. Indeed, we found that although anti–IL-17 blocks bacteria-driven innate colitis (Buonocore et al., 2010) and granulocyte recruitment, blockade of this pathway is not sufficient to reverse dysplasia and invasive adenocarcinoma. Instead, we observed a nonredundant role for IL-22 in this process.
Although IL-22 is host protective in acute intestinal infection and inflammation (Satoh-Takayama et al., 2008; Sugimoto et al., 2008), it is also implicated in the pathogenesis of chronic colitis through epithelial hyperproliferation (Kamanaka et al., 2011). We have also found a pathogenic role for IL-22 in development of Hh-induced colitis in immune-deficient and lymphocyte-replete mice in the presence of IL-10 deficiency (unpublished data). The reasons for the dual properties of IL-22 in terms of intestinal homeostasis are not known but may relate to the nature of microbial stimulation and that sustained production in response to chronic bacterial stimulation, such as that provided by the pathobiont Hh eventually becomes pathological. A recent study on IL-22 and its regulation by IL-22BP further supports this dual function of IL-22 and its role in inflammation and colorectal tumorigenesis but does not study tumor maintenance (Huber et al., 2012).
The presence of other cytokines also influences IL-22 functions. Thus, dual production of IL-17 and IL-22 by ILC led to immune pathology in a model of lung inflammation through recruitment of neutrophils (Sonnenberg et al., 2010). Granulocytes may contribute to carcinogenesis through production of reactive oxygen species; however, blockade of IL-22 or IL-17 did not significantly reduce granulocyte accumulation in the colon despite the former having anti-cancer properties, suggesting that other functions of IL-22, particularly its direct action on IEC, could be more relevant for tumor maintenance.
The main pathway of IL-22 action is via ligation of IL-22R on epithelial cells, leading to Stat3 activation. The role of Stat3 in colon cancer is well known but this has been attributed to activation by IL-6 and IL-11 (Ernst et al., 2008; Grivennikov et al., 2009). Surprisingly, neutralization of IL-22, but not IL-6 or IL-17, almost completely abrogated Stat3 phosphorylation in the epithelium, as well as epithelial proliferation and cyclin D1 expression. This indicates that IL-22 is the main activator of the Stat3 pathway in this model and could facilitate cancer perpetuation via proliferative effects on the epithelium.
IL-22 may also promote cancer through direct effects on intestinal epithelial stem cells, previously shown to be key precursors for intestinal cancer (Barker et al., 2009). IL-22R expression was recently demonstrated in intestinal stem cells and IL-22 had protective effects on these cells in a graft-versus-host disease model (Hanash et al., 2012). A persistent IL-22 signal during chronic inflammation could therefore directly act on the “master regulator” of epithelial regeneration, leading to hyperproliferation and later to tumor maintenance. Such a mechanism could also explain the therapeutic effect of anti–IL-22 treatment on established tumors.
It is also possible that through the induction of antimicrobial peptides, IL-22 could induce a shift in the intestinal microbiota, favoring the outgrowth of “inflammatory allies” that support inflammation and cancer in conjunction with Hh. The composition of the gut microbiota influences cancer susceptibility as germ-free mice do not develop cancer (Uronis et al., 2009). Furthermore, certain species like the anaerobic Fusobacterium nucleatum were found to be associated with human CRC tissue (Castellarin et al., 2012; Kostic et al., 2012). Further studies are required to assess the role of microbiota changes in IL-22–driven CAC.
Although we found that IL-22 blockade very efficiently reduced tumor maintenance, a role for other cILC-produced factors in tumorigenesis is not excluded. Besides IL-22, cILC can produce other molecules involved in epithelial repair. It was recently shown that lung ILC, phenotypically similar to nuocytes and natural helper cells, produced the EGFR ligand Areg in response to influenza virus-induced tissue damage in the lung (Monticelli et al., 2011). Administration of Areg in ILC-depleted mice was able to restore lung function upon influenza virus infection. In our cancer model, Areg is increased in aberrant crypt areas of Hh+AOM-treated mice. Furthermore, we found that cILCs express Areg message (unpublished data) and this could contribute to deregulated epithelial proliferation and tumor development. Further transcriptional profiling of cILC could lead to the identification of other factors involved in tissue repair and cancer.
In summary, our results show that innate immune responses intended to protect the host from invading pathogens and to initiate tissue repair can become deregulated and chronic in genetically susceptible hosts, paving the way for cancer development. Together, our results identify a novel role for IL-22 in maintenance of colon carcinogenesis in chronic inflammation. The finding that established colon tumors in this model system are not cell autonomous but require the immune mediator IL-22 for their perpetuation raises the possibility that targeting the IL-22 axis may have therapeutic utility in human CRC.
MATERIALS AND METHODS
Human study subjects.
CRC patients were recruited from the colorectal surgery department at the Churchill Hospital in Oxford. Colon specimens were collected from patients undergoing surgery for severe disease. Diagnosis of CRC was confirmed by a pathologist. Matched tumor and normal samples were selected from the same patients by a pathologist. Ethical approval was obtained from the Oxfordshire Research Ethics Committee (Reference numbers: 11/YH/0020 and 09/H0606/5) and informed written consent was given by all study patients.
Mice.
129SvEv.RAG−/− (129SvEvS6.Rag2−/−) mice were maintained in an accredited specific pathogen-free facility and experiments were conducted in accordance with the UK Scientific Procedures Act (1986). Mice were routinely screened for the absence of pathogens and Helicobacter spp.
Hh CAC.
Hh NCI-Frederick isolate 1A (strain 51449) was grown as described previously (Maloy et al., 2003). Mice were fed 3× on consecutive days with Hh 1A (1.0 × 108 CFU) by oral gavage and injected with AOM (Sigma-Aldrich) i.p. once a week at a dose of 10 mg/kg for 5 wk, starting 6 wk after Hh infection (Nagamine et al., 2008). For experiments where animals were treated with blocking or depleting antibodies, treatment groups were mixed in cages to minimize cage effects and animals were treated at weeks 12–19 with the respective antibodies and analyzed at week 20. In some experiments, a preinjection group was analyzed at week 12.
Hh colonization of mice was quantified in cecal contents, which were collected upon sacrifice. DNA was isolated using DNA Stool kit (QIAGEN) and QPCR with Hh-specific primers against the cdtB gene was performed as previously described (Maloy et al., 2003).
For analysis of precarcinogenic aberrant crypts, colons were fixed in 10% formalin, stained in 2% methylene blue, and analyzed using a dissection microscope and sliding caliper. For histological analysis, 200–300 sections of paraffin-embedded Swiss-rolled colons and cecums were prepared, and every 10th section was H&E-stained and blindly graded by a pathologist using criteria for grading dysplasia in human IBD as previously described (Boulard et al., 2012). Tumors were staged as per TNM classification of human CRC.
In vivo antibody treatment.
Therapeutic administration of different antibodies was started at week 12 of Hh+AOM treatment in 129SvEv.RAG−/− mice, when inflammation was already fully developed, and was continued until the end of the experiment at week 20. To deplete Thy1+ cells, 1 mg of depleting rat anti-Thy1 (YTS154.7.7.10) or isotype control (YKIX 337.217.1) was administered i.p. once a week. To specifically block different cytokines in vivo, 0.375 mg of IL-17 blocking Ab (UCB Celltech) or 1 mg of IFN-γ blocking Ab (clone AN18) was administered i.p. twice a week. A blocking mAb against IL-22 (Genentech, clone 8E11) or isotype control (GP120 10E7.1D2) was administered i.p. three times per week at 0.15 mg. For short-term treatment, anti–IL-22, anti–IL-17, anti-IL6R, or anti-Thy1 were administered to 5-mo Hh+AOM mice three times within 1 wk.
Histological assessment of colitis and typhlitis.
Colonic and cecal inflammation in Hh+AOM mice was assessed upon sacrifice at 3 or 5 mo. H&E-stained samples, prepared as described above, were analyzed for inflammation in the proximal, mid, and distal part of the colon as well as the cecum. Scoring of inflammation was performed using previously published criteria (Izcue et al., 2008). In brief, each sample was graded semiquantitatively from 0 to 3 for the four following criteria: degree of epithelial hyperplasia and goblet cell depletion, leukocyte infiltration in the lamina propria, area of tissue affected, and presence of markers of severe inflammation (such as crypt abscesses, submucosal inflammation, and ulcers). Scores for each criterion were added to give a final quantification for intestinal inflammation ranging from 0 to 12. For colitis scores, a mean value of the three sections (proximal, mid, and distal colon) was calculated.
Cell isolation and FACS staining.
For cLP cell isolation, mouse colon pieces were incubated (3×) in RPMI 1640 with 10% FCS and 5 mM EDTA at 37°C to remove epithelial cells. Tissue was then digested with 300 U/ml of type VIII collagenase for 45 min at 37°C. The isolated cells were layered on a 30/40/75% Percoll gradient (GE Healthcare), which was centrifuged for 20 min at 600 g, and the 40/75% interface, containing mostly leukocytes, was recovered. cLPs were stained with a fixable viability dye (eBioscience) and antibodies against CD45.2 and CD4 (RM4-5; both BD), a lineage mix (CD11b [M1/70], Ly6G [Gr1, RB6-8C5], and B220 [RA3-6B2]), Thy1 (53–2.1), Sca1 (D7), Nkp46 (29A1.4), and IL-7Rα (A7R34; all from eBioscience).
For intracellular cytokine staining, cLPs were either directly cultivated for 3 h in the presence of Golgistop-Monensin (BD) or stimulated overnight with 10 ng/ml of recombinant IL-23 (R&D Systems), and then for the last 3 h PMA and Ionomycin were added. Cells were stained intracellularly with antibodies against IL-22 (1H8PWSR) and IL-17 (17B7; both eBioscience) or isotype control antibodies using the Cytofix/Cytoperm kit (BD). cLPs were analyzed by flow cytometry using an LSRII flow cytometer (BD).
For the isolation of epithelial cells colons were incubated for 5 min in 5 mM EDTA at 37°C. Cells in suspension were collected, immediately washed with cold PBS, and centrifuged. Flow cytometry was performed using a fixable viability dye and antibodies against EpCAM (G8.8; BioLegend), CD45.2, and Ki-67 (both BD). Colonic stromal cells were collected from the 30/40% Percoll interface during cLP isolation.
Human cLPs from tumors and normal tissue were isolated as previously described using enzymatic and mechanical disruption by GentleMACS (Miltenyi Biotec; Geremia et al., 2011). Cells were stained for the following lineage markers: CD1a (HI149), CD2 (RPA-2.10), CD19 (HIB19; all eBioscience), CD11c (AbD Serotec), CD14 (BD), and CD16 (3G8; BioLegend); CD56 (HCD56), CD45 (HI30; both BioLegend), CD3 (OKT3), and CD4 (RPA-T4; both eBioscience); and intracellular using Cytofix/Cytoperm for IL-22 (R&D Systems) and IL-17A (BL168; BioLegend).
Cytokine detection.
For detection of cytokines by the Flowcytomix Multiplex system (eBioscience), 105 isolated cLPs were cultured overnight in 100 µl RPMI-1640 with 10% FCS at 37°C. Cytokines in the supernatant were detected using Simplex kits for mouse IL-17, IL-22, IFN-γ, IL-1β, IL-6, and TNF. Samples were analyzed on a FACSCalibur (BD).
Quantification of mRNA levels by real-time PCR.
cLPs, stromal cells, or epithelial cells were lysed in RLT buffer (QIAGEN) with β-mercaptoethanol. RNA isolation was performed using the RNeasy kit (QIAGEN) including a DNase I digest step. Content and purity of RNA was controlled with a Nanodrop spectrophotometer (Thermo Fisher Scientific). cDNA synthesis was performed using the Superscript III reverse transcription kit (Life Technologies). Quantitative real-time PCR for the candidate genes was done using SYBR green chemistry (SensiMix; Bioline) or the TaqMan system. cDNA samples were analyzed in triplicate using the CFX96 detection system (Bio-Rad Laboratories) and values were normalized to Hprt. Analysis was performed according to the Δ-Ct method. The following primer pairs were used for the quantification of gene expression in mouse tissue with SYBR green. QuantiTect Primer Assay for Hprt (QIAGEN): Mmp2, 5′-CACCACCGAGGACTATGACC-3′ and 5′-TCCTTGGTCAGGACAGAAGC-3′; Mmp9, 5′-TTCGACACTGACAAGAAGTGG-3′ and 5′-CCACGACCATACAGATACTGG-3′; Areg, 5′-GACTCACAGCGAGGATGACA-3′ and 5′-GGCTTGGCAATGATTCAACT-3; Vegfa, 5′-TTACTGCTGTACCTCCACC-3′ and 5′-ACAGGACGGCTTGAAGATG-3′; Hif1a, 5′-CGACACCATCATCTCTCTGG-3′ and 5′-TGATTCAGTGCAGGATCAGC-3′; and Ptgs2 (Cox2), 5′-CAGTCAGGACTCTGCTCACG-3′ and 5′-TTGACATGGATTGGAACAGC-3′. Primer pairs and probes for Taqman: TaqMan Gene Expression Assays (Life Technologies) for mouse IL-22, IL-17, IL-23, Ccnd1, Bcl2, Bclxl, Cxcl1, Cxcl2, Reg3g, Reg3b, S100A8, and SOCS3: HPRT, 5′-GACCGGTCCCGTCATGC-3′, 5′-TCATAACCTGGTTCATCATCGC-3′, and VIC/TAMRA probe 5′-ACCCGCAGTCCCAGCGTCGTC-3′. For human tissue, the following TaqMan Gene Expression Assays (Life Technologies) were used: IL-22, IL-17, IFN-γ, IL-10, and bAct.
Immunohistochemistry, immunofluorescence, and immunoblot.
For immunohistochemistry, subsequent 5-µm tissue paraffin sections were deparaffinized with Histo-Clear (National Diagnostics) and rehydrated. Antigen retrieval was performed using 10 mM citrate buffer, pH 6, and then intrinsic peroxidase activity was blocked (peroxidase block from Dako EnVision kit) as well as unspecific interactions of the secondary antibody using PBS with 2% BSA and 2% goat serum. Mouse tissue slides were stained with pStat3Y705 (Cell Signaling Technology), Cyclin D1 (NeoMarkers), IL-22R (R&D Systems), or rabbit isotype control (Zymed, Life Technologies). Human tissue slides were stained for Ki-67 (Abcam) or IL-22 (eBioscience). Staining was visualized using the appropriate secondary HRP-coupled antibodies and the EnVision System and slides were counterstained with hematoxylin.
For immunofluorescence in mouse, cryosections were fixed with 2% formaldehyde and for the nuclear ROR-γ stain additionally with Fix/Perm (eBioscience). Endogenous peroxidase activity was blocked with 3% H2O2 and 2% NaN3 and unspecific binding with 10% donkey serum. Sections were either incubated with anti–IL-7Rα (A7R34; eBioscience) in 10% donkey serum in PBS or with ROR-γ in Perm buffer (eBioscience), followed by donkey anti–rat-HRP (Jackson ImmunoResearch Laboratories). Tyramide signal amplification was performed (Perkin Elmer). Then sections were stained with E-cadherin–FITC (612130; BD) and with DAPI.
For immunofluorescence of human tumor and normal sections, tissue was frozen in OCT, cut into 7 µm sections using a cryostat, and fixed with 1% paraformaldehyde for 5 min. Sections were stained for CD3 (Abcam) with Cy3 Perkin Elmer tyramide amplification, reblocked, stained for IL-22 (eBioscience) with FITC Perkin Elmer tyramide amplification, and mounted with Vectashield containing DAPI. Images were collected using a 710 microscope (Carl Zeiss).
Western blots were performed using Nupage gels (Life Technologies) and standard immunoblot technique. Invitrolon (Life Technologies) membranes were treated with pStat3-Y705 (9145) and Stat3 (both from Cell Signaling Technology) and developed using ECL (Thermo Fisher Scientific).
Statistical analysis.
Statistical analysis and graphical representations were done using Prism5 software (GraphPad Software). For comparison of Q-PCR results and histological analysis, the nonparametric Mann-Whitney test and unpaired Student’s t test were used, and for multiple samples, one-way ANOVA with Bonferroni’s post-tests was applied. For analysis of dysplasia and CRC versus normal, the Fisher square test was used.
Acknowledgments
We would like to thank R. Stillion for histology; C. Pearson, S. Buonocore, and M. Huhn for technical advice and discussion; C. Arancibia for critically reading the manuscript; A. Schaupp for technical assistance; W. Ouyang (Genentech) for the blocking anti–IL-22 antibody and isotype controls; S. Cobbold and H. Waldman for the Thy1 and isotype control hybridoma and J. Langhorne for the AN18 hybridoma; and UCB Celltech for providing the blocking anti–IL-17. We acknowledge the contribution to this study made by the Oxford Centre for Histopathology Research and the Oxford Radcliffe Biobank, which are supported by the NIHR Oxford Biomedical Research Centre.
S. Kirchberger was supported by the European Commission research program Infla-Care (EC contract No. 223151) and an Austrian FWF Schroedinger Fellowship; D.J. Royston was supported by The Royal College of Pathologists and the Jean Shanks Foundation; O. Boulard was supported by Cancer Research UK grants A7297 and A11663; and F. Powrie was supported by the Wellcome Trust.
The authors declare no competing financial interests.
S. Kirchberger and O. Boulard planned and performed experiments. D.J. Royston graded and scored cancer pathology. R.L. Szabady, E. Thornton, O. Harrison, and F. Franchini were involved in experiments. F. Powrie directed the research. S. Kirchberger and F. Powrie wrote the manuscript.
Footnotes
Abbreviations used:
- ACF
- aberrant crypt foci
- AOM
- 2-azoxymethane
- CAC
- colitis-associated cancer
- cILC
- colonic innate lymphoid cell
- cLP
- colonic lamina propria leukocyte
- CRC
- colorectal cancer
- Hh
- Helicobacter hepaticus
- IBD
- inflammatory bowel disease
- IEC
- intestinal epithelial cell
References
- Barker N., Ridgway R.A., van Es J.H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A.R., Sansom O.J., Clevers H. 2009. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 457:608–611 10.1038/nature07602 [DOI] [PubMed] [Google Scholar]
- Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. ; NIDDK IBD Genetics Consortium; Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 40:955–962 10.1038/ng.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollrath J., Phesse T.J., von Burstin V.A., Putoczki T., Bennecke M., Bateman T., Nebelsiek T., Lundgren-May T., Canli O., Schwitalla S., et al. 2009. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 15:91–102 10.1016/j.ccr.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Boulard O., Kirchberger S., Royston D.J., Maloy K.J., Powrie F.M. 2012. Identification of a genetic locus controlling bacteria-driven colitis and associated cancer through effects on innate inflammation. J. Exp. Med. 209:1309–1324 10.1084/jem.20120239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S.T., Otte J.M., Diepolder H., Marquardt A., Jagla W., Popp A., et al. 2006. IL-22 is increased in active Crohn’s disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am. J. Physiol. Gastrointest. Liver Physiol. 290:G827–G838 10.1152/ajpgi.00513.2005 [DOI] [PubMed] [Google Scholar]
- Buonocore S., Ahern P.P., Uhlig H.H., Ivanov I.I., Littman D.R., Maloy K.J., Powrie F. 2010. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 464:1371–1375 10.1038/nature08949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin M., Warren R.L., Freeman J.D., Dreolini L., Krzywinski M., Strauss J., Barnes R., Watson P., Allen-Vercoe E., Moore R.A., Holt R.A. 2012. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22:299–306 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M., Fuchs A., Vermi W., Facchetti F., Otero K., Lennerz J.K., Doherty J.M., Mills J.C., Colonna M. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 457:722–725 10.1038/nature07537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr R.H., Taylor K.D., Brant S.R., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., et al. 2006. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 314:1461–1463 10.1126/science.1135245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaden J.A., Abrams K.R., Mayberry J.F. 2001. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 48:526–535 10.1136/gut.48.4.526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenring M., vom Berg J., Kristiansen G., Saller E., Becher B. 2010. IL-12 initiates tumor rejection via lymphoid tissue-inducer cells bearing the natural cytotoxicity receptor NKp46. Nat. Immunol. 11:1030–1038 10.1038/ni.1947 [DOI] [PubMed] [Google Scholar]
- Ernst M., Najdovska M., Grail D., Lundgren-May T., Buchert M., Tye H., Matthews V.B., Armes J., Bhathal P.S., Hughes N.R., et al. 2008. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J. Clin. Invest. 118:1727–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremia A., Arancibia-Cárcamo C.V., Fleming M.P., Rust N., Singh B., Mortensen N.J., Travis S.P., Powrie F. 2011. IL-23–responsive innate lymphoid cells are increased in inflammatory bowel disease. J. Exp. Med. 208:1127–1133 10.1084/jem.20101712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S., Karin E., Terzic J., Mucida D., Yu G.Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., Karin M. 2009. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 15:103–113 10.1016/j.ccr.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S.I., Wang K., Mucida D., Stewart C.A., Schnabl B., Jauch D., Taniguchi K., Yu G.Y., Osterreicher C.H., Hung K.E., et al. 2012. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 491:254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanash A.M., Dudakov J.A., Hua G., O’Connor M.H., Young L.F., Singer N.V., West M.L., Jenq R.R., Holland A.M., Kappel L.W., et al. 2012. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 37:339–350 10.1016/j.immuni.2012.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S., Gagliani N., Zenewicz L.A., Huber F.J., Bosurgi L., Hu B., Hedl M., Zhang W., O’Connor W., Jr, Murphy A.J., et al. 2012. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 491:259–263 10.1038/nature11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue S., Ahern P., Buonocore S., Kullberg M.C., Cua D.J., McKenzie B.S., Powrie F., Maloy K.J. 2006. Interleukin-23 drives innate and T cell–mediated intestinal inflammation. J. Exp. Med. 203:2473–2483 10.1084/jem.20061099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izcue A., Hue S., Buonocore S., Arancibia-Cárcamo C.V., Ahern P.P., Iwakura Y., Maloy K.J., Powrie F. 2008. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity. 28:559–570 10.1016/j.immuni.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Tan Z., Deng L., Chen Y., Xia Y., Gao Y., Wang X., Sun B. 2011. Interleukin-22 promotes human hepatocellular carcinoma by activation of STAT3. Hepatology. 54:900–909 10.1002/hep.24486 [DOI] [PubMed] [Google Scholar]
- Kamanaka M., Huber S., Zenewicz L.A., Gagliani N., Rathinam C., O’Connor W., Jr, Wan Y.Y., Nakae S., Iwakura Y., Hao L., Flavell R.A. 2011. Memory/effector (CD45RBlo) CD4 T cells are controlled directly by IL-10 and cause IL-22–dependent intestinal pathology. J. Exp. Med. 208:1027–1040 10.1084/jem.20102149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortylewski M., Xin H., Kujawski M., Lee H., Liu Y., Harris T., Drake C., Pardoll D., Yu H. 2009. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell. 15:114–123 10.1016/j.ccr.2008.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A.D., Gevers D., Pedamallu C.S., Michaud M., Duke F., Earl A.M., Ojesina A.I., Jung J., Bass A.J., Tabernero J., et al. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22:292–298 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I., Wei S., Szeliga W., Vatan L., Zou W. 2009. Endogenous IL-17 contributes to reduced tumor growth and metastasis. Blood. 114:357–359 10.1182/blood-2008-09-177360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langowski J.L., Zhang X., Wu L., Mattson J.D., Chen T., Smith K., Basham B., McClanahan T., Kastelein R.A., Oft M. 2006. IL-23 promotes tumour incidence and growth. Nature. 442:461–465 10.1038/nature04808 [DOI] [PubMed] [Google Scholar]
- Liang S.C., Tan X.Y., Luxenberg D.P., Karim R., Dunussi-Joannopoulos K., Collins M., Fouser L.A. 2006. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203:2271–2279 10.1084/jem.20061308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Duan Y., Cheng X., Chen X., Xie W., Long H., Lin Z., Zhu B. 2011. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochem. Biophys. Res. Commun. 407:348–354 10.1016/j.bbrc.2011.03.021 [DOI] [PubMed] [Google Scholar]
- Maloy K.J., Salaun L., Cahill R., Dougan G., Saunders N.J., Powrie F. 2003. CD4+CD25+ T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J. Exp. Med. 197:111–119 10.1084/jem.20021345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Orozco N., Muranski P., Chung Y., Yang X.O., Yamazaki T., Lu S., Hwu P., Restifo N.P., Overwijk W.W., Dong C. 2009. T helper 17 cells promote cytotoxic T cell activation in tumor immunity. Immunity. 31:787–798 10.1016/j.immuni.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L.A., Sonnenberg G.F., Abt M.C., Alenghat T., Ziegler C.G., Doering T.A., Angelosanto J.M., Laidlaw B.J., Yang C.Y., Sathaliyawala T., et al. 2011. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12:1045–1054 10.1038/ni.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranski P., Boni A., Antony P.A., Cassard L., Irvine K.R., Kaiser A., Paulos C.M., Palmer D.C., Touloukian C.E., Ptak K., et al. 2008. Tumor-specific Th17-polarized cells eradicate large established melanoma. Blood. 112:362–373 10.1182/blood-2007-11-120998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaiyan G., Saha B. 2009. Protumor vs antitumor functions of IL-17. J. Immunol. 183:4169–4175 10.4049/jimmunol.0901017 [DOI] [PubMed] [Google Scholar]
- Nagamine C.M., Rogers A.B., Fox J.G., Schauer D.B. 2008. Helicobacter hepaticus promotes azoxymethane-initiated colon tumorigenesis in BALB/c-IL10-deficient mice. Int. J. Cancer. 122:832–838 10.1002/ijc.23175 [DOI] [PubMed] [Google Scholar]
- Numasaki M., Fukushi J., Ono M., Narula S.K., Zavodny P.J., Kudo T., Robbins P.D., Tahara H., Lotze M.T. 2003. Interleukin-17 promotes angiogenesis and tumor growth. Blood. 101:2620–2627 10.1182/blood-2002-05-1461 [DOI] [PubMed] [Google Scholar]
- Pearson C., Uhlig H.H., Powrie F. 2012. Lymphoid microenvironments and innate lymphoid cells in the gut. Trends Immunol. 33:289–296 10.1016/j.it.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Pickert G., Neufert C., Leppkes M., Zheng Y., Wittkopf N., Warntjen M., Lehr H.A., Hirth S., Weigmann B., Wirtz S., et al. 2009. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206:1465–1472 10.1084/jem.20082683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N., Vosshenrich C.A., Lesjean-Pottier S., Sawa S., Lochner M., Rattis F., Mention J.J., Thiam K., Cerf-Bensussan N., Mandelboim O., et al. 2008. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 29:958–970 10.1016/j.immuni.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Sonnenberg G.F., Nair M.G., Kirn T.J., Zaph C., Fouser L.A., Artis D. 2010. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. J. Exp. Med. 207:1293–1305 10.1084/jem.20092054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg G.F., Monticelli L.A., Elloso M.M., Fouser L.A., Artis D. 2011. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 34:122–134 10.1016/j.immuni.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Ogawa A., Mizoguchi E., Shimomura Y., Andoh A., Bhan A.K., Blumberg R.S., Xavier R.J., Mizoguchi A. 2008. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 118:534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C.L., Plummer S.J., Tucker T.C., Casey G., Li L. 2010. Interleukin-22 genetic polymorphisms and risk of colon cancer. Cancer Causes Control. 21:1165–1170 10.1007/s10552-010-9542-5 [DOI] [PubMed] [Google Scholar]
- Tosolini M., Kirilovsky A., Mlecnik B., Fredriksen T., Mauger S., Bindea G., Berger A., Bruneval P., Fridman W.H., Pagès F., Galon J. 2011. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 71:1263–1271 10.1158/0008-5472.CAN-10-2907 [DOI] [PubMed] [Google Scholar]
- Trinchieri G. 2012. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu. Rev. Immunol. 30:677–706 10.1146/annurev-immunol-020711-075008 [DOI] [PubMed] [Google Scholar]
- Uronis J.M., Mühlbauer M., Herfarth H.H., Rubinas T.C., Jones G.S., Jobin C. 2009. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE. 4:e6026 10.1371/journal.pone.0006026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte E., Witte K., Warszawska K., Sabat R., Wolk K. 2010. Interleukin-22: a cytokine produced by T, NK and NKT cell subsets, with importance in the innate immune defense and tissue protection. Cytokine Growth Factor Rev. 21:365–379 10.1016/j.cytogfr.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Wu S., Rhee K.J., Albesiano E., Rabizadeh S., Wu X., Yen H.R., Huso D.L., Brancati F.L., Wick E., McAllister F., et al. 2009. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15:1016–1022 10.1038/nm.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Chen Y., Wei H., Zheng C., Sun R., Zhang J., Tian Z. 2008. Antiapoptotic activity of autocrine interleukin-22 and therapeutic effects of interleukin-22-small interfering RNA on human lung cancer xenografts. Clin. Cancer Res. 14:6432–6439 10.1158/1078-0432.CCR-07-4401 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., Ouyang W. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14:282–289 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]
- Ziesché E., Bachmann M., Kleinert H., Pfeilschifter J., Mühl H. 2007. The interleukin-22/STAT3 pathway potentiates expression of inducible nitric-oxide synthase in human colon carcinoma cells. J. Biol. Chem. 282:16006–16015 10.1074/jbc.M611040200 [DOI] [PubMed] [Google Scholar]