Abstract
Although extensive investigation has been made on miR-29a in relation to malignancies, only a little information has been provided about the angiogenic property of this miRNA so far. Herein, we sought to investigate the role of miR-29a in regulating cell cycle and angiogenic phenotype of endothelial cells. The results showed that miR-29a is highly expressed and upregulated by hypoxia-mimicking reagents in human umbilical vein endothelial cells (HUVEC). Consistent with this preliminary finding, introduction of exogenous agomiR-29a, or antagomir-29a altered cell cycle progression and promoted, or repressed the proliferation and tube formation of HUVEC, respectively. Furthermore, by using luciferase reporter assay, the expression of HBP1, a suppressor transcription factor was directly regulated by miR-29a through 3′-UTR. Increased or decreased HBP1 protein level was associated with the inhibition or overexpression of miR-29a, respectively. We conclude that miR-29a has a significant role in regulating cell cycle, proliferation and angiogenic properties of HUVEC, and this function is likely mediated through HBP1 protein at the post-transcriptional level. As a novel molecular target, miR-29a may have a potential value for the treatment of angiogenesis-associated diseases such as cardiovascular diseases and cancers.
Keywords: miRNA-29a, angiogenesis, endothelial cell, HBP1, hypoxia
1. Introduction
MiRNAs (miRNAs) are highly conserved, single-stranded, non-coding small RNAs, which regulate gene expression at the post-transcriptional level by inhibiting protein translation from mRNA or by promoting degradation of mRNA. Some specific miRNAs are related to hypoxia and involved in hypoxia-dependent modification of angiogenic property of endothelium [1,2]. Accumulating evidence indicates that miRNAs can modulate various aspects of angiogenesis, such as proliferation, migration, and morphogenesis of endothelial cells [3]. Although extensive investigation has been made on miR-29a in relation to malignancies [4–7], only a little information has been provided about the angiogenic property of this miRNA so far. MiR-29a has been found to be highly expressed in endothelium [8], however, its functions in endothelium and angiogenesis are not clearly clarified. Therefore, the roles of miR-29a in angiogenesis deserve to further study. Recently, a clinical study showed that plasma miR-29a levels of myocardial infarct patients were significantly upregulated during the first 1~5 days [9]. As a novel molecular target, miR-29a may have a potential value for treatment of angiogenesis-associated diseases.
Tumor suppressor protein HBP1 (HMG box-containing protein-1) is a transcriptional repressor that binds to the promoter region of target genes. HBP1 inhibited Wnt signaling and regulated cell proliferation in breast cancer cells [10], and it was upregulated in miRNA-deficient endothelial cells and predicted to be a target of miR-29a by using computational prediction algorithm such as TargetScan, Pictar and miRbase [11].
In this study, we investigate: (1) whether miR-29a expression in endothelium is regulated by hypoxia; (2) whether miR-29a directly targets HBP1 expression and regulates angiogenic properties of human endothelial cells.
2. Methods
2.1 Cell culture
Human umbilical venous endothelial cells (HUVEC) were purchased from KeyGEN Biotech (Nanjing, China), and were maintained in RPMI-1640 medium supplemented with 2% FBS and antibiotics [12].
2.2 Hypoxia inducement
Hypoxia inducement was carried out by incubating cells with Cobalt chloride (CoCl2, Yeyuan company, Shanghai, China) or Deferoxamine Mesylate (DFO; Calbiochem, Merk, Germany) at different concentration for 20 hours[2]. Then, cells were harvested and total RNA was isolated for miRNA quantification analysis.
2.3 Plasmid construction
pmiR-RB-Report-HBP1 which contains the hRluc (Renilla luciferase) gene subject to regulation of 3′-UTR (1132 bp) of HBP1 mRNA was created by inserting the whole length 3′-UTR of human HBP1 mRNA into pmiR-RB-Report double luciferase report vector (Ribobio Co, Guangzhou, China). Briefly, the human HBP1 3′-UTR fragment was obtained from the human genome DNA using PCR technique with primers (template: GenBank accession no. NM_012257):
HBP1-F: 5′-CCGCTCGAGACCAGGATGCTTATGTTCTT-3′
HBP1-R: 5′-GAATGCGGCCGCCAATCAGTAGACGTCAGAGAT-3′.
After the sequence was verified by restriction enzyme digestion and sequencing, the fragment was subcloned into pmiR-RB-Report.
Another reporter plasmid pmiR-RB-Report-VEGF which contains whole length 3′-UTR (1901bp) of VEGFA mRNA was also created with similar procedure. The human HBP1 3′-UTR fragment was obtained from the human genome DNA using PCR technique with primers (template: GenBank accession no. NM_001171630):
VEGFA-F: 5′ CCGCTCGAGGCCGGGCAGGAGGAAGGA 3′
VEGFA-R: 5′ GAATGCGGCCGCTTTAAGATATATCTGTATTTCTTTG 3′
2.4 Transfection
For overexpression of miR-29a, Agomir-29a (Ribobio Co, Guangzhou, China), a kind of thio-, cholesterol-modified miRNA mimic (double-stranded RNAs) was used to transfect HUVEs, and Agomir-Co, which was similar to Agomir-29a but with a scramble seeding sequence, was used as control.
Agomir-29a sequences used are:hsa-29a-Agomir sequence(Agomir-29a):
UAGCACCAUCUGAAAUCGGUUA
hsa-miR-29a-Agomir scramble control (Agomir-Co):
UGACAACCUCUGAAAUCGGUUA
For inhibition of miR-29a, a specific inhibitor, Antagomir-29a (Ribobio Co, Guangzhou, China) is used and Antagomir Negative Control (Antagomir-Co) was also used as control.
Briefly, cells in wells of 96-well/6-well plates were grow to 50~70% confluence. Agomir/Antagomir at final concentration of 100 nM were used to transfect the cells in the presence of Lipofectin Reagent (Invitrogen) according to the manufacturer’s protocol. Transfection efficiency in our system was evaluated by Cy3-labeled Agomir-Control (Ribobio Co, Guangzhou, China).
2.5 RNA isolation and miRNA Microarray analysis
Cells were collected, washed and lysed with 700μl Qiazol lysis reagent (Qiagen). Total RNA isolation, quality control, labeling, hybridization and miRNA microarray analysis were performed by LC Sciences (Houston, TX, USA). Data were analyzed by first subtracting the background and then normalizing to the signals using a LOWESS filter.
2.6 RNA Purification and qRT-PCR assay for miRNA expression
Total RNAs were purified using RNAiso Plus (TaKara) as manufacturer’s protocols. miRNAs were assayed by real-time reverse transcriptase polymerase chain reaction (qRT-PCR). The cDNAs were produced from total RNA with Reverse Transcription Kit (Takara, Dalian, China) and miRNA specific Bulge-loop™ miRNA RT primers (Ribobio Co, Guangzhou, China). Human U6 snRNA was used as endogenous control for data normalization. Real-time PCR was performed on the ABI Prism 7500 Sequence Detection System (Applied Biosystems) using an SYBR Green I Real-Time PCR kit (TaKara, Dalian, China). The relative expression levels of miRNAs were calculated and quantified by using the 2−Ct method after normalization by expression of endogenous control [13].
2.7 Enzyme-Linked Immunosorbent Assay (ELISA) for HIF1αprotein
Levels of HIF1α protein in HUVECs were determined using the Human HIF-1α ELISA Assay Kit (Shanghai Yili Co, Shanghai, China) according the manufacturer’s instructions. The Means of OD values in 450 nm were used as represents of HIF1α protein expression.
2.8 Cell proliferation assay
Cell proliferation was assessed using Cell Counting Kit-8 (Dojindo Laboratories, Japan) as the manufacturer’s protocol. At each time point of 24, 48, 64 and 72 h after Agomir/Antagomir transfection, 10μl of cell proliferation reagent WST-8 solution was added to each well of a 96-well plate and the cells were incubated 0.5~1 hour in a CO2 incubator. Absorbance at 450 nm (O.D. value) was measured by a microplate reader, and absorbance at 630 nm was used as reference. The average OD values were used as represents of the total cell numbers of each group.
2.9 In vitro tube network formation assay
For tube network formation assay, each well of 96-well plates was pre-coated with 50μl of Matrigel (BD Biosciences Discovery Labware, Bedford, MA, USA) and allowed to polymerize for 30 min at 37°C. Then, cells were seeded on Matrigel-coated wells at a density of 2×104 cells per well in RPMI-1640 medium containing 1% FBS at 37°C. Cells started to form tubes at 4 h. Tube formation was optimal after 8 h. The tube images were taken at 7~8 h with a digital camera attached to an inverted phase-contrast microscope. Total tube length in each well was measured and calculated using image software (IPP).
2.10 Cell cycle analysis
HUVECs in wells of 6-well plate were transfected with Agomir-29a/Antagomir-29a and incubated for 48h. Then, the cells were harvested for Flow Cytometry analysis. Briefly, Cells were fixed in 70% ethanol, suspended in PBS, and incubated with RNase A (0.5μg/μl) for 30min at 37°C. Then, the cells were stained with propidium iodide on ice for 1 h and subsequently measured with FACScan Flow Cytometry (Becton Dickinson, San Jose, Calf, USA). The percentage of cells in the G1, S, and G2 phases were analyzed by using the Motif LT Softwre.
2.11 Luciferase reporter assay
Cells in wells of a 96-well plate were grown to 50~60% confluence and co-transfected with 0.1 μg pmiR-RB-Report-HBP1 (or pmiR-RB-Report-VEGF) and 100 nM Agomir-29a/Antagomir-29a in the presence of Lipofectin Reagent (Invitrogen). Then, cells were lysed for luciferase assay using Dual-Luciferase Reporter Assay System (Promega). The ratio of Renilla luciferase activity to Firefly luciferase activity in each sample was served as a normalized luciferase activity and expressed as fold-induction.
2.12 Western blot for HBP1 protein
HUVECs in 6-well plates were trypsinized and washed with PBS. Then, the cells were lysed by 70 μl of ice-cold RIPA Cell Lysis Buffer. Cell lysates (10 μg protein) were separated on 12% SDS–PAGE gel and electroblotted onto a nitrocellulose membrane. Membrane was blocked and detected with rabbit anti-HBP1 polyclonal antibody (ab83402, Abcam Inc, USA) and a horseradish peroxidase-conjugated secondary antibody. β-actin was used as endogenous control.
2.13 Statistical analysis
Data are expressed as Mean±SE. Comparison of the means was performed using unpaired Student’s t-test and P<0.05 was considered statistically significant.
3. Results
3.1 MiR-29a expression was abundant in HUVECs
To investigate the role of miRNAs in angiogenesis, the miRNA expression profile in HUVECs was examined by miRNAs array, and 68 relative high-expressed miRNAs were shown (Table). Expressions of 15 miRNAs were further evaluated by qRT-PCR. The results showed that miR-21, miR-720, miR-29a, miR-16, miR-92a, miR-320 and let-7 family are abundant in HUVECs (Fig. 1A). Among them, miR-29a was predicted to target several mRNAs of important angiogenesis-related gene, including HBP1 and VEGF, etc.
Table 1.
Table MiRNAs microarray profile of HUVECs
The expression of 694 miRNA transcripts were detected in cultured HUVECs, and 68 relative highly expressed miRNAs (signal intensity larger than 50) were shown in the table.
| Probe_ID | Averaged Signal |
|---|---|
| let-7a | 1,080.38 |
| let-7b | 854.26 |
| let-7c | 403.75 |
| let-7d | 418.33 |
| let-7e | 399.88 |
| let-7f | 578.61 |
| let-7g | 63.92 |
| let-7i | 812.77 |
| miR-100 | 295.60 |
| miR-103 | 114.16 |
| miR-106a | 83.58 |
| miR-106b | 77.32 |
| miR-107 | 128.29 |
| miR-125a-5p | 295.34 |
| miR-125b | 294.53 |
| miR-1268 | 70.42 |
| miR-1275 | 63.53 |
| miR-1280 | 1,300.27 |
| miR-1308 | 130.42 |
| miR-146a | 199.31 |
| miR-151-3p | 66.17 |
| miR-151-5p | 291.11 |
| miR-155 | 83.74 |
| miR-15b | 116.65 |
| miR-16 | 134.27 |
| miR-17 | 74.25 |
| miR-181a | 63.89 |
| miR-182 | 812.22 |
| miR-1826 | 13,060.51 |
| miR-183 | 116.02 |
| miR-185 | 55.40 |
| miR-191 | 361.51 |
| miR-1915 | 139.00 |
| miR-1974 | 2,712.38 |
| miR-1975 | 4,701.08 |
| miR-1977 | 98.78 |
| miR-1978 | 70.86 |
| miR-1979 | 12,216.99 |
| miR-200b | 88.92 |
| miR-20a | 58.60 |
| miR-21 | 3,517.42 |
| miR-210 | 65.99 |
| miR-22 | 62.60 |
| miR-221 | 296.81 |
| miR-222 | 803.28 |
| miR-23a | 486.17 |
| miR-23b | 150.80 |
| miR-24 | 199.34 |
| miR-25 | 299.40 |
| miR-26a | 638.97 |
| miR-29a | 316.15 |
| miR-30b | 54.01 |
| miR-30c | 53.51 |
| miR-30d | 50.56 |
| miR-320a | 1,828.82 |
| miR-320b | 774.10 |
| miR-320c | 1,275.44 |
| miR-320d | 252.32 |
| miR-361-5p | 173.16 |
| miR-423-5p | 277.46 |
| miR-638 | 714.68 |
| miR-663 | 113.07 |
| miR-720 | 2,287.82 |
| miR-762 | 107.19 |
| miR-92a | 1,219.34 |
| miR-92b | 245.87 |
| miR-93 | 102.15 |
| miR-99b | 160.51 |
Fig. 1.
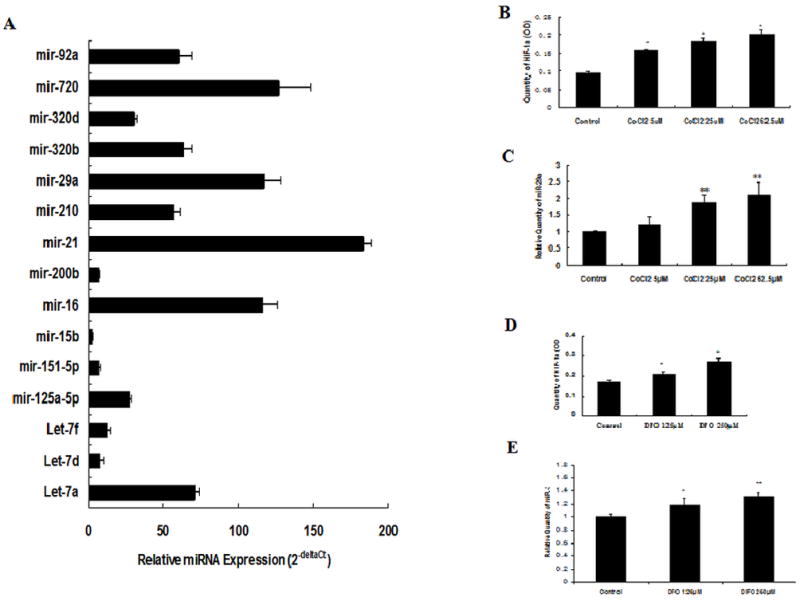
Expressions of 15 relatively high-expressed miRNAs in HUVECs(A) and the effect of hypoxia on miR-29a expression in HUVECs(B→E)
Expressions of 15 relatively high-expressed miRNAs in HUVECs were assayed by qRT-PCR. Data represent Means of value in 3 samples. To study the effect of hypoxia on miR-29a expression in endothelial cells, miR-29a and HIF-1α expression in HUVECs with or without chemical mimics CoCl2 (0, 5, 25, 62.5μM) or DFO (0, 125, 250μM) treatment were analyzed by qRT-PCR and ELISA method. CoCl2 or DFO treatment noticeably increased expression of both HIF-1α and miR-29a (Fig. 1 B→E). Data represents Mean ± SE of three independent experiments, *P <0.05, **P <0.01 vs. Control.
3.2 MiR-29a expression was upregulated by hypoxia treatment in HUVECs
To study the effect of hypoxia on miR-29a expression in endothelial cells, we performed qRT-PCR analysis using HUVECs with or without DFO or CoCl2 treatment. DFO or CoCl2 treatment noticeably increased both miR-29a and HIF-1α expression in a dose-dependent manner (Fig. 1B→E). These data suggest that miR-29a expression can be stimulated by hypoxia.
3.3 MiR-29a regulated cell cycle in HUVECs
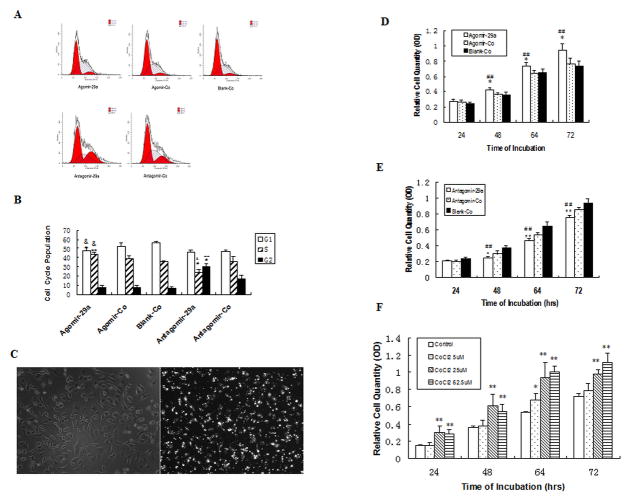
To investigate whether miR-29a regulated cell cycle of endothelial cells, Cell cycles of Agomir-29a or Antagomir-29a-transfected HUVECs were analyzed by Flow Cytometry fluorescence activated cell sorter (FACS). The results showed that G1 phase population of Agomir-29a-transfected cells was decreased, while S phase population was increased, compared with that of Agomir-Co groups, as well as Blank-Control groups (Fig. 2A, B), while inhibition of miR-29a by Antagomir-29a decreased S phase population and increased G2 phase population, compared with that of Antagomir-Control and Blank-Control groups. These data suggested that elevated miR-29a expression might accelerate G1 to S phase shift in endothelial cells, while inhibition of miR-29a might lead to a G2 phase arrest.
Fig. 2.
Effect of miR-29a on cell cycle(A→C) and proliferation(C→F) of HUVECs.
HUVECs were transfected with Agomir-29a or Antagomir-29a and subjected to Propidium iodide (PI) Flow Cytometry analysis. Overexpression of miR-29a by agomir-29a accelerated G1 to S cell cycle transition, while inhibition of miR-29a by antagomir-29a increase G2 population and reduced S phase population. A: Representative Flow Cytometry images of three independent experiments. B: Comparison of cell cycle populations in each group. Data represents Mean ± SE of three independent experiments, * P <0.05, ** P <0.01 vs. Blank-Co. & P <0.05 vs. Agomir-Co, # P <0.05 vs. Antagomir-Co. To show transfection efficiency of Agomir, Cy3-labeled Agomir-Control was used to transfect HUVECs by lipofectin and observed under phase-control microscope (C, left) or fluorescence microscope (C, right).
Proliferation of Agomir-29a/Antagomir-29a transfected cells or CoCl2 treated cells were analyzed by CCK-8 at different time point during 24~72 hours. D: Relative cell proliferation levels of HUVECs transfected with Agomir-29a, Agomir-control (Agomir-Co) and Blank-control (Blank-Co). E: Relative cell proliteration levels of HUVECs after transfection of Antagomir-29a, Antagomir-control (Antagomir-Co) or Blank-control (Blank-Co). * P<0.05, ** P<0.01 vs. Agomir-Co; # P<0.05, # # P<0.01 vs. Blank-Co. F: Relative cell proliteration levels of HUVECs treated with different concentration of CoCl2, * P<0.05, ** P<0.01 vs. Control of the same time point.
To determine transfection efficiency of Agomir-29a (or Antagomir-29a) in our system, Cy3-labeled Agomir-Control was used to transfect the endothelial cells in the presence or absence of Lipofectin. The results showed relatively high transfection efficiency of Agomir (or Antagomir-29a) in HUVEC (Fig. 2C).
3.4 MiR-29a regulated proliferation of HUVECs
To determine whether miR-29a overexpression mimics hypoxia-promoted proliferation and angiogenesis in endothelial cells, cell proliferations of Agomir-29a-transfected cells or treated with CoCl2 were assayed by CCK8 assay. Either miR-29a overexpression by agomir-29a or treated with CoCl2 significantly promoted cell proliferation of HUVECs at 48–72h (Fig. 2D, F). To further confirm the effect of miR-29a on cell proliferation in HUVECs, cell proliferation of antagomir-29a-transfected cell was also assessed by CCK8 assay (Fig. 2E). MiR-29a inhibition by antagomir-29a significantly suppressed cell proliferation of HUVECs at 48–72h.
3.5 MiR-29a promoted tube network formation of HUVECs
To investigate the influence of miR-29a on angiogenic properties of endothelial cell, the tube network formation on Matrigel of agomir-29a/antangomir-29a-transfected HUVECs was examined. The results showed that overexpression of miR-29a promoted tube network formation of HUVECs (Fig. 3A, B), It was suggested that elevated miR-29a in endothelial cells might activate endothelial cells and promote angiogenesis.
Fig. 3.
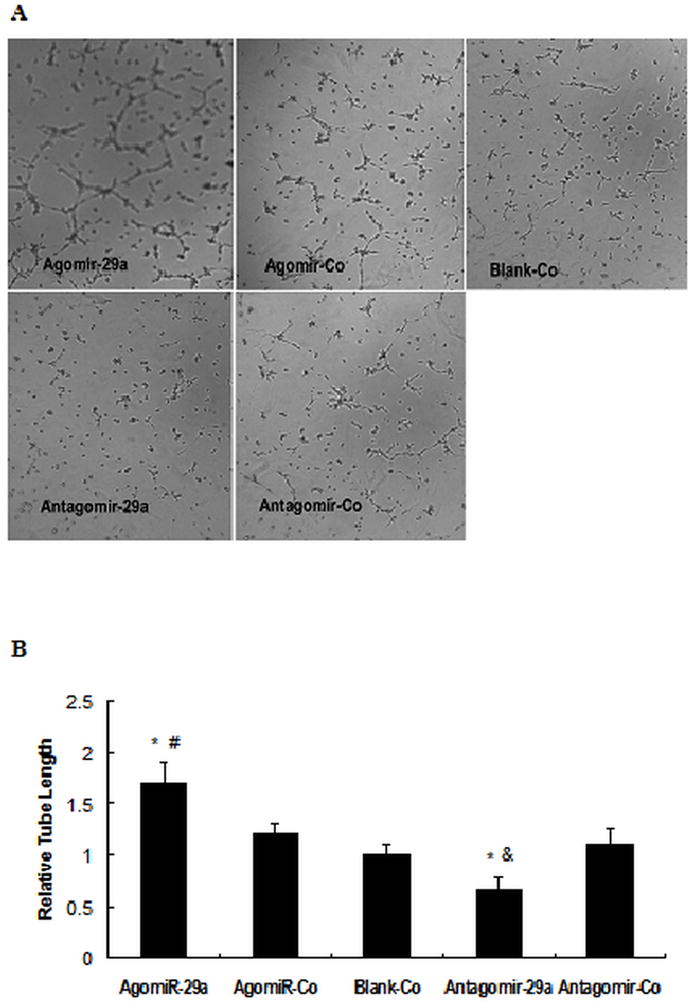
Effect of miR-29a on tube network formation in HUVECs.
HUVECs were transfected with Agomir-29a/Agomir-Co or Antagomir-29a /Antagomir-control. After 48 hrs, cells in suspension were seeded on wells that precoated with Matrigel. Tube network formations were measured after 6~8h. A: Representative images of three independent experiments. B. Total tube length was measured with image analysis software and normalized to that of Blank-Co group. * P<0.01 vs Blank-Co, # P<0.01 vs. Agomir-Co, & P<0.01 vs. Antagomir-Co.
3.6 HBP1 expression was directly regulated by miR-29a
To determine whether HBP1 is a direct target of miR-29a in endothelial cells, pmiR-RB-Report-HBP1 luciferase reporter which contains 3′-UTR of HBP1 mRNA was used to examine the effect of miR-29a on mRNA expression of HBP1. In the meantime, HBP1 protein expression was also assayed by Western blot. Overexpression of miR-29a by agomir-29a led to about remarkable inhibition of both HBP1-3′UTR reporter luciferase expression and HBP1 protein expression (Fig. 4 A,B,C), while inhibition of miR-29a by antagmir-29a induced remarkable increase of both HBP1-3′UTR reporter luciferase expression and HBP1 protein expression (Fig. 4. D,E,F). Meanwhile, over-expression or inhibition of miR-29a by agomir-29a/ antagmir-29a did not remarkably influence luciferase express of VEGF--3′UTR reporter (data not show). These results suggested that miR-29a might directly regulate HBP1 expression at the post-transcriptional level.
Fig. 4.
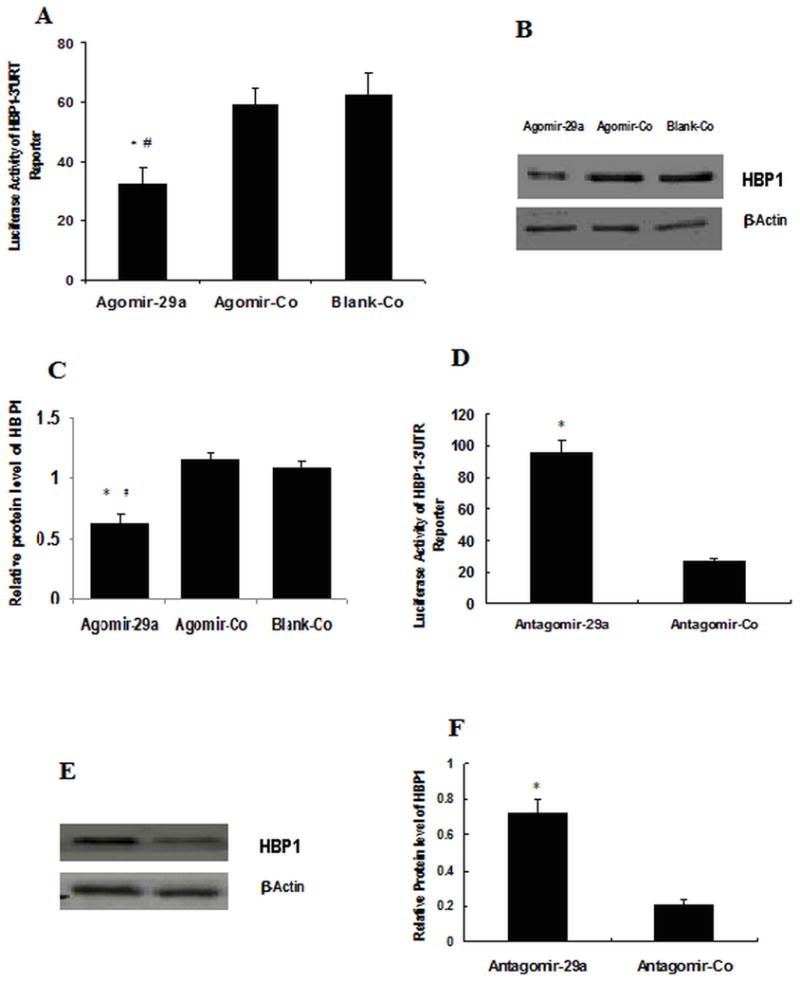
miR-29a target identification.
Luciferase reporter constructs (pmiR-RB-Report-HBP1) containing the 3′-UTR of HBP1 mRNA and Agomir-29a or Antagomir-29a were used to transfect HUVECs for 48 hs, and the cells were lysed for luciferase assay. Some groups of cells transfected with only Agomir-29a/Agomir-Co or Antagomir-29a/Antagomir-Co were lysed and subjected to western blot analysis. A. Relative luciferase activities of Agomir-29a-transfected cells. Agomir-29a: cells transfected with Agomir-29a; Agomir-Co: transfected with Agomir-control, Blank-Co: medium only; * P<0.01 vs. Agomir-29a; # P<0.01 vs. Blank-Co. Data represent Mean±SE of three independent experiments; B: HBP1 protein level of cells transfected with Agomir-29a, Agomir-Co and Blank-Co; C and F: Intensities of each protein band were quantified by Quantity One Software. The results were standardized against the levels of β-actin and presented as the relative intensity. The experiments were repeated there times; * P<0.05 vs. Agomir-Co (or Antagomir-Co), # P<0.01 vs. Blank-Co. D: Relative luciferase activities of Antagomir-29a-transfected cells. Antagmir-29a: cells transfected with Antagomir-29a; Antagmir-Co: transfected with Antagmir-control; ** P<0.01 vs. 29a-Antagmir; E: HBP1 protein level of cells transfected with Antagomir-29a, Antagomir-Co.
4. Discussion
As a crucial pathogenic component of major cardiovascular diseases and tumor microenvironment, hypoxia plays an important role in activation of endothelial cells and angiogenesis [14]. Endothelium-related miRNAs such as miR-210 have been reported to be upregulated by hypoxia and involved in modification of angiogenic property of endothelium [15]. Our results showed that miR-29a expression can be induced by a hypoxia-mimicking reagent in a dose-dependent manner in HUVECs. These data support the novel notion that miR-29a may be stimulated by hypoxia in endothelium.
We further investigated the effects of alteration of miR-29a expression on cell cycle, proliferation and angiogenic properties in endothelium. The results showed that G1 to S cell cycle transition of HUVECs can be accelerated by miR-29a overexpression. Enforced expression of miR-29a in endothelium remarkably promoted cell proliferation and tube network formation on Matrigel. Previous study showed that miR-29a promoted progenitor cell proliferation by expediting G1 to S cell cycle transition [16], and miR-29a expression was upregulated in a metastatic breast cancer cell line [17]. MiR-29a transgenic mice led to expanded CD5+ B-cell population and development of indolent CLL (chronic lymphocytic leukemia) phenotype [18]. Our results indicated that the functions of miR-29a in endothelial cells were similar to that of hematopoietic progenitor cells. In human osteoblast, miR-29a increased Wnt signaling by targeting some negative regulators of Wnt signaling [19], which also supported the promotive effect of miR-29a on cell proliferation, at least in some types of cells.
HBP1 is a transcriptional repressor and has been shown to negatively regulate the cell cycle. In human breast cancer, a correlation between HBP1 under-expression and poor prognosis has been described [10]. HBP1 represses some growth regulatory genes such as cyclin D1, c-myc, etc, and is an inhibitor of G1 progression [20]. Previous studies have demonstrated. that HBP1 negatively regulates p47phox of the NADPH oxidase complex, a signaling messengers in driving angiogenesis [11]. Lowering of cellular miRNA content by Dicer knockdown can lead to elevated HBP1 expression and impaired angiogenic response in endothelial cells, while knockdown of HBP1 restored the angiogenic response of miRNA-deficient endothelial cells [11]. Our study showed that inhibition of miR-29a by specific antagomir led to remarkable increase of both 3′UTR reporter luciferase and protein expression of HBP1, while overexpression of miR-29a remarkably suppressed HBP1 expression, leading to a shift of G1 to S phase population, which suggested that the role of miR-29a in regulating angiogenic properties of endothelium was partially attributed to targeting HBP1. Angiogenesis is essential in the development, progression and metastasis of solid tumors. MiR-29a has been proved to be associated to metastasis of some human carcinomas such as breast carcinoma, colorectal carcinoma [17, 21]. Other targets may be involved in regulative role of miR-29a in angiogenesis such as tristetraprolin (TTP), which can inhibit Ras-dependent tumor angiogenesis by inducing VEGF mRNA degradation. Overexpression of miR-29a suppressed the expression of TTP and led to EMT (epithelial-mesenchymal transition) and metastasis [17]. In view of the important role of VEGF in angiogenesis, we also analyzed the regulation effect of miR-29a on 3′UTR of VEGF, however, our results did not showed any significant influence of miR-29a on VEGF 3′UTR reporter expression.
In different kinds of cancer cells, miR-29a can act as oncogenic miRNA or tumor suppressor, depending on the context [4]. As a single miRNA can bind to multiple targets whereas single gene may be regulated by multiple miRNAs, further studies need to clarify the complex interactions between the endothelium-related miRNAs and their targets during angiogenesis. The role and mechanism of mir-29a in tumor angiogenesis in vivo need to be clarified further.
In conculusion, miR-29a has a significant role in regulating cell cycle, proliferation and angiogenic properties of HUVEC, and this function is likely mediated through HBP1 protein at the post-transcriptional level.
Highlights.
miR-29a may be stimulated by hypoxia in HUVEC.
miR-29a regulates cell cycle, proliferation and tube network formation of HUVEC.
HMG box-containing protein-1(HBP1) is a direct target of miR-29a.
miR-29a has a potential value for treating angiogenesis-associated diseases.
Acknowledgments
This work was supported by the National 863 Program of China (Grant No. 2008AA02Z109, Dr. Wu), National Natural Science Foundation of China (Grant No.81272354, Dr. Wu), Zhejiang Provincial Natural Science Foundation of China (Grant No.2090107, Dr. Yang), and in part by USA National Institutes of Health grant HL087990 (GL)
Abbreviations used
- miRNA
microRNA
- miR-29a
microRNA-29a
- HUVEC
human umbilical vein endothelial cells
- HBP1
HMG box-containing protein-1
- DFO
deferoxamine mesylate
- CCK-8
cell counting Kit-8
- UTR
untranslated regions
- FACS
fluorescence-activated cell sorting
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ivan M, Harris AL, Martelli F, Kulshreshtha R. Hypoxia response and microRNAs: no longer two separate worlds. Cell Mol Med. 2008;12(5A):1426–1431. doi: 10.1111/j.1582-4934.2008.00398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi H, Chen L, Wang H, Zhu S, Dong C, et al. Synergistic induction of miR-126 by hypoxia and HDAC inhibitors in cardiac myocytes. Biochem Biophys Res Commun. 2013;430:827–32. doi: 10.1016/j.bbrc.2012.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F, Yang Z, Li G. Role of specific microRNAs for endothelial function and angiogenesis. Biochem Biophys Res Commun. 2009;386:549–553. doi: 10.1016/j.bbrc.2009.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pekarsky Y, Croce CM. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget. 2010;1:224–227. doi: 10.18632/oncotarget.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui Y, Su WY, Xing J, Wang YC, Wang P, et al. MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation of p42.3 in human gastric cancer. PLoS One. 2011;6:e25872. doi: 10.1371/journal.pone.0025872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teichler S, Illmer T, Roemhild J, Ovcharenko D, Stiewe T, et al. MicroRNA29a regulates the expression of the nuclear oncogene Ski. Blood. 2011;118:1899–1902. doi: 10.1182/blood-2010-09-306258. [DOI] [PubMed] [Google Scholar]
- 7.Kong G, Zhang J, Zhang S, Shan C, Ye L, et al. Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS One. 2011;6:e19518. doi: 10.1371/journal.pone.0019518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 9.Zile MR, Mehurg SM, Arroyo JE, Stroud RE, DeSantis SM, et al. Relationship between the temporal profile of plasma microRNA and left ventricular remodeling in patients after myocardial infarction. Circ Cardiovasc Genet. 2011;4:614–619. doi: 10.1161/CIRCGENETICS.111.959841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulson KE, Rieger-Christ K, McDevitt MA, Kuperwasser C, Kim J, et al. Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res. 2007;67:6136–6145. doi: 10.1158/0008-5472.CAN-07-0567. [DOI] [PubMed] [Google Scholar]
- 11.Shilo S, Roy S, Khanna S, Sen CK. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:471–477. doi: 10.1161/ATVBAHA.107.160655. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z, Li JC. Stimulation of endothelin-1 gene expression by insulin via phosphoinositide-3 kinase-glycogen synthase kinase-3beta signaling in endothelial cells. Life Sci. 2008;82:512–518. doi: 10.1016/j.lfs.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 14.Nagasawa H. Pathophysiological response to hypoxia - from the molecular mechanisms of malady to drug discovery: drug discovery for targeting the tumor microenvironment. J Pharmacol Sci. 2011;115:446–452. doi: 10.1254/jphs.10r25fm. [DOI] [PubMed] [Google Scholar]
- 15.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han YC, Park CY, Bhagat G, Zhang J, Wang Y, et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400–405. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santanam U, Zanesi N, Efanov A, Costinean S, Palamarchuk A, et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci USA. 2010;107:12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. 2010;285:25221–25231. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escamilla-Powers JR, Daniel CJ, Farrell A, Taylor K, Zhang X, et al. The tumor suppressor protein HBP1 is a novel c-myc-binding protein that negatively regulates c-myc transcriptional activity. J Bio l Chem. 2009;285:4847–4858. doi: 10.1074/jbc.M109.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2011;36:e61–7. doi: 10.1016/j.canep.2011.05.002. [DOI] [PubMed] [Google Scholar]