Mutations in the transcription factor NF-E2 in patients with myeloproliferative neoplasms result in a truncated protein that enhances the function of wild-type NF-E2 and causes erythrocytosis and throbocytosis in a mouse model.
Abstract
The molecular etiology of myeloproliferative neoplasms (MPNs) remains incompletely understood, despite recent advances incurred through the discovery of several different mutations in MPN patients. We have recently described overexpression of the transcription factor NF-E2 in MPN patients and shown that elevated NF-E2 levels in vivo cause an MPN phenotype and predispose to leukemic transformation in transgenic mice. We report the presence of acquired insertion and deletion mutations in the NF-E2 gene in MPN patients. These result in truncated NF-E2 proteins that enhance wild-type (WT) NF-E2 function and cause erythrocytosis and thrombocytosis in a murine model. NF-E2 mutant cells acquire a proliferative advantage, witnessed by clonal dominance over WT NF-E2 cells in MPN patients. Our data underscore the role of increased NF-E2 activity in the pathophysiology of MPNs.
Myeloproliferative neoplasms (MPNs) constitute a group of clonal, malignant hematopoietic disorders, which includes polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). Patients present clinically with erythrocytosis and/or thrombocytosis (PV and ET, respectively) or with pan-cytopenia (PMF) and display a propensity to transform to acute leukemia. Despite the discovery of a variety of mutations in MPN patients (Vainchenker et al., 2011), most notably an activating point mutation in the JAK2 kinase (JAK2V617F; Baxter et al., 2005; James et al., 2005; Kralovics et al., 2005), the pathophysiology of these disorders remains incompletely understood. Consequently, at present, no curative treatment exists besides bone marrow transplantation, which in this patient population is associated with considerable morbidity and mortality.
We have shown that expression of the hematopoietic transcription factor nuclear factor erythroid-2 (NF-E2) is elevated in MPN patients irrespective of the presence or absence of the JAK2V617F mutation (Goerttler et al., 2005; Wang et al., 2010). Based on this observation, we have generated a novel murine model for MPN by overexpressing NF-E2 in transgenic mice (Kaufmann et al., 2012). NF-E2–overexpressing mice display many MPN features including thrombocytosis, leukocytosis, Epo-independent colony formation, characteristic bone marrow histology, expansion of stem and progenitor compartments, and spontaneous transformation to acute myeloid leukemia (AML; Kaufmann et al., 2012). During AML transformation, NF-E2 transgenic mice acquire recurrent genetic alterations, particularly a gain of genetic material corresponding to human chromosome 8, which is also frequently observed in MPN patients and is associated with a higher rate of transformation to acute leukemia (Hussein et al., 2009). These data have established a role for aberrant NF-E2 expression in the pathophysiology of MPN. Here we test the hypothesis that alterations in the NF-E2 gene may contribute to the development of MPN.
RESULTS
To further investigate the role of NF-E2 in the pathophysiology of MPN, we sequenced the NF-E2 gene in a cohort of MPN patients (144 PV, 120 ET and 192 MF patients), as well as in patients with MDS (n = 57), CMML (n = 67), secondary, post-MPN AML (n = 39), and healthy controls (n = 65; Table S1). Seven different insertions and deletions leading to frameshift mutations were detected in the NF-E2 coding sequence in eight patients with MPNs, three with PV and five with MF, either PMF or secondary post-MPN MF (Table 1 and Fig. S1). Two mutations, c.782-785delAGAG and c.662_663insG, were found in two different patients each, and one patient harbored two separate mutations (Table 1). The frameshifts introduce premature stop codons in the open reading frame, leading to truncations in the NF-E2 protein (Fig. 1 A). One 12 bp deletion, c.889-900del, causes an in-frame deletion of four amino acids within the leucine zipper heterodimerization domain, causing the two proximal leucine to lose the seven amino acid spacing typical of a leucine zipper (O’Shea et al., 1989).
Table 1.
NF-E2 Mutations detected in MPN patients
| UPN | Diagnosis | Variant cDNA NM_006163.1 | Variant protein | Genetics | JAK2V617F | MplW515L | ASXL1 (exon 12) | TET2 | EZH2 |
| 202 | PV | c.782-785delAGAG | p.E261AfsX3 | Acquired | + | − | − | - | − |
| 209 | PV | c.782-785delAGAG | p.E261AfsX3 | Acquired | + | − | − | c.3818G>A p.C1273Y | − |
| 241 | Post PV-MF | c.732_733insG | p.L245VfsX5 | + | − | − | c.285-309del p.K95NfsX10 | − | |
| 409 | PV | c.236delCc.234delT | p.P79LfsX32p.P79LfsX32 | Acquired | + | − | − | c.1276-1277delCT p.L426EfsX16 | − |
| 442 | Post PV-MF | c.662_663insG | p.E221GfsX7 | + | − | − | - | − | |
| 532 | PMF | c.889-900del | p.E297-R300del | Acquired | + | − | − | - | − |
| 980 | Post ET-MF | c.780_781insA | p.E261RfsX44 | Acquired | + | − | − | - | − |
| 2,836 | Post ET-MF | c.662_663insG | p.E221GfsX7 | n.d. | − | − | - | − |
Figure 1.
NF-E2 mutations in MPN patients cause truncations and loss of DNA binding. (A) Schematic representation of the NF-E2 protein (top) and the truncations resulting from mutations detected in MPN patients (bottom). Stippled bars indicate the changed amino acid sequence resulting from the frameshift mutations in p.E261RfsX44 and p.P79LfsX32. A gap marks the four amino acids deleted in p.E297-R300del. (B and C) EMSA of WT NF-E2 and NF-E2 mutants. (B) Nuclear extracts of HEK-293 cells transduced with expression vectors encoding WT NF-E2 (lane 2), MafG (lane 3), or both (lanes 4–8) were incubated with a 32P-labeled oligonucleotide containing an NF-E2 binding site (Mignotte et al, 1989). In lanes 5 and 6, a 100× excess of a nonradioactive oligonucleotide, either consensus NF-E2 (lane 5) or a negative control, NF-κB (lane 6), was added. Alternatively, an antibody to NF-E2 (lane 7) or a control NF-κB antibody (lane 8) was added. (C) Nuclear extracts of HEK-293 cells transduced with expression vectors encoding either WT NF-E2 (lane 1) or the indicated NF-E2 mutants (lanes 2–4) together with MafG. (B and C) A filled arrowhead indicates the specific NF-E2–DNA complex. The open circle shows nonspecific binding to the DNA probe. Variable binding of nonspecific bands is frequently observed in EMSAs (Mueller and Pahl, 2000). (D) Protein expression in the cell extracts used for EMSA in B and C. Cell lysates were subjected to SDS-PAGE and interrogated for NF-E2 (top) and β-actin (bottom) expression by Western blotting. A filled arrowhead demarks full-length WT NF-E2, whereas an open arrowhead points to the smaller, truncated NF-E2 proteins. B–D is representative of three independent experiments each. (E) Protein extracts of a healthy control (HC), as well as two patients carrying NF-E2 truncation mutations (UPN 202: 262 aa truncation; and UPN 241: 248 aa truncation, see Table 1) were interrogated for NF-E2 (top) and β-actin (bottom) expression by Western blotting. The outer right lane serves as a positive control and depicts lentivirally transduced CB3 cells expressing both WT and the 262aa truncated NF-E2. A filled arrowhead demarks full-length WT NF-E2, an open arrowhead points to the smaller, truncated NF-E2 proteins, and an asterisk demarks a nonspecific band. A representative blot of three independent experiments is shown.
Insertion and deletion mutations in NF-E2 were detected exclusively in PV and MF patients (3 of 144 patients, 2.1%; and 5 of 192 patients, 2.6%, respectively). These mutations were not observed in patients with ET, MPN-U, MDS, secondary post-MPN AML and CMML, or in healthy controls. Constitutive DNA, obtained from buccal swabs or T cells, was available from five patients with insertions or deletions, and in all cases, we were able to demonstrate that the mutations were acquired (Table 1).
Because the truncated NF-E2 proteins contain neither the DNA binding domain nor the leucine zipper required for dimerization to small Maf proteins or, in one case, contain a deletion in the leucine zipper, we investigated whether NF-E2 mutants retain DNA binding activity in an electrophoretic mobility shift assay (EMSA). As previously demonstrated (Igarashi et al., 1994), NF-E2 requires interaction with a small Maf protein, here MafG, to bind DNA (Fig. 1 B, compare lane 2 and lane 4). Specificity of the NF-E2/MafG heterodimer binding to its cognate DNA sequence was verified by competition and super-shift experiments (Fig. 1 B, lanes 5–8).
In contrast to WT NF-E2, two truncated NF-E2 mutants (p.L245VfsX5, here called 248aa, and p.E261AfsX3, here called 262aa) were unable to bind DNA even in the presence of MafG (Fig. 1 C, compare lanes 1 and 2–3). Furthermore, the NF-E2 mutant carrying the 4 aa deletion, here called Δ297-300, was likewise unable to bind DNA (Fig. 1 C, compare lanes 1 and 4). Protein expression of all mutants was verified by Western blotting (Fig. 1 D). Importantly, analysis of protein extracts from primary cells of MPN patients also demonstrates expression of the truncated, mutant protein in MPN patient cells (Fig. 1 E).
Subsequently, we investigated whether the NF-E2 mutants retained transactivation potential in reporter gene assays. Two different reporter gene constructs were used, one containing a known NF-E2 binding site from the β-globin promoter (Blank et al., 1997), and a second containing an NF-E2 binding site in the β1-tubulin promoter, recently characterized by us (unpublished data). Both reporter constructs yielded similar results (Figs. 2, A and B); all three NF-E2 mutants tested were no longer able to evoke reporter gene activity above the background levels observed with MafG alone.
Figure 2.
Transactivating activity of mutant NF-E2 proteins and their effect on WT NF-E2 activity. Plasmids encoding a β-globin promoter-luciferase construct (A; Igarashi et al., 1994) or a reporter construct encoding 5.2 kb of the β1-tubulin promoter coupled to the luciferase reporter gene (B) were cotransfected into HEK-293 cells either with expression vectors for MafG (white bars), WT NF-E2 (black bars), or various NF-E2 mutants (gray bars) as indicated. Luciferase activity was measured 16 h after transfection and normalized for transfection efficiency by determination of Renilla luciferase activity from a cotransfected vector. Activity of MafG alone was set at 1 and fold activity relative to this control is depicted. Bar graphs represent the mean ± SEM of four independent experiments, each performed in duplicate. Statistical analysis was done by one-way ANOVA with Bonferroni’s post-hoc multiple comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) CB3 cells, which lack endogenous NF-E2, were transfected with WT NF-E2 or various NF-E2 mutants as indicated. 72 h after transfection, RNA was harvested and assayed for β-globin and β-2-microglobulin housekeeping gene expression by qRT-PCR. Results represent the mean ± SEM of four independent experiments and are reported as relative expression levels setting β-globin expression in WT NF-E2 transfected CB3 cells at 100. Data were analyzed for statistical significance by one-way ANOVA with Bonferroni’s post-hoc multiple comparison test. ***, P < 0.001. (D) Experimental design. CB3 cells were transduced with pLeGO-iG-NF-E2, sorted for GFP expression, and a single clone (CB3-NF-E2wt), which displays WT NF-E2 expression, was selected. Subsequently, CB3-NF-E2wt cells were transduced with either an empty pLeGO-iT2 vector or with pLeGO-iT2 vectors encoding the indicated NF-E2 mutants. (E) Double-positive cells were FACS sorted and assayed for β-globin and β-2-microglobulin housekeeping gene expression by qRT-PCR. Results represent the mean ± SEM of four independent experiments and are reported as relative expression levels setting β-globin expression in empty pLeGO-iT2 transduced CB3-NF-E2wt cells at 1. Data were analyzed for statistical significance by one-way ANOVA with Bonferroni’s post-hoc multiple comparison test. **, P < 0.01. (F) Peripheral blood of transplanted mice expressing the indicated NF-E2 mutants and of control mice was FACS sorted for CD71, Ter-119 double-positive cells. RNA from sorted cells was assayed for β-globin and β-2-microglobulin housekeeping gene expression by qRT-PCR. Results represent the mean ± SEM of four to five animals in each group and are reported as relative expression levels setting β-globin expression in a mouse transduced with empty pLeGO-iG at 1. Data were analyzed for statistical significance by Student’s t tests. *, P < 0.05; **, P < 0.01.
As transcription off a plasmid DNA does not reflect physiological conditions within the chromatin-bound DNA, we used CB3 cells to test the mutants under physiological conditions. CB3 cells carry a homozygous viral insertion in the NF-E2 locus and therefore express neither NF-E2 nor its target β-globin (Ben-David et al., 1992). Upon reintroduction of NF-E2, β-globin is robustly expressed (Li et al., 2001). We introduced WT NF-E2 or the three NF-E2 mutants tested above into CB3 cells and measured β-globin mRNA expression by qRT-PCR. Although transduction of WT NF-E2 caused a 100-fold increase in β-globin mRNA expression, all three NF-E2 mutants were unable to activate β-globin expression (Fig. 2 C).
Because all NF-E2 mutations were observed in a heterozygous state, hence unfolding their effect in the presence of WT NF-E2, we investigated the effect of coexpression of WT NF-E2 and NF-E2 mutants in CB3 cells. CB3 cells stably transfected with WT NF-E2 were infected with an empty pLeGO-iT2 virus (Weber et al., 2008), or viruses expressing the two truncation mutants, 248aa and 262aa, or the 4aa deletion Δ297-300, and assayed for β-globin expression (Fig. 2, D and E). Although transduction with empty pLeGO-iT2 had no effect on β-globin mRNA expression in WT NF-E2–expressing CB3 cells (Fig. 2 E), transduction with the three mutants, which on their own retain no transactivation activity (Fig. 2 C), enhances WT NF-E2 activity between three- and ninefold, with the Δ297-300 mutant displaying the strongest effect (Fig. 2 E). Thus, although unable to transactivate transcription on their own, the NF-E2 mutants significantly enhance WT NF-E2 activity.
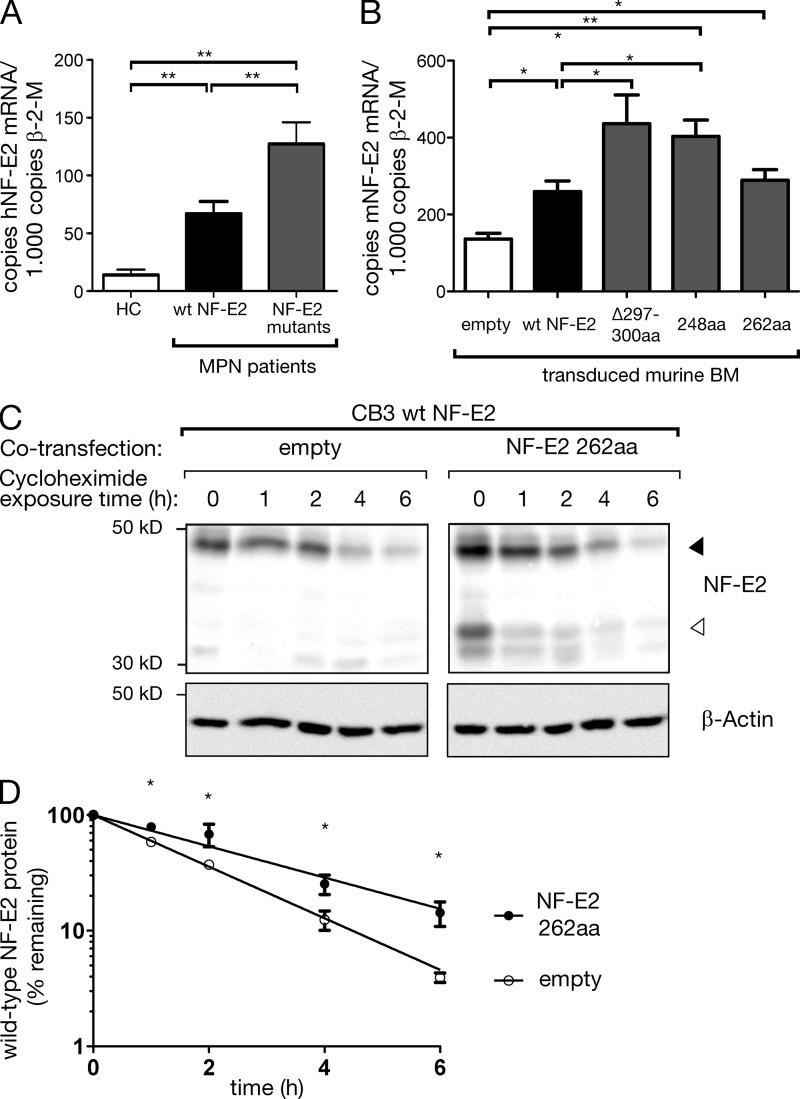
We investigated the molecular mechanism by which mutant NF-E2 enhances the activity of WT NF-E2. NF-E2 mRNA levels were determined by qRT-PCR in five patients carrying NF-E2 mutations, seven MPN patients with WT NF-E2, and three healthy controls (Fig. 3 A). As we have previously published, NF-E2 is overexpressed 2–10-fold in MPN patients expressing WT NF-E2 (Fig. 3 A; Goerttler et al., 2005). However, expression of NF-E2 mRNA is further significantly augmented in patients carrying NF-E2 mutations, reaching a median 2.5-fold of that observed in WT NF-E2 patients (Fig. 3 A). The presence of mutant NF-E2 therefore significantly increases NF-E2 mRNA levels.
Figure 3.
Effect of mutant NF-E2 on WT NF-E2 mRNA levels and on protein stability. (A) RNA was isolated from purified granulocytes of five patients carrying NF-E2 mutations, seven MPN patients with WT NF-E2, and three healthy controls as indicated and subjected to quantitative RT-PCR analysis for human NF-E2 and β-2-microglobulin housekeeping gene expression. A standard curve with known copy numbers of NF-E2 was included on each plate. Sample copy numbers of NF-E2 and β-2-microglobulin were determined from the standard curve and are expressed as relative ratios (copy number NF-E2 per 103 β-2-microglobulin molecules). Mean and SEM are shown. *, P < 0.05; **, P < 0.01 (B) Murine bone marrow of mice expressing the indicated NF-E2 mutants and of control mice was assayed for murine NF-E2 and β-2-microglobulin housekeeping gene expression by qRT-PCR. Results represent the mean ± SEM of four to five animals in each group (n = 2 empty) and are reported as relative ratios (copy number NF-E2 per 103 β-2-microglobulin molecules). Data were analyzed for statistical significance by Student’s t tests. *, P < 0.05; **, P < 0.01. (C and D) CB3 cells expressing WT NF-E2 were cotransfected either with a virus expressing the 262aa NF-E2 truncation mutant or with an empty control virus. Cells were treated with 20 µg/ml cycloheximide to inhibit de novo protein biosynthesis and cell extracts assayed for NF-E2 protein levels by Western blotting for up to 6 h, as indicated. The blot is representative of n = 4 independent experiments. (D) A nonlinear best-fit regression model was used to calculate the half-life of WT NF-E2 protein in the presence or absence of the 262aa truncation mutant. Graphs depict mean and SEM of n = 4 independent experiments. *, P < 0.05.
The qRT-PCR assay used does not distinguish between RNA encoding WT or mutant, truncated NF-E2. We therefore investigated NF-E2 protein levels in primary cells from MPN patients carrying NF-E2 mutations. Corresponding to the observed increase in NF-E2 mRNA, we detected elevated NF-E2 protein levels in patients expressing NF-E2 mutants (Fig. 1 E). Interestingly, the expression of full-length, WT NF-E2 was strongly increased, whereas the levels of truncated, mutant protein were low. Thus, presence of mutant NF-E2 results in elevated levels of WT NF-E2. These data provide one possible mechanism by which mutant NF-E2 augments the activity of its WT counterpart.
In a second series of experiments, we compared the stability of WT NF-E2 protein in the presence or absence of mutant NF-E2. The CB3 cells expressing WT NF-E2 were cotransfected either with a virus expressing the 262aa NF-E2 truncation mutant or with an empty control virus. The levels of WT and truncated, mutant NF-E2 were determined by Western blotting and shown to be comparable in their relative ratio to those observed in MPN patients; mutant NF-E2 was present at much lower levels than WT NF-E2 (compare Fig. 3 C, right, lane 1 with Fig. 1 E). Cells were treated with 20 µg/ml cycloheximide to inhibit de novo protein biosynthesis and assayed for NF-E2 protein levels by Western blotting for up to 6 h after the start of treatment (Fig. 3 C). The half-life of WT NF-E2, which is 1.35 h in control cells, is extended 1.65-fold to 2.23 h by the presence of truncated NF-E2 (Fig. 3 D). Expression of mutant NF-E2 therefore not only increases mRNA levels but, in addition, increases protein stability. Both mechanisms are likely to contribute to the increase in NF-E2 protein levels seen in NF-E2 mutant carrying MPN patients (Fig. 1 E).
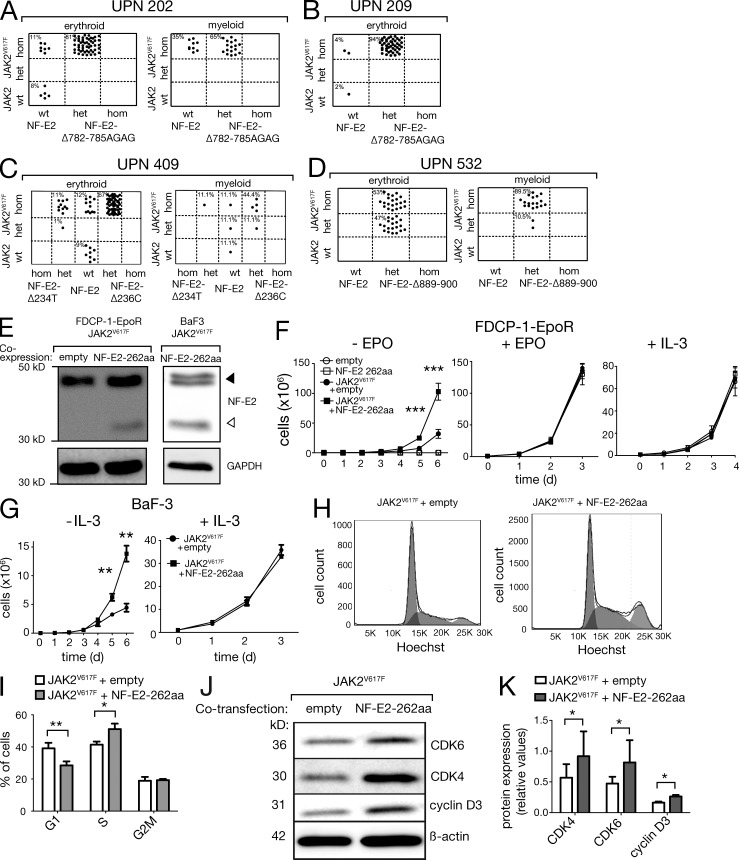
MPN patients carrying NF-E2 mutations were also tested for the presence of the JAK2V617F mutation as well as mutations in cMpl, ASXL1, TET2, and EZH2. Three patients displayed mutations in TET2, whereas all seven patients assayed carried both a NF-E2 mutation and the JAK2V617F mutation (Table 1). Analysis of individual hematopoietic colonies grown in methylcellulose revealed that in three patients, 202, 209, and 409, the NF-E2 mutation was acquired subsequent to the JAK2V617F mutation, as some colonies carrying only the JAK2V617F mutation but not the NF-E2 mutation were found and all colonies positive for the NF-E2 mutation also carried JAK2V617F (Fig. 4, A-C). Interestingly, these assays revealed a proliferative advantage of cells carrying the NF-E2 mutation over cells carrying only JAK2V617F (Fig. 4 A-C). In all three patients, cells carrying both a NF-E2 mutation and the JAK2V617F mutation represent the vast majority of the colonies analyzed (81, 94, and 79% of the erythroid colonies, respectively), demonstrating that the JAK2V617F-positive cell that acquired a NF-E2 mutation outcompeted cells carrying the JAK2V617F mutation alone.
Figure 4.
NF-E2 mutations confer a proliferative advantage on the MPN clone by enhancing cell proliferation. (A–D) Mononuclear cells of the MPN patients indicated were seeded in methylcellulose and individual erythroid and myeloid colonies harvested after 14 d. Colonies were assayed for presence of the JAK2V617F and the NF-E2 mutations by BsaXI restriction of PCR-amplified DNA (Baxter et al., 2005) and by GeneScan analysis (patients 202, 209, and 532) or direct sequencing (patients 209 and 409), respectively. Genotypes of the individual colonies, each represented by a single dot, are depicted. (E) FDCP-1-EpoR cells (left) and BaF3 cells (right) carrying the JAK2V617F mutation were transduced either with a vector expressing the 262aa NF-E2 truncation mutant or with an empty control vector. Total cell extracts were interrogated with an antibody that recognizes both human and murine NF-E2. Equal loading was assured by reprobing with an antibody against GAPDH. (F and G) FDCP-1-EpoR cells (F) and BaF3 cells (G) carrying the JAK2V617F mutation and expressing either the 262aa NF-E2 truncation mutant or carrying an empty control vector were cultured in the absence of growth factors (left) or in the presence of 1 IU/ml Epo or 5 ng/ml IL-3, as indicated. Mean and SEM of four independent experiments are shown. **, P < 0.01; ***, P < 0.001. (H) FDCP-1-EpoR cells carrying an empty control vector (left) or expressing the 262aa NF-E2 truncation mutant (right) were stained with Hoechst 33342 and analyzed for DNA content by FACS. Representative blots of n = 6 independent experiments are shown. (I) Statistical analysis of n = 6 independent experiments. *, P < 0.05; **, P < 0.01 by Student’s t test. (J) Cell extracts of FDCP-1-EpoR cells carrying an empty control vector (left lanes) or expressing the 262aa NF-E2 truncation mutant (right lanes) were interrogated with antibodies against CDK4, CDK6, and cyclin D3, as indicated. Equal loading was assured by reprobing with an antibody against β-actin. Representative blots of n = 6 independent experiments are shown. (K) Statistical analysis of n = 6 independent Western blot experiments. Mean and SEM are shown. *, P < 0.05 by Wilcox matched pairs test.
Patient 409 has acquired two separate and mutually exclusive NF-E2 mutations, c.236delC and c.234delT (Fig. 4 C and Table 1). The vast majority of both myeloid and erythroid colonies carry a homozygous JAK2V617F mutation. However, 67% of the erythroid and 55% of the myeloid colonies carry the c.236delC, whereas 12% of the erythroid and 11% of the myeloid colonies carry a c.234delT mutation (Fig. 4 C). The remaining colonies are WT for NF-E2, demonstrating that both mutations are acquired and again suggesting that both NF-E2 mutations confer a growth advantage, as they outcompete the JAK2V617F NF-E2 WT cells. The presence of both heterozygous and homozygous JAK2V617F clones containing either the c.234delT or the c.236delC NF-E2 mutation implies that in this patient, uniparental disomy (loss of heterozygosity) has occurred in two independent clones, a rare situation previously reported in individual MPN patients (Schaub et al., 2010; Wang et al., 2011).
In patient 532, it cannot be determined for sure whether the NF-E2 or the JAK2V617F mutation was incurred first. However, again, the presence of both mutations clearly provided a proliferative advantage over either of the mutations alone, as all colonies carry both mutations (Fig. 4 D). In addition, in the context of an NF-E2 mutation, as in WT NF-E2 patients, homozygous JAK2V617F cells outgrow heterozygous JAK2V617F cells.
As acquisition of the NF-E2 mutation consistently conferred a selective advantage, so that cells carrying both mutations outnumbered those carrying JAK2V617F alone (Fig. 4, A–D), we sought to determine the mechanism mediating this effect. JAK2V617F has been shown to confer growth factor independence on the Epo- and IL-3–dependent cell lines FDCP-1-EpoR and BaF3, respectively (James et al., 2005). We transduced FDCP-1-EpoR and BaF3 cells carrying the JAK2V617F mutation either with a vector expressing the 262aa NF-E2 truncation mutant or with an empty control vector. The levels of endogenous, WT NF-E2 and truncated, mutant NF-E2 were determined by Western blotting and shown to be comparable in their relative ratio to those observed in MPN patients; mutant NF-E2 was present at much lower levels than WT NF-E2 (compare Fig. 4 E with Fig. 1 E).
FDCP-1-EpoR and BaF3 cells were starved of growth factors and cell numbers recorded in the absence of Epo or IL-3, respectively (Fig. 4, F and G, left). As published previously (James et al., 2005), cells carrying the JAK2V617F mutation alone display growth in the absence of cytokines. Consistently, in the absence of cytokines, cells expressing mutant NF-E2 in addition to the mutant JAK2V617F kinase grew faster than cells expressing only JAK2V617F, reaching statistically significantly higher cell numbers (Fig. 4, F and G, left). In the presence of Epo or IL-3, all cells proliferated equally (Fig. 4 F and G, middle and right). The presence of mutant NF-E2 therefore enhanced cytokine independence and conferred a proliferative advantage in the absence of cytokines beyond that conferred by JAK2V617F alone. This was true for both the Epo independence of FDCP-1-EpoR cells and the independence from IL-3 in BaF3 cells (Fig. 4, F and G).
The proliferative advantage conferred by mutant NF-E2 was further investigated by conducting cell cycle analysis on FDCP-1-EpoR cells expressing both JAK2V617F and the NF-E2 262aa mutant as well as FDCP-1-EpoR cells expressing JAK2V617F alone (Fig. 4 H). Cells carrying mutant NF-E2 displayed a significant increase in the proportion of cells in S-phase and a concomitant significant decrease in the proportion of cells in G1-phase (Fig. 4 I). Expression of mutant NF-E2 therefore enhances cell division, causing increased proliferation.
To investigate the molecular basis for the proliferative advantage conferred by mutant NF-E2, we conducted microarray analysis comparing FDCP-1-EpoR cells expressing JAK2V617F alone to those carrying both JAK2V617F and the NF-E2 262aa mutant. We combined these results with ChIP-seq data available in silico, depicting in vivo NF-E2 DNA binding in K562 cells (Consortium, 2011). A series of candidates was selected whose RNA expression was increased in cells carrying both JAK2V617F and the NF-E2 262aa mutant compared with cells expressing JAK2V617F alone and whose promoter or enhancer regions displayed NF-E2 binding, suggesting that this transcription factor is involved in regulating their expression. Several candidate genes encode cell cycle regulators. Protein expression of three candidates, cyclin D3, CDK4 and CDK6, was examined by Western blotting (Fig. 4, J and K). Cells carrying both JAK2V617F and the NF-E2 262aa mutant expressed higher levels of all three of these cell cycle regulators. These data provide a molecular mechanism for the enhanced proliferation conferred by mutant NF-E2: by increasing expression of cell cycle regulators that promote the G1-S transition (Sherr, 1995; Herzinger and Reed, 1998), mutant NF-E2 causes an increase in the number of cells entering S-phase, hence promoting proliferation.
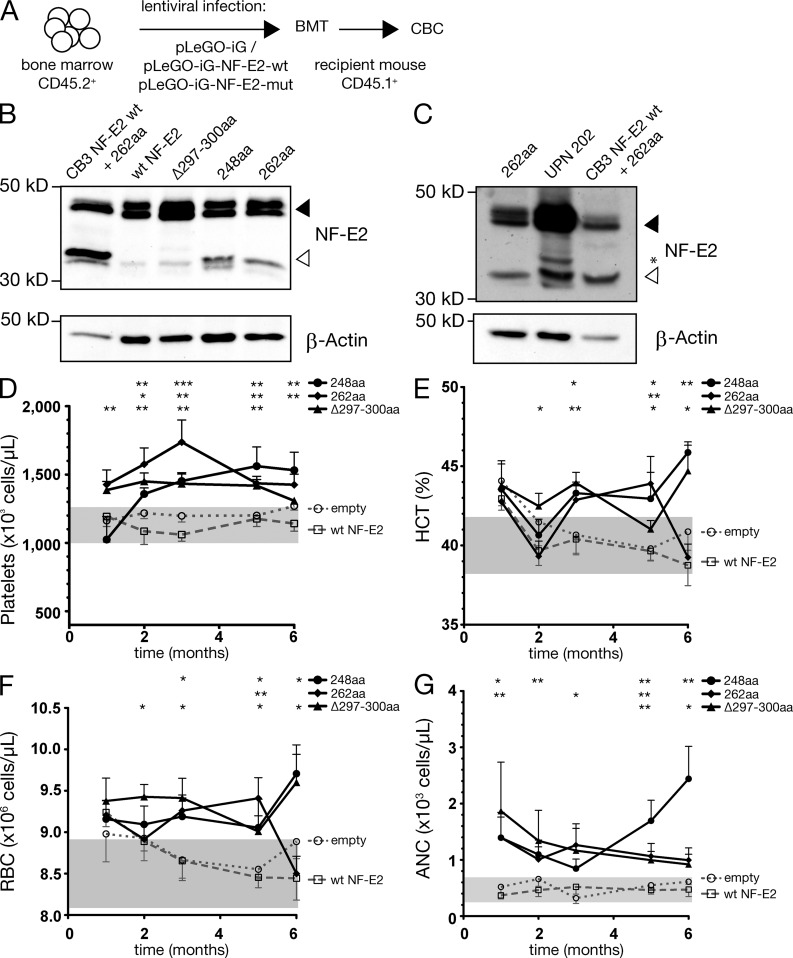
To test whether the NF-E2 mutants also confer an MPN phenotype in vivo, a murine bone marrow transplant model was used. Donor bone marrow was transduced with empty lentiviruses or viruses encoding WT NF-E2, the 248aa, the 262aa, or the Δ297-300 mutants and transplanted into recipient mice (Fig. 5 A). Donor engraftment exceeded 90% in all mice.
Figure 5.
Hematological parameters of bone marrow transplanted mice. (A) Experimental design. FVB/N mice (45.1) were transplanted intrafemorally with lentivirally transduced FVB/N-45.2 bone marrow, infected either with an empty vector, or with a vector expressing WT NF-E2 or the indicated NF-E2 mutants (n = 4–11 mice/construct, as indicated). 12 wk after transplantation, engraftment exceeded 90% in all cases. (B) Total cell extracts of murine bone marrow transduced with the indicated NF-E2 constructs was interrogated with an antibody that recognizes both human and murine NF-E2. Equal loading was assured by reprobing with an antibody against β-actin. A representative blot of n = 3 independent experiments is shown. (C) Total cell extracts of murine bone marrow transduced with the NF-E2-262aa mutant (left lane), as well as protein extracts of a patient carrying the NF-E2-262aa truncation mutations (UPN 202) were interrogated for NF-E2 (top) and β-actin (bottom) expression by Western blotting. The outer right lane serves as a positive control and depicts lentivirally transduced CB3 cells expressing both WT and the 262aa truncated NF-E2. A filled arrowhead demarks full-length WT NF-E2, an open arrowhead points to the smaller, truncated NF-E2 proteins, and an asterisk demarks a nonspecific band. A representative blot of two independent experiments is shown. (D–G) Peripheral blood was obtained by retro-orbital puncture and analyzed on an Advia 120 system at the indicated time points (n = 4–11 mice per genotype). Mean and SEM are shown. (D) Platelet counts. (E) Hematocrit. (F) RBC count. (G) ANC. Statistical analysis was conducted using Student’s t tests., *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus WT.
We determined NF-E2 mRNA and protein expression in this novel BMT model. Bone marrow was harvested by aspiration and NF-E2 protein levels assayed by Western blotting. Similar to what we observed in MPN patients (Fig. 1 E), truncated, mutant NF-E2 is expressed at low levels in our murine model, equivalent to those observed in patients (Fig. 5, B and C; in Fig. 5 C, mutant NF-E2 is 26% of WT in the transplanted mouse and 29% of WT in patient 202), demonstrating that the mouse model faithfully represents the pathophysiologic situation in MPN patients.
NF-E2 mRNA levels were determined by qRT-PCR. As noted above (Fig. 3 A), MPN patients carrying NF-E2 mutations display significantly elevated NF-E2 mRNA levels compared with patients with WT NF-E2. A similar effect was observed in mice carrying NF-E2 mutations. Two of the three mutants likewise express significantly elevated levels of murine NF-E2 mRNA compared with control mice expressing WT NF-E2 (Fig. 3 B).
To determine the effect of mutant NF-E2 on the transcription of a well characterized target gene, we measured β-globin expression in cells from control and NF-E2 mutant mice. Cells at equivalent stages of erythroid differentiation were isolated by FACS sorting of CD71, Ter-119 double-positive cells. β-globin mRNA expression was determined by qRT-PCR in this sorted population. As depicted in Fig. 2 F, β-globin expression is significantly elevated in NF-E2 mutant carrying mice compared with control mice. Moreover, the degree of β-globin overexpression in mice carrying the three different NF-E2 mutations precisely mirrors that previously observed in transiently transfected cells (Fig. 2, compare E and F). Therefore, the truncated mutant NF-E2 protein is expressed and functional in the BMT model.
After transplantation, peripheral blood was analyzed at monthly intervals. Compared with mice transplanted with empty vector or with vector expressing WT NF-E2, which, similar to NF-E2 transgenic mice in the first year of life, display normal platelet values (Kaufmann et al., 2012), mice transplanted with all three mutants displayed a statistically significant elevation in platelet number beginning 1–2 mo after transplantation (Fig. 5 D). Moreover, although WT NF-E2 transplanted mice, like NF-E2 transgenic mice, display normal hematocrit and normal RBCs (Kaufmann et al., 2012), mice transplanted with NF-E2 mutants displayed an elevation in hematocrit and in RBC number (Fig. 5, E and F). Furthermore, the absolute neutrophil count (ANC) is elevated in NF-E2 mutant mice beginning as early as 1 mo after transplantation (Fig. 5 G). These data demonstrate that presence of mutant NF-E2 is sufficient to cause thrombocytosis, erythrocytosis, and neutrophilia in a murine model.
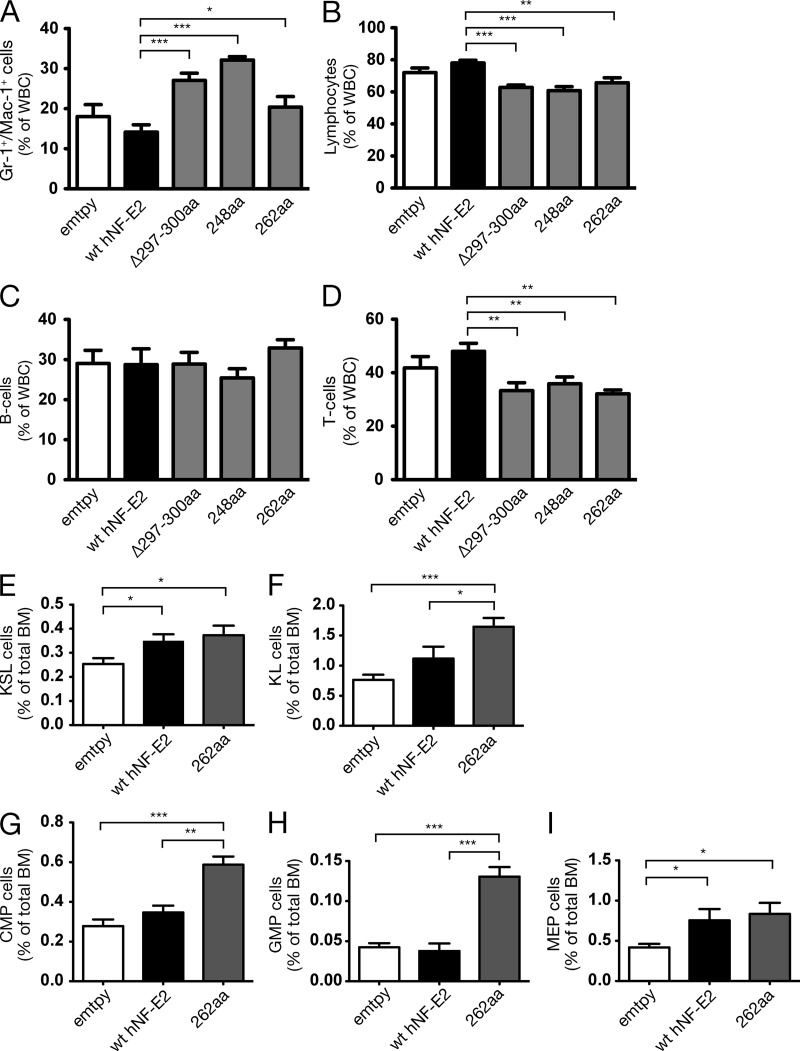
We investigated whether expression of mutant NF-E2 alters the proportion of myeloid and lymphoid cells in the peripheral blood. FACS analysis of peripheral blood leukocytes demonstrates that the proportion of Gr-1+-Mac-1+ myeloid cells is statistically significantly increased in mice carrying NF-E2 mutations (Fig. 6 A and Fig. S1). Consequently, the proportion of lymphocytes is statistically significantly reduced (Fig. 6 B). Among the lymphocytes, the proportion of B cells remained constant, whereas the proportion of T cells was significantly reduced (Fig. 6, C and D; and Fig. S1). In MPN patients, the B cells are frequently part of the malignant clone (Raskind et al., 1985), whereas the T cells often are not (el-Kassar et al., 1997). Therefore, mutant NF-E2 causes a lineage bias and increases the production of those lineages affected in MPN patients.
Figure 6.
Effect of NF-E2 mutants on lineage commitment and hematopoietic stem and progenitor populations. (A–D) Peripheral blood leukocytes were stained with antibodies against Gr-1, Mac-1, B220, and CD3 and analyzed by FACS. (A) Percentage of Gr-1+/Mac-1+ double-positive cells. (B) Percentage of lymphocytes (CD3+ and B220+). (C and D) Lymphocyte subpopulations. (C) B220+ B cells. (D) CD3+ T cells. (E–I) BM cells were stained with antibodies against a cocktail of lineage markers as well as against c-kit, Sca-1, CD34, Fc-gammaR, and Flt3/Flk2. Lineage-negative, c-kit–positive, Sca-1–positive (KSL) cells (E) and lineage-negative, c-kit-positive, Sca-1-negative (KL) cells (F) are depicted as a percentage of total BM cells. (G–I) CMP, GMP, and MEP are depicted as a percentage of total BM cells. (A–I) n = 5–17 per genotype, as indicated. Mean and SEM are shown.*, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine the origin of the observed lineage bias, we analyzed the stem and progenitor compartments in NF-E2 mutant-bearing mice. Compared with control mice, NF-E2 mutant-expressing mice show a significant expansion of KSL cells (Kit+, Sca+, lin− cells; Fig. 6 E and Fig. S2). Moreover, the size of the KL compartment (Kit+, Sca−, lin− cells) is more than doubled in NF-E2 mutant mice, compared with empty control transduced mice (Fig. 6 F). This expansion is the result of an increase in the numbers of all three types of KL progenitors, megakaryocyte/erythroid progenitor (MEP), granulocyte/monocyte progenitor (GMP), and common myeloid progenitor (CMP; Fig. 6, G–I; and Fig. S2). An increase of the latter three cell types in PV patients has previously been reported by Jamieson et al. (2006).
We have recently shown that transgenic mice overexpressing WT NF-E2 contain elevated numbers of KSL and KL cells (Kaufmann et al., 2012). In addition, WT NF-E2 transgenic mice display increased numbers of CMP and MEP but not GMP (Kaufmann et al., 2012). In contrast, NF-E2 mutant-bearing mice contain an expansion of all three progenitor types, CMP, MEP, and GMP, and show a highly significant increase in the number of KL cells, CMP, and GMP over WT NF-E2–expressing mice (Fig. 6, F–I). Therefore, mutant, truncated NF-E2 increases and augments the myeloid lineage bias already evident in NF-E2 WT transgenic mice. In the murine model, as in the cell lines, mutant NF-E2 enhances and extends the effect of WT NF-E2.
Histopathological analysis was conducted in mice sacrificed at 6–7 mo of age. As shown in Fig. 5, mice expressing WT NF-E2, similar to NF-E2 transgenic mice (Kaufmann et al., 2012), did not show changes in blood counts in the first 6 mo after transplant (Fig. 5, D–G). Accordingly, bone marrow of mice expressing WT NF-E2 is histologically normal and similar to control bone marrow (Fig. 7, F and G). Mice expressing mutant NF-E2, in contrast, display an increased ANC (Fig. 5 G) as well as a myeloid lineage bias (Fig. 6). Correspondingly, bone marrow histologies of mutant NF-E2–expressing mice show increased granulopoiesis and display a right shift, manifested by the increased presence of mature forms, which can be found extending to the trabecula (Fig. 7, H–J).
Figure 7.
Histopathology of mutant NF-E2–expressing mice and control animals. H&E staining of formalin-fixed and paraffin-embedded organ sections. (A–E) Spleen. (A) Empty. (B) WT NF-E2. (C) NF-E2-248aa. (D) NF-E2-262aa. (E) NF-E2-Δ-297-300. Bars: (left) 50 µm; (right) 100 µm. (F–J) Bone marrow. (F) Empty. (G) WT NF-E2. (H) NF-E2-248aa. (I) NF-E2-262aa. (J) NF-E2-Δ-297-300. Bars: (left) 100 µm; (right) 200 µm, respectively. (K–N) Lung. (K) WT NF-E2. (L) NF-E2-248aa. (M) NF-E2-262aa. (N) NF-E2-Δ-297-300. Bars, 50 µm. (O–R) Liver. (O) WT NF-E2. (P) NF-E2-248aa. (Q) NF-E2-262aa. (R) NF-E2-Δ-297-300. Bars, 100 µm.
Similar to what we have shown in WT NF-E2 transgenic animals (Kaufmann et al., 2012), animals transplanted with WT NF-E2 show an increase in siderin deposits in the spleen (Fig. 7 B), as well as an expanded red pulp, compared with control mice (Fig. 7 A). Mice expressing mutant NF-E2 show changes in splenic architecture very similar to those observed in WT NF-E2 transduced mice (Fig. 7, C–E).
The livers and lungs of NF-E2 WT and mutant NF-E2–expressing mice display normal histology in all animals, notably an absence of myeloid infiltrates (Fig. 7, K–N and O–R). We wished to determine whether the cooperation between JAK2V617F and mutant NF-E2 observed in MPN patients and in vitro models (Fig. 4) can be recapitulated in vivo.
To correctly model the order of acquisition of the two mutations, the following experiment was conducted (Fig. 8 A). Because the patients first acquire JAK2V617F and subsequently mutant NF-E2 (Fig. 4, A–C), we infected murine bone marrow from balb/c mice with JAK2V617F and transplanted recipient mice. This JAK2V617F model is well documented in the literature (Bumm et al., 2006; Wernig et al., 2006; Zaleskas et al., 2006). After 10 wk, the transplanted bone marrow was harvested and infected either with a lentivirus expressing mutant NF-E2 (pLeGO-iC2-NF-E2-262aa) or an empty LeGO-iC2 control. Cells expressing both alleles, JAK2V617F and either mutant NF-E2 or empty control, were sorted by FACS. Subsequently, 50,000 double-positive cells were transplanted, together with 300,000 bone marrow support cells, into lethally irradiated recipient mice (Fig. 8 A).
Figure 8.
Effect of coexpression of JAK2V617F and mutant NF-E2 in vivo. (A) Experimental design. Murine bone marrow was retrovirally transduced with a vector expressing JAK2V617F and transplanted into lethally irradiated balb/c animals. 10 wk after transplantation, bone marrow was harvested and infected with an empty control lentivirus, pLeGO-iC2, or with a lentivirus expressing the 262aa NF-E2 truncation mutant (pLeGO-iC2-NF-E2-262aa). Cells expressing both JAK2V617F and the 262aa NF-E2 mutant or empty control virus were FACS sorted. Subsequently, 50,000 double-positive cells were transplanted together with 300,000 bone marrow support cells into lethally irradiated recipient mice. (B–E) Peripheral blood was obtained by retro-orbital puncture and analyzed on an Advia 120 system. (n = 3–7 mice per genotype). Mean and SEM are shown. (B) Hemoglobin. (C) Mean corpuscular volume (MCV). (D) Mean corpuscular hemoglobin (MCH). (E) WBC count. Statistical analysis was conducted using Student’s t tests. *, P < 0.05; ***, P < 0.001. (F–I) H&E staining of formalin-fixed and paraffin-embedded organ sections. (F and G) Mice expressing JAK2V617F alone (with the empty pLeGo-iC2 control). (H and I) Mice expressing both JAK2V617F and the NF-E2-262aa mutant. Bars: (F and H) 100 µm; (G and I) 50 µm.
As previously published, mice expressing JAK2V617F alone (with the empty pLeGo-iC2 control) display an increased hemoglobin of 17 g/dl (Bumm et al., 2006; Wernig et al., 2006; Zaleskas et al., 2006). Mice expressing both JAK2V617F and mutant NF-E2 display a further, statistically significant increase in hemoglobin (Fig. 8 B). Furthermore, addition of mutant NF-E2 significantly increased the MCV, also previously shown to be elevated in the JAK2V617F BMT model (Lacout et al., 2006), and increased MCH, as well as the WBC count, above that affected by JAK2V617F alone (Fig. 8, C–E). These data demonstrate that in vivo mutant NF-E2 cooperates with JAK2V617F to augment the MPN phenotype.
We evaluated bone marrow histologies of mice expressing JAK2V617F alone (with the empty pLeGo-iC2 control) and mice expressing both JAK2V617F and mutant NF-E2. Compared with JAK2V617F-expressing mice, mice expressing both mutations show increased granulopoiesis and display a right shift, similar to mice expressing mutant NF-E2 alone (Fig. 8, F–I). Histological analysis thus supports the data from peripheral blood counts in that the presence of mutant NF-E2 confers a phenotype beyond the changes affected by the expression of JAK2V617F alone.
DISCUSSION
Based on our previous data documenting a role for aberrant NF-E2 expression in the pathophysiology of MPNs (Kaufmann et al., 2012), we hypothesized that MPN patients may harbor alterations in the NF-E2 gene. Here, we report the presence of insertion and deletion mutations in patients with PV and PMF, which lead to the expression of truncated NF-E2 proteins (Fig. 1). Although these truncation mutants demonstrate neither DNA binding nor transcriptional transactivating activity on their own (Fig. 1 C; and Fig. 2, A–C), in the presence of WT NF-E2, they augment the effect of this transcription factor, both in vitro and in vivo (Fig. 2, E and F; Fig. 3, A and B; Fig. 5, D–G; and Fig. 6).
We demonstrate two distinct effects of truncated NF-E2 on the expression of its WT counterpart. First, mutant NF-E2 increases the steady-state levels of WT NF-E2 mRNA (Fig. 3, A and B). Second, the presence of mutant NF-E2 increases protein stability of WT NF-E2 (Fig. 3, C and D). How the truncated protein causes these effects mechanistically remains to be investigated. Increased amounts of steady-state mRNA can arise from an increased rate of transcription, from delayed mRNA degradation, or from a combination of both effects. Run-on transcription assays, as well as the determination of mRNA decay, will be used to investigate by which mechanisms truncated NF-E2 proteins increase WT NF-E2 mRNA levels.
The presence of mutant NF-E2 augments the levels and activity of WT NF-E2 protein (Fig. 2, E and F; Fig. 3 C; Fig. 5, D–G; and Fig. 6). Because the truncated proteins lack a DNA binding domain, they most likely exert their effect via protein–protein interactions. However, as the leucine zipper domain required for interactions with other β-zip proteins is also deleted or inactivated, these protein–protein interactions must take place within the remaining portion of the protein, the N-terminal transactivating domain, which extends from amino acids 1 to 206. The shortest truncating mutant detected in our cohort retains only the first 79 amino acids in frame (Fig. 1 A and Table 1). Interestingly, amino acids 1–79 constitute the proline-rich domain, which contains two heme binding sites (Moore et al., 2006).
NF-E2 has been reported to interact with several partners through sites within its transactivating domain. These include the ubiquitin ligases ITCH and NEDD4, which bind the NF-E2 PY domain (aa 79–83) through their WW domains (Gavva et al., 1997; Chen et al., 2001; Lee et al., 2008). This interaction domain is retained in all truncation mutants except for the shortest p.P79LfsX32 protein. In addition, NF-E2 binds the transcriptional coactivators YAP and CBP (Gavva et al., 1997; Hung et al., 2001) as well as a member of the TFIID transcription complex, TAFII130 (Amrolia et al., 1997).
Revealing a potential role as both an activator and a repressor, NF-E2 also binds the transcriptional repressor NAPP2, which acts epigenetically by recruiting HDAC1 and causing histone deacetylation (Gavva et al., 2002). NF-E2 is known to exert further epigenetic functions. It has been shown to remodel chromatin at the β-globin locus, establishing two chromatin marks, histone 3 trimethylated lysine 4 and trimethylated lysine 79 (Kiekhaefer et al., 2004). NF-E2 is required for recruitment of the MLL2 (mixed-lineage leukemia 2) and the G9a methyltransferase complexes to the β-globin gene and interacts physically with several of their members (Demers et al., 2007; Chaturvedi et al., 2009).
Presence of truncated NF-E2 proteins, which may still be able to bind their interaction partners but lack additional domains required for full function, could perturb any of the protein–protein interactions detailed above. We are currently investigating which interactions are retained by the truncated NF-E2 mutants and whether these alter the functions of the resulting complexes. Alternatively or in addition, mutant protein could serve as a decoy, occupying binding sites in regulatory proteins, thereby preventing the degradation of WT NF-E2 leading to its accumulation. These hypotheses are currently being addressed experimentally.
In summary, our data identify NF-E2 as a novel mutational target in MPN patients. Based on our studies, these alterations are rare in MPN patients, affecting ∼2% of cases. We report truncating mutations of NF-E2 that enhance WT NF-E2 function, promote erythrocytosis and thrombocytosis in a murine model, and cooperate with JAK2V617F to augment the MPN phenotype. Our data underscore the role of elevated NF-E2 activity in the pathophysiology of MPNs.
MATERIALS AND METHODS
MPN patients and healthy controls.
Peripheral blood samples were obtained from PV (n = 144), ET (n = 120), MF (n = 192, including 139 PMF), 53 post-PV or post-ET secondary MF patients), MPN-U (n = 49), MDS (n = 57), post-MPN-sAML (n = 39), and CMML (n = 67) patients, all fulfilling the WHO criteria for diagnosis (Swerdlow et al., 2008). Patient characteristics are summarized in Table S1. Buffy coats of healthy volunteer blood donors were obtained from the University Hospital Freiburg Center for Blood Transfusion and from Sanquin Bloodsupply, Nijmegen. Isolated granulocytes, obtained by dextran sedimentation and Ficoll gradient centrifugation as previously described (Temerinac et al., 2000), were used for DNA preparation and subsequent NF-E2 sequence analysis. Buccal swabs or isolated T cells (Nikoloski et al., 2010) were used to obtain DNA for germline analysis. The study protocol was approved by the local ethics committees (University Hospital Freiburg, University Hospital Ulm, Radboud University Nijmegen Medical Centre, as well as the Member Institutions of the MPD-RC) and informed consent was obtained from all patients. Each patient was assigned a unique patient number (UPN), which was used thereafter for the protection of privacy. All patients save one were tested for the presence of the JAK2V617F mutation by qRT-PCR or LNA PCR as previously described (Steimle et al., 2007).
NF-E2 sequencing.
Exons 2 and 3 of the NF-E2 locus were amplified by PCR and subjected to direct sequencing according to standard protocols. Primer sequences are available upon request. Sequence variations were confirmed by an independent PCR amplification of the original genomic DNA and subsequent sequence analysis. Constitutive DNA, obtained either from buccal swabs (patients 532 and 980) or isolated T cells (patient 209), was subjected to PCR amplification and sequencing to determine the acquired nature of the detected mutations. Detected mutations were described according to the consensus nomenclature (http://www.hgvs.org/mutnomen; den Dunnen and Antonarakis, 2001).
EMSA analysis.
HEK-293 nuclear extracts were prepared as previously described (Schreiber et al., 1989). For EMSA analysis, 1–2 µg of nuclear extracts were added to a binding reaction containing 2 µg poly dI-dC, 2 µl 10× binding buffer (10 mM Hepes, pH 7.9, 5 mM MgCl2, 30 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 12% glycerol), and 0.5 ng 32P-labeled oligonucleotide. The oligonucleotide containing the NF-E2 consensus binding site at bp −160 of the human porphobilinogen deaminase gene promoter (Mignotte et al., 1989) was synthesized by Eurofins MWG Operon and the NF-κB consensus oligonucleotide was purchased from Promega. When indicated, a 100-fold excess of nonradioactive oligonucleotide, an NF-E2 antibody (Sigma-Aldrich), or an NF-κB p65 antibody (Santa Cruz Biotechnologies, Inc.) were added. The reaction was incubated at room temperature for 15 min. For supershift assays, extracts and antibodies were preincubated for 10 min at room temperature before addition of the radioactive nucleotide.
Immunoblotting.
Murine bone marrow, peripheral blood MNCs from MPN patients, CB3 cells, or HEK-293 cells, transduced with expression vectors encoding WT NF-E2 or mutants, were lysed in SC buffer (50 mM Tris/HCl, pH 8.0, 170 mM NaCl, 0.1% vol/vol NP-40, 50 mM NaF, 2 mM Na3VO4, 0.2 mM DTT, 20% vol/vol glycerol, and 1 µg/ml BSA) supplemented with 1× Complete (Roche). Cell lysates were subjected to SDS-PAGE-gel-electrophoresis and Western blotting. Primary antibodies against NF-E2 (Goerttler et al., 2005) were used as described. FDCP-1-EpoR cells expressing JAK2V617F as well as the 262aa NF-E2 truncation mutant or carrying an empty control vector were analyzed for CDK4 (clone DCS 156; Cell Signaling Technology), CDK6 (clone DCS 83; Cell Signaling Technology), and Cyclin D3 (clone DCS 22; Cell Signaling Technology) expression.
The blots were stripped and reprobed against β-actin (Clone AC-15; Sigma-Aldrich) or GAPDH (Clone 71.1; Sigma-Aldrich) to control for equal loading. Immune complexes were detected using chemiluminescence Western Blotting Reagents (GE Healthcare).
Plasmids: NF-E2 reporter constructs and lentiviral vectors.
A 5.2 kb region, spanning bp −1 to bp −5221 of the β1 tubulin gene, was amplified by PCR from human genomic DNA and inserted into the pGL3basic luciferase reporter vector (Promega), resulting in the β1-tubulin −5.2 kb-luc construct. The pRBGP2-Luciferase (pRBGP2-Luc) reporter plasmid (Igarashi et al., 1994), which contains three copies of the NF-E2 binding site from the chicken β-globin enhancer in a TATA-luciferase reporter vector, was a gift of M. Yamamoto (Tsukuba University, Tsukuba, Japan).
Insertion and deletion mutations detected in MPN patients were inserted into the NF-E2 cDNA by site-directed mutagenesis using the GeneTailor site-directed mutagenesis system (Invitrogen). WT and mutant NF-E2 cDNAs were cloned into pLeGO-iG, pLeGO-iT2, pLeGo-iC2 (Weber et al., 2008), and pRc/CMV vectors by restriction enzyme digestion. The MafG-pCMV6-XL4 expression vector was purchased from OriGene.
Transient transfections and luciferase assays.
HEK-293 cells were transiently transfected with 0.2 µg of either the pRBGP2-Luc or the β1-tubulin-Luciferase reporter gene construct, together with 1.36 µg of either pRc/CMV-NF-E2 WT or a pRc/CMV-NF-E2 mutant expression vector or the pRc/CMV empty control vector as well as 0.34 µg of a MafG-pCMV6-XL4 expression vector (OriGene). In addition, 0.1 µg of a TK-Renilla plasmid (Promega) were cotransfected as an internal control.
Cells were harvested 16 h after transfection, and luciferase activity was determined using the Dual Luciferase Reporter Assay System (Promega). Luciferase activity was normalized to the Renilla internal control to compensate for variations in transfection efficiency.
Lentiviral transductions.
CB3 cells, a kind gift of V. Blank (McGill University, Montreal, Quebec, Canada) and E. Bresnick (University of Wisconsin, Madison, WI), were transduced with empty pLeGO-iG, pLeGO-iT, or pLeGO-iC2 containing WT NF-E2 or NF-E2 mutants as previously described (Roelz et al., 2010) using an MOI between 1 and 8. GFP-expressing cells were sorted to obtain pure populations. CB3-NF-E2wt cells were obtained by transducing CB3 cells with pLeGO-iG-WT-NF-E2 and subsequently cloning single transduced cells by limiting dilution. A stable clone was selected and used for coexpression of NF-E2 mutants and variants. CB3-NF-E2wt cells were transduced with either pLeGO-iT2 or pLeGO-iC2 containing mutant and variant NF-E2 cDNAs or the empty control vector using an MOI between 0.5 and 9. Tomato or Cherry and GFP-expressing cells were sorted to obtain pure populations.
FDCP-1-Epo-R and FDCP-1-EpoR-JAK2V617F cells were transduced either with pLeGO-iC2 expressing the 262aa NF-E2 truncation mutant or the empty control vector. BaF3 cells were transduced with both the pMIG retrovirus expressing JAK2V617F as well as with pLeGO-iC2 expressing the 262aa NF-E2 truncation mutant or the empty control pLeGo-iC2 vector. All cells were sorted to obtain pure populations. Cells were starved of growth factors and subsequently counted daily to assess the cumulated cell number. As a control, FDCP-1-Epo-R cells were cultured in 1 IU/ml Epo and both BaF3 and FDCP-1-Epo-R cells in 5 ng/ml mIL-3.
Measurement of NF-E2 protein stability.
CB3 cells stably transduced with WT NF-E2 were subsequently transduced with either pLeGO-iC2 expressing the 262aa NF-E2 truncation mutant or the empty control vector and sorted to obtain pure populations. Cells were treated with 20 µg/ml cycloheximide to arrest protein synthesis and protein extracts collected at 1, 2, 4, and 6 h thereafter. NF-E2 and GAPDH levels were detected by Western blotting as described above and quantitated densitometrically. A nonlinear best-fit regression model was used to calculate the half-life of WT NF-E2 protein in the presence or absence of the 262aa truncation mutant.
Cell cycle analysis.
FDCP-1-EpoR cells expressing JAK2V617F, as well as the 262aa NF-E2 truncation mutant, or carrying an empty control vector were analyzed for cell cycle stage by staining with Hoechst 33342 (Molecular Probes) and 1 µg/ml LIVE/DEAD Fixable Near-IR Dead Cell Stain (Molecular Probes) at the manufacturer’s recommendation and analyzed by FACS using FlowJo software (Tree Star).
Quantitative RT-PCR assays.
Quantitative RT-PCR measurements were performed using an Assay on Demand (Mm01611268_g1 Hbb-b1; Applied Biosystems) for analysis of murine β-globin as well as the following primer and probe sequences for murine β-2-Microglobulin: forward primer, 5′-CTTTCTGGTGCTTGTCTCACTGAC-3′; reverse primer, 5′-GGTGGCGTGAGTATACTTGAATTTG-3′; TaqMan Probe, 6-FAM-ATCCAGAAAACCCC-MGBNFQ. NF-E2 mRNA was quantitated as previously described (Kaufmann et al., 2012). Reverse transcription of total CB3 cell RNA was performed using the Multiscribe Reverse transcription (Applied Biosystems). Q-PCR assays were performed in duplicate in an ABI PRISM 7000 Cycler and analyzed using the ABI PRISM 7000 software (Applied Biosystems). β-globin expression was determined relative to expression of the β-2-Microglobulin house-keeping gene using the ΔΔCt method.
Colony assays and mutation analysis.
Cells were seeded in methylcellulose media (1,000 cells/ml) containing SCF, IL-3, GM-CSF, and EPO (H4434; Stem Cell Technologies) and incubated for 14 d at 37°C with 5% CO2. Individual colonies (erythroid burst-forming units), as well as granulocytic/macrophage colony-forming units, were harvested on day 14 and assayed for the presence of the JAK2V617F mutation as well as the NF-E2 mutation. Presence of the JAK2V617F mutation was assessed by PCR amplification and BsaXI digestion, as well as by a JAK2V617F mutant-specific PCR, whereas presence of the NF-E2 mutation was detected by GeneScan analysis (patients 202, 209, and 532) and by direct sequencing (patients 209 and 409).
GeneScan analysis of NF-E2 mutations.
GeneScan analysis of genomic DNA extracted from single colonies was performed if the NF-E2 mutation comprised a deletion of three or more nucleotides (patients 202, 209, and 532). Two different primer pairs were used: forward primer, 5′-GACTTTAATGAGCTATTGGCAAGGTA-3′, and reverse primer, 5′-ACAATGGTTTCCAGCTTCCTCTT-3′ (coupled to 6FAM); or forward primer, 5′-CCCTACTCACTCATGCCCAACT-3′, and reverse primer, 5′-AAGCCGCTCCCGTTCATT-3′ (coupled to 6FAM). GeneScan analysis was performed on a 3730 DNA Analyzer using the GeneScan 500 LIZ Size Standard (Life Technologies). The generated peaks were analyzed using the Peak Scanner Software (Life Technologies).
Assessment of the JAK2V617F burden by PCR amplification and BsaXI digestion.
Detection of the JAK2V617F mutation by BsaXI digestion was performed essentially as previously described (Baxter et al., 2005). In brief, the following primers, located in exon 14 and intron 14 of the JAK2 gene, were used for PCR of genomic DNA: forward primer, 5′-AGCAGCAAGTATGATGAGCAAG-3′; reverse primer, 5′-GAGAAAGGCATTAGAAAGCCTGTAG-3′. The DNA fragment generated was subjected to BsaXI restriction digestion. The c.2343 G>T transversion, which generates the V617F mutation, destroys a BsaXI recognition sequence. Therefore, DNA containing only WT JAK2 alleles is cut by the enzyme, resulting in smaller fragments. Whereas DNA heterozygous for the JAK2V617F mutation generates smaller fragments, visible as one band, as well as the uncut PCR amplicon, the homozygous JAK2V617F DNA shows only the uncut PCR amplicon. Genomic DNA from the JAK2 WT EBV-DUZ cell line and from HEL cells, which harbor 10 copies of the JAK2V617F allele, was used as a negative and a positive control, respectively.
Assessment of the JAK2V617F burden by mutant-specific PCR.
Detection of the JAK2V617F mutation by a mutant-specific PCR assay was performed essentially as described previously (Sidon et al., 2006). Genomic DNA was amplified using the following primers: a forward primer recognizing both WT and mutant JAK2 alleles, 5′-TTATGGACAACAGTCAAACAACAATTC-3′; and a mutant-specific reverse primer, 5′-CTTACTCTCGTCTCCACAAAA-3′. The mutant-specific reverse primer contains one additional mutation to enhance specificity. Genomic DNA from the EBV-DUZ and HEL cell lines was used as a negative and a positive control, respectively.
Murine bone marrow transplantation.
FVB/N-45.2 donor mice (Guibal et al., 2009) were treated with 150 mg/kg body weight 5-fluorouracil (5-FU) 4 d before bone marrow harvest. Donor bone marrow was either infected with empty lentiviral particles (pLeGO-iG) or with lentiviruses expressing WT or mutant NF-E2 at an MOI of 10 as previously described (Roelz et al., 2010). FVB/N mice (45.1) were transplanted intrafemorally with lentivirally transduced FVB/N-45.2 bone marrow, as previously described (Roelz et al., 2010; Kaufmann et al., 2012). 12 wk after transplantation, engraftment exceeded 90% in all cases.
Bone marrow isolated from 5-FU–treated balb/c mice was infected with pMIG-JAK2V617F, as previously described (Bumm et al., 2006; Zaleskas et al., 2006) and transplanted into lethally irradiated recipients. 10 wk after transplantation, bone marrow was harvested and transduced with either the empty pLeGO-iC2 lentivirus or a lentivirus expressing the 262aa NF-E2-262 truncation mutant (pLEGO-iC2-NF-E2-262aa). Double-positive cells, expressing both GFP and Cherry, were FACS sorted. 50,000 double-positive cells were transplanted intrafemorally together with 300,000 support bone marrow cells into lethally irradiated recipients. Peripheral blood was obtained by retro-orbital puncture and analyzed on an Advia 120 system (Siemens).
FACS analysis of murine peripheral blood and bone marrow.
Murine peripheral blood was stained with antibodies against Gr1 (clone RB6-8C5; BioLegend), Mac-1 (clone M1/70; BioLegend), B220 (clone RA3-6B2,103212; BioLegend), and CD3ε (clone 145-2C11, 100308; BioLegend) and analyzed by FACS. For FACS sorting of populations to determine β-globin expression, antibodies against Ter-119 (clone Ter-119, 22155234; ImmunoTools) and CD71 (clone R17217; Leinco) were used.
Bone marrow was stained with a cocktail for lineage markers (CD3, Mac-1/CD11b, B220, Ter-119, and Gr1/Ly6G/6C; BioLegend) and lineage-negative cells (Lin−) were analyzed for c-Kit/CD117 (clone 2B8; eBioscience) and Sca-1 (clone D7; BioLegend) expression as previously described (Akashi et al., 2000; Christensen and Weissman, 2001; Passegué et al., 2003). Progenitor and stem cell subpopulations were identified as described by staining with the following additional markers: CD34 (clone MEC14.7; BioLegend), Fc-gamma-II/III-R (clone 93; eBioscience), as well as Flt3-biotin (clone A2F10; BioLegend) detected with streptavidin-PerCP (405213, BioLegend). Positively staining cells, as well as negative cells, were determined by fluorescence-minus-one (FMO) stainings, as previously published (Kaufmann et al., 2012).
Histology.
Organs were fixed in 4% formalin, femurs subsequently decalcified in 10% buffered ethylene-diamine tetra-acetic acid (EDTA), pH 7.2, and all organs paraffin embedded. Sections were stained with H&E for histological analysis.
Data analysis.
Paired or unpaired Student’s t tests (two sided or one sided) and the Mann-Whitney Rank sum test were used to determine whether a significant (P < 0.05) difference existed between two groups. When comparing more than two groups, a one- or two-way ANOVA was used. These analyses were performed using SigmaPlot 11.0 (Systat) or Prism 5.0 (GraphPad Software) software.
Online supplemental material.
Fig. S1 depicts the quantification of peripheral blood leukocytes (granulocytes, B and T cells), in PB of transplanted mice. Fig. S2 shows the quantification of hematopoietic stem and precursor (KL, KSL, CMP, GMP, and MEP) cells in BM of transplanted mice. Table S1 shows the clinical characteristics of the MPN, MDS, sAML, and CMML patients studied. Table S2 depicts the clinical features of MPN patients carrying NF-E2 mutations and Table S3 lists variants in NF-E2 that could not be verified as somatic. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120521/DC1.
Supplementary Material
Acknowledgments
The authors sincerely thank Martina de Groot for expert technical assistance, Sandra Kaiser and Titiksha Basu for lending a helping hand, and Prof. Dr. Torsten Haferlach, MLL, München for providing patient samples and gratefully acknowledge support from the MPD Research Consortium (MPD-RC) Tissue Bank, which also provided tissue samples. CB3 cells were kindly provided by Dr. Blank. The pRBGP2-Luciferase (pRBGP2-Luc) reporter plasmid was a kind gift of Dr. Masayuki Yamamoto (Tsukuba University, Tsukuba, Japan). FDCP-1-Epo-R and FDCP-1-EpoR-JAK2V617F cells were generous gifts of Dr. Stefan Constantinescu, Brussels. We gratefully acknowledge the gift of FVB/N-45.2 mice by Dr. Daniel G. Tenen, Harvard Medical School through Prof. Dr. Steffen Koschmieder, University Hospital Aachen. The pLeGO-iG, pLeGO-iT2, and pLeGO-iC2 vectors were generously provided by Prof. Dr. B. Fehse, Hamburg.
This work was supported by grants from the National Cancer Institute (PO1 CA108671 to H.L. Pahl), the Deutsche Forschungsgemeinschaft (Pa 611/6-1 to H.L. Pahl), and the Netherlands Institute for Regenerative Medicine to J.H. Jansen. H.L. Pahl is a member of the MPD Research Consortium. R. Bogeska was funded by the Excellence Initiative of the German Federal and States Governments (GSC-4, Spemann Graduate School).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- ANC
- absolute neutrophil count
- CMP
- common myeloid progenitor
- EMSA
- electrophoretic mobility shift assay
- ET
- essential thrombocythemia
- GMP
- granulocyte-monocyte progenitor
- MEP
- myeloid erythroid progenitor
- MPN
- myeloproliferative neoplasm
- NF-E2
- nuclear factor erythroid-2
- PMF
- primary myelofibrosis
- PV
- polycythemia vera
- UPN
- unique patient number
- WBC
- white blood cell
References
- Akashi K., Traver D., Miyamoto T., Weissman I.L. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- Amrolia P.J., Ramamurthy L., Saluja D., Tanese N., Jane S.M., Cunningham J.M. 1997. The activation domain of the enhancer binding protein p45NF-E2 interacts with TAFII130 and mediates long-range activation of the alpha- and beta-globin gene loci in an erythroid cell line. Proc. Natl. Acad. Sci. USA. 94:10051–10056 10.1073/pnas.94.19.10051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter E.J., Scott L.M., Campbell P.J., East C., Fourouclas N., Swanton S., Vassiliou G.S., Bench A.J., Boyd E.M., Curtin N., et al. ; Cancer Genome Project 2005. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 365:1054–1061 [DOI] [PubMed] [Google Scholar]
- Ben-David Y., Bani M.R., Chabot B., De Koven A., Bernstein A. 1992. Retroviral insertions downstream of the heterogeneous nuclear ribonucleoprotein A1 gene in erythroleukemia cells: evidence that A1 is not essential for cell growth. Mol. Cell. Biol. 12:4449–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank V., Kim M.J., Andrews N.C. 1997. Human MafG is a functional partner for p45 NF-E2 in activating globin gene expression. Blood. 89:3925–3935 [PubMed] [Google Scholar]
- Bumm T.G., Elsea C., Corbin A.S., Loriaux M., Sherbenou D., Wood L., Deininger J., Silver R.T., Druker B.J., Deininger M.W. 2006. Characterization of murine JAK2V617F-positive myeloproliferative disease. Cancer Res. 66:11156–11165 10.1158/0008-5472.CAN-06-2210 [DOI] [PubMed] [Google Scholar]
- Chaturvedi C.P., Hosey A.M., Palii C., Perez-Iratxeta C., Nakatani Y., Ranish J.A., Dilworth F.J., Brand M. 2009. Dual role for the methyltransferase G9a in the maintenance of beta-globin gene transcription in adult erythroid cells. Proc. Natl. Acad. Sci. USA. 106:18303–18308 10.1073/pnas.0906769106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Wen S., Fukuda M.N., Gavva N.R., Hsu D., Akama T.O., Yang-Feng T., Shen C.K. 2001. Human ITCH is a coregulator of the hematopoietic transcription factor NF-E2. Genomics. 73:238–241 10.1006/geno.2001.6512 [DOI] [PubMed] [Google Scholar]
- Christensen J.L., Weissman I.L. 2001. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. USA. 98:14541–14546 10.1073/pnas.261562798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium E.P.; ENCODE Project Consortium 2011. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9:e1001046 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers C., Chaturvedi C.P., Ranish J.A., Juban G., Lai P., Morle F., Aebersold R., Dilworth F.J., Groudine M., Brand M. 2007. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol. Cell. 27:573–584 10.1016/j.molcel.2007.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen J.T., Antonarakis S.E. 2001. Nomenclature for the description of human sequence variations. Hum. Genet. 109:121–124 10.1007/s004390100505 [DOI] [PubMed] [Google Scholar]
- el-Kassar N., Hetet G., Brière J., Grandchamp B. 1997. Clonality analysis of hematopoiesis in essential thrombocythemia: advantages of studying T lymphocytes and platelets. Blood. 89:128–134 [PubMed] [Google Scholar]
- Gavva N.R., Gavva R., Ermekova K., Sudol M., Shen C.J. 1997. Interaction of WW domains with hematopoietic transcription factor p45/NF-E2 and RNA polymerase II. J. Biol. Chem. 272:24105–24108 10.1074/jbc.272.39.24105 [DOI] [PubMed] [Google Scholar]
- Gavva N.R., Wen S.C., Daftari P., Moniwa M., Yang W.M., Yang-Feng L.P., Seto E., Davie J.R., Shen C.K. 2002. NAPP2, a peroxisomal membrane protein, is also a transcriptional corepressor. Genomics. 79:423–431 10.1006/geno.2002.6714 [DOI] [PubMed] [Google Scholar]
- Goerttler P.S., Kreutz C., Donauer J., Faller D., Maiwald T., März E., Rumberger B., Sparna T., Schmitt-Gräff A., Wilpert J., et al. 2005. Gene expression profiling in polycythaemia vera: overexpression of transcription factor NF-E2. Br. J. Haematol. 129:138–150 10.1111/j.1365-2141.2005.05416.x [DOI] [PubMed] [Google Scholar]
- Guibal F.C., Alberich-Jorda M., Hirai H., Ebralidze A., Levantini E., Di Ruscio A., Zhang P., Santana-Lemos B.A., Neuberg D., Wagers A.J., et al. 2009. Identification of a myeloid committed progenitor as the cancer-initiating cell in acute promyelocytic leukemia. Blood. 114:5415–5425 10.1182/blood-2008-10-182071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzinger T., Reed S.I. 1998. Cyclin D3 is rate-limiting for the G1/S phase transition in fibroblasts. J. Biol. Chem. 273:14958–14961 10.1074/jbc.273.24.14958 [DOI] [PubMed] [Google Scholar]
- Hung H.L., Kim A.Y., Hong W., Rakowski C., Blobel G.A. 2001. Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J. Biol. Chem. 276:10715–10721 10.1074/jbc.M007846200 [DOI] [PubMed] [Google Scholar]
- Hussein K., Van Dyke D.L., Tefferi A. 2009. Conventional cytogenetics in myelofibrosis: literature review and discussion. Eur. J. Haematol. 82:329–338 10.1111/j.1600-0609.2009.01224.x [DOI] [PubMed] [Google Scholar]
- Igarashi K., Kataoka K., Itoh K., Hayashi N., Nishizawa M., Yamamoto M. 1994. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature. 367:568–572 10.1038/367568a0 [DOI] [PubMed] [Google Scholar]
- James C., Ugo V., Le Couédic J.P., Staerk J., Delhommeau F., Lacout C., Garçon L., Raslova H., Berger R., Bennaceur-Griscelli A., et al. 2005. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 434:1144–1148 10.1038/nature03546 [DOI] [PubMed] [Google Scholar]
- Jamieson C.H., Gotlib J., Durocher J.A., Chao M.P., Mariappan M.R., Lay M., Jones C., Zehnder J.L., Lilleberg S.L., Weissman I.L. 2006. The JAK2 V617F mutation occurs in hematopoietic stem cells in polycythemia vera and predisposes toward erythroid differentiation. Proc. Natl. Acad. Sci. USA. 103:6224–6229 10.1073/pnas.0601462103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K.B., Gründer A., Hadlich T., Wehrle J., Gothwal M., Bogeska R., Seeger T.S., Kayser S., Pham K.-B., Jutzi J.S., et al. 2012. A novel murine model of myeloproliferative disorders generated by overexpression of the transcription factor NF-E2. J. Exp. Med. 209:35–50 10.1084/jem.20110540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiekhaefer C.M., Boyer M.E., Johnson K.D., Bresnick E.H. 2004. A WW domain-binding motif within the activation domain of the hematopoietic transcription factor NF-E2 is essential for establishment of a tissue-specific histone modification pattern. J. Biol. Chem. 279:7456–7461 10.1074/jbc.M309750200 [DOI] [PubMed] [Google Scholar]
- Kralovics R., Passamonti F., Buser A.S., Teo S.S., Tiedt R., Passweg J.R., Tichelli A., Cazzola M., Skoda R.C. 2005. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352:1779–1790 10.1056/NEJMoa051113 [DOI] [PubMed] [Google Scholar]
- Lacout C., Pisani D.F., Tulliez M., Gachelin F.M., Vainchenker W., Villeval J.L. 2006. JAK2V617F expression in murine hematopoietic cells leads to MPD mimicking human PV with secondary myelofibrosis. Blood. 108:1652–1660 10.1182/blood-2006-02-002030 [DOI] [PubMed] [Google Scholar]
- Lee T.L., Shyu Y.C., Hsu T.Y., Shen C.K. 2008. Itch regulates p45/NF-E2 in vivo by Lys63-linked ubiquitination. Biochem. Biophys. Res. Commun. 375:326–330 10.1016/j.bbrc.2008.07.164 [DOI] [PubMed] [Google Scholar]
- Li Y.J., Higgins R.R., Pak B.J., Shivdasani R.A., Ney P.A., Archer M., Ben-David Y. 2001. p45(NFE2) is a negative regulator of erythroid proliferation which contributes to the progression of Friend virus-induced erythroleukemias. Mol. Cell. Biol. 21:73–80 10.1128/MCB.21.1.73-80.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignotte V., Eleouet J.F., Raich N., Romeo P.H. 1989. Cis- and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc. Natl. Acad. Sci. USA. 86:6548–6552 10.1073/pnas.86.17.6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A., Merad Boudia M., Lehalle D., Massrieh W., Derjuga A., Blank V. 2006. Regulation of globin gene transcription by heme in erythroleukemia cells: analysis of putative heme regulatory motifs in the p45 NF-E2 transcription factor. Antioxid. Redox Signal. 8:68–75 10.1089/ars.2006.8.68 [DOI] [PubMed] [Google Scholar]
- Mueller J.M., Pahl H.L. 2000. Assaying NF-kappa B and AP-1 DNA-binding and transcriptional activity. Methods Mol. Biol. 99:205–216 [DOI] [PubMed] [Google Scholar]
- Nikoloski G., Langemeijer S.M., Kuiper R.P., Knops R., Massop M., Tönnissen E.R., van der Heijden A., Scheele T.N., Vandenberghe P., de Witte T., et al. 2010. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 42:665–667 10.1038/ng.620 [DOI] [PubMed] [Google Scholar]
- O’Shea E.K., Rutkowski R., Kim P.S. 1989. Evidence that the leucine zipper is a coiled coil. Science. 243:538–542 10.1126/science.2911757 [DOI] [PubMed] [Google Scholar]
- Passegué E., Jamieson C.H., Ailles L.E., Weissman I.L. 2003. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc. Natl. Acad. Sci. USA. 100:11842–11849 10.1073/pnas.2034201100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué E., Wagers A.J., Giuriato S., Anderson W.C., Weissman I.L. 2005. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 202:1599–1611 10.1084/jem.20050967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskind W.H., Jacobson R., Murphy S., Adamson J.W., Fialkow P.J. 1985. Evidence for the involvement of B lymphoid cells in polycythemia vera and essential thrombocythemia. J. Clin. Invest. 75:1388–1390 10.1172/JCI111840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelz R., Pilz I.H., Mutschler M., Pahl H.L. 2010. Of mice and men: human RNA polymerase III promoter U6 is more efficient than its murine homologue for shRNA expression from a lentiviral vector in both human and murine progenitor cells. Exp. Hematol. 38:792–797 10.1016/j.exphem.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Schaub F.X., Looser R., Li S., Hao-Shen H., Lehmann T., Tichelli A., Skoda R.C. 2010. Clonal analysis of TET2 and JAK2 mutations suggests that TET2 can be a late event in the progression of myeloproliferative neoplasms. Blood. 115:2003–2007 10.1182/blood-2009-09-245381 [DOI] [PubMed] [Google Scholar]
- Schreiber E., Matthias P., Müller M.M., Schaffner W. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419 10.1093/nar/17.15.6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. 1995. D-type cyclins. Trends Biochem. Sci. 20:187–190 10.1016/S0968-0004(00)89005-2 [DOI] [PubMed] [Google Scholar]
- Sidon P., Heimann P., Lambert F., Dessars B., Robin V., El Housni H. 2006. Combined locked nucleic acid and molecular beacon technologies for sensitive detection of the JAK2V617F somatic single-base sequence variant. Clin. Chem. 52:1436–1438 10.1373/clinchem.2006.066886 [DOI] [PubMed] [Google Scholar]
- Steimle C., Lehmann U., Temerinac S., Goerttler P.S., Kreipe H., Meinhardt G., Heimpel H., Pahl H.L. 2007. Biomarker analysis in polycythemia vera under interferon-alpha treatment: clonality, EEC, PRV-1, and JAK2 V617F. Ann. Hematol. 86:239–244 10.1007/s00277-006-0214-1 [DOI] [PubMed] [Google Scholar]
- Swerdlow S., Campo E., Harris N. 2008. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IRAC Press Lyon, France; 439 pp [Google Scholar]
- Temerinac S., Klippel S., Strunck E., Röder S., Lübbert M., Lange W., Azemar M., Meinhardt G., Schaefer H.E., Pahl H.L. 2000. Cloning of PRV-1, a novel member of the uPAR receptor superfamily, which is overexpressed in polycythemia rubra vera. Blood. 95:2569–2576 [PubMed] [Google Scholar]
- Vainchenker W., Delhommeau F., Constantinescu S.N., Bernard O.A. 2011. New mutations and pathogenesis of myeloproliferative neoplasms. Blood. 118:1723–1735 10.1182/blood-2011-02-292102 [DOI] [PubMed] [Google Scholar]
- Wang K., Swierczek S., Hickman K., Hakonarson H., Prchal J.T. 2011. Convergent mechanisms of somatic mutations in polycythemia vera. Discov. Med. 12:25–32 [PMC free article] [PubMed] [Google Scholar]
- Wang W., Schwemmers S., Hexner E.O., Pahl H.L. 2010. AML1 is overexpressed in patients with myeloproliferative neoplasms and mediates JAK2V617F-independent overexpression of NF-E2. Blood. 116:254–266 10.1182/blood-2009-11-254664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Bartsch U., Stocking C., Fehse B. 2008. A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol. Ther. 16:698–706 10.1038/mt.2008.6 [DOI] [PubMed] [Google Scholar]
- Wernig G., Mercher T., Okabe R., Levine R.L., Lee B.H., Gilliland D.G. 2006. Expression of Jak2V617F causes a polycythemia vera-like disease with associated myelofibrosis in a murine bone marrow transplant model. Blood. 107:4274–4281 10.1182/blood-2005-12-4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaleskas V.M., Krause D.S., Lazarides K., Patel N., Hu Y., Li S., Van Etten R.A. 2006. Molecular pathogenesis and therapy of polycythemia induced in mice by JAK2 V617F. PLoS ONE. 1:e18 10.1371/journal.pone.0000018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.