Tonsil-resident BDCA1+ DCs, BDCA3+ DCs, and pDCs all cross-present antigen efficiently.
Abstract
Dendritic cells (DCs) represent a heterogeneous population of antigen-presenting cells that initiate and orient immune responses in secondary lymphoid organs. In mice, lymphoid organ–resident CD8+ DCs are specialized at cross-presentation and have developed specific adaptations of their endocytic pathway (high pH, low degradation, and high export to the cytosol). In humans, blood BDCA3+ DCs were recently shown to be the homologues of mouse CD8+ DCs. They were also proposed to cross-present antigens more efficiently than other blood DC subsets after in vitro activation, suggesting that in humans cross-presentation is restricted to certain DC subsets. The DCs that cross-present antigen physiologically, however, are the ones present in lymphoid organs. Here, we show that freshly isolated tonsil-resident BDCA1+ DCs, BDCA3+ DCs, and pDCs all cross-present soluble antigen efficiently, as compared to macrophages, in the absence of activation. In addition, BDCA1+ and BDCA3+ DCs display similar phagosomal pH and similar production of reactive oxygen species in their phagosomes. All three DC subsets, in contrast to macrophages, also efficiently export internalized proteins to the cytosol. We conclude that all freshly isolated lymphoid organ–resident human DCs, but not macrophages, display high intrinsic cross-presentation capacity.
DCs are a rare and heterogeneous population of antigen-presenting cells. Murine DCs can be divided into plasmacytoid DCs (pDCs) and conventional DCs, which include lymphoid organ–resident DCs (present in all lymphoid organs where they spend their entire life cycle) and migratory DCs (which arise in peripheral tissues then migrate into the lymph nodes). Murine DC subsets display functional specializations, including in antigen presentation (Heath and Carbone, 2009). Among resident DCs, CD8+ DCs are superior to CD8− DCs for the presentation of exogenous antigens on MHC class I molecules, a process known as cross-presentation (Joffre et al., 2012). This differential ability lies both on the selective expression of phagocytic receptors (for dead cells for example) and on the specialization of their endocytic pathway. In CD8+ DCs, proteolysis in the endocytic pathway is limited, which preserves potential epitopes from destruction. This occurs through different mechanisms, including the inhibition of acidification in endosomes and phagosomes through the production of reactive oxygen species (ROS; Savina et al., 2009). In addition, transfer of exogenous proteins into the cytosol, a key step for cross-presentation through the proteasome-dependent pathway (Segura and Villadangos, 2011), is more efficient in CD8+ than in CD8− DCs (Lin et al., 2008). Therefore, at steady-state, mouse DC subsets have different intrinsic capacity for cross-presentation.
This does not mean that CD8− DCs cannot cross-present in any circumstance. Antigens complexed to antibodies are cross-presented by both DC types (den Haan and Bevan, 2002). In addition, when equal amounts of antigen are delivered to murine CD8+ and CD8− DCs through the forced expression of a human receptor, both DC types can cross-present (Kamphorst et al., 2010). When antigen is delivered through endogenously expressed murine receptors, however, CD8+ DCs are more efficient for cross-presentation than CD8− DCs (Dudziak et al., 2007). CD8− DCs also cross-present Saccharomyces cerevisiae-derived antigens more efficiently than CD8+ DCs in vitro (Backer et al., 2008). The intracellular mechanisms involved in cross-presentation in CD8− DCs have not been explored so far. Therefore, although CD8+ DCs are better at cross-presentation than CD8− DCs in a vast majority of experimental systems in vivo and in vitro, a few exceptions have been reported.
Human DCs can be divided into pDCs and resident BDCA1+ DCs and BDCA3+ DCs that are found in the blood, spleen, tonsil, and lymph nodes (Dzionek et al., 2000; McIlroy et al., 2001; Lindstedt et al., 2005; Segura et al., 2012). In the skin, three DC subsets have been described: Langerhans cells, CD1a+ dermal DCs, and CD14+ dermal DCs (Nestle et al., 1993; Klechevsky et al., 2008; Haniffa et al., 2009), which all migrate into skin-draining lymph nodes (Segura et al., 2012).
In humans, antigen cross-presentation has been analyzed on DC subsets isolated from the skin, blood, and spleen. Among skin DCs, Langerhans cells and CD1a+ DCs cross-present antigens efficiently, whereas CD14+ DCs do not (Klechevsky et al., 2008). Blood pDCs can also cross-present efficiently soluble or particulate antigens (Hoeffel et al., 2007; Di Pucchio et al., 2008). Blood BDCA3+ DCs were recently proposed to be the homologues of murine resident CD8+ DCs (Bachem et al., 2010; Crozat et al., 2010; Jongbloed et al., 2010; Poulin et al., 2010) based on phenotypic and transcriptomic evidence. In these studies, blood BDCA3+ DCs, after activation by poly:IC (a TLR3 ligand), were more efficient than blood BDCA1+ DCs for antigen cross-presentation, suggesting that BDCA3+ DCs, like mouse CD8+ DCs, are specialized in antigen cross-presentation. Blood DCs, however, may actually be a precursor form of resident DCs in transit to lymphoid organs (Ziegler-Heitbrock et al., 2010; Segura et al., 2012). Finally, splenic DC subsets were shown to cross-present an influenza virus-derived antigen, although in this study donors had been treated with a glucocorticosteroid, which alters DC properties (Mittag et al., 2011).
Because antigen cross-presentation in vivo occurs in secondary lymphoid organs and not in the blood, we decided to investigate the specialization of human lymphoid organ–resident DC subsets for cross-presentation. We measured cross-presentation and phagosomal functions in the three DC subsets and macrophages directly purified from tonsils of healthy donors. We show that cross-presentation is a functional specialization of all freshly isolated human lymphoid tissue-resident DCs, as opposed to macrophages, which fail to cross-present antigen efficiently.
RESULTS
Human tonsils contain three populations of resident DCs
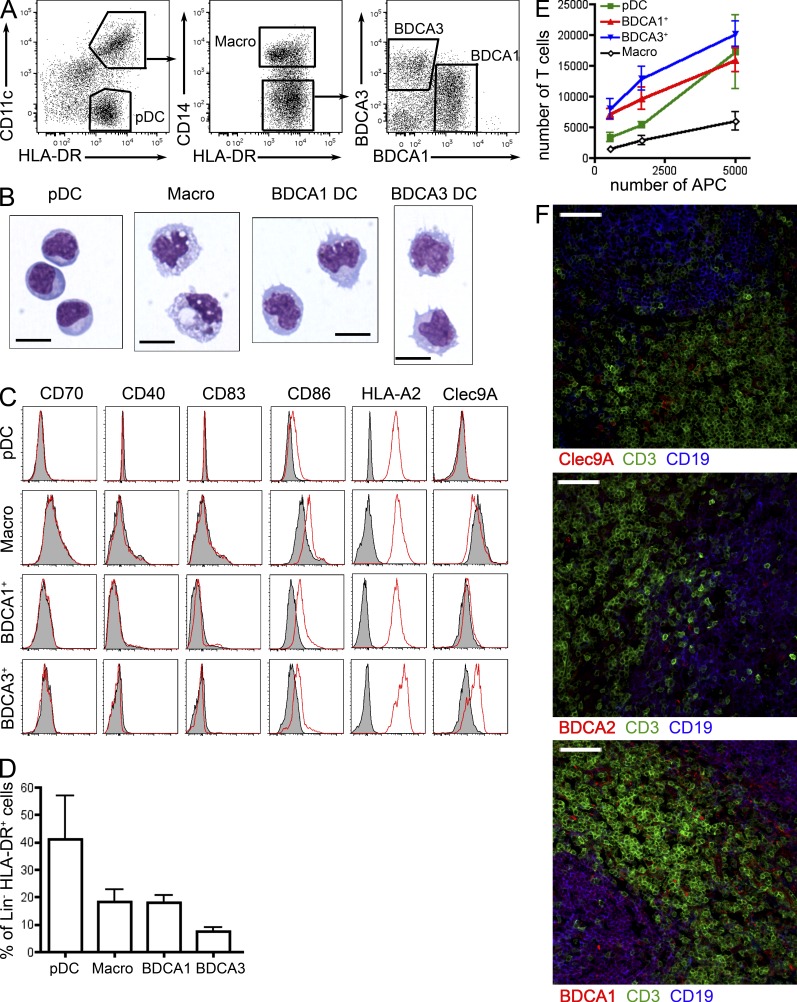
To analyze cross-presentation in human lymphoid organ–resident DCs, we isolated DC subsets from tonsils of healthy donors. Phenotypical (Fig. 1 A) and morphological (Fig. 1 B) analysis showed that human tonsils contain three populations of DCs: pDCs (CD11c−HLA-DR+) displaying a lymphoid-like morphology, BDCA1+ DCs (CD11c+HLA-DR+CD14−BDCA1+), and BDCA3+ DCs (CD11c+HLA-DR+CD14−BDCA3+) showing a typical DC morphology with numerous dendrites. Tonsils also contain CD11c+HLA-DR+CD14+ cells, which have previously been shown to be macrophages rather than DCs (Segura et al., 2012). In addition, tonsil macrophages bear large characteristic phagocytic vacuoles (Fig. 1 B). BDCA3+ DCs correspond to Clec9A+ DCs (Fig. 1 C). Phenotypical analysis showed that none of these populations expressed co-stimulatory molecules, except for low levels of CD86, indicating that tonsil DC subsets are not mature (Fig. 1 C). In addition, all DC types expressed similar levels of HLA-A2 MHC class I molecules. BDCA3+ DCs is the rarest subset, whereas pDCs represent the largest population of tonsil DCs (Fig. 1 D). In an allogeneic mixed leukocyte reaction, BDCA1+ and BDCA3+ DCs were the most efficient for stimulating naive CD4+ T cell proliferation (Fig. 1 E), whereas pDCs only activated naive CD4+ T cells at a high DC/T cell ratio. As expected, macrophages did not stimulate naive CD4+ T cell proliferation efficiently.
Figure 1.
Human tonsils contain three populations of dendritic cells. (A) Tonsil cells were enriched for DCs and stained for HLA-DR, CD11c, CD14, BDCA1, and BDCA3. Representative results of 12 independent experiments. (B) Purified cell populations were submitted to cytospin and Giemsa/May-Grünwald staining. Representative images of four independent experiments. Bar, 10 µm. (C) HLA-A2+ tonsil cell populations were analyzed by flow cytometry for the expression of CD70, CD40, CD83, CD86, HLA-A2, or Clec9A. Gray shaded histograms represent control isotype staining. Representative results of five independent experiments. (D) Percentage of each population among lineage (Lin)− HLA-DR+ cells. Mean ± SD in 12 donors. (E) Different numbers of purified APCs were cultured for 6 d with allogeneic naive CD4 T cells. Mean ± SEM of five independent experiments. (F) Fixed tonsil sections were stained for CD3, CD19, and Clec9A, BDCA2 or BDCA1. Bar, 50 µm. Representative images of four donors.
In situ immunostainings showed that the three DC subsets were mostly found in T cell areas (Fig. 1 F). Some DCs were also observed in B cell areas (note that B cells also express BDCA1). Overall, there was no clear difference between BDCA1+ and BDCA3+ DCs, whereas pDC were found more often than the other two subsets in B cell areas.
Cross-presentation of necrotic cell-derived antigen by BDCA1+ and BDCA3+ DCs
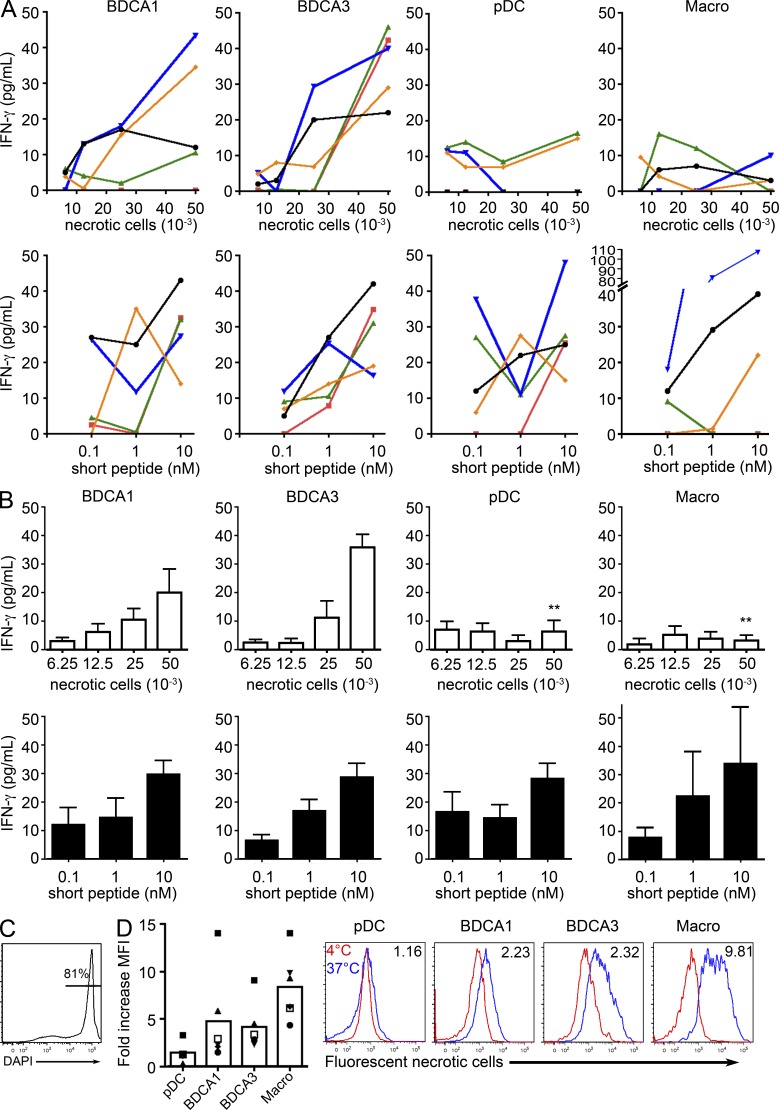
It has been suggested that blood BDCA3+ DCs are superior to blood BDCA1+ DCs in their ability to cross-present dead cell–derived antigens (Jongbloed et al., 2010). To investigate the efficiency of cross-presentation by pDCs, BDCA1+ and BDCA3+ DCs, and macrophages, we used a MelanA-specific CD8+ T cell clone (Dufour et al., 1997). As a control of the potency of activation of the T cell clone by the different DC subsets, we always use a preprocessed short peptide in our experiments, for all donors. We first analyzed cross-presentation of necrotic melanoma cell–derived MelanA antigen by tonsil DC subsets (Fig. 2). Necrotic melanoma cells expressing MelanA but not HLA-A2 were used (Fig. 2 C). BDCA3+ DCs from all donors cross-present necrotic cell–derived antigen efficiently, whereas BDCA1+ DCs from two out of five donors also did so (Fig. 2, A and B). Overall, the difference between BDCA3+ and BDCA1+ DCs was not statistically significant (unpublished data), but the number of donors may not be sufficient to reach significance. In contrast, pDCs and macrophages did not cross-present this form of antigen, although they could activate the CD8+ T cell clone when incubated with the short peptide (Fig. 2, A and B). The uptake of fluorescent necrotic cells by the different cell populations was evaluated by flow cytometry. BDCA1+ and BDCA3+ DCs took up the necrotic cells with similar efficiency, whereas pDCs failed to do so, probably accounting for their inability to cross-present antigen from dead cells (Fig. 2 D). Macrophages were by far the most efficient at taking up dead cells, indicating that the lack of necrotic cell cross-presentation in these cells is caused by postinternalization events.
Figure 2.
Cross-presentation of necrotic cell–derived antigens. (A and B) Purified DC subsets were incubated with different numbers of necrotic melanoma cells or different concentrations of MelanA short peptide. After washing, DCs were cultured for 24 h with antigen-specific CD8 T cell clones. IFN-γ secretion was assessed as a measure of T cell activation. Background level was subtracted. **, P < 0.01. (C) Mel888 melanoma cells were treated with oxaliplatin for 48 h, stained for DAPI, and analyzed by flow cytometry. Results representative of four independent experiments. (D) Necrotic melanoma cells were stained with a fluorescent dye and incubated with tonsil DC-enriched cell suspensions at 37°C or 4°C. Cells were analyzed by flow cytometry. The ratio of fluorescence for necrotic cells at 37°C versus at 4°C is depicted. Mean ± SEM of six independent experiments. Representative histograms of six independent experiments. Fluorescence ratio is indicated.
All tonsil DC types can cross-present soluble antigens
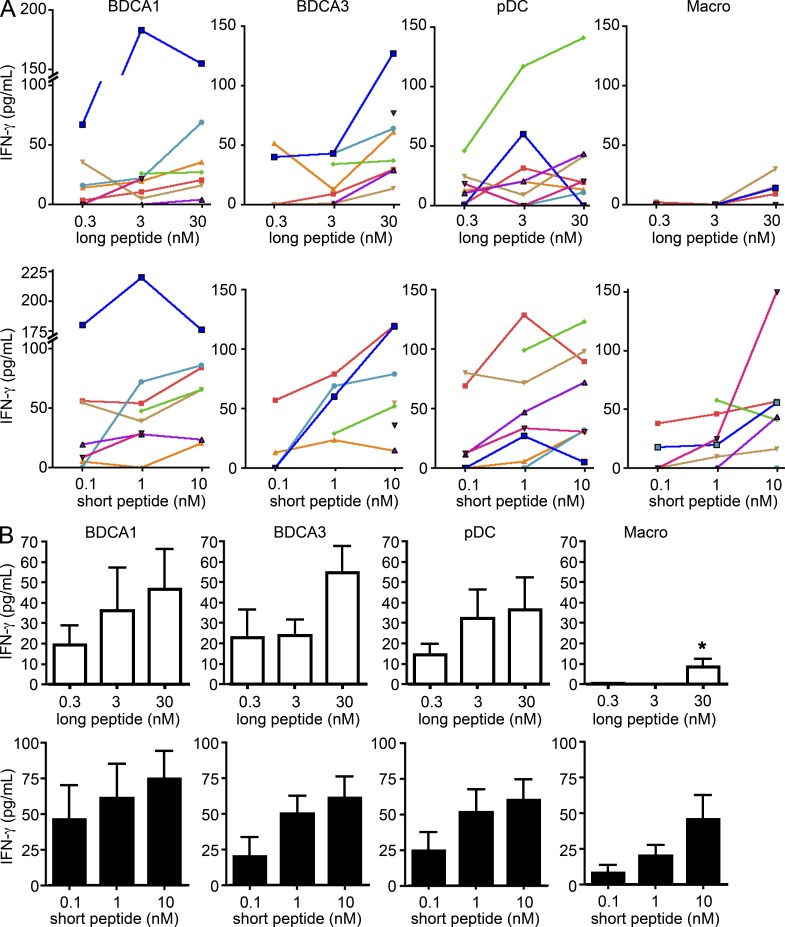
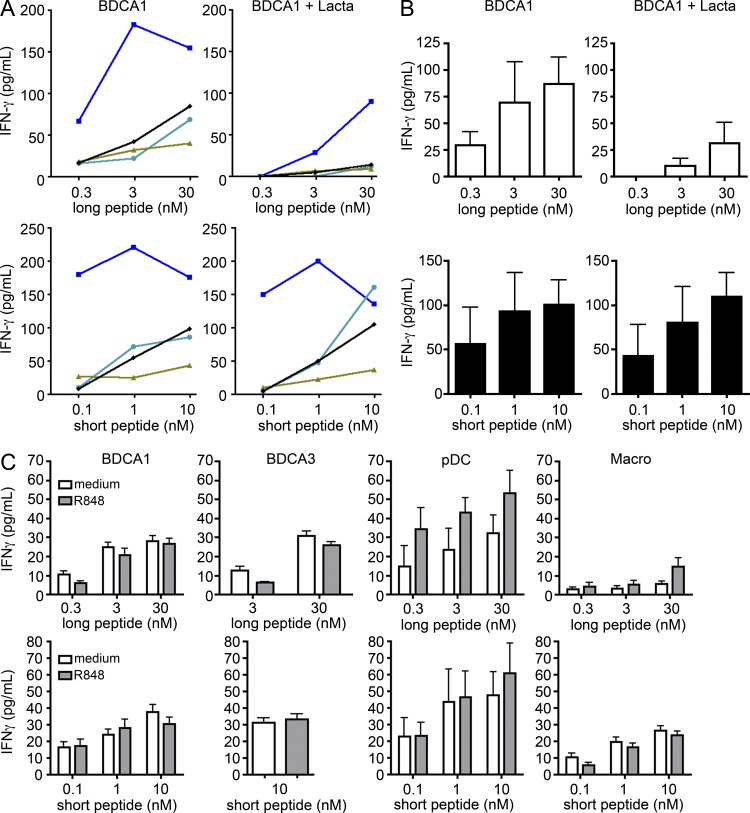
To bypass the effect of differential expression of dead cell–specific receptors on DC subsets, we next investigated cross-presentation of soluble antigen. Cross-presentation was measured after uptake of a 34-aa-long peptide that includes the epitope recognized by the MelanA-specific CD8+ T cell clone. Although we observed variability between donors (Fig. 3 A), pDCs, BDCA1+ DCs, and BDCA3+ DCs all efficiently cross-present the long peptide and present the short peptide with similar efficiency (Fig. 3, A and B). Overall, there was no clear hierarchy between the different DC subsets (Fig. 3 B). In contrast, macrophages did not cross-present the long peptide, but activated the T cell clone when incubated with the short peptide, although less efficiently than DCs.
Figure 3.
All tonsil DC subsets cross-present soluble MelanA antigen. (A and B) Purified DC subsets were incubated with different concentrations of MelanA long or short peptide. After washing, DCs were cultured for 24 h with antigen-specific CD8 T cell clones. IFN-γ secretion was assessed as a measure of T cell activation. Background level was subtracted. (A) Each color represents results from the same donor. (B) Mean ± SEM of eight independent experiments. *, P < 0.05.
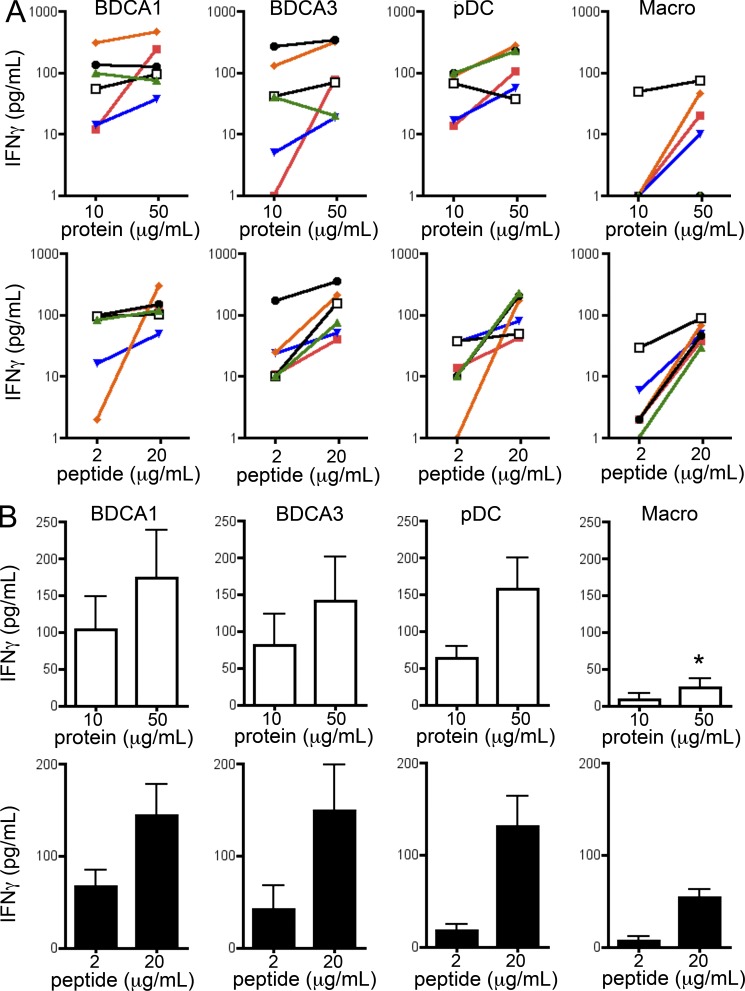
We sought to confirm these results using a different soluble antigen, the nonstructural 3 (NS3) antigen of hepatitis C virus, and a specific CD8+ T cell clone (Francavilla et al., 2004). Consistent with previous results, we found that pDC, BDCA1+ DCs, and BDCA3+ DCs all cross-present efficiently the NS3 antigen and present the corresponding preprocessed peptide with similar efficiency (Fig. 4). Macrophages did not cross-present the antigen, although they could activate the T cells in the presence of the NS3 minimal peptide (although less potently than DCs).
Figure 4.
All tonsil DC subsets cross-present soluble NS3 antigen. Purified DC subsets were incubated with different concentrations of NS3 protein or minimal peptide. After washing, DCs were cultured for 16 h with antigen-specific CD8 T cell clones. IFN-γ production was assessed in the supernatant by ELISA. Background level was subtracted. (A) Each color represents results from the same donor. (B) Mean ± SEM of six independent experiments. *, P < 0.05.
As in the case of monocyte-derived DCs (Faure et al., 2009), cross-presentation of the long MelanA, but not of the short peptide by tonsil BDCA1+ DCs was abolished in the presence of the proteasome inhibitor lactacystin (Fig. 5, A and B), suggesting that it requires export to the cytosol and processing by the proteasome. Because it has been shown that activation with TLR-ligands increases cross-presentation by blood DCs (Bachem et al., 2010; Crozat et al., 2010; Jongbloed et al., 2010; Mittag et al., 2011), we also investigated the effect of TLR-ligands on cross-presentation by tonsil DCs. R848, a TLR7/8 ligand, induced the activation of all DC subsets (as indicated by cytokine secretion; unpublished data) but did not affect the cross-presentation of the MelanA antigen by BDCA1+ and BDCA3+ DCs (Fig. 5 C). Cross-presentation by pDCs was slightly increased.
Figure 5.
Cross-presentation by tonsil DC is proteasome-dependent but remains unaffected by TLR-ligand activation. Purified DC subsets were incubated with different concentrations of MelanA long or short peptide in the presence or absence of lactacystin (Lacta) (A and B) or R848 (C). After washing, DCs were cultured for 24 h with antigen-specific CD8 T cell clones. IFN-γ secretion was assessed as a measure of T cell activation. Background level was subtracted. (A) Each color represents results from the same donor. (B) Mean ± SEM of 4 independent experiments. (C) Mean ± SEM of 6 independent experiments.
We conclude that all three tonsil DC subsets cross-present antigens with similar efficiency in the absence of further in vitro activation.
Antigen cross-presentation in in vitro-generated CD1a+ DCs and CD14+ DCs
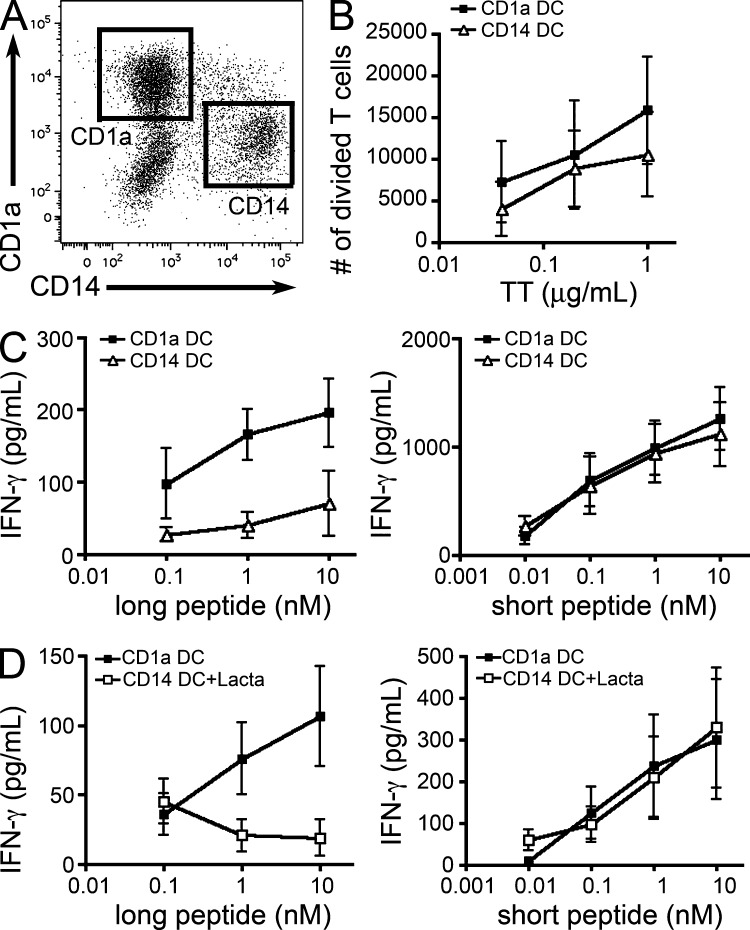
In mice, differences in phagosomal ROS production, pH, and export to the cytosol were shown to participate to the superior cross-presentation efficiency of CD8+ DCs, as compared with CD8− DCs (Lin et al., 2008; Savina et al., 2009). To investigate these phagosomal functions and their possible correlation to cross-presentation in human DCs, we first sought to validate the assays developed in mice using DC populations with clear differential cross-presentation ability. To do this, we used an in vitro culture model of CD1a+ and CD14+ DCs derived from CD34+ blood precursors (Caux et al., 1997; Fig. 6 A). Both DC types present antigens on their MHC class II molecules, as shown by the activation of autologous CD4+ T cells after incubation of DCs with Tetanus toxoid (Fig. 6 B). Similar to fresh skin CD1a+ and CD14+ DCs, in vitro-generated CD1a+ DCs are reportedly more efficient at cross-priming than CD14+ DCs (Klechevsky et al., 2008), suggesting that they are more efficient at cross-presenting antigen. In our MelanA assay, only CD1a+ DCs cross-present the long peptide efficiently (Fig. 6 C), whereas both DC types activate the T cell clone with similar efficiency when incubated with the short peptide. Similar to tonsil DCs, cross-presentation of the long, but not of the short, peptide by CD1a+ DCs was inhibited by lactacystin (Fig. 6 D). Therefore, as expected, CD1a+ DCs are more efficient for cross-presentation than CD14+ DCs.
Figure 6.
Only CD1a+ DC cross-present efficiently. (A) CD34+ cell–derived DCs were stained for CD1a and CD14. Representative result of 20 experiments. (B) Purified CD1a+ or CD14+ DC were incubated with different concentrations of tetanus toxoid (TT). After extensive washing, DCs were cultured for 6 d with CFSE-stained autologous blood CD4 T cells. T cell proliferation was assessed by flow cytometry. Mean ± SEM of five independent experiments. (C and D) Purified CD1a+ or CD14+ DCs were incubated with different concentrations of MelanA long or short peptide in the absence or presence of lactacystin (Lacta). After washing, DCs were cultured for 24 h with antigen-specific CD8 T cell clones. IFN-γ secretion was assessed as a measure of T cell activation. Background level was subtracted. Mean ± SEM of five (C) or three (D) independent experiments.
Phagosomal pH and ROS production in human DC subsets
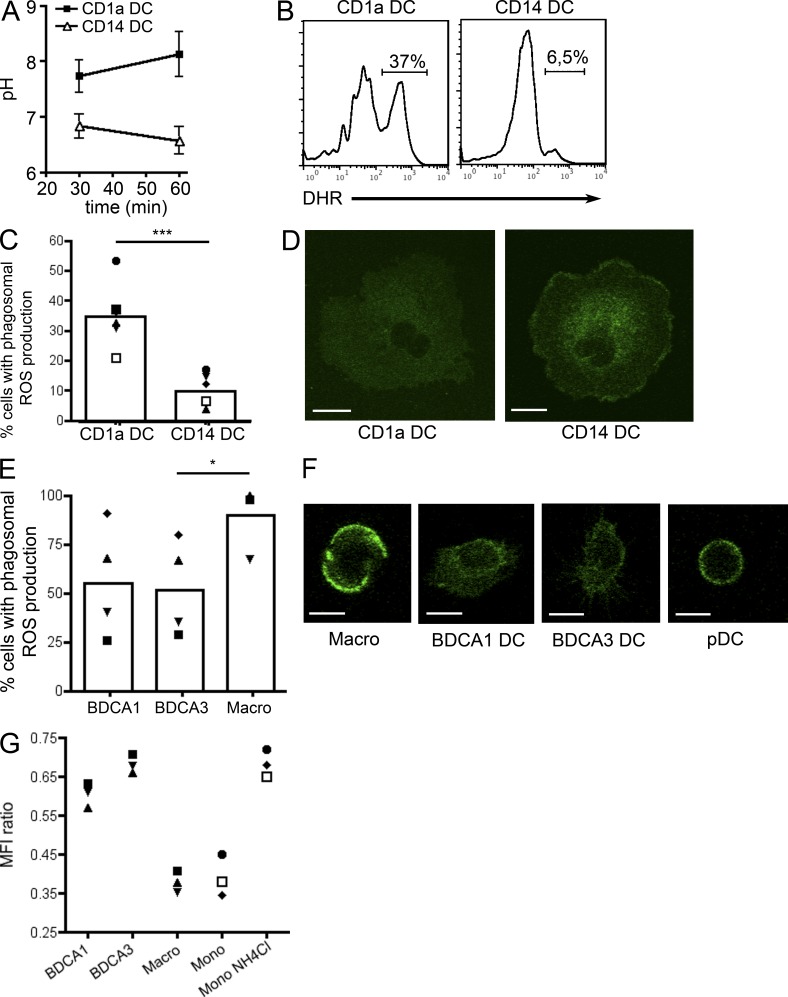
We first measured the pH in DC phagosomes using ratiometric fluorescence measurements in cells having phagocytosed latex beads coated with pH-sensitive and pH-insensitive fluorochromes (Savina et al., 2006). CD1a+ DCs phagosomes have an alkaline pH, whereas CD14+ DC phagosomes have a slightly acidic pH (Fig. 7 A), similar to mouse CD8+ and CD8− DCs, respectively (Savina et al., 2009). In murine CD8+ DCs, high phagosomal pH requires the production of ROS by NOX2 within phagosomes (Savina et al., 2009). ROS dismutate into peroxides, a reaction that consumes protons and thereby alkalinizes the phagosomal lumen. Consistently, production of ROS, as detected by the oxidation of dihydrorhodamine (DHR)-coated beads, was higher in the phagosomes of CD1a+ DCs than in the phagosomes of CD14+ DCs (Fig. 7, B and C). In addition, the subcellular localization of gp91-phox, a NOX2 subunit, differed between the two subtypes (Fig. 7 D). In CD1a+ DCs, a diffuse gp91-phox staining was observed in the cytoplasm, whereas in CD14+ DCs gp91-phox was enriched at the plasma membrane, similar to what is observed in mouse CD8+ and CD8− DCs, respectively (Savina et al., 2009). We conclude that human in vitro–generated CD1a+ and CD14+ DCs display phagosome characteristics similar to mouse CD8+ and CD8− DCs, and consistent with their differential abilities to cross-present antigens.
Figure 7.
Cross-presenting DCs maintain an alkaline pH in their phagosomes. (A) Purified CD1a+ or CD14+ DCs were incubated with beads coated with pH-sensitive and -insensitive dyes. The pH in phagosomes was determined by comparison with a standard curve. Mean ± SEM of four independent experiments. (B and C) Purified CD1a+ or CD14+ DC were incubated with beads coated with DHR. ROS production in the phagosomes was measured by flow cytometry. (B) Representative histograms of six independent experiments. (C) Symbols represent results from the same donor. Mean is shown (n = 6). ***, P < 0.001. (D) Purified CD1a+ or CD14+ DC were coated on microscopy slides, fixed, permeabilized, and stained for gp91-phox. One representative image out of four independent experiments. Bar, 10 µm (E) Purified tonsil cell subsets were incubated with beads coated with DHR. ROS production in the phagosomes was measured by flow cytometry. Symbols represent results from the same donor. Mean is shown (n = 4). *, P < 0.05. (F) Purified tonsil cell subsets were coated on microscopy slides, fixed, permeabilized, and stained for gp91-phox. One representative image out of three independent experiments. Bar, 10 µm (G) Tonsil cell subsets or blood monocytes were incubated with beads coated with pH-sensitive and -insensitive dyes for 30 min. Monocytes were also incubated with NH4Cl during 5 min before analysis. Symbols represent results from the same donor (n = 3).
We next performed a similar analysis on tonsil DC subsets. BDCA1+ and BDCA3+ DCs both oxidize DHR-coated beads at similar levels (Fig. 7 E). ROS production in macrophage phagosomes was very high, consistent with what has been observed in in vitro–generated human macrophages (Mantegazza et al., 2008). Results in pDCs were inconclusive due to very inefficient uptake of beads (unpublished data). The subcellular localization of gp91-phox was similar in BDCA1+ and BDCA3+ DCs, with a diffuse cytoplasmic staining (Fig. 7 F), similar to cross-presenting human CD1a+ DCs and mouse CD8+ DCs. In contrast, gp91-phox was enriched at the plasma membrane in macrophages. In pDCs, staining was inconclusive due to the very small size of the cells (Fig. 7 F). These results show that both BDCA1+ and BDCA3+ DCs resemble mouse CD8+ DCs in terms of gp91-phox distribution and ROS production in phagosomes.
We next sought to measure the phagosomal pH in the human DC subsets. Because of the limited number of cells purified from human tonsils, we could not perform measurements for a standard curve (which allows calculating the absolute pH for each cell type, from the observed ratio of fluorescence between the pH-sensitive and -insensitive dyes). We therefore compared the ratio of mean fluorescence intensity for the pH-sensitive and -insensitive fluorochromes in the different cell populations. We also included blood monocytes (which have been shown to acidify their phagosomes strongly; Horwitz and Maxfield, 1984) for comparison. As expected, both monocytes and macrophages presented low ratios, indicating acidic pH, which could be neutralized by incubation with NH4Cl (a weak base that neutralizes the pH). Results in pDCs were inconclusive as a result of very inefficient uptake of beads (unpublished data). In BDCA1+ and BDCA3+ DCs, the ratios were similar, and higher than in macrophages and monocytes, suggesting similar high pH levels in the phagosomes of the two DC subsets (Fig. 7 G). We conclude that in both human BDCA1+ and BDCA3+ DCs, similar to murine CD8+ DCs, phagosome acidification is very inefficient.
Cross-presenting human DCs transfer exogenous proteins into their cytosol
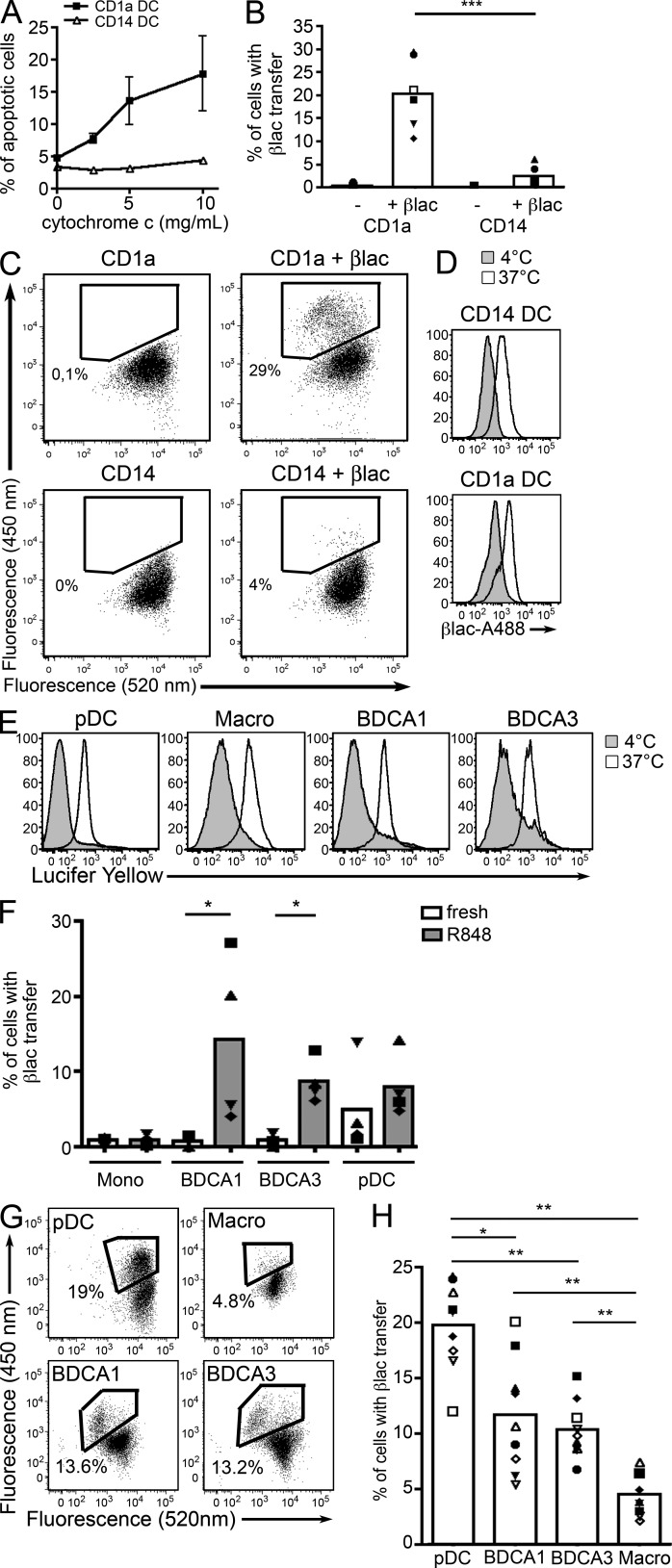
Transfer of exogenous proteins into the cytosol is a key step of proteasome-dependent cross-presentation in murine DCs. Indeed, CD8+ DCs were shown to export antigens to their cytosol more efficiently than CD8− DCs (Lin et al., 2008). We therefore investigated the transfer of exogenous proteins into the cytosol of CD1a+ and CD14+ DCs using two different assays. In the first assay, the cells are incubated with horse cytochrome C, which induces caspase-dependent apoptosis upon translocation to the cytosol. In mice, only cross-presenting DCs are sensitive to cytochrome C–induced apoptosis in vitro and in vivo (Lin et al., 2008). As expected, we found that only CD1a+ DCs become apoptotic after incubation with cytochrome C (Fig. 8 A). The second assay is based on a cytosolic β-lactamase–sensitive FRET probe that is digested upon transfer of the exogenous enzyme into the cytosol (Ray et al., 2010; Cebrian et al., 2011). After loading with the cytosolic substrate, in the absence of β-lactamase, the fluorescence of the probe remained unchanged (Fig. 8 C). When DCs were incubated with β-lactamase, a change in fluorescence, indicating cleavage of the probe, was observed only in CD1a+ DCs (Fig. 8, B and C), although both DC types internalized β-lactamase efficiently (Fig. 8 D). We conclude that similar to murine CD8+ DCs, human cross-presenting CD1a+ DCs, but not CD14+ DCs, efficiently deliver internalized cargo to the cytosol.
Figure 8.
Cross-presenting DCs transfer exogenous antigens into their cytosol. (A) CD34+ cell–derived DCs were cultured with different concentrations of cytochrome c and apoptosis was measured 24 h later. Mean ± SEM of four independent experiments. ***, P < 0.001. (B and C) Purified CD1a+ or CD14+ DC were loaded with a FRET-sensitive, β-lactamase–cleavable probe and incubated in the presence or absence of β-lactamase (βlac). After 3 h, change in fluorescence was measured by flow cytometry. (B) Symbols represent results from the same donor. Mean is shown (n = 6). (C) Representative results of six independent experiments. (D) CD1a+ or CD14+ DC were incubated with Alexa Fluor 488–conjugated β-lactamase at 4°C or 37°C. Representative results of three independent experiments. (E) Tonsil cells were incubated for 3 h with Lucifer Yellow at 4°C or 37°C and analyzed by flow cytometry. Representative results of four independent experiments. (F) Freshly isolated or R848-activated blood DC subsets or CD14+ monocytes were loaded with a FRET-sensitive, β-lactamase–cleavable probe and incubated in the presence or absence of β-lactamase. After 3 h, change in fluorescence was measured by flow cytometry. Symbols represent results from the same donor. Mean is shown (n = 4). *, P < 0.05. (G and H) Purified tonsil DC subsets were loaded with a FRET-sensitive, β-lactamase–cleavable probe and incubated in the presence or absence of β-lactamase. After 3 h, change in fluorescence was measured by flow cytometry. (F) Symbols represent results from the same donor. Mean is shown (n = 9). *, P < 0.05; **, P < 0.01. (G) Representative results of nine independent experiments.
We next investigated the ability of blood DCs, with CD14+ monocytes as a comparison, to transfer internalized proteins into their cytosol. Freshly isolated blood DCs did not transfer β-lactamase into their cytosol (Fig. 8 F). In contrast, R848-activated DC subsets, but not monocytes, were capable of transferring β-lactamase into their cytosol with overall similar efficiency. We conclude that blood DCs can transfer exogenous proteins from endosomes to cytosol efficiently only after activation.
Finally, we analyzed the transfer of exogenous proteins into the cytosol of tonsil DC subsets and macrophages using the β-lactamase assay. All cell types captured soluble proteins efficiently, as shown by the efficient internalization of Lucifer yellow (Fig. 8 E). All DC subsets, in contrast to macrophages, translocated β-lactamase into their cytosol, with pDC being the most efficient and BDCA1+ and BDCA3+ DCs being overall similar (Fig. 8, G and H). We conclude that the three resident DC subsets present in human tonsils efficiently deliver internalized cargo to their cytosol.
DISCUSSION
In this work, we show that the essential mechanisms of cross-presentation are conserved between mouse and human: only cross-presenting DCs maintain an alkaline pH and produce ROS in their endocytic compartments, and only cross-presenting DCs transfer internalized proteins into their cytosol. We also show that, whereas in the mouse among resident DCs only CD8+ DCs are specialized at cross-presentation in vivo, in humans all freshly isolated lymphoid organ–resident DC subsets have the intrinsic ability to cross-present antigens.
Comparative transcriptional and functional studies suggested that human BDCA3+ DCs are homologous to murine CD8+ DCs (Hoeffel et al., 2007; Robbins et al., 2008; Bachem et al., 2010; Crozat et al., 2010; Jongbloed et al., 2010 ; Poulin et al., 2010). This conclusion is further supported by the shared requirement for the transcription factor Batf3 for the differentiation of both mouse CD8+ DCs and human BDCA3+ DCs (Hildner et al., 2008; Poulin et al., 2012). It was also suggested, by analogy to mouse CD8+ DCs, that human BDCA3+ DCs are more efficient than BDCA1+ DCs for antigen cross-presentation (Bachem et al., 2010; Crozat et al., 2010; Jongbloed et al., 2010). This conclusion is based on the analysis of cross-presentation of two forms of antigen, soluble and dead cell–associated, by blood DC subsets in vitro. In the case of dead cell–associated antigen, these studies show superior cross-presentation efficiency of BDCA3+ over BDCA1+ DCs. Consistent with these results, we found that in three out of five donors, tonsil-resident BDCA3+ DCs were more efficient than BDCA1+ DCs for cross-presentation of dead tumor cell–associated antigen.
In the case of soluble antigen, two of these studies used HCMV-derived pp65 protein and antigen-specific CD8+ T cell clones. Bachem et al. (2010) showed that blood BDCA3+ DCs cross-present pp65, whereas BDCA1+ DCs did not. In contrast, Jongbloed et al. (2010) show that in the absence of poly:IC activation, blood BDCA1+ DCs are at least as efficient for cross-presentation as BDCA3+ DCs. Similar observations were made in a subsequent study with a virus-derived antigen (Mittag et al., 2011). The evidence for nonactivated BDCA3+ DCs cross-presenting better than BDCA1+ DCs is therefore not compelling. After activation by poly:IC, cross-presentation is enhanced in BDCA3+ DCs, but not in BDCA1+ DCs. As a result, after poly:IC activation blood BDCA3+ DCs are more efficient than BDCA1+ DCs for cross-presentation. Poly:IC, however, does not activate BDCA1+ DCs as well as BDCA3+ DCs (Jongbloed et al., 2010). Indeed, TLR3 is expressed at lower levels in BDCA1+ DCs than in BDCA3+ DCs (Jongbloed et al., 2010).
Using two different soluble antigens, we failed to evidence any major difference in the efficiency of cross-presentation by freshly isolated tonsil DCs. This is consistent with the results obtained with DCs from the spleen of glucocorticosteroid-treated donors (Mittag et al., 2011) and with BDCA1+ and BDCA3+ DCs from lymph nodes of untreated cancer patients (Segura et al., 2012), suggesting that this property is common to resident DCs from all lymphoid organs. To circumvent the inherent limitations of such indirect biological assays, which can sometimes lead to opposite conclusions in different laboratories, we sought to gain further insight by analyzing the cell biology of cross-presentation in human DCs. With regards to control of phagosomal pH and ROS production, as well as export of internalized proteins into the cytosol, we could not find any major difference between lymphoid organ–resident BDCA1+ and BDCA3+ DCs. In addition, we found that human pDC export internalized cargo into their cytosol very efficiently. In mouse DCs, these phagosomal functions are restricted to CD8+ DCs. These results suggest that the endocytic features that promote cross-presentation are evolutionarily conserved between humans and mice, but independently of DC ontogeny. In addition, we found that blood DC only transfer efficiently exogenous proteins into their cytosol after activation. This result suggests that blood DCs are not fully functional, as they only export antigen into their cytosol and cross-present efficiently after a final step of differentiation, most likely taking place in lymphoid organs, that can be mimicked or bypassed in vitro by TLR-mediated activation.
It has been suggested that limited degradation in endocytic compartments favors cross-presentation over MHC class II-restricted presentation. Indeed, in addition to specialized phagosomal functions, mouse resident CD8+ DCs and migratory CD103+ DCs exhibit lower levels of lysosomal proteases as compared with their CD8− or CD11b+ counterparts (Dudziak et al., 2007; Helft et al., 2012). Whether human DC subsets bear different proteases in their endocytic compartments remains open for future investigation.
While all freshly isolated human resident DC subsets have the ability to cross-present antigens, it is possible that in vivo one subset may become dominant depending on the form of antigen, the environment or the type of pathogen encountered. For instance, although excelling at transferring exogenous antigen into their cytosol, pDCs have limited ability to phagocytose large particles and take up dead cells, therefore limiting their efficiency for cross-presentation of particulate or necrotic cell-derived antigens. Certain forms of antigen can also profoundly impact the cross-presentation ability of DC subsets, as it is the case for mouse CD8− DCs which are generally poor at cross-presentation but cross-present efficiently immune complexes (den Haan and Bevan, 2002). Differential expression of cell surface receptors is also likely to impact cross-presentation of different forms of antigens by the DC subsets. Although both BDCA1+ and BDCA3+ DCs capture necrotic cells and can cross-present necrotic cell-derived antigens, BDCA3+ DCs seem more efficient, which is probably due to the selective expression of Clec9A, a receptor for dead cells that favors cross-presentation through diversion of internalized dead cell material into recycling endosomes (Sancho et al., 2009; Zelenay et al., 2012).
Our results have broad implications for the design of new vaccination strategies. DC targeting for the in vivo delivery of vaccines has proved a powerful tool in the mouse (Bonifaz et al., 2002; Bonifaz et al., 2004; Tacken et al., 2007; Caminschi et al., 2009). Special emphasis is put on the targeting of cross-presenting DCs to elicit strong cytotoxic T cell responses (Sancho et al., 2008; Idoyaga et al., 2011). To apply this strategy to humans, it is essential to determine which DC subsets to target. Our results suggest that pDCs, BDCA1+ and BDCA3+ DCs are all potential targets when aiming for cross-presentation of vaccine antigen. Our data further indicates that our knowledge of murine DC biology cannot always be directly translated to humans and that further studies are required to better understand human DC biology.
MATERIALS AND METHODS
Samples and cell isolation.
Tonsils from healthy patients undergoing tonsillectomy for obstructive sleep apnea were obtained from Hôpital Necker (Paris, France) in accordance with hospital ethical guidelines. Samples were cut into small fragments, digested with 0.1 mg/ml Liberase TL (Roche) in the presence of 0.1 mg/ml DNase (Roche) for 40 min at room temperature before addition of 10 mM EDTA. Cells were filtered on a 40-µm cell strainer (Falcon; BD) and washed. Light density cells were isolated by centrifugation on a Ficoll gradient (Lymphoprep; Greiner Bio-One). DCs were enriched by depletion of cells expressing CD3, CD15, CD19, CD56, and CD235a using antibody-coated magnetic beads (Miltenyi Biotec). Cell subsets were further isolated by cell sorting on a FACSAria instrument (BD). Buffy coats from healthy donors were obtained from Etablissement Français du Sang. PBMCs were isolated after centrifugation on a Ficoll gradient. Blood CD14+ monocytes were isolated from PBMC by positive selection using anti-CD14–coated magnetic beads according to manufacturer’s instructions (Miltenyi Biotec). Blood DCs were enriched by positive selection of cells expressing BDCA4, BDCA1, and BDCA3 using antibody-coated magnetic beads (Miltenyi Biotec). All human samples were anonymized, and therefore in accordance with Institut National de la Santé et de la Recherche Médicale ethical guidelines, informed consent was not necessary.
Cell culture.
Blood CD34+ cells were isolated from PBMCs by positive selection using anti-CD34-coated magnetic beads and magnetic columns according to manufacturer’s instructions (Miltenyi). CD34+ cells were cultured for 9–10 d in Yssel medium supplemented with 10% fetal calf serum (FCS), penicillin/streptomycin and 50 ng/ml GM-CSF (AbCys), 100 ng/ml Flt3-liter (R&D), and 10 ng/ml TNF (R&D). DC subsets were isolated by cell sorting on a FACSAria instrument after staining for CD1a and CD14. HLA-A2− Mel888 cells were cultured in RPMI 1640-Glutamax (Gibco) supplemented with penicillin/streptomycin and 10% FCS. Cell death was induced by addition of 10 µg/ml Oxaliplatin (Sanofi-Aventis). After 48 h, >95% of cells were necrotic as assessed by flow cytometry with DAPI (Invitrogen) and Annexin V (BD Biosciences) staining.
Flow cytometry.
Nonspecific binding was blocked using TruStain (BioLegend). Cells were stained with FITC or Pe/Cy5 anti-CD1a (BD), FITC or PE anti-CD14 (BD Biosciences), Alexa Fluor 700 anti-CD14 (BioLegend), PerCP-eFluor710 anti-BDCA1 (eBioscience), APC-eFluor780 anti-HLA-DR (eBioscience), Pe/Cy7 anti-CD11c (BioLegend), APC anti-BDCA4 (Miltenyi), PE or APC anti-BDCA3 (Miltenyi Biotec), FITC anti-CD86 (BD), PE anti-CD40 (BD), PE anti-CD83 (BD), FITC anti-CD70 (BD), PE anti-HLAA2 (BD), biotin anti-Clec9A (Miltenyi Biotec), or isotype control antibodies where indicated. Cells were analyzed on a LSR II (BD), FACSVerse (BD), or MACSQuant (Miltenyi Biotec) instrument. Data were analyzed with FlowJo (Tree Star).
Mixed leukocyte reaction.
Naive CD4+ T cells were isolated from healthy donors’ PBMC by negative selection using magnetic beads (Miltenyi Biotec). Different numbers of antigen-presenting cells were cultured with naive CD4+ T cells (1.5 × 104 cells/well) for 6 d in Yssel medium supplemented with 10% FCS. The number of live cells was evaluated by flow cytometry.
Cross-presentation assay.
Purified HLA-A2+ APCs were incubated (104 cells/well) in V-bottom 96-well plates (Corning) with different concentrations of MelanA long peptide (KGHGHSYTTAEEAAGIGILTVILGVL), MelanA short peptide (EAAGIGILTV), or different numbers of necrotic Mel888 cells for 3–4 h in Yssel medium in the presence or absence of 2.5 µg/ml lactacystin (EMD Millipore) or in the presence or absence of 1 µg/ml R848 (InvivoGen). After extensive washing, APCs were cultured for 24 h with CD8 T cell LT12 clones (Dufour et al., 1997; 2 × 104 cells/well) in Yssel medium supplemented with 10% FCS. Supernatants were collected and kept at −20°C until measurement of IFN-γ concentration by ELISA (BD). Background levels (APCs cultured with LT12 cells without peptide) were subtracted for each cell type. Background levels were typically 20–25 ng/ml for CD1a+, CD14+, BDCA1+ DCs, BDCA3+ DCs, and pDCs, and 70–100 pg/ml for macrophages. For assays with NS3 antigen, purified HLA-A2+ APCs were incubated (104 cells/well) in V-bottom 96-well plates with different concentrations of NS3 c33c subtype 1a recombinant protein (Meridian Life Science) or NS31073-1081 peptide (CINGVCWTV) for 4 h in Yssel medium. After extensive washing, APCs were cultured for 16 h with NS31073-1081-specific CD8 T cell clones (a gift from V. Barnaba, Sapienza Universita di Rome, Rome, Italy; Francavilla et al., 2004; 2 × 104 cells/well) in Yssel medium supplemented with 10% FCS.
MHC II–restricted antigen presentation.
Purified DC populations (104 cells/well) were incubated with different concentrations of Tetanus Toxoid (EMD Millipore) for 3 h in Yssel medium. Autologous blood CD4 T cells were labeled with 5 µM 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (CFSE, Invitrogen). After extensive washing, DCs were cultured for 6 d with CFSE-labeled autologous CD4 T cells (105 cells/well). T cell proliferation was assessed by flow cytometry.
Cytosolic translocation assays.
For the cytochrome c killing assay, cells (2 × 105 cells/ml) were cultured with different concentrations of horse cytochrome c (Sigma-Aldrich) for 24 h. After washing, apoptosis was measured with Annexin V (BD). For the β-lactamase assay, cells were incubated with 0.5 µg/ml CCF4-AM (Invitrogen) for 30 min at room temperature in loading buffer (120 mM NaCl, 7 mM KCl, 1.8 mM CaCl2, 0.8 mM MgCl2, 5 mM glucose, and 25 mM Hepes, pH 7.3) containing solution B (dilution 1/20, LiveBLAzer FRET-B/G loading kit, Invitrogen), and 1 mM probenicid (Invitrogen). After washing, cells were incubated in loading buffer containing 1 mM probenicid in the presence or absence of β-lactamase (Penicillinase from Bacillus cereus; Sigma-Aldrich) for 3 h at 37°C. After washing, cells were analyzed on a LSR II Instrument or FACSVerse by measuring the 450 and 520 nm fluorescence channels. Cell viability was assessed using Topro-3 (Invitrogen). For blood cell populations, cells were tested either directly after isolation or after 16 h of culture with 0.5 µg/ml R848 and 25 ng/ml GM-CSF.
Phagosomal ROS production assay.
1 µm aminobeads (Polysciences) were covalently coupled to 1 mg/ml DHR 123 (Invitrogen) and 1 mg/ml FluoProbes 647 (Interchim) in NaHCO3 buffer (pH 8.5) for 2 h at room temperature. After washing, beads were resuspended in PBS. Cells were pulsed with DHR-coated beads in CO2-independent medium (Gibco) for 15 min at 37°C, and then extensively washed in a large volume of cold PBS. Cells were further incubated for 20 min at 37°C in Yssel medium and analyzed on a LSR II (BD) or MACSQuant (Miltenyi Biotec) instrument.
Phagosomal pH assay.
pH in phagosomes was determined as previously described (Savina et al., 2006). In brief, 1 µm aminobeads were covalently coupled to 1 mg/ml FITC (Sigma-Aldrich) and 1 mg/ml FluoProbes 647 in NaHCO3 buffer (pH 8.5) for 2 h at room temperature. After washing, beads were resuspended in PBS. Cells were pulsed with beads in CO2-independent medium for 15 min at 37°C and then extensively washed in a large volume of cold PBS. Cells were further incubated for 30 or 60 min at 37°C in Yssel medium and analyzed by FACS. In some experiments, cells were incubated with 4 mM NH4Cl for 5 min before analysis. The ratio of the mean fluorescence intensity emission between the two dyes was determined. For in vitro–derived DCs, this ratio was compared with a standard curve obtained by analyzing cells incubated with beads for 30 min and placed for 8 min in a solution at a fixed pH (ranging from pH 6.0 to 8) and containing 0.1% Triton X-100.
Immunofluorescence analysis on cryosections.
Tonsils were fixed in 4% paraformaldehyde, washed with PBS, incubated overnight in 30% sucrose at 4°C, embedded in tissue-tek OCT (Sakura), and stored at −80°C. 5-µm-thick sections were cut with a CM 1850 UV cryomicrotome (Leica). For staining, sections were incubated for 30 min in TNB blocking buffer (0.1 M Tris HCl, 0.15 M NaCl, and 0.5% blocking reagent; Perkin Elmer) and incubated overnight with primary antibodies in TNB at 4°C. After five washes in PBS, sections were incubated for 1 h with fluorochrome-conjugated secondary antibodies or fluorochrome-conjugated streptavidin and DAPI (4′,6-diamidine-2′-phenylindole dihydrochloride; Roche) in TNB. Samples were mounted on coverslip with fluorescent mounting medium (Dako) and visualized using a Zeiss LSM710 confocal microscope using a 25×/0.8 numerical aperture objective. Images were analyzed with ImageJ (National Institutes of Health). Primary antibodies used were as follows: biotinylated or pure anti–human CD19 (HIB19; eBioscience), anti–human CD3 (UCHT1; Invitrogen), biotinylated anti–human BDCA1 (Miltenyi Biotec), anti–human BDCA2 (Miltenyi Biotec), and anti–human Clec9A (Miltenyi Biotec). The corresponding streptavidin conjugate and secondary antibodies (all from Molecular Probes) were as follows: Alexa Fluor 488 anti–rabbit, Alexa Fluor 568 or 647 anti–mouse, and Alexa Fluor 568 or 647 streptavidin.
Confocal microscopy.
Cells were coated on Poly-Lysine–treated glass slides, fixed with 2% paraformaldehyde, and permeabilized in PBS containing 0.05% saponin and 10% bovine serum. Cells were stained with anti-gp91phox (a gift from J El Benna, INSERM U773, Paris, France) followed by staining with Alexa Fluor 488–coupled anti–mouse (Invitrogen) and DAPI. Slides were analyzed on a LSM 510 confocal microscope (Carl Zeiss).
Morphological analysis.
Cells were subjected to cytospin and colored with May-Grünwald/Giemsa staining. Images were acquired with a CFW-1308C color digital camera (Scion Corporation) on a Leica DM 4000 B microscope.
Necrotic cells capture assay.
Oxaliplatin-treated Mel888 cells were stained with 1 µM Dye eFluor670 (eBioscience) and incubated with tonsil DC-enriched cell suspensions for 4 h at 37°C or 4°C at a ratio of 3/1. After washing, cells were stained for HLA-DR, CD11c, CD14, BDCA1, BDCA3, and DAPI, and analyzed by flow cytometry.
Statistical analysis.
Wilcoxon matched paired test or Mann-Whitney test were performed using Prism (GraphPad Software).
Acknowledgments
The authors wish to thank the Institut Curie flow cytometry facility, the imaging platform PICT-IBiSA@BDD, and V. Barnaba for the gift of NS3-specific CD8 T cell clones.
This work was supported by the European Research Council (2008-AdG n°233062 PhagoDC , INCa/DGOS 2011-189, Labex DCBIOL, INSERM) and Ligue contre le Cancer. E. Segura is a fellow of Association pour la Recherche sur le Cancer.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- DHR
- dihydrorhodamin
- NS3
- nonstructural 3
- pDC
- plasmacytoid DC
- ROS
- reactive oxygen species
References
- Bachem A., Güttler S., Hartung E., Ebstein F., Schaefer M., Tannert A., Salama A., Movassaghi K., Opitz C., Mages H.W., et al. 2010. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207:1273–1281 10.1084/jem.20100348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer R., van Leeuwen F., Kraal G., den Haan J.M. 2008. CD8- dendritic cells preferentially cross-present Saccharomyces cerevisiae antigens. Eur. J. Immunol. 38:370–380 10.1002/eji.200737647 [DOI] [PubMed] [Google Scholar]
- Bonifaz L., Bonnyay D., Mahnke K., Rivera M., Nussenzweig M.C., Steinman R.M. 2002. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J. Exp. Med. 196:1627–1638 10.1084/jem.20021598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaz L.C., Bonnyay D.P., Charalambous A., Darguste D.I., Fujii S., Soares H., Brimnes M.K., Moltedo B., Moran T.M., Steinman R.M. 2004. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J. Exp. Med. 199:815–824 10.1084/jem.20032220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminschi I., Lahoud M.H., Shortman K. 2009. Enhancing immune responses by targeting antigen to DC. Eur. J. Immunol. 39:931–938 10.1002/eji.200839035 [DOI] [PubMed] [Google Scholar]
- Caux C., Massacrier C., Vanbervliet B., Dubois B., Durand I., Cella M., Lanzavecchia A., Banchereau J. 1997. CD34+ hematopoietic progenitors from human cord blood differentiate along two independent dendritic cell pathways in response to granulocyte-macrophage colony-stimulating factor plus tumor necrosis factor alpha: II. Functional analysis. Blood. 90:1458–1470 [PubMed] [Google Scholar]
- Cebrian I., Visentin G., Blanchard N., Jouve M., Bobard A., Moita C., Enninga J., Moita L.F., Amigorena S., Savina A. 2011. Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell. 147:1355–1368 10.1016/j.cell.2011.11.021 [DOI] [PubMed] [Google Scholar]
- Crozat K., Guiton R., Contreras V., Feuillet V., Dutertre C.A., Ventre E., Vu Manh T.P., Baranek T., Storset A.K., Marvel J., et al. 2010. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 207:1283–1292 10.1084/jem.20100223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Haan J.M., Bevan M.J. 2002. Constitutive versus activation-dependent cross-presentation of immune complexes by CD8(+) and CD8(-) dendritic cells in vivo. J. Exp. Med. 196:817–827 10.1084/jem.20020295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pucchio T., Chatterjee B., Smed-Sörensen A., Clayton S., Palazzo A., Montes M., Xue Y., Mellman I., Banchereau J., Connolly J.E. 2008. Direct proteasome-independent cross-presentation of viral antigen by plasmacytoid dendritic cells on major histocompatibility complex class I. Nat. Immunol. 9:551–557 10.1038/ni.1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudziak D., Kamphorst A.O., Heidkamp G.F., Buchholz V.R., Trumpfheller C., Yamazaki S., Cheong C., Liu K., Lee H.W., Park C.G., et al. 2007. Differential antigen processing by dendritic cell subsets in vivo. Science. 315:107–111 10.1126/science.1136080 [DOI] [PubMed] [Google Scholar]
- Dufour E., Carcelain G., Gaudin C., Flament C., Avril M.F., Faure F. 1997. Diversity of the cytotoxic melanoma-specific immune response: some CTL clones recognize autologous fresh tumor cells and not tumor cell lines. J. Immunol. 158:3787–3795 [PubMed] [Google Scholar]
- Dzionek A., Fuchs A., Schmidt P., Cremer S., Zysk M., Miltenyi S., Buck D.W., Schmitz J. 2000. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 165:6037–6046 [DOI] [PubMed] [Google Scholar]
- Faure F., Mantegazza A., Sadaka C., Sedlik C., Jotereau F., Amigorena S. 2009. Long-lasting cross-presentation of tumor antigen in human DC. Eur. J. Immunol. 39:380–390 10.1002/eji.200838669 [DOI] [PubMed] [Google Scholar]
- Francavilla V., Accapezzato D., De Salvo M., Rawson P., Cosimi O., Lipp M., Cerino A., Cividini A., Mondelli M.U., Barnaba V. 2004. Subversion of effector CD8+ T cell differentiation in acute hepatitis C virus infection: exploring the immunological mechanisms. Eur. J. Immunol. 34:427–437 10.1002/eji.200324539 [DOI] [PubMed] [Google Scholar]
- Haniffa M., Ginhoux F., Wang X.N., Bigley V., Abel M., Dimmick I., Bullock S., Grisotto M., Booth T., Taub P., et al. 2009. Differential rates of replacement of human dermal dendritic cells and macrophages during hematopoietic stem cell transplantation. J. Exp. Med. 206:371–385 10.1084/jem.20081633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath W.R., Carbone F.R. 2009. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat. Immunol. 10:1237–1244 10.1038/ni.1822 [DOI] [PubMed] [Google Scholar]
- Helft J., Manicassamy B., Guermonprez P., Hashimoto D., Silvin A., Agudo J., Brown B.D., Schmolke M., Miller J.C., Leboeuf M., et al. 2012. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J. Clin. Invest. 122:4037–4047 10.1172/JCI60659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G., Ripoche A.C., Matheoud D., Nascimbeni M., Escriou N., Lebon P., Heshmati F., Guillet J.G., Gannagé M., Caillat-Zucman S., et al. 2007. Antigen crosspresentation by human plasmacytoid dendritic cells. Immunity. 27:481–492 10.1016/j.immuni.2007.07.021 [DOI] [PubMed] [Google Scholar]
- Horwitz M.A., Maxfield F.R. 1984. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99:1936–1943 10.1083/jcb.99.6.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idoyaga J., Lubkin A., Fiorese C., Lahoud M.H., Caminschi I., Huang Y., Rodriguez A., Clausen B.E., Park C.G., Trumpfheller C., Steinman R.M. 2011. Comparable T helper 1 (Th1) and CD8 T-cell immunity by targeting HIV gag p24 to CD8 dendritic cells within antibodies to Langerin, DEC205, and Clec9A. Proc. Natl. Acad. Sci. USA. 108:2384–2389 10.1073/pnas.1019547108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffre O.P., Segura E., Savina A., Amigorena S. 2012. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 12:557–569 10.1038/nri3254 [DOI] [PubMed] [Google Scholar]
- Jongbloed S.L., Kassianos A.J., McDonald K.J., Clark G.J., Ju X., Angel C.E., Chen C.J., Dunbar P.R., Wadley R.B., Jeet V., et al. 2010. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 207:1247–1260 10.1084/jem.20092140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst A.O., Guermonprez P., Dudziak D., Nussenzweig M.C. 2010. Route of antigen uptake differentially impacts presentation by dendritic cells and activated monocytes. J. Immunol. 185:3426–3435 10.4049/jimmunol.1001205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klechevsky E., Morita R., Liu M., Cao Y., Coquery S., Thompson-Snipes L., Briere F., Chaussabel D., Zurawski G., Palucka A.K., et al. 2008. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. 29:497–510 10.1016/j.immuni.2008.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.L., Zhan Y., Proietto A.I., Prato S., Wu L., Heath W.R., Villadangos J.A., Lew A.M. 2008. Selective suicide of cross-presenting CD8+ dendritic cells by cytochrome c injection shows functional heterogeneity within this subset. Proc. Natl. Acad. Sci. USA. 105:3029–3034 10.1073/pnas.0712394105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstedt M., Lundberg K., Borrebaeck C.A. 2005. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J. Immunol. 175:4839–4846 [DOI] [PubMed] [Google Scholar]
- Mantegazza A.R., Savina A., Vermeulen M., Pérez L., Geffner J., Hermine O., Rosenzweig S.D., Faure F., Amigorena S. 2008. NADPH oxidase controls phagosomal pH and antigen cross-presentation in human dendritic cells. Blood. 112:4712–4722 10.1182/blood-2008-01-134791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy D., Troadec C., Grassi F., Samri A., Barrou B., Autran B., Debré P., Feuillard J., Hosmalin A. 2001. Investigation of human spleen dendritic cell phenotype and distribution reveals evidence of in vivo activation in a subset of organ donors. Blood. 97:3470–3477 10.1182/blood.V97.11.3470 [DOI] [PubMed] [Google Scholar]
- Mittag D., Proietto A.I., Loudovaris T., Mannering S.I., Vremec D., Shortman K., Wu L., Harrison L.C. 2011. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J. Immunol. 186:6207–6217 10.4049/jimmunol.1002632 [DOI] [PubMed] [Google Scholar]
- Nestle F.O., Zheng X.G., Thompson C.B., Turka L.A., Nickoloff B.J. 1993. Characterization of dermal dendritic cells obtained from normal human skin reveals phenotypic and functionally distinctive subsets. J. Immunol. 151:6535–6545 [PubMed] [Google Scholar]
- Poulin L.F., Salio M., Griessinger E., Anjos-Afonso F., Craciun L., Chen J.L., Keller A.M., Joffre O., Zelenay S., Nye E., et al. 2010. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J. Exp. Med. 207:1261–1271 10.1084/jem.20092618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin L.F., Reyal Y., Uronen-Hansson H., Schraml B., Sancho D., Murphy K.M., Hakansson U.K., Moita L.F., Agace W.W., Bonnet D., Reis E.S.C. 2012. DNGR-1 is a specific and universal marker of mouse and human Batf3-dependent dendritic cells in lymphoid and non-lymphoid tissues. Blood. 119:6952–6962 10.1182/blood-2012-01-406967 [DOI] [PubMed] [Google Scholar]
- Ray K., Bobard A., Danckaert A., Paz-Haftel I., Clair C., Ehsani S., Tang C., Sansonetti P., Tran G.V., Enninga J. 2010. Tracking the dynamic interplay between bacterial and host factors during pathogen-induced vacuole rupture in real time. Cell. Microbiol. 12:545–556 10.1111/j.1462-5822.2010.01428.x [DOI] [PubMed] [Google Scholar]
- Robbins S.H., Walzer T., Dembélé D., Thibault C., Defays A., Bessou G., Xu H., Vivier E., Sellars M., Pierre P., et al. 2008. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9:R17 10.1186/gb-2008-9-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho D., Mourão-Sá D., Joffre O.P., Schulz O., Rogers N.C., Pennington D.J., Carlyle J.R., Reis e Sousa C. 2008. Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin. J. Clin. Invest. 118:2098–2110 10.1172/JCI34584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho D., Joffre O.P., Keller A.M., Rogers N.C., Martínez D., Hernanz-Falcón P., Rosewell I., Reis e Sousa C. 2009. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 458:899–903 10.1038/nature07750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I.C., Lennon-Duménil A.M., Seabra M.C., Raposo G., Amigorena S. 2006. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 126:205–218 10.1016/j.cell.2006.05.035 [DOI] [PubMed] [Google Scholar]
- Savina A., Peres A., Cebrian I., Carmo N., Moita C., Hacohen N., Moita L.F., Amigorena S. 2009. The small GTPase Rac2 controls phagosomal alkalinization and antigen crosspresentation selectively in CD8(+) dendritic cells. Immunity. 30:544–555 10.1016/j.immuni.2009.01.013 [DOI] [PubMed] [Google Scholar]
- Segura E., Villadangos J.A. 2011. A modular and combinatorial view of the antigen cross-presentation pathway in dendritic cells. Traffic. 12:1677–1685 10.1111/j.1600-0854.2011.01254.x [DOI] [PubMed] [Google Scholar]
- Segura E., Valladeau-Guilemond J., Donnadieu M.H., Sastre-Garau X., Soumelis V., Amigorena S. 2012. Characterization of resident and migratory dendritic cells in human lymph nodes. J. Exp. Med. 209:653–660 10.1084/jem.20111457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacken P.J., de Vries I.J., Torensma R., Figdor C.G. 2007. Dendritic-cell immunotherapy: from ex vivo loading to in vivo targeting. Nat. Rev. Immunol. 7:790–802 10.1038/nri2173 [DOI] [PubMed] [Google Scholar]
- Zelenay S., Keller A.M., Whitney P.G., Schraml B.U., Deddouche S., Rogers N.C., Schulz O., Sancho D., Reis e Sousa C. 2012. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. J. Clin. Invest. 122:1615–1627 10.1172/JCI60644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Heitbrock L., Ancuta P., Crowe S., Dalod M., Grau V., Hart D.N., Leenen P.J., Liu Y.J., MacPherson G., Randolph G.J., et al. 2010. Nomenclature of monocytes and dendritic cells in blood. Blood. 116:e74–e80 10.1182/blood-2010-02-258558 [DOI] [PubMed] [Google Scholar]