SUMMARY
Many bacteria contain an ortholog of the Ro autoantigen, a ring-shaped protein that binds noncoding RNAs (ncRNAs) called Y RNAs. In the only studied bacterium, Deinococcus radiodurans, the Ro ortholog Rsr functions in heat stress-induced rRNA maturation and starvation-induced rRNA decay. However, the mechanism by which this conserved protein and its associated ncRNAs act has been obscure. We report that Rsr and the exoribonuclease polynucleotide phosphorylase (PNPase) form an RNA degradation machine that is scaffolded by Y RNA. Single-particle electron microscopy, followed by docking of atomic models into the reconstruction, suggests that Rsr channels single-stranded RNA into the PNPase cavity. Biochemical assays reveal that Rsr and Y RNA adapt PNPase for effective degradation of structured RNAs. A Ro ortholog and ncRNA also associate with PNPase in Salmonella Typhimurium. Our studies identify a new ribonucleoprotein machine and demonstrate that ncRNA, by tethering a protein cofactor, can alter the substrate specificity of an enzyme.
Noncoding RNAs (ncRNAs) are involved in an enormous variety of cellular processes. Many ncRNAs assemble with proteins and function as ribonucleoprotein (RNP) complexes. These RNPs include small nuclear RNPs that function in pre-mRNA splicing, small nucleolar RNPs that modify pre-rRNAs, the telomerase RNP that maintains chromosome ends, and the microRNA/Argonaute complexes that modulate mRNA translation and stability (Hannon et al., 2006). For each of these RNPs, the ncRNA moiety basepairs with nucleic acid targets to direct enzymatic activity to specific RNA and DNA sequences. For other RNPs, the ncRNA influences the function of bound proteins. For example, in the signal recognition particle (SRP), which mediates targeting of secretory and membrane proteins to plasma membranes, the ncRNA increases the interaction of SRP with its receptor and stimulates the GTPase activities of the SRP-receptor complex (Ataide et al., 2011).
In contrast to the many well-characterized RNPs, the function of Ro RNPs has been mysterious since their discovery (Lerner et al., 1981). The major protein component, the ring-shaped Ro 60 kDa autoantigen, is a clinically important target of the immune response in patients with the rheumatic diseases systemic lupus erythematosus and Sjögren’s syndrome (Sim and Wolin, 2011). Ro orthologs are present in most metazoans and approximately 5% of sequenced bacterial genomes (Perreault et al., 2007; Sim and Wolin, 2011). Bacteria containing likely Ro orthologs are present in most phyla, and include the radiation-resistant Deinococcus radiodurans and the human pathogens Bacteroides thetaiotaomicron and Salmonella enterica serovar Typhimurium (S. Typhimurium) (Sim and Wolin, 2011).
In all studied organisms, Ro binds ncRNAs called Y RNAs. These ncRNAs, which are ~ 100 nt in length, fold into secondary structures consisting of one or more large internal loops and a long stem that contains the Ro binding site (Teunissen et al., 2000; Sim and Wolin, 2011). Many species contain between two and four distinct Y RNAs that differ largely in the sequences and sizes of the internal loops (Sim and Wolin, 2011). Ro binding stabilizes Y RNAs from degradation (Labbe et al., 1999). Conversely, Y RNAs block a nuclear accumulation signal on Ro, retaining Ro in the cytoplasm (Sim et al., 2009).
Because the Ro protein also binds misfolded ncRNAs in some animal cell nuclei, it is proposed to function in ncRNA quality control (O’Brien and Wolin, 1994; Shi et al., 1996; Chen et al., 2003; Hogg and Collins, 2007). Structural and biochemical studies revealed that Ro binds misfolded RNAs that contain both a 3′ single-stranded end and adjacent protein-free helices (Fuchs et al., 2006). The 3′ ends of these RNAs insert through the Ro ring, while helices contact the Ro outer surface. Because the binding of Ro to misfolded ncRNAs is largely sequence non-specific, Ro may scavenge aberrant RNAs that fail to assemble with their correct RNA-binding proteins (Fuchs et al., 2006). However, both the mechanism by which Ro affects misfolded RNA metabolism and whether Y RNAs contribute to this function are unknown.
To have a genetically tractable system, we characterized Ro and a Y RNA in the first sequenced bacterium with a Ro ortholog, D. radiodurans (Chen et al., 2000). These studies revealed that the ortholog Rsr (Ro sixty-related) functions with 3′ to 5′ exoribonucleases during some types of environmental stress. Rsr and two exoribonucleases, RNase II and RNase PH, are required for efficient 23S rRNA maturation during heat stress (Chen et al., 2007). In stationary phase, Rsr and the ring-shaped exoribonuclease polynucleotide phosphorylase (PNPase) are important for rRNA degradation (Wurtmann and Wolin, 2010). Rsr and PNPase are found together in immunoprecipitates, and the sedimentation of PNPase with ribosomal subunits in stationary phase requires Rsr (Chen et al., 2007; Wurtmann and Wolin, 2010). However, the components of the putative Rsr/PNPase complex, the way in which Rsr influences PNPase activity, and whether Y RNA is involved have not been addressed.
To understand how a Ro protein can influence the function of an exoribonuclease, we purified the Rsr/PNPase complex from D. radiodurans and examined its composition, molecular architecture and activity. We report that Y RNA tethers Rsr to PNPase to form an RNA degradation machine. Single particle electron microscopy (EM), followed by docking Ro and PNPase atomic structures into the three-dimensional reconstruction, is consistent with a model in which single-stranded RNA threads from the Rsr ring into the PNPase cavity. Biochemical experiments demonstrate that Rsr and Y RNA specialize PNPase for effective degradation of structured RNAs. Notably, a Ro ortholog and a ncRNA also associate with PNPase in an evolutionarily distant bacterium, S. Typhimurium. We discuss the similarities between the Rsr/Y RNA/PNPase RNP and other RNA degradation machines.
RESULTS
An Rsr/Y RNA/PNPase RNP
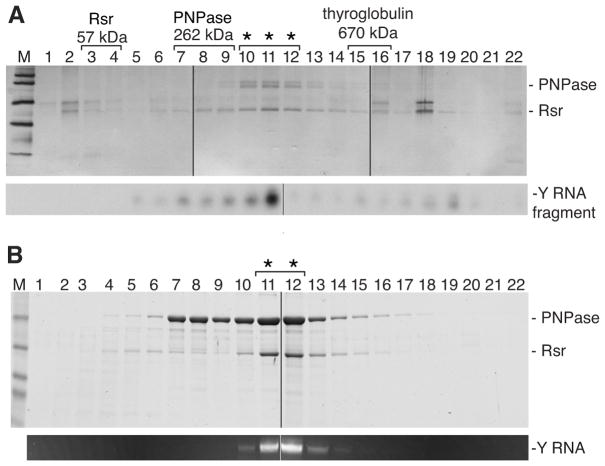
To identify components of the Rsr/PNPase complex, we subjected a D. radiodurans strain in which Rsr and PNPase each carried a distinct epitope tag to two rounds of affinity purification. Rsr was fused to the IgG-binding domains of Staphylococcus aureus Protein A, while PNPase was fused to 3 copies of FLAG. After applying extracts to IgG Sepharose, Rsr-containing complexes were eluted with TEV protease and applied to anti-FLAG agarose to isolate the Rsr/PNPase complex. Fractionation of the final eluate on glycerol gradients, followed by silver staining, revealed that the complex migrated at ~300–450 kD and that the major components were Rsr and PNPase (Figure 1A). Northern analyses revealed that a Y RNA degradation fragment co-migrated with the two proteins (Figure 1A).
Figure 1. Rsr, Y RNA and PNPase form an RNP.
(A) After purification from D. radiodurans, the complex was fractionated in glycerol gradients and proteins detected by silver staining (top). Protein identities were confirmed by immunoblotting. By subjecting RNA from the fractions to Northern analysis, a ~30 nt Y RNA fragment was detected (bottom). Positions of Rsr, PNPase and thyroglobulin run in parallel gradients are shown. Asterisks, fractions containing complex. Extra bands in lanes 2 and 18 are keratins.
(B) After E. coli expression, the purified complex was fractionated in glycerol gradients. Proteins were visualized with Coomassie blue (top). Extracted RNA was visualized with ethidium bromide (bottom). Asterisks, complex. In both (A) and (B), samples were analyzed in multiple gels that were joined at the lines.
Because the apparent molecular size suggested the complex consisted of Rsr (57 kD), one PNPase trimer (262 kD), and a Y RNA (42 kD), we attempted a reconstitution by co-expressing these components in E. coli, a bacterium that lacks a Ro ortholog. Epitope-tagged forms of Rsr and PNPase were expressed in E. coli in the presence or absence of Y RNA. Complex formation was only detected when Y RNA was co-expressed. Following purification, the complex formed in E. coli migrated identically in glycerol gradients to the complex purified from D. radiodurans (Figure 1B). Thus, Rsr and PNPase form a stable complex that also contains Y RNA.
Y RNA tethers Rsr to PNPase
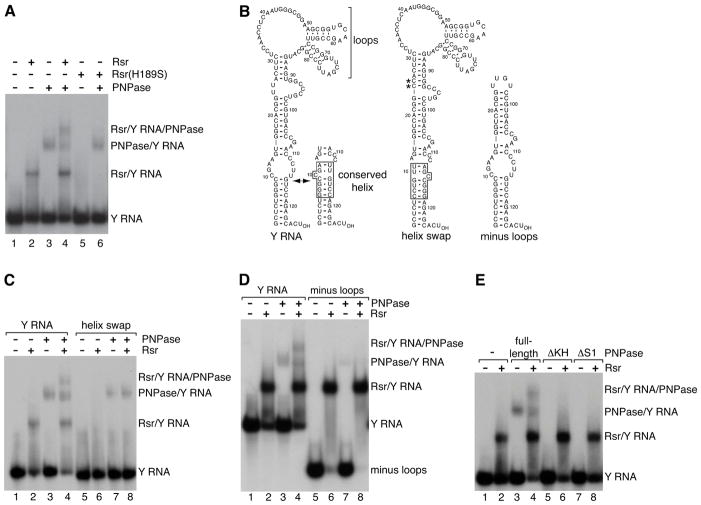
Since Rsr and PNPase only associated when Y RNA was present, we hypothesized that Y RNA scaffolds the complex. To test this idea, we established an assay to evaluate how Rsr and PNPase interact via Y RNA. Rsr and PNPase were mixed with 32P-labeled Y RNA and the RNPs separated from unbound RNA in nondenaturing gels. Rsr and PNPase each formed discrete RNPs with Y RNA (Figure 2A). When both Rsr and PNPase were present, a new RNP appeared that migrated more slowly than the two binary complexes (Figure 2A). As expected if Y RNA tethers Rsr to PNPase to form a ternary complex, this slowest migrating RNP failed to form in the presence of a mutant Rsr (Rsr-H189S) that does not bind Y RNA (Chen et al., 2007) (Figure 2A). By excising the bands, fractionating the proteins in SDS-PAGE and performing Western blotting, we confirmed that the slowest migrating RNP contained both Rsr and PNPase (Figures S1A and S1B).
Figure 2. Y RNA is a scaffold.
(A) 32P-labeled Y RNA was mixed with no protein (lane 1), Rsr (lane 2), PNPase (lane 3) or Rsr and PNPase (lane 4) and RNPs separated in native gels. Rsr-H189S was assayed in lanes 5–6.
(B) Proposed secondary structures for wild-type and mutant Y RNAs. A fraction of the RNA forms a conformer in which the nucleotides in the conserved helix (box) are basepaired (Green et al., 1998). In the helix swap mutant, the basepairs are reversed. To reduce alternative structures, Us at positions 24 and 25 were converted to Cs (asterisks). These changes do not affect Rsr binding. In the truncated RNA, nts are numbered according to positions in the full-length RNA.
(C and D) Wild-type and mutant Y RNAs were incubated with Rsr and PNPase as indicated.
(E) PNPase and mutants lacking the KH or S1 domain were incubated with Y RNA in the absence or presence of Rsr.
See also Figure S1.
If Y RNA is a scaffold, then Rsr and PNPase should bind separate sites on the RNA. In vertebrates, Ro makes specific contacts with the 5′ strand of a conserved helix in a stem formed by basepairing the 5′ and 3′ Y RNA ends (Stein et al., 2005) (Figure 2B). As expected if Rsr interacts similarly, a mutant Y RNA in which the conserved basepairs were reversed (Figure 2B, helix swap) did not bind Rsr or form the Rsr/Y RNA/PNPase RNP, although PNPase binding to the mutant RNA was unaffected (Figure 2C).
A second feature of Y RNAs is the presence of one or more large internal loops (Teunissen et al., 2000; Sim and Wolin, 2011) (Figure 2B). A Y RNA lacking these loops exhibited reduced PNPase binding and did not form the Rsr/Y RNA/PNPase RNP, although Rsr bound the truncated RNA indistinguishably from the full-length RNA (Figure 2D). The Y RNA loops likely interact with the KH and S1 domains of PNPase, since PNPases lacking either of these single-stranded RNA binding domains (Lunde et al., 2007) failed to bind Y RNA (Figure 2E), although the mutant proteins formed trimers similar to full-length PNPase as judged by fractionation in native gels and electron microscopy (Figure S1). Since mutations that reduce binding of either Rsr or PNPase to Y RNA impair complex formation, we conclude that Y RNA scaffolds the complex.
Molecular architecture of the Rsr/Y RNA/PNPase RNP
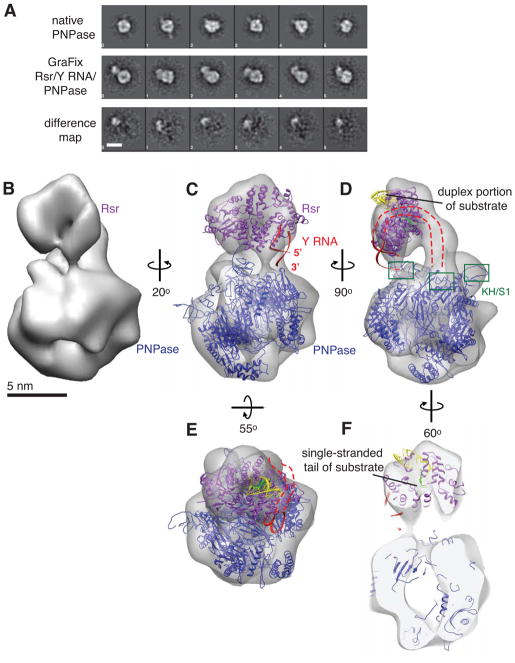
We examined the structure of the Rsr/Y RNA/PNPase RNP by negative stain electron microscopy (EM). PNPase appeared as a homogeneous monodisperse globular particle with a central cavity and ~100 Å diameter in the longest direction in raw micrographs (Figure S1F). Two-dimensional class averages showed a large ring-shaped particle containing three major domains of density with 3-fold symmetry (Figure 3A). Since the Rsr/Y RNA/PNPase RNP dissociated under the conditions used for negative stain EM, with particles corresponding to PNPase and smaller ring-like particles the size and shape of Rsr (Ramesh et al., 2007) easily recognizable in raw micrographs, we used the GraFix method (Kastner et al., 2008) to improve recovery. The use of GraFix, which involves a final purification step of sedimenting the complex through a double gradient of glutaraldehyde and glycerol, improved the homogeneity of the RNP dramatically. Large globular particles similar to PNPase but containing a marked additional density and a total longest diameter of ~140–150 Å appeared (Figure S2A). The majority of the two-dimensional class averages of the Rsr/Y RNA/PNPase RNP appeared as a small ring connected to a larger ring (Figures 3A and S2B). No differences were observed between native PNPase and PNPase in the GraFix-prepared RNP (Figure 3A), indicating the GraFix method does not affect PNPase structure.
Figure 3. Architecture of the Rsr/Y RNA/PNPase RNP.
(A) Representative reference-free two-dimensional class averages of native PNPase (top row) compared with those of the GraFix-prepared complex (middle row). Class averages of PNPase and Rsr/Y RNA/PNPase in the same column are aligned in or to the same orientation. The difference maps between each pair of images are shown in the bottom row. Scale bar, 10 nm.
(B) Rsr/Y RNA/PNPase single particle reconstruction from negatively stained complexes.
(C–F) The atomic models of S. antibioticus PNPase (PDB 1E3P) and X. laevis Ro complexed with Y RNA (PDB 1YVP) and misfolded RNA (PDB 2I91) fragments were docked into the EM density to obtain a model of the RNP. PNPase, blue; Ro, purple; Y RNA, red; duplex portion of substrate, yellow; single-stranded portion of substrate, light green. The PNPase KH/S1 domains, which were only partly visualized in S. antibioticus PNPase, are boxed in (D). The Y RNA fragment is oriented such that in the full-length RNA, the 5′ and 3′ ends point towards PNPase (C–D), and the part of the Y RNA that is absent from the crystal structure is depicted as a dashed red line (C–E). Front and back sections of the reconstruction were removed from the view to show the single-stranded end of the substrate threading through Rsr towards the PNPase cavity in (F). Scale bar, 5 nm.
See also Figure S2.
Using single particle reconstruction methods, we obtained a three-dimensional reconstruction of the Rsr/Y RNA/PNPase RNP at ~25 Å resolution based on the 0.5 Fourier Shell Correlation (FSC) criterion from ~9,000 particle images (Figures 3B and S2). In agreement with the reference-free two-dimensional class averages, the reconstruction showed a small elongated ring-shaped density positioned at an angle almost directly above the central channel of the larger ring-shaped structure. The smaller and larger densities have the sizes and shapes of Rsr (Ramesh et al., 2007) and PNPase (Symmons et al., 2000) respectively, and reference-free two-dimensional class averages of Rsr/Y RNA/PNPase complexes labeled with an antibody against the His-tag of Rsr confirm the identity of the smaller ring as Rsr (Figure S2D and S2E). There is an additional rod-shaped density that forms the major connection between the two rings along one side, which may represent Y RNA.
By docking atomic models of Rsr and PNPase into the EM map, we obtained a model for the complex (Figure 3C). The smaller ring accommodates the crystal structure of Rsr (Ramesh et al., 2007) or X. laevis Ro (Stein et al., 2005). The Rsr structure (PDB 2NVO) more closely resembles the size and shape of the smaller ring-shaped density and could be docked into the map with the highest cross-correlation coefficient (0.939). Because a X. laevis Ro structure included a Y RNA fragment (Stein et al., 2005), we used this structure (PDB 1YVP) for final modeling by superimposing its coordinates onto the Rsr structure already deposited into the EM density. This orientation places one end of the Y RNA fragment, corresponding to the 5′ and 3′ termini of the intact RNA, in close proximity to one of the KH and S1 domains of the PNPase trimer (Figure 3C). The Y RNA fragment used for crystallization (Stein et al., 2005) contains only the part of the stem encompassing the conserved helix, and we propose that the remainder of the stem and the distal loops continue along the Ro surface and across the rod-shaped density to allow the loops to contact a second KH/S1 domain of PNPase, thus anchoring the two protein components (Figures 3D and 3E).
To examine how the Rsr/Y RNA/PNPase RNP could interact with substrates, we superimposed the coordinates of X. laevis Ro bound to a misfolded RNA fragment [PDB 2I91; (Fuchs et al., 2006)] onto the structure of Ro deposited into the EM density. In this structure, the single-stranded 3′ end of the misfolded RNA passes through the Ro cavity, while a duplex binds on the outer surface (Fuchs et al., 2006). Docking of this substrate (Figures 3D–3F) places the duplex on the Rsr outer face (colored yellow) with the single-stranded end (colored green) passing through the Rsr cavity directly above the PNPase channel. The Streptomyces antibioticus PNPase structure (PDB 1E3P; Symmons et al., 2000) can fit into the larger ring of the reconstruction in two opposite orientations with similar cross-correlation coefficients (0.835 and 0.841); however the docking result that places the KH and S1 domains facing Rsr and the protruding single-stranded RNA is most consistent with the biochemical data (Figures 2 and S1). In this orientation, RNA could thread from Rsr into the PNPase cavity (Figure 3F).
Rsr and Y RNA specialize PNPase for degrading structured RNAs
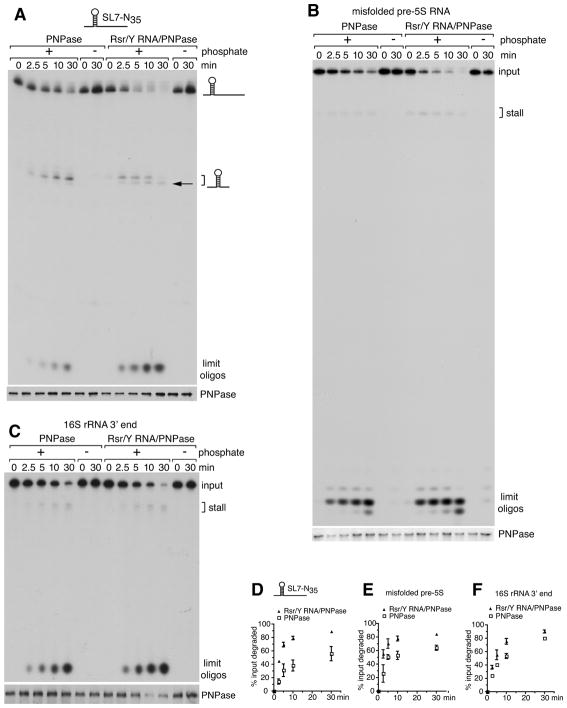
Our model predicts that Rsr and Y RNA should influence the activity of PNPase in the RNP complex. By testing a substrate containing a 7GC basepair stemloop and a 35 nt 3′ extension (SL7-N35 RNA; Figure S3A), we determined that both PNPase and the purified RNP exhibit processive phosphate-dependent degradation (Figure 4A). As reported for E. coli PNPase (Spickler and Mackie, 2000), both PNPase and the RNP stalled 7–8 nt 3′ to the stemloop (Figure 4A, bracket; Figure S3B). Importantly, the RNP degraded the substrate more effectively than PNPase alone, as measured by both increased degradation of the input RNA and the accumulation of limit oligonucleotides (Figures 4A, 4D and S3B). Consistent with a role in unwinding RNA, the RNP complex also stalled slightly closer to the stemloop than PNPase (Figures 4A and S3B, arrows). Enhanced RNA degradation was not detected when purified Rsr, Y RNA and PNPase were added to the reaction under conditions that result in inefficient complex assembly, indicating that RNP formation is required (Figure S3B).
Figure 4. Enhanced degradation of structured RNAs by the RNP.
(A–C) The activity of PNPase and the RNP was compared on 5′-labeled SL7-N35 RNA (A), misfolded X. laevis pre-5S rRNA (B) and the D. radiodurans 16S rRNA 3′ end substrate (C). At intervals, aliquots were removed and RNAs fractionated in denaturing gels (top panels). Brackets, sites of enzyme stalling. Compared to PNPase, the RNP stalls closer to the stemloop of the SL7-N35 RNA (A, arrow). The gel in (B) was run further than those in (A) and (C). Aliquots were subjected to immunoblotting to confirm that equal amounts of PNPase were present (bottom panels).
(D–F) Data from the degradation assays were plotted. Data are represented as mean values from three replicates ± SEM.
See also Figure S3.
We next examined the activity of the Rsr/Y RNA/PNPase RNP on substrates that resemble most cellular RNAs in consisting of short helices separated by bulges and loops. One substrate consisted of a misfolded pre-5S rRNA that is bound by X. laevis Ro (Fuchs et al., 2006)(Figure S3A). Since Rsr and PNPase function in rRNA degradation (Wurtmann and Wolin, 2010), we also tested a 160 nt substrate resembling the D. radiodurans 16S rRNA 3′ end (Figure S3A). The RNP was more active than PNPase in degrading both these RNAs (Figures 4B, 4C, 4E and 4F).
To identify the specific RNA features that result in more effective degradation by the RNP, we assayed a series of duplex-containing substrates similar to those used to characterize other exoribonucleases (Cheng and Deutscher, 2005; Lorentzen et al., 2008). In addition to a GC-rich 17 bp duplex, the substrates contained poly(U) 3′ extensions of varying length (Figure S4A). Both PNPase and the Rsr/Y RNA/PNPase RNP degraded substrates with 3′ overhangs of at least 20 nt, with more efficient decay occurring when the overhang contained 30 nt (Figure S4B). As reported for E. coli PNPase (Cheng and Deutscher, 2005), neither D. radiodurans PNPase nor the RNP could degrade the 17 bp duplex; however both digested through a 13 bp duplex (Figure S4E).
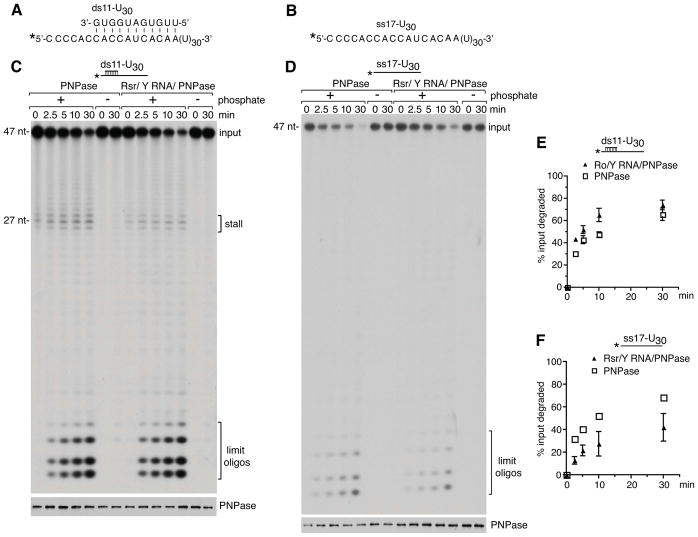
Importantly, when we tested substrates containing shorter duplexes, the RNP degraded the RNAs more effectively than PNPase alone. The difference was evident with a substrate containing an 11 bp duplex and a 30 nt overhang, as measured by increased degradation of the input RNA and decreased stalling 3′ to the duplex (Figures 5A, 5C and 5E). Although the enhancement of PNPase activity was less than that observed on other structured RNAs (Figure 4), this may be because disruption of the first few bps of the short duplex by either PNPase or the RNP results in dissociation of the remaining helix. Notably, the RNP was less active than PNPase in degrading a single-stranded RNA corresponding to the overhang strand of the duplex (Figures 5B, 5D and 5F), most likely because the S1 and KH domains that contribute to single-stranded RNA binding by PNPase are less accessible in the RNP (Figure 3D). Similar effects were seen with duplex and single-stranded substrates containing poly(A) tails (data not shown). Together with the increased activity of the Rsr/Y RNA/PNPase RNP we observed on stemloop-containing substrates (Figure 4), we conclude that Rsr and Y RNA increase the activity of PNPase in degrading structured RNAs.
Figure 5. The RNP degrades a duplex more effectively than PNPase but is less active on single-stranded RNA.
(A and B) Sequences of ds11-U30 and ss17-U30 substrates. The ss17-U30 RNA corresponds to the overhang strand of the ds11-U30 duplex. In both substrates the 5′ end of the ss17-U30 RNA was labeled with [γ-32P]ATP.
(C and D) The activity of PNPase and the RNP was compared on the ds11-U30 (C) and ss17-U30 (D) substrates. Aliquots were removed at intervals and fractionated in denaturing gels to visualize the labeled RNA (top panels). Aliquots were also assayed by immunoblotting to confirm that equal amounts of PNPase were present (bottom panels). Notably, although more of the RNP may have been used in the ss17-U30 assay (D, bottom panel), the RNP is less active than PNPase on this substrate (top panel).
(E and F) Degradation reactions using the ds11-U30 and ss17-U30 substrates were performed in triplicate. Data are represented as mean values from three replicates ± SEM. In (F), error bars for PNPase are too small to be visible.
See also Figure S4.
In addition to its role as an exoribonuclease, PNPase adds A-rich tails to RNA using nucleoside diphosphates as precursors (Mohanty and Kushner, 2011). No differences were detected between the RNP and PNPase when assayed for polyadenylation activity using a structured RNA that is a substrate for other PNPases (Sohlberg et al., 2003)(Figures S3C–S3E). To determine if the RNP contributes to tail synthesis in vivo, we compared the A-rich tails in RNA isolated from wild-type D. radiodurans and strains lacking PNPase, Rsr or the Y RNA. This revealed that PNPase, but not Rsr or the Y RNA, is required for tail formation (Figure S3F). As we do not detect effects of Rsr and the Y RNA on A-tail addition, we conclude that Rsr and Y RNA sculpt PNPase for effective degradation of structured RNAs.
A Ro ortholog and a ncRNA associate with PNPase in S. Typhimurium
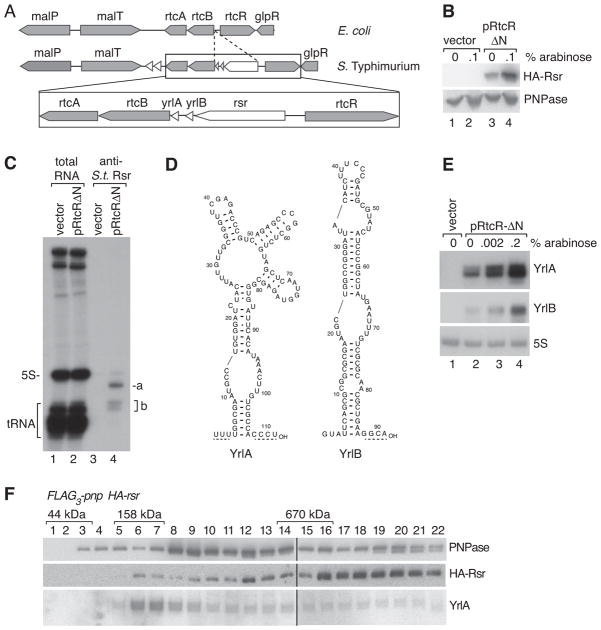
To determine if the role we identified for Rsr in D. radiodurans could be conserved in an evolutionarily distant species, we examined S. Typhimurium, an enteric bacterium closely related to E. coli. S. Typhimurium Rsr is encoded within the rtcBA operon, whose E. coli orthologs encode the RNA ligase (rtcB) and RNA cyclase (rtcA) components of a σ54-regulated RNA repair operon (Figure 6A) (Genschik et al., 1998; Tanaka and Shuman, 2011). In E. coli, this locus is tightly regulated by the adjacent rtcR transcriptional activator, as rtcBA transcription is not detected during standard growth, but becomes detectable when a N-terminally truncated, constitutively active form of RtcR is overexpressed (Genschik et al., 1998). Consistent with similar regulation, we could only detect S. Typhimurium Rsr when a parallel version of the truncated RtcR was expressed from a plasmid (Figure 6B).
Figure 6. Rsr and ncRNA associate with PNPase in S. Typhimurium.
(A) Maps of the S. Typhimurium rsr locus and the corresponding E. coli region.
(B) S. Typhimurium HA-rsr cells carrying empty vector or the truncated RtcR under control of the arabinose-inducible promoter (pRtcRΔN) were grown in the presence or absence of arabinose. Lysates were subjected to immunoblotting with anti-HA (top) and anti-PNPase antibodies (bottom panel). Expression in the absence of arabinose (lane 3) is due to promoter leakiness.
(C) Lysates from cells carrying the empty vector or pRtcRΔN were subjected to immunoprecipitation with anti-S. Typhimurium Rsr antibodies. Total RNA (lanes 1–2) and RNAs within immunoprecipitates (lanes 3–4) were visualized by end-labeling.
(D) Possible secondary structures of YrlA and YrlB.
(E) RNA from cells carrying the indicated plasmids was subjected to Northern blotting to detect YrlA and YrlB.
(F) FLAG3-pnp HA-rsr lysates were subjected to affinity purification using anti-FLAG. After fractionating eluates in glycerol gradients, HA-Rsr and PNPase were detected by immunoblotting. YrlA was detected by Northern blotting. Samples were analyzed in multiple gels that were joined at the lines. Positions of ovalbumin (44 kD), bovine γ-globulin (158 kD) and thyroglobulin (670 kD) run in parallel gradients are shown.
See also Figure S5.
Immunoprecipitation with an antibody against S. Typhimurium Rsr, followed by end-labeling of associated RNAs, revealed two novel ncRNAs encoded 3′ to rsr (Figures 6A, 6C and 6D). Both ncRNAs are only detected when the truncated RtcR is overexpressed (Figures 6C and 6E). Thus, similar to D. radiodurans Rsr and Y RNA, which are upregulated following several forms of environmental stress (Sim and Wolin, 2011), transcription of S. Typhimurium Rsr and its associated ncRNAs may be regulated in response to an external stimulus. We call the S. Typhimurium ncRNAs YrlA (Y RNA-like) and YrlB.
To examine if S. Typhimurium Rsr associates with PNPase, we fused PNPase to 3 copies of FLAG and carried out affinity purification using anti-FLAG agarose. Fractionation of the eluate in glycerol gradients, followed by immunoblotting, revealed that PNPase sedimented both at the expected size for the homotrimer (Figure 6F, fractions 8–9) and in heavier fractions. Rsr was also present in the eluate. However, while some Rsr and PNPase migrated similarly in the gradient to the D. radiodurans complex (Figure 6F, fractions 11–12), Rsr was present predominantly in heaver fractions (fractions 16–22). Using Northern blotting, we detected YrlA, but not YrlB, in the gradient fractions. Although some YrlA was degraded during sedimentation, making it difficult to assess where it peaked, we detected YrlA in all of the gradient fractions that contained Rsr (Figure 6F). The presence of Rsr and YrlA in the eluate was dependent on PNPase, as both components were greatly reduced when the affinity purification was performed from an untagged strain (Figure S5). Thus, while the larger size of the S. Typhimurium Rsr/YrlA/PNPase complex suggests that it may contain additional components or differ in stoichiometry from the minimal D. radiodurans RNP, the association of PNPase with Rsr and a ncRNA is conserved.
DISCUSSION
The function of the Ro protein and its associated ncRNAs have been enigmatic for over thirty years. By studying a bacterial Ro protein, we identified a ribonucleoprotein machine in which the ortholog Rsr is tethered via Y RNA to the ring-shaped exoribonuclease PNPase. Single particle electron microscopy of the RNP revealed a double ring architecture, suggesting that Rsr channels RNA into the PNPase cavity for degradation. Biochemical studies demonstrated that Rsr and Y RNA specialize PNPase for degrading structured RNAs. This work has identified a novel role for ncRNA and revealed a bacterial RNA degradation machine that resembles the archaeal and eukaryotic exosomes.
A ncRNA functions as a tether
We showed that Y RNA tethers D. radiodurans Rsr to PNPase to form a RNP machine. Since proteins with RNA binding domains that could interact with Y RNA loops are widespread, Y RNAs could potentially tether Ro orthologs to additional RNA remodeling proteins, such as helicases and RNA chaperones. In this case, Y RNAs and their associated Ro rings would function as trans-acting modules that can be attached to multiple distinct proteins to augment their handling of structured RNAs. Conversely, Y RNAs could tether proteins that affect the activity or subcellular location of Ro. The use of loops to tether interacting proteins ensures that Y RNAs will not be substrates for enzymes that require an RNA end for activity, such as exoribonucleases and some helicases. The role of Y RNAs as tethers is supported by reports of proteins that interact with mammalian Ro proteins through binding Y RNAs (Bouffard et al., 2000; Fabini et al., 2001; Fouraux et al., 2002; Hogg and Collins, 2007; Sim et al., 2012). Although in most cases the significance is unknown, binding of the zipcode-binding protein ZBP1 to a mouse Y RNA allows nuclear export of the RNP (Sim et al., 2012).
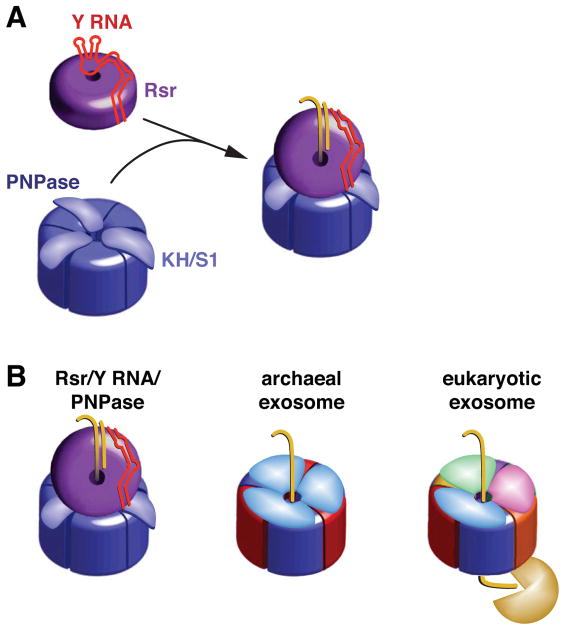
Because the binding sites of Y RNAs and misfolded ncRNAs overlap on the Ro surface (Fuchs et al., 2006), it has been unclear how Y RNAs could be repositioned to allow substrates to access the Ro cavity. We propose that binding of proteins such as PNPase to Y RNA loops removes this portion of Y RNA from the Rsr ring, rendering the cavity accessible (Figure 7A). An appealing feature of this model is that in addition to its role as a scaffold, Y RNA could function as a gate to ensure that RNA substrates enter the Rsr cavity only when PNPase or another remodeling enzyme is available to act on them.
Figure 7. Model for Rsr/Y RNA/PNPase RNP function.
(A) Formation of the Rsr/Y RNA/PNPase RNP. Residues on the Rsr outer surface contact conserved bases in the Y RNA stem, while the rest of the RNA interacts with basic residues on the Rsr surface, preventing binding of other RNAs (Stein et al., 2005). When PNPase is present, Y RNA loops bind the PNPase KH and S1 domains, repositioning the RNA so that substrates can access the Rsr cavity.
(B) The Rsr/Y RNA/PNPase RNP resembles archaeal and eukaryotic exosomes. All three complexes contain rings with six RNase PH domains. PNPase is a trimer in which each monomer contains two RNase PH domains. The archaeal ring contains three copies of each of two proteins containing single RNase PH domains, while in eukaryotes six distinct proteins form the ring. Both exosomes contain RNA-binding caps formed by KH and S1 domain proteins. We propose that, similar to the archaeal exosome, RNA passes through an RNA-binding ring (Rsr) to undergo phosphorolytic degradation in the catalytic ring (PNPase). As yeast and human exosomes have a catalytically inactive RNase PH domain ring, RNA passes through the nine subunit Exo1–9 ring to reach a hydrolytic exonuclease.
Although Y RNA-mediated tethering of Rsr is needed for the enhanced activity of PNPase on the tested RNAs, Rsr may be able to assist some enzymes as a Y RNA-free protein. This is suggested by the requirement for Rsr in heat stress-induced 23S rRNA maturation, which involves RNase II and RNase PH (Chen et al., 2007). In this case, either deleting the Y RNA or increasing the levels of Y RNA-free Rsr results in constitutive 23S rRNA maturation, implying that Rsr may assist 3′ end trimming by some exonucleases without a Y RNA tether. If so, the ability of Rsr and other Ro proteins to act both as free proteins and as Ro/Y RNA modules would increase their functional repertoires.
The Rsr/Y RNA/PNPase RNP: a bacterial exosome?
Our characterization of the Rsr/Y RNA/PNPase RNP expands the inventory of RNA degradation machines and reveals a new way in which exonucleases such as PNPase can be adapted to degrade structured RNAs. Previously, the only described bacterial machine was the degradosome, which in E. coli contains PNPase, the scaffolding endonuclease RNase E, an RNA helicase and the metabolic enzyme enolase (Gorna et al., 2011). Although the Rsr/Y RNA/PNPase RNP may function with helicases or endonucleases in vivo, these enzymes are not required to enhance the activity of PNPase in degrading at least some RNAs. Moreover, as some bacteria could have both an Rsr/Y RNA/PNPase RNP and a degradosome, there may be selective advantages associated with each assembly. We note that in many bacteria the degradosome contains different exonucleases, endonucleases, helicases and metabolic enzymes than those present in the E. coli complex (Gorna et al., 2011). If the Rsr/Y RNA/PNPase RNP diverges similarly in composition (as suggested by the apparent size differences between the D.radiodurans and S. Typhimurium complexes), Rsr and Y RNA could associate with additional nucleases or remodeling proteins in some bacteria.
Although the use of ncRNA to scaffold an RNA degradation machine is unique, the D. radiodurans Rsr/Y RNA/PNPase RNP exhibits structural and functional similarities to the multiprotein nuclease complexes known as exosomes. Like the archaeal exosome (Evguenieva-Hackenberg, 2011), the Rsr/Y RNA/PNPase RNP contains an RNA-binding ring atop an RNase PH domain-containing ring that catalyzes both phosphorolytic degradation of RNA and 3′ tail addition (Figure 7B). Similar to the trimeric RNA-binding ring of the archaeal exosome (Evguenieva-Hackenberg et al., 2008), the Rsr ring is important for efficient RNA degradation but is not needed for synthesis of A-rich tails. Also, the conformational flexibility present in both the archaeal RNA binding ring and Rsr should allow both rings to interact with diverse substrates (Ramesh et al., 2007; Evguenieva-Hackenberg, 2011). However, while the exosome RNA-binding ring is coaxial to the degradation ring, Rsr sits at an angle above the PNPase cavity, due to the asymmetric bulk of the ncRNA tether.
Notably, while the RNA-binding ring of the archaeal exosome enhances the activity of the RNase PH ring in degrading both single-stranded and structured RNAs (Evguenieva-Hackenberg et al., 2008), the Rsr/Y RNA/PNPase RNP is less active than PNPase on single-stranded RNAs (Figure 5). A likely explanation for the decreased activity of the RNP on single-stranded RNA is that Y RNA-mediated tethering of Rsr to the S1/KH domains of PNPase obstructs one or more of these single-stranded RNA binding surfaces. In this model, the RNA-binding surface of PNPase would be largely replaced with that of Rsr (Figure 7B). If the RNA-binding platform of Rsr, which is composed of HEAT repeats, resembles X. laevis Ro in preferring substrates that contain both a single-stranded tail and adjacent helices (Fuchs et al., 2006), the presence of Rsr would specialize PNPase for binding structured RNAs.
As described for the yeast exosome, in which RNA is predicted to thread through the catalytically inactive Exo1–9 ring to reach the Exo11 RNase (Bonneau et al., 2009) (Figure 7B), Rsr may contribute to ATP-independent unwinding of structured RNAs. Such a role is consistent with our finding that Rsr did not contribute to polyadenylation by PNPase, since addition of nucleotides to an already accessible end may be less dependent on RNA unwinding than exonucleolytic decay. We note that our experiments do not allow us to determine if Rsr and/or Y RNA dissociate after PNPase engages a substrate, or whether they remain on the substrate throughout degradation. Moreover, as crystal structures of Ro and Rsr have revealed considerable flexibility in the organization of the HEAT repeat ring and in the size of the central hole (Stein et al., 2005; Ramesh et al., 2007), the Rsr ring could open to allow RNA to enter the channel and/or Rsr to disengage from the substrate. As remains the case for the eukaryotic exosome, atomic resolution structures of the Rsr/Y RNA/PNPase RNP complexed with substrates will be required to determine the actual paths of RNA substrates. Nonetheless, the crystal structures showing single-stranded RNA traversing the X. laevis Ro cavity (Stein et al., 2005; Fuchs et al., 2006), the juxtaposition of the Rsr and PNPase rings observed with EM, and the increased activity of the RNP in degrading structured RNA are all consistent with a mechanism in which RNA passes from the Rsr ring into the PNPase cavity. Finally, the homology between bacterial and vertebrate Ro proteins and Y RNAs, together with the finding that vertebrate Ro proteins bind misfolded ncRNAs (O’Brien and Wolin, 1994; Chen et al., 2003), suggests that the RNA remodeling function that we identified for Rsr could be conserved in eukaryotes.
EXPERIMENTAL PROCEDURES
Purification of the Rsr/Y RNA/PNPase RNP from D. radiodurans
Strain PTR17 expressing Protein A-TEV-Rsr and PNPase-FLAG3 was grown to OD600 = 0.8, resuspended in buffer A (40 mM Tris-HCl pH 7.5, 100 mM NaCl, 10% glycerol, 0.1% NP-40, 3 mM MgCl2, 3 mM MnCl2, 1 mM DTT) with 0.5 mM PMSF, 1 mM EGTA, 0.1% diethyl pyrocarbonate, 2 mM vanadyl ribonuclease complex (VRC), 1X protease inhibitor cocktail (Roche Applied Science) and lysed with a French press. After clearing in a Beckman Type 50.2Ti rotor at 20,000 rpm for 30 min, the supernatant was mixed with IgG-Sepharose (GE Healthcare) for 2 hr at 4°C. After washing with 40 ml buffer A containing 0.5 mM PMSF and 20 ml TEV buffer (10 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% NP-40, 1 mM MgCl2, 1 mM MnCl2, 1X protease inhibitor cocktail, 0.5 mM PMSF), the beads were incubated with 100 units TEV protease for 16 hr at 4°C. The eluate was mixed with anti-FLAG agarose (Sigma-Aldrich) for 2 hr at 4°C, washed with buffer B (40 mM Tris-HCl pH 7.4, 100 mM NaCl, 0.1% NP-40, 1 mM MgCl2, 1 mM MnCl2), and eluted with 1 mg/ml 3XFLAG peptide in buffer B containing 0.02% NP-40. This eluate was layered on a 10–40% glycerol gradient in 20 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM MgCl2, 1 mM MnCl2, 2 mM β-mercaptoethanol, and sedimented at 36,000 rpm in a Beckman SW41 rotor for 20 hr at 4°C. Fractions were subjected to SDS-PAGE and silver staining. To detect Y RNA, extracted RNA was separated in 6% polyacrylamide/8.3 M urea gels, transferred to Zeta-Probe GT (Bio-Rad) and probed with a [α-32P]rCTP-labeled RNA complementary to the Y RNA.
Electrophoretic Mobility Shift Assays (EMSA)
To visualize RNP formation, 9.1 pmol of unlabeled Y RNA was mixed with 86 fmol of Y RNA that was transcribed in the presence of [α-32P]rATP, heated to 95°C in binding buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 2 mM MnCl2), frozen on dry ice and thawed on ice. The refolded RNA was incubated with 17.4 pmol Rsr for 30 min at 4°C and 30 min at 30°C. This mixture was incubated with 6.8 pmol PNPase in a final volume of 5 μl (20 mM Tris-HCl pH 7.5, 50 mM NaCl, 1.25 mM MnCl2, 2 mM DTT, 5% glycerol) for 30 min at 4°C and 30 min at 30°C. Next, 1 μl of binding buffer containing 10% glycerol was added and the reaction fractionated in 4% polyacrylamide gels (80:1, acrylamide:bisacrylamide) in 25 mM Tris, 25 mM boric acid, 1 mM EDTA. Gels were run at 4°C for 1.5 hr at 250 V.
Purification of PNPase and the Rsr/Y RNA/PNPase RNP Following E. coli Expression
Because attempts to separate in vitro assembled RNPs from unassembled PNPase were unsuccessful, we used co-expression of tagged components in E. coli, followed by purification, to visualize the complex by EM and assay its activity. For PNPase, E. coli expressing D. radiodurans PNPase carrying an N-terminal Strep-tag (Schmidt and Skerra, 2007) were lysed with a French press in 20 mM Hepes pH 7.5, 200 mM NaCl, 5% glycerol, 2 mM β-mercaptoethanol, 0.5 mM PMSF, 1 mM Pefabloc, and 1× protease inhibitor cocktail. Strep-PNPase was purified on Strep-Tactin Superflow Plus (Qiagen), eluted with 5 mM d-desthiobiotin, 20 mM Hepes pH 7.5, 200 mM NaCl, 2 mM β-mercaptoethanol, 5% glycerol and concentrated with Amicon Ultra centrifugal filter units to 0.8 ml. Next, 0.3 ml of Strep-PNPase was applied to 11 ml 10–40% glycerol gradients in 20 mM Hepes, pH 7.5, 50 mM NaCl, 1 mM MgCl2, 1 mM MnCl2, 1 mM Pefabloc, 2 mM β-mercaptoethanol, and sedimented at 36,000 rpm in a Beckman SW41 rotor for 20 hr at 4°C.
To express the Rsr/Y RNA/PNPase RNP, Rsr containing an N-terminal His6-tag and Strep-PNPase were cloned into pRSFDuet-1 (Novagen), while Y RNA, followed by a hammerhead ribozyme and a T7 terminator, was cloned into pETDuet-1. To purify the Rsr/Y RNA/PNPase RNP, E. coli carrying both plasmids were grown in LB with ampicillin and kanamycin (100 μg/ml each), induced with IPTG and lysed by passing through a French Press in buffer C [20 mM Hepes pH 7.5, 50 mM NaCl, 5% glycerol, 1 mM MgCl2, 1 mM MnCl2, 2 mM β-mercaptoethanol, 0.5 mM PMSF, 1.25 mM VRC, 1 mM Pefabloc, 1X protease inhibitor cocktail]. After clearing at 20,000 rpm in a Beckman 50.2Ti rotor for 30 min, lysate was incubated with Ni-NTA agarose (Qiagen) for 2 hr, washed with buffer C and Rsr-containing complexes eluted with 160 mM imidazole in buffer C. The eluate was incubated with Strep-Tactin resin (Qiagen) for 2 hr, washed with buffer C lacking VRC and eluted from the resin with 0.3 ml of 5 mM d-desthiobiotin, 20 mM Hepes pH 7.5, 50 mM NaCl, 2 mM β-mercaptoethanol, 5% glycerol, 1 mM MgCl2, 1 mM MnCl2, 0.5% PMSF. This eluate was fractionated in 11 ml 10–40% glycerol gradients in 20 mM Hepes pH 7.5, 50 mM NaCl, 2 mM β-mercaptoethanol, 1 mM MgCl2, 1 mM MnCl2, 1 mM Petabloc as described for PNPase. One liter of E. coli yielded 44–106 pmols of RNP. Variations in which we used forms of Rsr and PNPase with cleavable tags, followed by tag cleavage, resulted in lower yields, probably due to the extra manipulations needed to remove the tags. Because the RNP dissociated upon freezing, fractions containing the complex were stored at 4°C and used within 3 weeks. To allow exact comparisons, PNPase was stored identically. For EM, 0.1% glutaraldehyde (EM grade, Sigma) was added to the 40% glycerol solution prior to gradient preparation (Kastner et al., 2008).
Negative Stain EM Sample Preparation and Data Collection
PNPase and the Rsr/Y RNA/PNPase RNP were diluted to ~25–50 nM and immediately applied to glow-discharged holey carbon grids with a thin layer of carbon over the holes. To form the RNP-antibody complex, 100 nM of the GraFix-prepared RNP and 50 nM of monoclonal anti-His (A00186, GenScript) were incubated at 0°C for 30 min in 10 μl prior to grid application. After 1 min, grids were stained consecutively in 3 droplets of 2% (w/v) uranyl formate solution and excess stain removed by blotting with filter paper.
For the two-dimensional analysis, samples were examined with an FEI Technai-12 electron microscope equipped with a LaB6 filament operated at 120 keV acceleration voltage using a nominal magnification of 42,000. Images were recorded on a 4k × 4k Ultrascan4000 CCD camera (Gatan) using low-dose mode with an exposure dose of 20–30 e−Å−2. The defocus used to collect the raw image was between −0.8 μm and −1.1 μm. Electron micrographs recorded using the Technai-12 electron microscope had a pixel size of 2.6 Å and were directly used for image processing.
For three-dimensional reconstruction of the Rsr/Y RNA/PNPase RNP, the sample was examined using a Technai 12 Bio-TWIN operated at 120 keV. The RNP was imaged at a nominal magnification of 49,000 (2.18 Å per pixel at specimen level) and data were acquired on an F416CMOS 4k × 4k CCD camera (TVIPS) using a dose of 20 e−Å−2. Tilt-pairs of images were recorded automatically using the RCT application (Yoshioka et al., 2007) within the LEGINON data collection software (Suloway et al., 2005). Briefly, the tilted (+60°) image was collected first using a defocus of approximately −1.5 μm, then the stage was tilted back to 0° and a second image was collected using a defocus range of −0.7 μm to −1.4 μm. Image processing and three-dimensional reconstruction are described in Extended Experimental Procedures.
Exonuclease Assays
Degradation assays contained 20 fmol of PNPase or Rsr/Y RNA/PNPase RNP and 94 fmol RNA in 20 mM Tris-HCl pH 7.5, 50 mM KCl, 1 mM MgCl2, 2 mM DTT, 10 mM phosphate at 30°C. At intervals, aliquots were added to equal volumes 90% formamide, 1 mM EDTA, 0.25% SDS, 0.03% bromophenol blue, 0.03% xylene cyanol and fractionated in 8% or 15% polyacrylamide/8.3 M urea gels. A PhosphorImager (Molecular Dynamics), followed by ImageQuant analysis, was used to quantify degradation. Aliquots were subjected to immunoblotting with anti-PNPase (Chen et al., 2007) to confirm equal amounts of PNPase were added. For details, see Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
A bacterial Ro protein (Rsr) is complexed with ncRNA and the PNPase exoribonuclease
ncRNA scaffolds this new RNA degradation machine
Single particle EM suggests that Rsr channels RNA into the PNPase cavity
Rsr and the ncRNA specialize PNPase for degrading structured RNA
Acknowledgments
We thank J. Button for assistance in preparing S. Typhimurium strains, A. Carpousis, N. Kucera, A. Ramesh, J. Sacchettini, S. Sim and E. Wurtmann for reagents, H. Zheng and K. Reinisch for assistance with baculovirus, G. Lander, M. Cianfrocco and P. Grob for electron microscopy assistance, E. Conti for advice, and S. Baserga, E. De La Cruz, A. Horwich, J. Steitz, E. Ullu and Y. Zuo for comments on the manuscript. We thank the Yale Center for Cellular and Molecular Imaging and E. Nogales for use of the EM facility at UC-Berkeley. D.W.T is a NSF Graduate Research Fellow. This work was supported by the Smith Family Awards Program for Excellence in Biomedical Research (to H.W.), NIH grants R01 AI079022 (to J.E.G.), R01 GM073863 (to S.L.W.) and Molecular Biophysics training grant 5 T32 GM008283 (D.W.T.).
Footnotes
X.C. identified the Rsr/Y RNA/PNPase RNP, reconstituted it in E. coli and performed all biochemical experiments. D.W.T. performed the single-particle electron microscopy and three-dimensional reconstruction. C.C.F. prepared S. Typhimurium strains and extracts. All authors contributed to planning experiments and analyzing data.
ACCESSION NUMBER
The EM map of the Rsr/Y RNA/PNPase RNP has been deposited into the Electron Microscopy Data Bank with accession number EMDB-5389.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ataide SF, Schmitz N, Shen K, Ke A, Shan SO, Doudna JA, Ban N. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- Bouffard P, Barbar E, Briere F, Boire G. Interaction cloning and characterization of RoBPI, a novel protein binding to human Ro ribonucleoproteins. RNA. 2000;6:66–78. doi: 10.1017/s1355838200990277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Quinn AM, Wolin SL. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev. 2000;14:777–782. [PMC free article] [PubMed] [Google Scholar]
- Chen X, Smith JD, Shi H, Yang DD, Flavell RA, Wolin SL. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr Biol. 2003;13:2206–2211. doi: 10.1016/j.cub.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Chen X, Wurtmann EJ, Van Batavia J, Zybailov B, Washburn MP, Wolin SL. An orthologue of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev. 2007;21:1328–1339. doi: 10.1101/gad.1548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Evguenieva-Hackenberg E. The archaeal exosome. Adv Exp Med Biol. 2011;702:29–38. doi: 10.1007/978-1-4419-7841-7_3. [DOI] [PubMed] [Google Scholar]
- Evguenieva-Hackenberg E, Roppelt V, Finsterseifer P, Klug G. Rrp4 and Csl4 are needed for efficient degradation but not for polyadenylation of synthetic and natural RNA by the archaeal exosome. Biochemistry. 2008;47:13158–13168. doi: 10.1021/bi8012214. [DOI] [PubMed] [Google Scholar]
- Fabini G, Raijmakers R, Hayer S, Fouraux MA, Pruijn GJ, Steiner G. The heterogeneous nuclear ribonucleoproteins I and K interact with a subset of Ro ribonucleoprotein-associated Y RNAs in vitro and in vivo. J Biol Chem. 2001;276:20711–20718. doi: 10.1074/jbc.M101360200. [DOI] [PubMed] [Google Scholar]
- Fouraux MA, Bouvet P, Verkaart S, van Venrooij WJ, Pruijn GJ. Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J Mol Biol. 2002;320:475–488. doi: 10.1016/s0022-2836(02)00518-1. [DOI] [PubMed] [Google Scholar]
- Fuchs G, Stein AJ, Fu C, Reinisch KM, Wolin SL. Structural and biochemical basis for misfolded RNA recognition by the Ro protein. Nat Struct Mol Biol. 2006;13:1002–1009. doi: 10.1038/nsmb1156. [DOI] [PubMed] [Google Scholar]
- Genschik P, Drabikowski K, Filipowicz W. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its sigma54-regulated operon. J Biol Chem. 1998;273:25516–25526. doi: 10.1074/jbc.273.39.25516. [DOI] [PubMed] [Google Scholar]
- Gorna MW, Carpousis AJ, Luisi BF. From conformational chaos to robust regulation: the structure and function of the multi-enzyme RNA degradosome. Q Rev Biophys. 2011:1–41. doi: 10.1017/S003358351100014X. [DOI] [PubMed] [Google Scholar]
- Green CD, Long KS, Shi H, Wolin SL. Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA. 1998;4:750–765. doi: 10.1017/s1355838298971667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Rivas FV, Murchison EP, Steitz JA. The expanding universe of noncoding RNAs. Cold Spring Harb Symp Quant Biol. 2006;71:551–564. doi: 10.1101/sqb.2006.71.064. [DOI] [PubMed] [Google Scholar]
- Hogg JR, Collins K. Human Y5 RNA specializes a Ro ribonucleoprotein for 5S ribosomal quality control. Genes Dev. 2007;21:3067–3072. doi: 10.1101/gad.1603907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- Labbe JC, Hekimi S, Rokeach LA. The levels of the RoRNP-associated Y RNA are dependent upon the presence of ROP-1, the Caenorhabditis elegans Ro60 protein. Genetics. 1999;151:143–150. doi: 10.1093/genetics/151.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Hardin JA, Steitz JA. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. 1981;211:400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR. Bacterial/archaeal/organellar polyadenylation. WIREs RNA. 2011;2:256–276. doi: 10.1002/wrna.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien CA, Wolin SL. A possible role for the 60 kd Ro autoantigen in a discard pathway for defective 5S ribosomal RNA precursors. Genes & Dev. 1994;8:2891–2903. doi: 10.1101/gad.8.23.2891. [DOI] [PubMed] [Google Scholar]
- Perreault J, Perreault JP, Boire G. Ro-associated Y RNAs in metazoans: evolution and diversification. Mol Biol Evol. 2007;24:1678–1689. doi: 10.1093/molbev/msm084. [DOI] [PubMed] [Google Scholar]
- Ramesh A, Savva CG, Holzenburg A, Sacchettini JC. Crystal structure of Rsr, an ortholog of the antigenic Ro protein, links conformational flexibility to RNA binding activity. J Biol Chem. 2007;282:14960–14967. doi: 10.1074/jbc.M611163200. [DOI] [PubMed] [Google Scholar]
- Schmidt TGM, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc. 2007;2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- Shi H, O’Brien CA, Van Horn DJ, Wolin SL. A misfolded form of 5S rRNA is associated with the Ro and La autoantigens. RNA. 1996;2:769–784. [PMC free article] [PubMed] [Google Scholar]
- Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, Wolin SL. The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell. 2009;20:1555–1564. doi: 10.1091/mbc.E08-11-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Wolin SL. Emerging roles for the Ro 60 kDa autoantigen in noncoding RNA metabolism. WIREs RNA. 2011;2:686–689. doi: 10.1002/wrna.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Yao J, Weinberg DE, Niessen S, Yates JR, 3rd, Wolin SL. The zipcode-binding protein ZBP1 influences the subcellular location of the Ro 60-kDa autoantigen and the noncoding Y3 RNA. RNA. 2012;18:100–110. doi: 10.1261/rna.029207.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlberg B, Huang J, Cohen SN. The Streptomyces coelicolor polynucleotide phosphorylase homologue, and not the putative poly(A) polymerase, can polyadenylate RNA. J Bacteriol. 2003;185:7273–7278. doi: 10.1128/JB.185.24.7273-7278.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickler C, Mackie GA. Action of RNase II and polynucleotide phosphorylase against stem-loops of defined structure. J Bacteriol. 2000;182:2422–2427. doi: 10.1128/jb.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: The Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–539. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;15:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem. 2011;286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunissen SW, Kruithof MJ, Farris AD, Harley JB, Venrooij WJ, Pruijn GJ. Conserved features of Y RNAs: a comparison of experimentally derived secondary structures. Nucleic Acids Res. 2000;28:610–619. doi: 10.1093/nar/28.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtmann EJ, Wolin SL. A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc Natl Acad Sci USA. 2010;107:4022–4027. doi: 10.1073/pnas.1000307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka C, Pulokas J, Fellmann D, Potter CS, Milligan RA, Carragher B. Automation of random conical tilt and orthogonal tilt data collection using feature-based correlation. J Struct Biol. 2007;159:335–346. doi: 10.1016/j.jsb.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.