Loss of Sirt1 causes increased Hoxa9 expression and expansion of HSPC subsets under hematopoietic stress, resulting in increased DNA damage and exhaustion of long-term progenitors.
Abstract
The (histone) deacetylase Sirt1 is a mediator of genomic and epigenetic maintenance, both of which are critical aspects of stem cell homeostasis and tightly linked to their functional decline in aging and disease. We show that Sirt1 ablation in adult hematopoietic stem and progenitor cells (HSPCs) promotes aberrant HSPC expansion specifically under conditions of hematopoietic stress, which is associated with genomic instability as well as the accumulation of DNA damage and eventually results in a loss of long-term progenitors. We further demonstrate that progenitor cell expansion is mechanistically linked to the selective up-regulation of the HSPC maintenance factor and polycomb target gene Hoxa9. We show that Sirt1 binds to the Hoxa9 gene, counteracts acetylation of its histone target H4 lysine 16, and in turn promotes polycomb-specific repressive histone modification. Together, these findings demonstrate a dual role for Sirt1 in HSPC homeostasis, both via epigenetic regulation of a key developmental gene and by promoting genome stability in adult stem cells.
Stem cell homeostasis is a central aspect of tissue maintenance and regeneration in response to injury or stress, which is perhaps best exemplified in hematopoietic stem and progenitor cells (HSPCs; Geiger and Rudolph, 2009; Wilson et al., 2009). Adult HSPCs show extensive self-renewal capacity and allow for long-term multilineage differentiation in the hematopoietic system. If deregulated, this process can result in a variety of disorders including myeloproliferative diseases and loss of immune function (Aggarwal et al., 2012). Recent evidence suggests that DNA damage accumulation or a defective response to genotoxic stress is a key contributor to aberrant expansion and concomitant functional exhaustion of HSPCs, which is further associated with altered expression of hematopoietic differentiation factors and age-associated stem cell decline (Rossi et al., 2005, 2007; Dykstra et al., 2011; Wang et al., 2012). Determining the factors that affect HSPC function with regard to both DNA damage accumulation and epigenetic deregulation is, thus, critical for our understanding of HSPC homeostasis in aging and disease.
The silent information regulator 2 (Sir2) family of proteins is a group of NAD+-dependent protein deacetylases initially discovered in yeast (Klar et al., 1979; Rine et al., 1979), which have been linked to longevity as well as a variety of physiological stress responses (Haigis and Sinclair, 2010; Chalkiadaki and Guarente, 2012; Yuan et al., 2012b). The closest mammalian Sir2 ortholog, Sirt1, is further involved in the transcriptional regulation of several key developmental regulators (Calvanese et al., 2010; Haigis and Sinclair, 2010; Lu et al., 2011) and was reported to associate with the stem cell–specific Polycomb group (PcG) repressive complex PRC4, preferentially under conditions of oxidative stress (Kuzmichev et al., 2005; O’Hagan et al., 2011). Consistent with a role for Sirt1 in stem cell homeostasis, Sirt1-deficient embryonic stem (ES) cells show increased sensitivity to oxidative stress, DNA damage accumulation, and genomic instability (Oberdoerffer et al., 2008; Wang et al., 2008). Moreover, Sirt1 has been linked to compromised differentiation of ES cells into cells of the hematopoietic lineage (Han et al., 2008; Ou et al., 2011).
Based on these observations, it is tempting to speculate that Sirt1 may also be a critical regulator of adult stem cell maintenance. The role of Sirt1 in HSPC homeostasis is, however, a current topic of debate. To date, no obvious HSPC-related phenotype has been detected in Sirt1-deficient mice, although in vitro analyses have implicated Sirt1 as either a positive or negative regulator of HSPC maintenance and/or differentiation after growth factor stimulation, suggesting that Sirt1 function in HSPCs may be susceptible to environmental cues (Narala et al., 2008; Leko et al., 2012; Matsui et al., 2012; Peled et al., 2012).
To unequivocally address the role of Sirt1 in HSPCs, we used two complementary approaches for conditional ablation of Sirt1: induced deletion in adult mice and constitutive deletion specifically in the hematopoietic lineage. In so doing, we uncovered an unexpected aspect of Sirt1 function that has the potential to reconcile previously conflicting reports: we found that Sirt1 ablation promotes aberrant expansion of HSPCs in vivo specifically in response to hematopoietic stress, as shown here for cytotoxic and genotoxic agents. Sirt1 ablation in HSPCs further led to a reduced ability to repair DNA damage, resulting in genomic instability and a progressive loss of long-term progenitors after stress-induced proliferative expansion or DNA damage. Through gene expression profiling and molecular characterization, we implicate epigenetic deregulation and inappropriate induction of the PcG target and HSPC maintenance factor Hoxa9 as the mechanism that promotes HSPC expansion in the absence of Sirt1. Together, these findings highlight the relevance of stress-related physiological changes with regard to Sirt1 function in HSPCs and uncover a critical role for Sirt1 in both genomic and epigenetic HSPC maintenance.
RESULTS
Induced Sirt1 ablation results in expansion of adult HSPCs
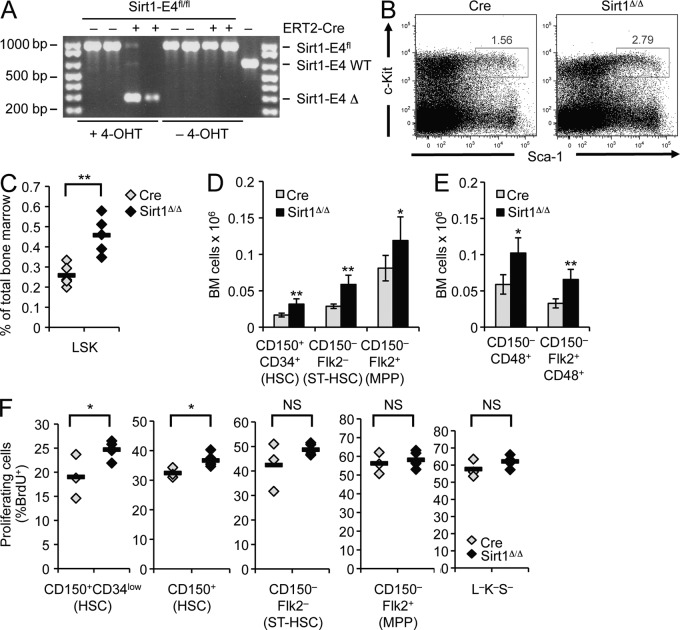
To determine the role of Sirt1 in the maintenance of adult HSPCs and, thereby, avoid possible developmental defects associated with conventional Sirt1 knockout mice (McBurney et al., 2003; Ou et al., 2011), we crossed mice homozygous for a conditional Sirt1 knockout allele in which Sirt1 exon 4 is flanked by two loxP sites (Sirt1-E4fl/fl) to the 4-hydroxy-tamoxifen (4-OHT)–inducible ERT2-Cre transgene (Ruzankina et al., 2007). Exon 4 was efficiently removed in the BM of mice fed with 4-OHT–supplemented chow, whereas no deletion was detected in the absence of 4-OHT (Fig. 1 A). Deletion of Sirt1 exon 4 was previously shown to result in a truncated protein, which lacks catalytic activity and phenocopies the Sirt1-null phenotype (Cheng et al., 2003).
Figure 1.
Induced loss of Sirt1 in adult mice causes HSPC expansion. (A) PCR was used to detect deletion of Sirt1 exon 4 using genomic DNA from BM of Sirt1-E4fl/fl mice with (+) or without (−) ERT2-Cre transgene in the presence or absence of a 4-OHT–supplemented diet. Δ: deletion of exon 4. (B) Representative FACS analysis of lineage marker-negative HSPCs based on expression of c-Kit and Sca-1 in 4-OHT–treated Sirt1-E4fl/fl ERT2-Cre (SIRT1Δ/Δ, n = 5) or ERT2-Cre control mice (Cre, n = 5). (C) Frequency of LSK subset in BM from Sirt1-E4fl/fl Cre (n = 5) and Sirt1Δ/Δ (n = 5) mice after 8–9 wk of 4-OHT administration. Each diamond represents an individual mouse, and results from two independent experiments were combined. P-values are based on a two-tailed Student’s t test. (D and E) Total number of indicated BM subsets in two independent experiments. BM cells were derived from two hind legs (femurs and tibias) of SIRT1Δ/Δ mice and Cre controls after 7–8 wk of 4-OHT administration. Subsets were identified based on CD34, Flk2, and CD150 (D; Cre n = 4, SIRT1Δ/Δ n = 5) or CD48 and Flk2 (E; Cre n = 4, SIRT1Δ/Δ n = 3). Error bars depict SD. See Fig. S1 A for representative gating strategies. (F) Fraction of cycling, BrdU+ HSPC subsets measured after 18 h of BrdU administration in CD150+CD34low, total CD150+, CD150−Flk2−, and CD150−Flk2+ progenitors and Lin−Sca-1−c-Kit− (L−K−S−) cells from mice in D: Cre (n = 3) and SIRT1Δ/Δ (n = 5), diamonds represent individual animals. Horizontal bars represent the mean of all data points. *, P ≤ 0.05; **, P ≤ 0.01; NS, not significant.
After 4-OHT treatment, we observed a pronounced increase in the HSPC-containing lineage-negative (Lin−), c-Kit+, Sca1+ (LSK) BM subset from Sirt1-E4fl/fl, ERT2-Cre (SIRT1Δ/Δ) mice when compared with ERT2-Cre transgenic (Cre) control mice (Fig. 1, B and C). Based on differential surface marker expression, LSK cells can be further subdivided into long-term hematopoietic stem cells (LT-HSCs: CD150+, CD34−, CD48−, Flk2−) and multipotent progenitor cell subsets, listed here by decreasing self-renewal and increasing proliferation capacity (based on Wilson et al., 2008): MPP1/2 (CD150+, CD34+, CD48−/+, Flk2−), MPP3 or short-term HSCs (CD150−, CD34+, CD48+, Flk2−), and MPP4 (CD150−, CD34+, CD48+, Flk2+). Sirt1 loss promoted an increase in HSPC subsets that was first apparent in CD150+CD34+ progenitors and continued throughout more differentiated CD150− MPP subsets (Fig. 1, D and E; Fig. S1 A). CD34-expressing CD150+ HSCs represent activated, early progenitors that are characterized by increased proliferative capacity as compared with quiescent HSCs (Wilson et al., 2008). We, thus, sought to determine if Sirt1 loss is associated with increased HSPC proliferation by analyzing BrdU incorporation in SIRT1Δ/Δ and Cre control mice. In so doing, we observed increased cycling in Sirt1-deficient, early (CD150+CD34low and total CD150+) progenitor subsets (Fig. 1 F; Fig. S1 B). More differentiated MPPs and Lin−c-Kit−Sca-1− non-HSPCs showed only minor changes in BrdU incorporation compared with Cre controls (Fig. 1 F), suggesting that the observed HSPC expansion in the absence of Sirt1 is, at least in part, the result of an early progenitor cell–specific increase in proliferation. Together, these data establish Sirt1 as a modulator of adult HSPC homeostasis in vivo.
HSPC expansion in the absence of Sirt1 depends on hematopoietic stress
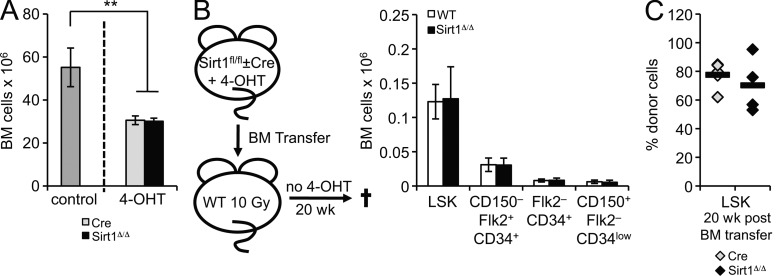
HSPC expansion upon 4-OHT–induced Sirt1 deletion was unexpected given that no change in HSPC subsets was found using conventional Sirt1 knockout mice (Leko et al., 2012). Notably, 4-OHT treatment caused a reduction in overall BM cellularity, which was comparable between SIRT1Δ/Δ mice and Cre controls (Fig. 2 A), and is consistent with general clastogenic cytotoxicity in response to 4-OHT (Vijayalaxmi and Rai, 1996). Both blood loss and cytotoxic agents represent common sources of hematopoietic stress that are capable of promoting activation of quiescent HSPCs (Passegué et al., 2005; Wilson et al., 2009). We, thus, hypothesized that HSPC expansion in the absence of Sirt1 may be the result of an exacerbated response to hematopoietic stress. To determine if altered HSPC maintenance is stress dependent, we transferred BM from 4-OHT–treated Sirt1-E4fl/fl, ERT2-Cre (SIRT1Δ/Δ) or Sirt1-E4fl/fl (WT) control donors into lethally irradiated, CD45-congenic WT hosts that were subsequently maintained in the absence of 4-OHT.
Figure 2.
Expansion of Sirt1-deficient HSPCs is dependent on hematopoietic stress. (A) Cell numbers are from two femurs and two tibias of untreated control mice (n = 5), 4-OHT–treated Sirt1-E4fl/fl, ERT2-Cre (SIRT1Δ/Δ, n = 3), and 4-OHT–treated ERT2-Cre controls (Cre, n = 3). Error bars represent SD. (B) Before BM transfer, WT and SIRT1Δ/Δ mice were treated with 4-OHT for 4 wk. After transfer, mice were kept on a 4-OHT–free diet for 20 wk. Sirt1 exon 4 deletion in BM chimeric mice was validated as described in Fig. 1 A (not depicted). At least five recipient mice were analyzed per group for indicated LSK subsets by FACS, and donor origin was confirmed using CD45.2 staining (>95%). Total BM cell numbers from two hind legs are shown. Error bars represent SD. (C) BM from 4-OHT–treated Sirt1-E4fl/fl, ERT2-Cre mice (SIRT1Δ/Δ, n = 4) and age-matched 4-OHT–treated ERT2-Cre controls (Cre, n = 5) was mixed with congenic CD45.1+ host BM at a 1:1 ratio. The frequency of CD45.2+ donor LSK in total BM was analyzed by FACS 20 wk after transfer, and diamonds depict individual recipient mice. Horizontal bars represent the mean of all data points. **, P ≤ 0.01.
In contrast to our findings in mice that were chronically exposed to 4-OHT (Fig. 1, D and E), we observed no difference in the total number of HSPC subsets between recipients of Sirt1-deficient and control BM in the absence of 4-OHT (Fig. 2 B), suggesting a role for hematopoietic stress in promoting expansion of Sirt1-deficient HSPCs. In further support of this notion, both SIRT1Δ/Δ and Cre control BM contributed equally to the LSK subset when competitively transferred with CD45-congenic WT BM in the absence of 4-OHT (Fig. 2 C).
The effect of Sirt1 on HSPC maintenance is cell autonomous and specific to stress-induced but not developmental expansion
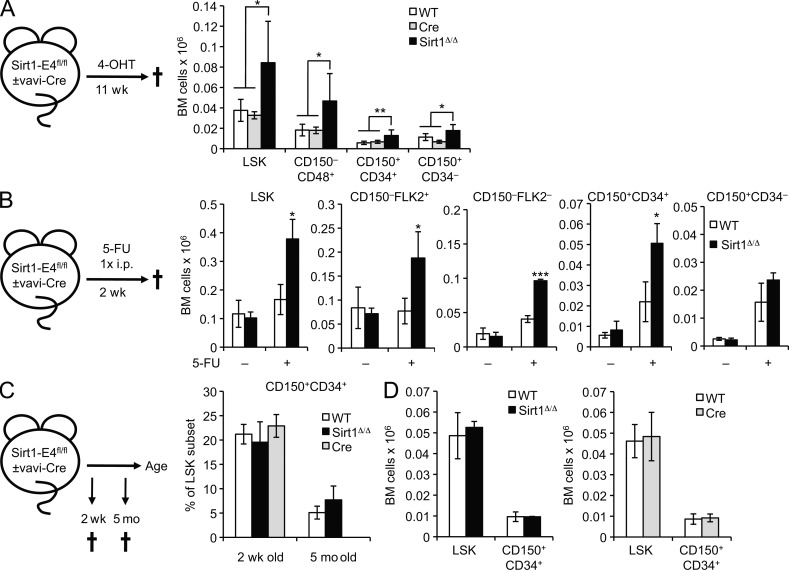
To validate the impact of Sirt1 loss on HSPC homeostasis in response to hematopoietic stress in a cell-autonomous manner, and to eliminate experimental limitations as a result of the dual use of 4-OHT as both ERT2-Cre inducer and a source of hematopoietic stress, we generated SIRT1-deficient mice using the vav-iCre transgene, which is predominantly expressed in the hematopoietic lineage (de Boer et al., 2003). Sirt1-E4fl/fl, vav-iCre (SIRT1Δ/Δ) mice, Sirt1-E4fl/fl (WT), and vav-iCre (Cre) controls were then exposed to 4-OHT for 11 wk and analyzed for HSPC subset distribution. We observed a significant increase in LSK subset numbers in the absence of Sirt1 compared with control mice, in agreement with our observations in the ERT2-Cre model (Fig. 3 A). We next sought to determine the consequences of Sirt1 loss in response to an independent, temporally controlled source of hematopoietic stress: treatment with the pyrimidine analogue 5-Fluorouracil (5-FU). A single injection of 5-FU selectively depletes proliferating cells, which in turn promotes the cycling of quiescent HSPCs and concomitant hematopoietic cell recovery (Harrison and Lerner, 1991). HSPC subset distribution and frequencies in Sirt1-E4fl/fl, vavi-Cre mice and Sirt1-E4fl/fl controls were analyzed both in the presence and absence of 5-FU. In agreement with previous observations using conventional Sirt1 knockout mice and our data from non–4-OHT–treated SIRT1Δ/Δ mice, we found no change in HSPC subsets between Sirt1-E4fl/fl, vav-iCre mice and littermate controls in the absence of 5-FU (Leko et al., 2012). 5-FU treatment, in contrast, caused a significant increase in HSPC subset frequencies and numbers, which was again first observed in CD150+CD34+ early progenitors, further corroborating the requirement for hematopoietic stress in driving the expansion of Sirt1-deficient HSPCs (Fig. 3 B).
Figure 3.
The effect of Sirt1 loss is cell-autonomous and linked to stress but not developmental HSPC expansion. (A) Hematopoietic lineage-specific Sirt1-E4fl/fl, vav-iCre mice (SIRT1Δ/Δ, n = 5), Sirt1-E4fl/fl (WT, n = 5), and vav-iCre (Cre, n = 2) control mice were treated with 4-OHT at 5 mo of age for 11 wk and the frequency of the indicated LSK subsets was analyzed by FACS. Cell numbers from two hind legs are shown. P-values are based on comparison of SIRT1Δ/Δ to combined controls. (B) Sirt1-E4fl/fl, vav-iCre mice (SIRT1Δ/Δ, n = 3) and Sirt1-E4fl/fl controls (WT, n = 3) were treated with 5-FU at 4–5 mo of age and analyzed 2 wk later as in A. Representative gating strategies are shown in Fig. S2. Total BM cell number from two hind legs is shown for the indicated subsets. (C) Frequency of CD150+CD34+ LSK progenitors and total LSKs in 2-wk-old Sirt1-E4fl/fl, vav-iCre mice (SIRT1Δ/Δ, n = 6), WT (n = 12), or vav-iCre (n = 3) controls. 5-mo-old mice from B are shown for comparison (n = 3 per group). (D) Total BM cell numbers of indicated HSPC subsets in 2-wk-old Sirt1-E4fl/fl, vav-iCre mice (SIRT1Δ/Δ) and WT or vav-iCre controls (Cre). Panels represent individual litters of either WT (n = 9) and SIRT1Δ/Δ (n = 3) or WT (n = 2) and Cre mice (n = 6). Error bars represent SD. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
To determine if the effect of Sirt1 loss is specific to conditions of hematopoietic stress or a general consequence of stem and progenitor cell expansion, we analyzed the impact of Sirt1 ablation on HSPC subsets during preadult hematopoietic development. Fetal and preadult HSCs are predisposed to stress-independent proliferative expansion, which is, in part, reflected in higher CD34 expression levels compared with adult, quiescent HSCs (Ito et al., 2000; Mikkola and Orkin, 2006; Wilson et al., 2008). Fittingly, we found an increase in activated, CD34+CD150+ early progenitors in preadult (2 wk old) BM LSK cells from both SIRT1Δ/Δ and control mice when compared with 5-mo-old adult mice (Fig. 3 C). However, in sharp contrast to Sirt1 ablation after hematopoietic stress (Fig. 3, A and B), Sirt1 loss had no effect on either CD150+CD34+ HSPC or total LSK frequency during developmental expansion (Fig. 3, C and D). This result likely reflects intrinsic differences in the regulation of preadult and adult hematopoietic progenitor maintenance and self-renewal (Mikkola and Orkin. 2006; see discussion). Together, these data demonstrate that Sirt1 is required for adult HSPC maintenance in a cell-autonomous manner, specifically under conditions of hematopoietic stress.
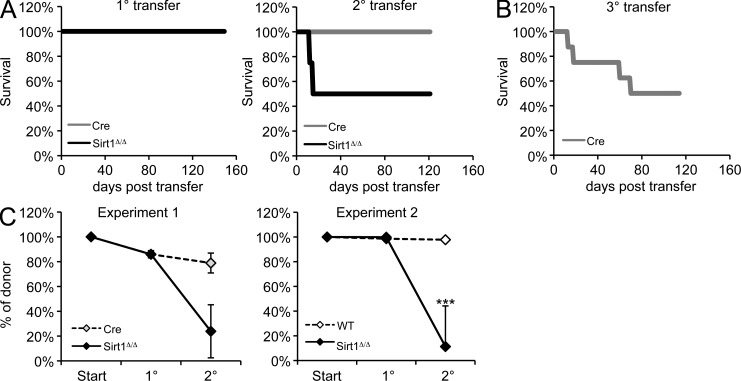
Chronic stress exposure promotes loss of long-term progenitor cells
Defects in HSPC maintenance have been associated with the functional exhaustion of stem and progenitor cells over time. This is perhaps best exemplified in aged HSPCs, which show significant expansion but a pronounced loss of long-term reconstitution potential (Rossi et al., 2007; Beerman et al., 2010). To examine the relationship between stress-associated HSPC maintenance defects after Sirt1 loss and HSC function, we performed serial BM transplantation experiments using Sirt1-E4fl/fl, ERT2-Cre, and ERT2-Cre or WT control donors that were exposed to 4-OHT for 6 wk before the first transfer. Serial transplantation poses a recurrent stress on LT-HSCs as they are forced to repeatedly expand and reconstitute the hematopoietic system. Recipients of Sirt1-deficient BM showed a pronounced decrease in survival compared with recipients of Cre control BM upon secondary transfer, and Sirt1-deficient hematopoietic cells were counter-selected in surviving recipients by host marrow that escaped irradiation-induced death (Fig. 4, A and C). Recipients of Cre control BM exhibited impaired survival only upon tertiary transfer (Fig. 4 B). Our data, thus, demonstrate that the sustained absence of Sirt1 causes the functional exhaustion of long-term progenitors in vivo.
Figure 4.
Sirt1-deficient BM shows serial transplantation defects. (A) Survival plots from serial transfer experiments using BM from 4-OHT–treated Sirt1-E4fl/fl, ERT2-Cre (SIRT1Δ/Δ) and ERT2-Cre (Cre) control mice. Surviving mice were subjected to secondary transfer. Recipient animal numbers three per group for primary transfer and four per group for secondary transfer. (B) Tertiary transfer of Cre control BM (n = 8). (C) Contribution of CD45.2+ donor cells to recipient BM at the end of 1 and 2° transfer in surviving mice from A (left) and from a second independent experiment with WT (n = 7) and SIRT1Δ/Δ (n = 9) mice (right). Error bars represent SD. ***, P ≤ 0.001.
SIRT1-deficient hematopoietic progenitors show increased DNA damage, p53 activation, and genomic instability
We next sought to determine the molecular basis for stress-associated loss of HSPC function in Sirt1-deficient progenitor cells. Functional decline of HSPCs has been previously linked to DNA damage accumulation after both (age-related) HSPC expansion and defective genome maintenance resulting from genetic manipulation (Rossi et al., 2007; Niedernhofer, 2008; Wang et al., 2012). Notably, Sirt1 has been implicated in genome maintenance of ES cells by us and others (Oberdoerffer et al., 2008; Wang et al., 2008) and is involved in several DNA repair processes ranging from homologous recombination to the repair of oxidative lesions in a variety of cell types including leukemic tumor cell lines (Haigis and Sinclair, 2010; Wang et al., 2013). We, therefore, reasoned that Sirt1 ablation may result in increased DNA damage accumulation and/or genomic instability in HSPCs after stress-induced expansion.
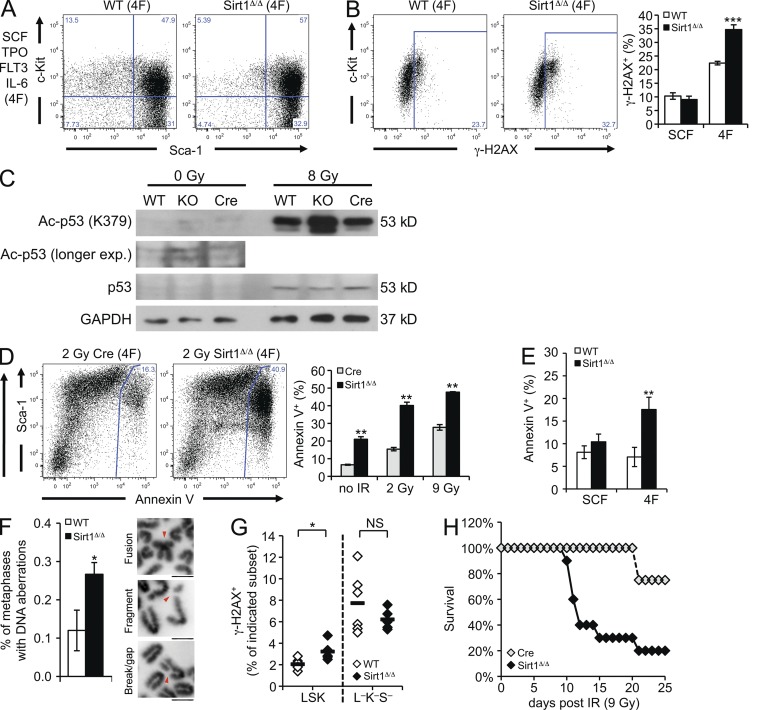
To determine DNA damage accumulation under conditions of proliferative stress in the presence or absence of Sirt1, we cultured Sirt1-E4fl/fl WT and Sirt1-E4fl/fl, ERT2-Cre BM cells using stem cell factor (SCF), Flk-2/Flt3 ligand (FLT3L), thrombopoietin (TPO), and IL-6. This and related four factor (4F) cytokine combinations were reported previously to promote selective survival and proliferation of multipotent hematopoietic progenitor cells in vitro (Zhang and Lodish, 2008). Surface staining confirmed that the majority of both SIRT1Δ/Δ and WT cells coexpressed the LSK markers Sca-1 and c-Kit after up to 12 d in culture (Fig. 5 A). Culture in SCF alone supports survival, but has a limited effect on HSPC expansion and served as a control. Sirt1 was deleted before culture in vivo or by transient 4-OHT–mediated Cre induction in vitro, thereby allowing for BM culture in the absence of continued Cre activity. To assess the frequency of cells with DNA double-strand breaks, we measured the accumulation of H2AX phosphorylated on S139 (γ-H2AX), an early marker of the DSB-induced DNA damage response. Consistent with proliferation-induced damage accumulation, we found an increase in γ-H2AX+ cells upon treatment with all four factors compared with SCF alone. Notably, the frequency of γ-H2AX+ cells was further increased in the absence of Sirt1 (Fig. 5 B), indicating a synergistic role for proliferation and Sirt1 loss in promoting the accumulation of DNA damage.
Figure 5.
Loss of Sirt1 causes increased sensitivity to DNA damage. (A) FACS analysis of c-Kit and Sca-1 expression in Sirt1-E4fl/fl (WT) or Sirt1-E4fl/fl, ERT2-Cre (SIRT1Δ/Δ) BM cells cultured for 10 d in SCF, TPO, FLT3L, and IL-6 (4F). One of at least three representative experiments is shown. (B) BM cultures from SIRT1Δ/Δ mice and WT controls maintained in SCF alone or 4F were analyzed in triplicate for the fraction of γ-H2AX+ cells after 11 d in culture. A representative staining is shown. Error bars represent SD. (C) SIRT1Δ/Δ, WT, and ERT2-Cre control BM cultured in the presence of 4F for 11 d was subjected to IR (8 Gy) or left untreated (0 Gy) and analyzed 6 h thereafter for p53 expression and p53-K379 acetylation. GAPDH served as loading control. One of three representative experiments is shown. (D) FACS analysis of Annexin V+ cells in Sirt1-E4fl/fl, vav-iCre (SIRT1Δ/Δ) or vav-iCre (Cre) control BM after 11 d of 4F culture. Cultures were left untreated or exposed to irradiation (2 Gy or 9 Gy) and analyzed 6 h thereafter. Samples were analyzed in triplicate for the total fraction of Annexin V+ cells. A representative staining is shown. Error bars represent SD. (E) FACS analysis of Annexin V+ cells cultured for 11 d in the presence of SCF alone or 4F. Analysis was performed as in D. Error bars represent SD. (F) Metaphase analysis from Sirt1-E4fl/fl (WT) and Sirt1-E4fl/fl, ERT2-Cre (SIRT1Δ/Δ) 4F BM cultures. Sirt1 was deleted in vivo before culture. The frequency of total metaphases containing DNA aberrations is shown. DAPI images depict examples of individual aberrations. Bar, 10 µm. See Table 1 for details. Mean and SD are based on BM cultures from three animals per group. 50 metaphases were analyzed per sample. (G) Frequency of γ-H2AX+ LSK and Lin−c-Kit−Sca-1− non-HSPC cells in vivo from mice in Fig. 2 B. Recipients of Sirt1-deficient (SIRT1Δ/Δ, n = 5) or control (WT, n = 6) BM were analyzed 20 wk after transfer. Diamonds represent individual recipient mice. Horizontal bars represent the mean of all data points. (H) Sirt1-E4fl/fl, vav-iCre mice (SIRT1Δ/Δ, n = 10) and age-matched vav-iCre controls (Cre, n = 4) were exposed to sublethal IR (9 Gy) and survival was monitored over 1 mo. P = 0.045 (Wilcoxon Test); P = 0.049 (Log-rank). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; NS, not significant.
Sirt1 was previously shown to counteract activation of the proapoptotic tumor suppressor p53 by deacetylating lysine 379 in response to genotoxic stress (Luo et al., 2001; Vaziri et al., 2001). Consistent with the increased DNA damage accumulation in Sirt1-deficient hematopoietic progenitor cells cultured in all four factors, we observed a modest increase in p53 acetylation and more Annexin V+ apoptotic cells upon Sirt1 loss (Fig. 5, C and D). No change in the frequency of Annexin V+ cells was observed when cells were cultured with SCF alone, in agreement with comparable levels of DNA damage after SCF treatment (Fig. 5, B and E). These findings support the notion that increased activation of the DNA damage response in Sirt1-deficient cells directly correlates with hematopoietic progenitor cell expansion.
To determine possible functional consequences of DNA damage accumulation in the absence of Sirt1, we analyzed metaphase spreads from 4F cultures and found that Sirt1 loss caused an increase in DSB-related aberrations, including centromeric chromosome fusions, chromosome fragments, gaps, and breaks (Fig. 5 F). The frequency of individual aberrations observed in cultures from three SIRT1Δ/Δ and three WT mice is listed in Table 1. Together, these findings implicate Sirt1 in the genomic maintenance of hematopoietic progenitor cells.
Table 1.
DNA aberrations in in vitro-expanded hematopoietic progenitor cells.
| Genotype | Break | Gap | Frag | DC | CF | Trans | Radial | 30-39 | 40 | 41+ | Cells | Sex |
| WT | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 10 | 40 | 0 | 50 | F |
| WT | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 7 | 43 | 0 | 50 | M |
| WT | 5 | 3 | 0 | 0 | 1 | 0 | 0 | 12 | 38 | 0 | 50 | F |
| SIRT1Δ/Δ | 3 | 9 | 0 | 0 | 1 | 0 | 0 | 13 | 37 | 0 | 50 | F |
| SIRT1Δ/Δ | 4 | 10 | 0 | 0 | 1 | 0 | 0 | 11 | 39 | 0 | 50 | M |
| SIRT1Δ/Δ | 3 | 5 | 1 | 0 | 3 | 0 | 0 | 7 | 42 | 1 | 50 | F |
BM from three Sirt1-E4fl/fl, ERT2-Cre mice (SIRT1Δ/Δ) and three Sirt1-E4fl/fl controls (WT) was cultured in the presence of SCF, FLT3L, IL-6, and TPO, and metaphases were harvested after 10 d. Chromosomal DNA aberrations were scored for 50 metaphases per sample. Frag: DNA fragment; DC: dicentric chromosome; CF: centric fusion; Trans: translocation. Number of chromosomes per cell and gender are indicated.
Sirt1 protects HSPCs from DNA damage in vivo
To determine the consequences of Sirt1 loss on DNA damage accumulation in vivo, we analyzed HSPCs from Sirt1-E4fl/fl, ERT2-Cre animals and littermate controls for the presence of γ-H2AX. To exclude non–cell-autonomous effects as well as continued activation of Cre, SIRT1Δ/Δ and WT BM from 4-OHT–treated mice was transferred into CD45-congenic recipients (Fig. 2 B). In agreement with our in vitro findings, we observed an increase in γ-H2AX+ SIRT1Δ/Δ HSPCs. Consistent with the HSPC-specific effect of Sirt1 loss on proliferation (Fig. 1 F), no increase in γ-H2AX+ cells was observed in Lin−c-Kit−Sca-1− non-HSPCs (Fig. 5 G; Fig. S3).
We next sought to determine if increased susceptibility to DNA damage affects HSPC function in Sirt1-deficient mice. To do so, we measured the ability of Sirt1-deficient BM to recover from acute genotoxic stress after γ-irradiation (IR), which depends on the survival of HSPCs, and short-term progenitors in particular, to allow for recovery of the hematopoietic system. It is of note that the effect of irradiation may be exacerbated by damage to the intestine or other radiation-sensitive tissues. To rule out possible nonhematopoietic cell-specific effects of Sirt1 loss in response to IR, we used Sirt1-E4fl/fl, vav-iCre mice for our analysis. We found that, in the absence of Sirt1, mice succumb to sublethal irradiation (9 Gy) with significantly higher frequency than vav-iCre controls (Fig. 5 H). Similar results were obtained with ERT2-Cre, Sirt1-E4fl/fl mice (unpublished data). To gain mechanistic insight into Sirt1-mediated protection of HSPCs from genotoxic stress, we next analyzed acetyl-p53 and total p53 levels in in vitro–expanded hematopoietic progenitor cells after IR. Consistent with the known antiapoptotic role of Sirt1 via p53 deacetylation (Luo et al., 2001; Vaziri et al., 2001), we observed a pronounced increase in p53 acetylation and concomitant induction of apoptosis in Sirt1-deficient cells, suggesting that activation of the p53 pathway at least in part contributes to DNA damage-induced HSPC loss in the absence of Sirt1 (Fig. 5, C and D). Together, these data demonstrate that Sirt1-deficient HSPCs are more sensitive to genotoxic stress in vivo and support the notion that accumulated DNA damage promotes the loss of pluripotent HSCs after stress-induced deregulation of HSPC homeostasis.
Sirt1 ablation promotes hyperproliferation and increased Hoxa9 expression in vitro
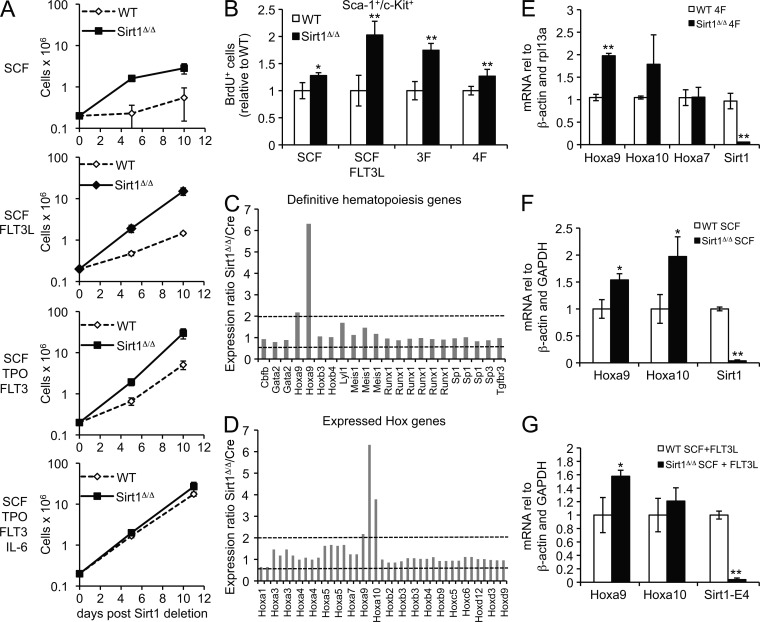
Having shown that Sirt1 loss promotes stress-associated expansion of hematopoietic progenitors, we next sought to dissect the molecular basis for this phenomenon using the in vitro culture approach described above. Consistent with our in vivo observations, Sirt1 loss caused an increase in cell number, which was most notable under suboptimal growth factor conditions, such as SCF alone, SCF and FLT3L, or SCF, TPO, and FLT-3 (Fig. 6 A). Only addition of all four cytokines (SCF, TPO, FLT3L, and IL-6) was able to consistently promote efficient expansion of WT cells. BrdU labeling confirmed that Sirt1 loss promotes a significant increase in cycling (BrdU+) c-Kit+Sca-1+ hematopoietic progenitor cells, which is most pronounced in suboptimal growth conditions (Fig. 6 B). Together, these findings demonstrate that ablation of Sirt1 results in proliferative expansion of hematopoietic progenitor cells under conditions of cytokine-induced proliferative stress in vitro.
Figure 6.
Induced Sirt1 deletion promotes hematopoietic progenitor cell expansion and deregulation of Hoxa9 in vitro. (A) BM culture of Sirt1-E4fl/fl, ERT2-Cre (SIRT1Δ/Δ) and Sirt1-E4fl/fl (WT) cells in indicated growth factors after 4-OHT–induced Sirt1 ablation. Cell numbers were counted in duplicate at indicated passage time points, and similar results were obtained in three independent experiments. (B) Fraction of BrdU+, c-Kit+, Sca-1+ SIRT1Δ/Δ, and WT cells cultured in the indicated growth conditions. BrdU incorporation was measured after 5 d of culture after 6 h of BrdU administration. The fraction of BrdU+ cells was normalized to WT for each culture condition, and samples were analyzed in triplicate. (C) Gene expression array analysis of FACS-sorted c-Kit+Sca-1+ERT2-Cre (Cre) control and SIRT1Δ/Δ cells cultured in the presence of SCF, TPO, FLT3L, and IL-6. Relative expression of SIRT1Δ/Δ compared with Cre is shown for probe sets representing definitive hematopoiesis genes. Dashed lines depict twofold expression changes (up or down). (D) Hox gene expression analysis of microarray samples from C. (E) Analysis of expression levels of Hoxa9, Hoxa10, Hoxa7, and Sirt1 exon 4 in unsorted WT and SIRT1Δ/Δ cells from an independent 4F culture. RNA was isolated 5 d after Sirt1 deletion and subjected to qRT-PCR, and samples were analyzed in triplicate and normalized to β-actin and Rpl13a. (F and G) Analysis of expression levels of Hoxa9, Hoxa10, and Sirt1 exon 4 in WT and SIRT1Δ/Δ cells cultured in the presence of SCF (F) or SCF and FLT3L (G). RNA was isolated 5 d after Sirt1 deletion, and samples were analyzed by qRT-PCR in triplicate and normalized to Gapdh and β-actin. Error bars represent SD. *, P ≤ 0.05; **, P ≤ 0.01.
To identify molecular changes caused by the loss of Sirt1, we performed gene expression profiling of in vitro expanded c-Kit+Sca-1+ cells from 4-OHT–treated Sirt1-E4fl/fl, ERT2-Cre and ERT2-Cre control mice. BM was cultured in the presence of all four cytokines, which resulted in the most homogeneous progenitor phenotype and further ensured comparable cell numbers between SIRT1Δ/Δ and control BM (Fig. 5 A and Fig. 6 A). Microarray analysis of RNA isolated from these cultures revealed that <2% of total probe sets were altered greater than twofold upon Sirt1 loss (Table S1), suggesting selective transcriptional deregulation in the absence of Sirt1. Given the HSPC phenotype described above, we focused on HSPC maintenance and hematopoiesis-related genes (Gene ontology group definitive hematopoiesis, GO:0060216). Of 11 genes found to be expressed within this GO group, only Hoxa9 showed a greater than twofold change in expression in Sirt1-deficient HSPCs (Fig. 6 C). Moreover, of all expressed Hox genes, only Hoxa9 and its neighbor, Hoxa10 were up-regulated greater than twofold in the absence of Sirt1, suggesting locus-specific regulation by Sirt1 (Fig. 6 D). Transcriptional up-regulation of Hoxa9 was validated by qRT-PCR in an independent experiment, Hoxa7 is shown as a control (Fig. 6 E). Hoxa9 expression was also increased in Sirt1-deficient cells cultured in SCF or SCF and FLT-3L, and Hoxa10 was up-regulated to varying degrees under different growth factor conditions (Fig. 6, E–G). Together, these data implicate Sirt1 in the transcriptional repression of the Hoxa9 locus. Hoxa9 was previously shown to be a key regulator of HSPC function and maintenance in mice (Thorsteinsdottir et al., 2002; Lawrence et al., 2005) and is expressed in both early and more differentiated HSPC progenitor subsets (Sauvageau et al., 1994). Moreover, Hoxa9 was shown to be required for efficient proliferation of hematopoietic progenitors in vitro (Thorsteinsdottir et al., 2002; Lawrence et al., 2005), suggesting Hoxa9 deregulation as a critical mediator of HSPC expansion in the absence of Sirt1.
Sirt1 physically associates with the Hoxa9 gene and promotes repressive histone marks
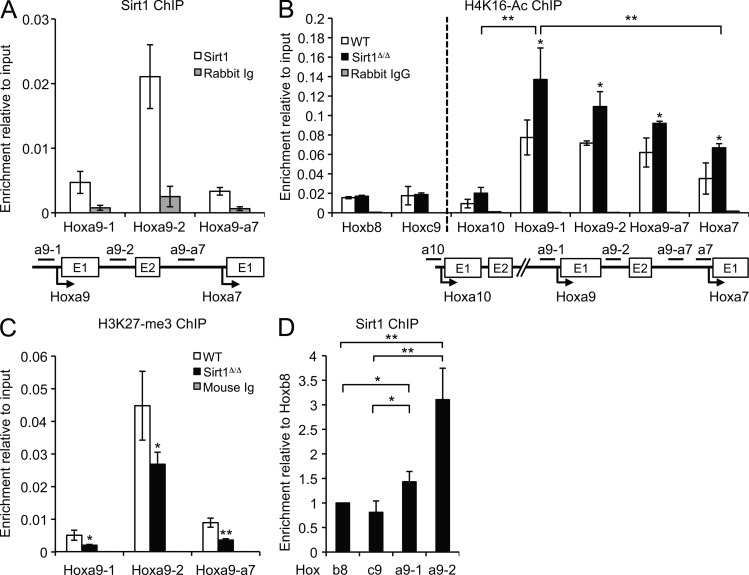
Given the increase in Hoxa9 mRNA levels in Sirt1-deficient hematopoietic progenitor cells, we next examined whether Sirt1 directly regulates Hoxa9 expression. Hoxa9 was previously shown to be repressed via PcG-mediated H3K27 trimethylation (Cao and Zhang, 2004). Notably, Sirt1 was found in complex with several PcG components in an interaction that is enhanced in response to stress (Kuzmichev et al., 2005; O’Hagan et al., 2011). To investigate if Sirt1 regulates Hoxa9 at the chromatin level, we performed chromatin immunoprecipitation (ChIP) for Sirt1 in in vitro–expanded hematopoietic progenitor cells and found Sirt1 enrichment at the Hoxa9 locus, which was most pronounced in the gene body (Fig. 7 A). Sirt1 deletion caused a significant increase in K16-acetylated histone H4 (H4K16-Ac), which is a modification associated with transcriptional activation and a known preferred enzymatic target for Sirt1 deacetylase activity (Fig. 7 B; Vaquero et al., 2004). H4K16-Ac levels were highest at the Hoxa9 promoter consistent with the robust effect of Sirt1 loss on Hoxa9 expression. Notably, a modest increase in H4K16 acetylation was also observed at two neighboring Hox-A cluster genes, Hoxa10 and Hoxa7. Although Hoxa10 showed variable up-regulation upon Sirt1 loss, Hoxa7 levels remained stable, suggesting an additional level of regulation downstream of Sirt1-mediated changes in chromatin accessibility. Importantly, no increase in H4AcK16 was observed at two distant Hox loci, Hoxb8 and Hoxc9, which further showed significantly reduced Sirt1 binding compared with Hoxa9, supporting a locus-specific effect of Sirt1-mediated Hox gene regulation (Fig. 7, B and D). Increased H4K16-Ac was previously found to be inversely correlated with PcG-associated repressive marks (O’Hagan et al., 2011), and consistent with this notion, H3K27 trimethylation was reduced in the absence of Sirt1. Moreover, we observed a strikingly similar pattern of enrichment for both H3K27-me3 and Sirt1 across the Hoxa9 gene (Fig. 7, B and C). Together, these data support a direct role for Sirt1 in the epigenetic regulation of the PcG target Hoxa9 and provide a molecular basis for Hoxa9 up-regulation and concomitant HSPC expansion in the absence of Sirt1.
Figure 7.
Sirt1 directly binds to the Hoxa9 locus and facilitates a repressive chromatin environment. ChIP analysis of 4F BM cultures. (A–C) Total cultures (B and D) or sorted c-Kit+Sca-1+ cells (A and C) were subjected to ChIP with the indicated antibodies. Enrichment over input is shown. Primer set locations are indicated, and values represent mean of triplicates. (D) Sirt1 ChIP at the indicated Hox loci. Enrichment over input is shown relative to the Hoxb8 locus. Error bars represent SD. *, P ≤ 0.05; **, P ≤ 0.01.
DISCUSSION
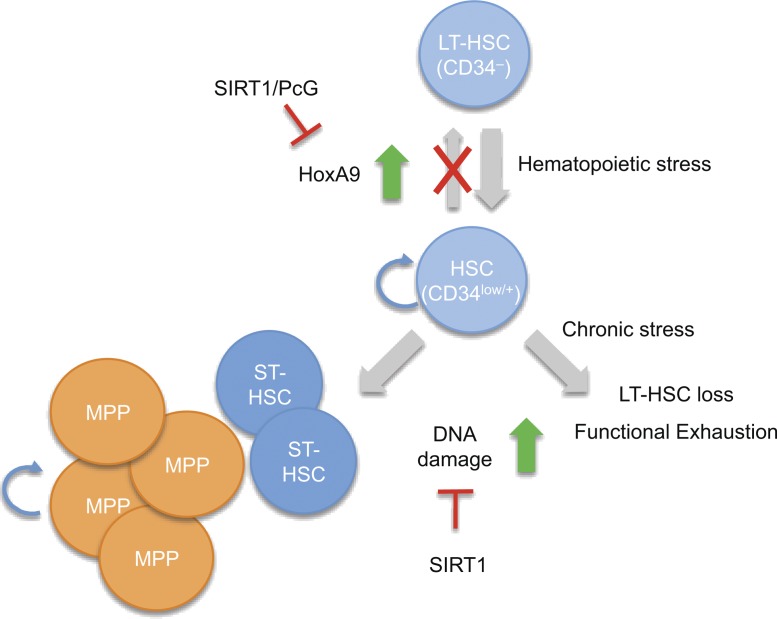
Here, we identify a dual role for Sirt1 in HSPC maintenance under conditions of hematopoietic stress. We find that Sirt1 negatively regulates the expression of a key developmental regulator of HSPC maintenance and differentiation, the PcG target gene Hoxa9. Consistent with the observation that Hoxa9 overexpression can promote HSPC expansion (Thorsteinsdottir et al., 2002), we show that Sirt1 loss promotes a stress-dependent increase in HSPC numbers and proliferation both in vivo and in vitro. Expansion of Sirt1-deficient HSPCs is further associated with the accumulation of DNA damage and genomic aberrations, which causes a loss of hematopoietic potential in response to serial transplantation or sublethal doses of irradiation. Together, these findings place Sirt1 at a central position in stress-associated HSPC homeostasis (see model in Fig. 8).
Figure 8.
Model for Sirt1 function in HSPC maintenance. Loss of Sirt1 causes an increase in Hoxa9 expression and expansion of HSPC subsets under conditions of hematopoietic stress, which results in increased DNA damage accumulation and exhaustion of Sirt1-deficient long-term progenitors over time.
Sirt1 was previously shown to interact with the PcG repressive complex PRC4, which is expressed specifically in stem and cancer cells (Kuzmichev et al., 2005). Notably, the formation of Sirt1-containing PcG complexes was enhanced in the presence of oxidative stress (O’Hagan et al., 2011); their physiological relevance, however, remained elusive. Our data now provide evidence that Sirt1 can directly modulate the expression of PcG target genes at the chromatin level, which may at least in part control maintenance of HSCs. In agreement with the limited expression pattern of PRC4, we found that HSPCs were more susceptible to the consequences of Sirt1 loss than more differentiated hematopoietic cell subsets (Figs. 1 F and 5 G). Fittingly, the effect of Sirt1 on Hox gene regulation was limited to the HSPC-associated Hox-A cluster genes Hoxa9 and Hoxa10, and increased H4K16 acetylation in the absence of Sirt1 appeared specific to the Hox-A cluster (Fig. 7 B). Notably, increased H4 acetylation was not sufficient to induce Hox-A gene expression, as indicated by the lack of Hoxa7 up-regulation despite a moderate increase in H4K16-Ac levels. Similar observations have been made for myc- or glucocorticoid receptor-associated H4 acetylation, which is necessary but not sufficient to induce transcription of target genes (Frank et al., 2001; Flavin et al., 2004). These findings suggest that loss of PcG-mediated silencing poises HSPC developmental genes for transcriptional deregulation in response to additional, environmental cues.
Consistent with this notion and in agreement with a previous study by Leko et al. (2012), we found that loss of Sirt1 by itself is not sufficient to promote HSPC expansion in vivo (Figs. 2 B and 3 B). Instead, we observed a significant increase in HSPCs specifically in response to hematopoietic stress. This observation is reminiscent of work by Chua et al. (2005) demonstrating that Sirt1-deficient MEFs have enhanced proliferative capacity under conditions of chronic stress. Notably, Sirt1 loss did not affect homeostasis of preadult hematopoietic progenitors, which are predisposed to proliferative expansion during HSPC development. Unlike preadult HSPCs, their adult counterparts rely on the maintenance of conservative self-renewal/quiescence, likely mediated by inherent differences in transcription factor requirements and gene expression profiles (Mikkola and Orkin, 2006; He et al., 2011). Interestingly, the PcG gene Bmi-1 is required specifically for adult hematopoiesis, consistent with the notion that the regulation for PcG target genes may differentially affect fetal and adult hematopoiesis (Park et al., 2003). We speculate that Sirt1-associated epigenetic changes may play a similar role to ensure controlled stress-associated HSPC expansion in the adult, a process which would likely be disadvantageous during developmental HSPC expansion. The finding that Sirt1 function in HSPCs is affected by both developmental state and hematopoietic stress may further help reconcile previously conflicting reports regarding the role of Sirt1 in HSPCs (Narala et al., 2008; Leko et al., 2012; Matsui et al., 2012; Peled et al., 2012).
In agreement with the known role for Sirt1 in genomic maintenance in ES and tumor cell lines, Sirt1 loss was further associated with the accumulation of DNA damage in HSPCs in vivo and in vitro (Fig. 5). Notably, this process was aggravated during HSPC expansion, reminiscent of DNA damage accumulation during HSPC expansion with age (Rossi et al., 2007). As a consequence, Sirt1-deficient HSPCs showed increased radiation sensitivity, p53 activation, and apoptosis and a decline in LT-HSC number and function after serial transplantation. These findings are in agreement with several recent studies demonstrating the detrimental impact of DNA damage accumulation on HSPC function. Mice with defects in various DNA repair pathways, as well as mouse models of telomere instability, show DNA damage accumulation, HSPC exhaustion, and concomitant BM failure (Rossi et al., 2007; Beier et al., 2012; Garaycoechea et al., 2012). Moreover, recent work by Wang et al. (2012) demonstrated that activation of DNA damage response pathways promotes HSPC differentiation and loss of self-renewal capacity. Increased sensitivity to DNA damage in the absence of Sirt1 appears to be independent of Hoxa9, as increased Hoxa9 levels were not sufficient to promote DNA damage in vitro (see SCF treatment, Fig. 5 B) and loss of Hoxa9 was previously shown to result in increased radiation-sensitivity (Lawrence et al., 2005). Together, these findings establish Sirt1 as a mediator of genomic stability in adult HSPCs, thereby extending previous findings in ES cells by us and others (Haigis and Sinclair, 2010; Palacios et al., 2010).
In addition to Hoxa9, Sirt1 was recently found to regulate several of its known enzymatic targets, including FoxOs, in HSPCs and HSPC-derived cell lines (Matsui et al., 2012; Yuan et al., 2012a). Interestingly, FoxO-deficient mice show increased LSK cycling and defects in long-term self-renewal in response to oxidative stress (Tothova et al., 2007), suggesting a synergistic role for FoxOs and Hoxa9 in Sirt1-regulated HSPC maintenance.
It is of note that HSPC homeostasis is not only a central aspect of normal hematopoiesis but is also closely linked to tumors of the hematopoietic lineage. Perturbed quiescence and aberrant expansion of HSPCs, as observed here in the absence of Sirt1, are key steps in malignant transformation of both acute and chronic myeloid leukemia (AML and CML), which has furthermore been linked to increased Hoxa9 expression (Argiropoulos and Humphries, 2007). In contrast, Sirt1 inhibition in chronic phase leukemia stem cells was found to enhance induction of apoptosis and blast crisis by promoting p53 acetylation (Li et al., 2012; Yuan et al., 2012a), consistent with the DNA damage-associated increase in p53 acetylation and apoptosis in Sirt1-deficient HSPCs observed here (Fig. 5 C). However, given that p53 expression, as well as its acetylation, were comparatively low in the absence of DNA damage, Sirt1 loss appears to promote stress-induced expansion rather than extensive cell death in nontransformed HSPCs in vivo. Together, these findings imply that Sirt1 may play a dual role in HSPC malignancies, where Sirt1 promotes survival in established CML tumors but protects primary HSPCs from aberrant proliferation and accumulation of mutagenic genomic defects. Our data, thus, add to the understanding of stress-induced changes in HSPC homeostasis with implications for HSPC maintenance with age as well as during malignant transformation.
MATERIALS AND METHODS
Mouse breeding and treatments.
Mice were housed under special pathogen-free conditions. Sirt1-E4fl/fl mice (Cheng et al., 2003) were backcrossed to C57BL/6 ERT2-Cre mice (Ruzankina et al., 2007) for at least six generations; all other strains were obtained on a C57BL/6 background. WT and Cre control mice were bred in house. To induce SIRT1 exon 4 deletion, 8–12-wk-old mice were injected i.p. with 1 mg 4-OHT, followed by administration of 4-OHT–supplemented chow for the indicated time periods (360 mg/kg tamoxifen citrate [Sigma-Aldrich] in AIN-93G purified rodent diet [Dyets Inc.]). PCR-based genotyping for exon 4 deletion was performed on peripheral blood lymphocytes isolated by retro-orbital bleeding after 2–6 wk of 4-OHT treatment. For tissue-specific deletion, Sirt1-E4fl/fl mice were crossed to vav-iCre transgenic C57BL/6 mice and deletion was confirmed by PCR as above. To determine radiation sensitivity, mice were subjected to sublethal doses of whole body irradiation (8 or 9 Gy) using a 137Cs irradiator. Animals were monitored daily for signs of morbidity and euthanized when moribund. Kaplan-Meier survival curves were generated and analyzed using JMP 5 software. For BM transfer experiments, recipient mice were subjected to lethal irradiation (10 Gy) before transfer. For 5-FU treatments, a single dose of 150 mg/kg was injected i.p. and mice were analyzed 2 wk thereafter. BrdU was administered by injecting a single dose of 100 mg/kg i.p., followed by exposure to 1 mg/ml BrdU-supplemented drinking water for 18 h.
BM transfers.
BM cells from Sirt1-E4fl/fl, ERT2-Cre mice and littermate controls were transplanted into lethally irradiated C/57BL6-Ly5.2 recipient mice (The Jackson Laboratory). For competitive transfer experiments, experimental CD45.2+ BM was mixed 1:1 with CD45.1+ recipient BM and 106 cells were injected by lateral tail vein injection. For noncompetitive transfers, 106 experimental BM cells were injected into CD45.1+ lethally irradiated recipients and mice were euthanized and analyzed at the indicated time points. For serial transplantation assays, 106 BM cells from pooled WT or SIRT1Δ/Δ donors were transferred into CD45.1+ congenic recipients, three recipients per group were used for primary transfers, four recipients per group for secondary transfers, and eight recipients for tertiary transfers. Survival was monitored daily over the indicated time period. Animals were euthanized when moribund.
BM culture.
BM cultures were performed as described previously (Suh et al., 2008). In brief, 2 × 105 cells/ml of total nucleated BM cells were cultured in IMDM with 10% serum in presence of the indicated growth factors, 100 ng/ml murine SCF (PeproTech), 50 ng/ml murine IL6 (Biosource), 100 ng/ml human TPO (PeproTech), and 100 ng/ml human FLT3L (PeproTech). Cells were passaged every 4–5 d and reseeded at 2 × 105 cells/well. For metaphase and microarray analysis, BM cells were isolated after 4–6 wk of 4-OHT administration in vivo and expanded in the presence of all four growth factors for two to three passages. To achieve acute ablation of Sirt1, exon 4 was deleted in vitro: 106 total BM cells were plated in 1 ml and treated with 0.5 µM 4-OHT for 48–60 h in serum-free media (StemSpan SFEM; STEMCELL Tech) in the presence of SCF and TPO (Fig. 3 A and Fig. 4 E). Cells were counted at every passage using a T4 cell counter (Nexcelom).
Immunophenotyping and cell sorting.
For immunostaining of BM, spleen, and thymus, single cell suspension was prepared and nucleated cells were isolated by Leukocyte Separation Media (MP Biomedical). For HSPCs stainings, lineage-positive cells were excluded using Ter119, CD4, CD8, Mac-1, Gr-1, B220, and IL-7R (BD). Abs used to identify hematopoietic cell subsets were: c-Kit, Sca-1, CD34, CD48, Flk2/FLT3, CD150, and CD45.2 (eBioscience). Apoptotic cells were stained using the Annexin V Apoptosis detection kit II (BD). To detect DNA damage, cells were stained for cell surface markers, fixed and permeabilized using Cytofix/Cytoperm solution (BD), followed by intracellular staining with α-γ-H2AX-FITC Ab (Cell Signaling Technology). To detect BrdU incorporation, BM cells were isolated and analyzed using the FITC BrdU Flow kit according to the manufacturer’s instructions (BD). FACS acquisition was performed on an LSRII flow cytometer (BD) and cell sorting on FACSAria (BD). FACS data were analyzed using FlowJo software (Tree Star).
ChIP analysis.
ChIP analysis was performed as previously described (Shukla et al., 2011). In brief, BM cells were subjected to Sirt1 exon 4 deletion in vitro and subsequent culture for one to two passages with four growth factors as described above. 5–10 million FACS-sorted c-Kit+Sca-1+ cells or unsorted cells from in vitro BM cultures were cross-linked for 10 min in 1% formaldehyde (Sigma-Aldrich) at room temperature. Nuclei were isolated and resuspended in sonication buffer (10 mM Tri-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% EGTA, 0.1% Na-deoxycholate, 0.5% N-lauryl sarcosine, and protease inhibitors). Sonication of DNA was performed using Bioruptor (Diagenode). Chromatin was immunoprecipitated after addition of 1% Triton X-100 using the following Abs: α-SIRT1, α-H3K27me3 (Abcam), α-H4K16Ac (Millipore), normal rabbit IgG (Cell Signaling Technology), and normal mouse IgG (Millipore). Purified ChIP DNA was analyzed for enrichment at the Hoxa9 locus using primers described previously (Table S2; Okada et al., 2005)).
Western blotting.
4F BM cultures were subjected to whole cell lysis in NETN buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Igepal CA-630, and 5% Glycerol) in the presence of 1× complete mini protease inhibitor mix (Roche), 5 mM Nicotinamide (Sigma-Aldrich), and 1 µM TSA (Sigma-Aldrich). Immunoblots were performed according to standard techniques. The following antibodies were used for immunoblotting: α-mouse Ac-p53(K379), α-mouse p53 (Cell Signaling Technology), and α-GAPDH (Abcam).
PCR and RT-PCR.
In vitro and in vivo Sirt1 deletion was checked in tail, blood, and BM cells by PCR using a set of two primers simultaneously detecting WT, loxP-flanked and deleted exon 4 (see Table S2). Mice with >80% deletion were used for experiments. For gene expression analysis, total RNA was extracted from cultured total BM cells and sorted LSK cells by RNeasy Mini kit (QIAGEN). Eluted RNA samples were reverse transcribed by ThermoScript RT-PCR system (Invitrogen) and expression of various genes was analyzed by RT-PCR using the LightCycler 480 II (Roche) and primers listed in Table S2.
Affymetrix microarray analysis.
Total RNA was isolated from sorted LSKs after in vitro culture in the presence of IL6, SCF, TPO, and FLT3L. RNA was isolated using RNeasy Micro kit (QIAGEN). RNA with a RIN quality score >9 was amplified and labeled using Ovation Whole Blood Solution kit (Nugen). 4.4 µg of labeled cDNA per sample was hybridized to Affymetrix mouse 430 2.0 GeneChip array at 45°C for 16 h. GeneChip arrays were washed and scanned on the GeneChip scanner 3000 (Affymetrix). Data were collected using Affymetrix AGCC software.
Metaphase aberration assay.
Metaphase spreads were prepared and analyzed as described previously (Biswas et al., 2011). In brief, BM cells cultured in the presence of four factors were treated with 10 µg/ml Colcemid (Invitrogen) for 90 min before harvest. Cells were collected and treated with hypotonic solution (KCl 0.075 M) for 15 min at 37°C and fixed with methanol: acetic acid (3:1). Chromosomes were stained with DAPI. Analyses were performed using an Axioplan 2 (Carl Zeiss) microscope. DAPI images of metaphase spreads were scored for the following DSB-related defects: fusions, gaps, broken chromosome arms, fragments, dicentric chromosomes, centromere fucions/robertsonian translocations, and radials. Same scoring standard were applied for all samples, metaphases were scored blindly, and 50 metaphases were analyzed per sample. The sum of all aberrations was divided by the number of metaphases to determine the mean frequency of aberrant metaphases.
Online supplemental material.
Fig. S1 shows FACS staining strategies for BM subset analyses. Fig. S2 shows gating strategy for LSK subset analysis in Fig. 3 based on CD150 versus Flk2 staining. Fig. S3 shows representative FACS graphs for g-H2AX and c-Kit in gated LSK and Lin2Sca-12c-Kit2 BM cells from mice shown in Fig. 5 G. Table S1 shows gene expression changes in Sirt1-deficient hematopoietic progenitor cell cultures. Only changes greater than twofold compared with Cre controls are listed. Table S2 shows PCR primer sequences.
Supplementary Material
Acknowledgments
We would like to thank T. Misteli, A. Nussenzweig, S. Oberdoerffer, D. Sinclair, and L. Tessarollo for helpful comments and/or critical reading of the manuscript, D. Swing and R. Koogle for animal support, X. Wu for microarray analysis, and R. Matthai and K. Noer for cell sorting. Animal work was approved by the NCI-Frederick Animal Care and Use Committee (ASP 09-066).
This work was supported by federal funds from the National Cancer Institute, The National Institutes of Health, The National Institutes of Health intramural research program, and under contract HHSN26120080001E.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- 4-OHT
- 4-hydroxy-tamoxifen
- 5-FU
- 5-Fluorouracil
- ES
- embryonic stem
- HSPC
- hematopoietic stem and progenitor cell
- LT-HSC
- long-term hematopoietic stem cell
- PcG
- Polycomb group
- SCF
- stem cell factor
- TPO
- thrombopoietin
References
- Aggarwal R., Lu J., Pompili V.J., Das H. 2012. Hematopoietic stem cells: transcriptional regulation, ex vivo expansion and clinical application. Curr. Mol. Med. 12:34–49 10.2174/156652412798376125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiropoulos B., Humphries R.K. 2007. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 26:6766–6776 10.1038/sj.onc.1210760 [DOI] [PubMed] [Google Scholar]
- Beerman I., Bhattacharya D., Zandi S., Sigvardsson M., Weissman I.L., Bryder D., Rossi D.J. 2010. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl. Acad. Sci. USA. 107:5465–5470 10.1073/pnas.1000834107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier F., Foronda M., Martinez P., Blasco M.A. 2012. Conditional TRF1 knockout in the hematopoietic compartment leads to bone marrow failure and recapitulates clinical features of dyskeratosis congenita. Blood. 120:2990–3000 10.1182/blood-2012-03-418038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K., Das R., Alter B.P., Kuznetsov S.G., Stauffer S., North S.L., Burkett S., Brody L.C., Meyer S., Byrd R.A., Sharan S.K. 2011. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood. 118:2430–2442 10.1182/blood-2010-12-324541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V., Lara E., Suárez-Alvarez B., Abu Dawud R., Vázquez-Chantada M., Martínez-Chantar M.L., Embade N., López-Nieva P., Horrillo A., Hmadcha A., et al. 2010. Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc. Natl. Acad. Sci. USA. 107:13736–13741 10.1073/pnas.1001399107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Zhang Y. 2004. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell. 15:57–67 10.1016/j.molcel.2004.06.020 [DOI] [PubMed] [Google Scholar]
- Chalkiadaki A., Guarente L. 2012. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol. 8:287–296 10.1038/nrendo.2011.225 [DOI] [PubMed] [Google Scholar]
- Cheng H.L., Mostoslavsky R., Saito S., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F. 2003. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA. 100:10794–10799 10.1073/pnas.1934713100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.F., Mostoslavsky R., Lombard D.B., Pang W.W., Saito S., Franco S., Kaushal D., Cheng H.L., Fischer M.R., Stokes N., et al. 2005. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2:67–76 10.1016/j.cmet.2005.06.007 [DOI] [PubMed] [Google Scholar]
- de Boer J., Williams A., Skavdis G., Harker N., Coles M., Tolaini M., Norton T., Williams K., Roderick K., Potocnik A.J., Kioussis D. 2003. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33:314–325 10.1002/immu.200310005 [DOI] [PubMed] [Google Scholar]
- Dykstra B., Olthof S., Schreuder J., Ritsema M., de Haan G. 2011. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 208:2691–2703 10.1084/jem.20111490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavin M., Cappabianca L., Kress C., Thomassin H., Grange T. 2004. Nature of the accessible chromatin at a glucocorticoid-responsive enhancer. Mol. Cell. Biol. 24:7891–7901 10.1128/MCB.24.18.7891-7901.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.R., Schroeder M., Fernandez P., Taubert S., Amati B. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069–2082 10.1101/gad.906601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea J.I., Crossan G.P., Langevin F., Daly M., Arends M.J., Patel K.J. 2012. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 489:571–575 10.1038/nature11368 [DOI] [PubMed] [Google Scholar]
- Geiger H., Rudolph K.L. 2009. Aging in the lympho-hematopoietic stem cell compartment. Trends Immunol. 30:360–365 10.1016/j.it.2009.03.010 [DOI] [PubMed] [Google Scholar]
- Haigis M.C., Sinclair D.A. 2010. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5:253–295 10.1146/annurev.pathol.4.110807.092250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.K., Song E.K., Guo Y., Ou X., Mantel C., Broxmeyer H.E. 2008. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2:241–251 10.1016/j.stem.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D.E., Lerner C.P. 1991. Most primitive hematopoietic stem cells are stimulated to cycle rapidly after treatment with 5-fluorouracil. Blood. 78:1237–1240 [PubMed] [Google Scholar]
- He S., Kim I., Lim M.S., Morrison S.J. 2011. Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes Dev. 25:1613–1627 10.1101/gad.2052911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Tajima F., Ogawa M. 2000. Developmental changes of CD34 expression by murine hematopoietic stem cells. Exp. Hematol. 28:1269–1273 10.1016/S0301-472X(00)00535-X [DOI] [PubMed] [Google Scholar]
- Klar A.J., Fogel S., Macleod K. 1979. MAR1-a Regulator of the HMa and HMalpha Loci in SACCHAROMYCES CEREVISIAE. Genetics. 93:37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Margueron R., Vaquero A., Preissner T.S., Scher M., Kirmizis A., Ouyang X., Brockdorff N., Abate-Shen C., Farnham P., Reinberg D. 2005. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc. Natl. Acad. Sci. USA. 102:1859–1864 10.1073/pnas.0409875102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence H.J., Christensen J., Fong S., Hu Y.L., Weissman I., Sauvageau G., Humphries R.K., Largman C. 2005. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 106:3988–3994 10.1182/blood-2005-05-2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leko V., Varnum-Finney B., Li H., Gu Y., Flowers D., Nourigat C., Bernstein I.D., Bedalov A. 2012. SIRT1 is dispensable for function of hematopoietic stem cells in adult mice. Blood. 119:1856–1860 10.1182/blood-2011-09-377077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang L., Li L., Wang Z., Ho Y., McDonald T., Holyoake T.L., Chen W., Bhatia R. 2012. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 21:266–281 10.1016/j.ccr.2011.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Li L., Lv X., Wu X.S., Liu D.P., Liang C.C. 2011. Modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment and activities on the chromatin. Cell Res. 21:1182–1195 10.1038/cr.2011.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. 2001. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 107:137–148 10.1016/S0092-8674(01)00524-4 [DOI] [PubMed] [Google Scholar]
- Matsui K., Ezoe S., Oritani K., Shibata M., Tokunaga M., Fujita N., Tanimura A., Sudo T., Tanaka H., McBurney M.W., et al. 2012. NAD-dependent histone deacetylase, SIRT1, plays essential roles in the maintenance of hematopoietic stem cells. Biochem. Biophys. Res. Commun. 418:811–817 10.1016/j.bbrc.2012.01.109 [DOI] [PubMed] [Google Scholar]
- McBurney M.W., Yang X., Jardine K., Hixon M., Boekelheide K., Webb J.R., Lansdorp P.M., Lemieux M. 2003. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 23:38–54 10.1128/MCB.23.1.38-54.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola H.K., Orkin S.H. 2006. The journey of developing hematopoietic stem cells. Development. 133:3733–3744 10.1242/dev.02568 [DOI] [PubMed] [Google Scholar]
- Narala S.R., Allsopp R.C., Wells T.B., Zhang G., Prasad P., Coussens M.J., Rossi D.J., Weissman I.L., Vaziri H. 2008. SIRT1 acts as a nutrient-sensitive growth suppressor and its loss is associated with increased AMPK and telomerase activity. Mol. Biol. Cell. 19:1210–1219 10.1091/mbc.E07-09-0965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L.J. 2008. DNA repair is crucial for maintaining hematopoietic stem cell function. DNA Repair (Amst.). 7:523–529 10.1016/j.dnarep.2007.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan H.M., Wang W., Sen S., Destefano Shields C., Lee S.S., Zhang Y.W., Clements E.G., Cai Y., Van Neste L., Easwaran H., et al. 2011. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 20:606–619 10.1016/j.ccr.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P., Michan S., McVay M., Mostoslavsky R., Vann J., Park S.K., Hartlerode A., Stegmuller J., Hafner A., Loerch P., et al. 2008. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 135:907–918 10.1016/j.cell.2008.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V.M., Su L., Xu G., Zhang Y. 2005. hDOT1L links histone methylation to leukemogenesis. Cell. 121:167–178 10.1016/j.cell.2005.02.020 [DOI] [PubMed] [Google Scholar]
- Ou X., Chae H.D., Wang R.H., Shelley W.C., Cooper S., Taylor T., Kim Y.J., Deng C.X., Yoder M.C., Broxmeyer H.E. 2011. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood. 117:440–450 10.1182/blood-2010-03-273011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios J.A., Herranz D., De Bonis M.L., Velasco S., Serrano M., Blasco M.A. 2010. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J. Cell Biol. 191:1299–1313 10.1083/jcb.201005160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 423:302–305 10.1038/nature01587 [DOI] [PubMed] [Google Scholar]
- Passegué E., Wagers A.J., Giuriato S., Anderson W.C., Weissman I.L. 2005. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 202:1599–1611 10.1084/jem.20050967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled T., Shoham H., Aschengrau D., Yackoubov D., Frei G., Rosenheimer G N., Lerrer B., Cohen H.Y., Nagler A., Fibach E., Peled A. 2012. Nicotinamide, a SIRT1 inhibitor, inhibits differentiation and facilitates expansion of hematopoietic progenitor cells with enhanced bone marrow homing and engraftment. Exp. Hematol. 40:342–355: e1 10.1016/j.exphem.2011.12.005 [DOI] [PubMed] [Google Scholar]
- Rine J., Strathern J.N., Hicks J.B., Herskowitz I. 1979. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics. 93:877–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D.J., Bryder D., Zahn J.M., Ahlenius H., Sonu R., Wagers A.J., Weissman I.L. 2005. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. USA. 102:9194–9199 10.1073/pnas.0503280102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D.J., Bryder D., Seita J., Nussenzweig A., Hoeijmakers J., Weissman I.L. 2007. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 447:725–729 10.1038/nature05862 [DOI] [PubMed] [Google Scholar]
- Ruzankina Y., Pinzon-Guzman C., Asare A., Ong T., Pontano L., Cotsarelis G., Zediak V.P., Velez M., Bhandoola A., Brown E.J. 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 1:113–126 10.1016/j.stem.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau G., Lansdorp P.M., Eaves C.J., Hogge D.E., Dragowska W.H., Reid D.S., Largman C., Lawrence H.J., Humphries R.K. 1994. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. USA. 91:12223–12227 10.1073/pnas.91.25.12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S. 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 479:74–79 10.1038/nature10442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H.C., Leeanansaksiri W., Ji M., Klarmann K.D., Renn K., Gooya J., Smith D., McNiece I., Lugthart S., Valk P.J., et al. 2008. Id1 immortalizes hematopoietic progenitors in vitro and promotes a myeloproliferative disease in vivo. Oncogene. 27:5612–5623 10.1038/onc.2008.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir U., Mamo A., Kroon E., Jerome L., Bijl J., Lawrence H.J., Humphries K., Sauvageau G. 2002. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 99:121–129 10.1182/blood.V99.1.121 [DOI] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., et al. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 128:325–339 10.1016/j.cell.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 16:93–105 10.1016/j.molcel.2004.08.031 [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 107:149–159 10.1016/S0092-8674(01)00527-X [DOI] [PubMed] [Google Scholar]
- Vijayalaxmi K.K., Rai S.P. 1996. Studies on the genotoxicity of tamoxifen citrate in mouse bone marrow cells. Mutat. Res. 368:109–114 10.1016/0165-1218(95)00101-8 [DOI] [PubMed] [Google Scholar]
- Wang J., Sun Q., Morita Y., Jiang H., Gross A., Lechel A., Hildner K., Guachalla L.M., Gompf A., Hartmann D., et al. 2012. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 148:1001–1014 10.1016/j.cell.2012.01.040 [DOI] [PubMed] [Google Scholar]
- Wang R.H., Sengupta K., Li C., Kim H.S., Cao L., Xiao C., Kim S., Xu X., Zheng Y., Chilton B., et al. 2008. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 14:312–323 10.1016/j.ccr.2008.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yuan H., Roth M., Stark J.M., Bhatia R., Chen W.Y. 2013. SIRT1 deacetylase promotes acquisition of genetic mutations for drug resistance in CML cells. Oncogene. 32:589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Trumpp A. 2009. Balancing dormant and self-renewing hematopoietic stem cells. Curr. Opin. Genet. Dev. 19:461–468 10.1016/j.gde.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Yuan H., Wang Z., Li L., Zhang H., Modi H., Horne D., Bhatia R., Chen W. 2012a. Activation of stress response gene SIRT1 by BCR-ABL promotes leukemogenesis. Blood. 119:1904–1914 10.1182/blood-2011-06-361691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Luo K., Liu T., Lou Z. 2012b. Regulation of SIRT1 activity by genotoxic stress. Genes Dev. 26:791–796 10.1101/gad.188482.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.C., Lodish H.F. 2008. Cytokines regulating hematopoietic stem cell function. Curr. Opin. Hematol. 15:307–311 10.1097/MOH.0b013e3283007db5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.