Abstract
Cytogenetic analyses have been historically limited in Waldenström’s macroglobulinemia (WM) by the difficulty to obtain tumor metaphases. Thus, few recurrent karyotypic abnormalities have been reported and the molecular consequences of these imbalances are largely unknown. We used an array-based comparative genomic hybridization approach to better characterize the recurrent chromosome abnormalities associated with WM pathogenesis and to compare them with the publicly available findings in other B-cell neoplasias. The majority of the recurrent chromosome abnormalities identified in WM were shared with marginal zone lymphomas (MZL), as deletions of 6q23 and 13q14 and gains of 3q13-q28, 6p and 18q. On the other hand, gains of 4q and 8q were recurrently identified in WM but have not been described as being common abnormalities in MZL. The genetic consequences of these specific abnormalities remain elusive and further studies are critical to refine the search and to precise the molecular pathways affected by these abnormalities.
Keywords: aCGH, Comparative genomic analysis
Introduction
Waldenström’s macroglobulinemia (WM) is an incurable B-lymphoproliferative disorder characterized by bone marrow infiltration with a clonal population of small B-lymphocytes, plasmacytoid lymphocytes and plasma cells that secrete monoclonal immunoglobulin M.1 Although it is considered a distinct clinicopathologic entity, the absence of morphologic, immunophenotypic or chromosomal disease-specific markers makes difficult the differential diagnosis of WM from other lowgrade B-cell neoplasias, as marginal zone lymphomas (MZL).
The identification of chromosome abnormalities by karyotypic analysis has not been very successful because the low proliferative rate and subsequent low yield of abnormal metaphases. Thus, few recurrent abnormalities have been described and the genetic basis of WM remains poorly defined. Deletion of 6q is the most frequently observed abnormality, identified in approximately half of the patients by fluorescence in situ hybridization (FISH).2 This abnormality usually involves chromosome bands 6q21-q23, but no tumor suppressor genes have been associated with the disease.3 Recently, trisomy of chromosome 4 has been identified in about 20% of patients.4 Deletions of 13q14 and 17p13 are not common at the time of diagnosis, but might be observed in up to 15% of patients at the time of disease progression.5 Unlike other B-cell lymphomas, WM does not have translocations involving the immunoglobulin heavy chain locus at chromosome 14. Originally, it was believed that about 50% of patients with WM/LPL contain the t(9;14)(p13;q32), but subsequent studies confirmed that this abnormality is restricted to cases without serum monoclonal protein and is not present in WM.2
Several studies were recently completed by using high-throughput transcriptional and proteomic profiles,6–8 but no high-resolution whole-genome approaches have been done in WM. Here, we showed the most recurrent chromosome copy-number changes identified in a WM cohort by using a comprehensive high-resolution approach. The main chromosomal findings were compared with data already published in other low-grade B-cell neoplasias, as MZL, IgM monoclonal gammopathy of undetermined significance (MGUS) and B-chronic lymphocytic leukemia (B-CLL).
Materials and Methods
Patients
Bone marrow samples from 42 symptomatic patients with WM patients (21 of whom previously treated) were collected after informed consent was obtained in accordance with the Declaration of Helsinki. The Mayo Clinic Institutional Review Board approved the study. DNA was obtained from tumor cells enriched by positive selection using anti-CD19+ or concomitant anti-CD19+ and CD138+ immunomagnetic beads (AutoMACS; Miltenyi-Biotec, Auburn, CA). Genomic DNA was obtained from all tumor samples using standard phenol-chloroform extraction methods.
Array-Based Comparative Genomic Hybridization
High-resolution array-based comparative genomic hybridization (aCGH) was performed with the Human Genome 244A microarray (Agilent Technologies; Palo Alto, CA), according to fabricant protocols, with minor modifications. Briefly, 1.2 μg of tumor and reference DNAs were digested with Bovine DNaseI (Ambion; Austin, TX) during 12 minutes at room temperature. Random primers and exo-Klenow fragment were used to differentially label tumor (Cy5) and reference (Cy3) genomic DNA samples (Agilent Technologies). Microarrays were scanned; data were extracted by feature extraction software and were analyzed by using CGH analytics software (Agilent Technologies).
Results and Discussion
Overall, 83% of cases (35 of 42) showed chromosomal imbalances. A total of 187 abnormalities were identified, ranging from 0 to 27 per patient (median of 3). Deletions were more frequently observed than amplifications (59% and 41%, respectively), but the size per deletion (median size of 2.5 Mb per abnormality and 53 Mb per karyotype, respectively) was considerably smaller than per amplifications (21.4 Mb and 78 Mb, respectively).
Eleven regions of recurrent copy number changes (5 deleted and 6 amplified regions) in > 10% of patients were identified (Table 1). Whole or partial deletion of 6q was identified in 40% of cases. Deletion of 6q is not specific of WM, being frequently found in several B-cell disorders as CLL, multiple myeloma (MM), mucosaassociated lymphoid tissue (MALT) lymphoma, diffuse large B-cell lymphoma, and follicular lymphoma.9–11 On the other hand, the presence of 6q deletion discriminates WM from IgM MGUS, as was previously demonstrated by Schop et al.3 Different minimal deletion regions (MDRs) have been defined in specific subtypes of lymphomas: on 6q21, associated with high-grade lymphomas; 6q23, associated with low-grade lymphomas; and 6q24-q27, associated with intermediate-grade lymphomas.12 In WM, we were not able to identify a unique MDR, but 2 non-overlapped regions were found in about 95% of cases each, covering 1.4 Mb and 3.4 Mb on cytobands 6q21-q22.1 (MDR1) and 6q23 (MDR2), respectively. Potential target genes localized inside those regions are PRDM1 (MDR1) and TNFAIP3 (MDR2), two tumor suppressor that have been associated with the pathogenesis of other B-cell neoplasias.13–15
Table 1.
Delineation of Minimal Deleted Regions and Minimal Amplified Regions Based on Abnormalities Identified in > 10% of Patients
| Genetic Abnormalities |
Cytoband | Number of Patients (%) |
Chromosome Position (bp) | Size (Mb) |
Number of Genes and MIRs |
Candidate Targets |
|---|---|---|---|---|---|---|
| Gains | ||||||
| 3q13.3-q28 | 3 (10) | chr3: 121238508-199379625 | 78.1 | 506 | – | |
| 4q13.1-q35.2 | 4 (12) | chr4: 65598026-191173837 | 125.6 | 580 | – | |
| 6p12.2-p25 | 7 (17) | chr6: 1-39874314 | 39.8 | 562 | – | |
| 8q | 3 (10) | chr8: 76726872-136133311 | 59.4 | 249 | – | |
| 18q | 7 (17) | chr18: 16793910-58926021 | 42.1 | 174 | – | |
| Xq27.1-q28 | 3 (10) | chrX: 149024400-154582473 | 5.6 | 123 | BGN, IRAK1, FLNA, F8, MTCP1, BRCC3 | |
| Losses | ||||||
| 6q16.1 | 14 (33) | chr6: 93266833-97766491 | 4.5 | 12 | MANEA | |
| 6q21-q22.1 | 16 (38) | chr6: 105811723-107183767 | 1.4 | 5 | PRDM1, AIM1 | |
| 6q23 | 16 (38) | chr6: 138093510-141518652 | 3.4 | 15 | TNFAIP3 | |
| 6q25.2-q25.3 | 14 (33) | chr6: 155012960-159967461 | 4.9 | 22 | – | |
| 13q14 | 3 (10) | chr13: 49414571-50454033 | 1 | 11 | MIRN15a, MIRN16-1 |
In regions smaller than 20 Mb were listed the CIG (cancer implicated genes) expressed in ≥ 50% of patients with Waldenström’s macroglobulinemia.
The remaining abnormalities were identified in < 20% of cases, mainly comprising whole chromosomes or entire chromosome arms. Gain of 6p was found in 17% of patients, and its presence was always associated with 6q loss, thus suggesting that 6p gain would be generated as a secondary event to 6q deletion. Gain of 6p has been also reported in nodal MZL (NMZL) in a similar rate than in WM, but its association with 6q deletion has not been confirmed.16
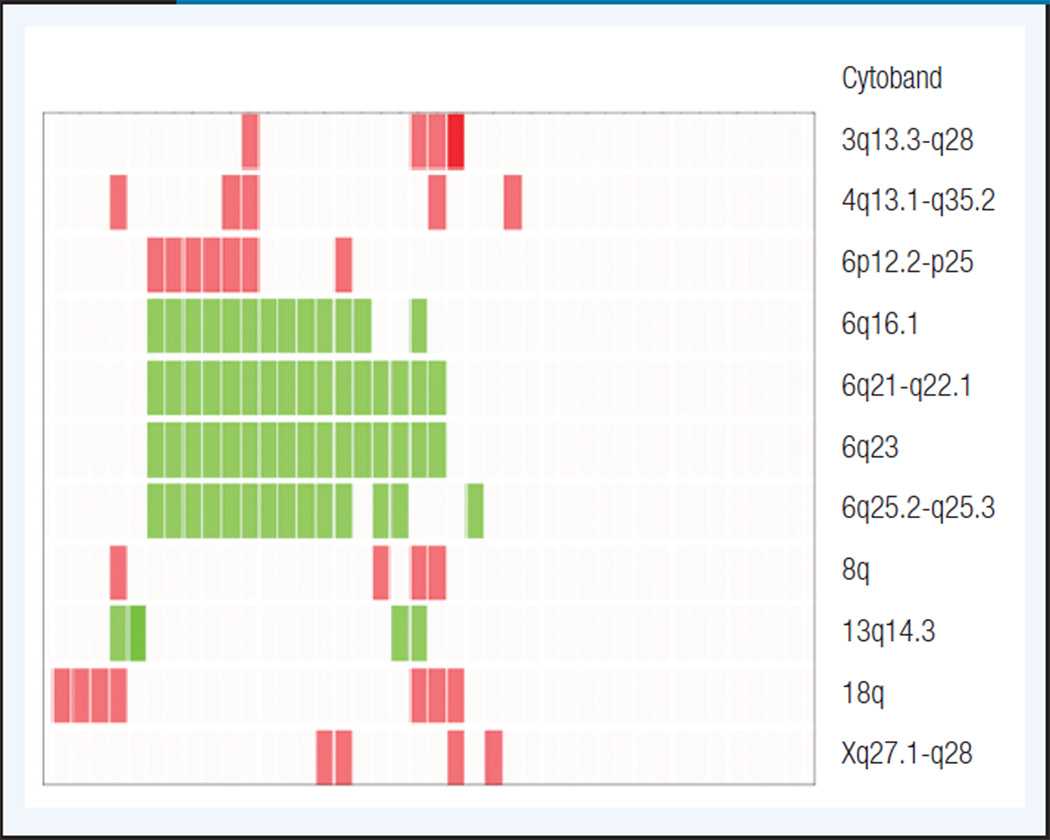
Waldenström macroglobulinemia shares with all the MZL subtypes the whole or partial gain of chromosomes 3 and 18,17,18 but conversely to MZL and CLL, no trisomies 12 were identified in WM. Gains of chromosomes 3 and 18 are very common in all the MZL entities ranging from 20% to 50% and their concomitant presence in the same karyotype is commonly observed. Here, we found gain of 3q13-q28 and 18q in 4 (10%) and 7 (17%) patients, respectively. Three of those cases showed a concomitant gain of both chromosomes and none of them showed 6q deletions (Figure 1).
Figure 1. Unsupervised Hierarchical Clustering of the Genomic Imbalances Detected in > 10% of Waldenström’s Macroglobulinemia Patients.
Red blocks represent chromosome gains; green blocks represent chromosome losses. Each column represents a patient.
Chromosome 13 imbalances were characterized by focal mono or biallelic deletions comprising the cytoband q14.3, thus delineating a MDR that includes the microRNAs MIRN15A and MIRN16-1. Both MIRN15A and MIR16-1 act as tumor suppressor genes in CLL and solid tumors, as prostate cancer, by controlling genes involved in the control of cell survival, proliferation, invasion, and apoptosis.19,20 In addition to WM and CLL, deletions affecting the same area were also recently described in splenic MZL (SMZL),16 thus suggesting that the dysregulation of this pathway is common to several low-grade B-cell neoplasias.
Karyotypic imbalances specific of WM seem to be gains on 4q and 8q arms, which were identified in 12% and 10% of WM cases, respectively, but they were absent on the remaining low-grade B-cell lymphomas. Gain of chromosome 4 was found with additional abnormalities in all but one karyotype, and its presence was irrespective of the 6q status (Figure 1). Even using this high-resolution approach, we were not able to delineate focal minimal amplified regions and no potential target genes were identified on 4q or 8q.
Overall, few recurrent abnormalities have been identified in WM and our data suggest a similar level of genomic instability than in other low-grade B-cell neoplasias. Indeed, WM shares several chromosomal imbalances with other closed-related entities as MZL and CLL, thus suggesting a comparable cytogenetic background with these malignancies. On the other hand, disease-specific recurrent abnormalities have been also identified, as whole or partial gains of chromosome 4 and 8q arm, helping to distinguish WM from other indolent B-cell malignancies. However, the genetic consequences of these specific-abnormalities remain elusive and further studies are critical to refine the search and to precise the molecular pathways affected by these abnormalities.
Acknowledgements
This work was funded by the International WaldenstrOm's Macroglobulinemia Foundation. We thank Chris Gooden and Michael Bittner for helpful assistance. E.B. is supported by IWMF 2S grant from the IWMF. J.J.K. is supported by a MMRF Fellowship and the Gene and Mary-Lou Kurtz fellowship in myeloma. P.L.B. is supported by R01-AG020686 and SPORE P50-CA100707-01. R.F. is supported by R01-CA83724-01, SPORE P50-CA100707-01 and P01-CA62242 from the National Cancer Institute, and the Donaldson Charitable Trust Fund. R.F. is a Clinical Investigator of the Damon Runyon Cancer Research Fund.
Support
E.B. is supported by IWMF 2S grant from the International Waldenström’ s Macroglobulinemia Foundation. J.J.K. is supported by a MMRF Fellowship and the Gene and Mary-Lou Kurtz fellowship in myeloma. P.L.B. is supported by R01-AG020686 and SPORE P50-CA100707-01. R.F. is supported by CA 972 74, R01- CA83724-01, SPORE P50-CA100707-01 and P01-CA62242 from the National Cancer Institute, and the Donaldson Charitable Trust Fund. R.F. is a Clinical Investigator of the Damon Runyon Cancer Research Fund.
Footnotes
Work was performed at Mayo Clinic, Scottsdale, AZ
Authorship Contributions
E.B., J.J.K., M.C., P.L.B., and R.F. designed the study; E.B., J.J.K., S.V.W., R.V., R.F.J.C., and M.B. performed the research; X.L., V.H.J.Z., T.P.T., and K.H. collected data; P.G., M.G., S.H., S.V.R., A.K.S., A.D., and I.G. contributed with patient samples; E.B. and R.F. wrote the article; all authors reviewed and gave final approval of the article.
Disclosures
The authors declare no relevant conflicts of interests.
References
- 1.Fonseca R, Hayman S. Waldenstrom macroglobulinaemia. Br J Haematol. 2007;138:700–720. doi: 10.1111/j.1365-2141.2007.06724.x. [DOI] [PubMed] [Google Scholar]
- 2.Schop RF, Kuehl WM, Van Wier SA, et al. Waldenström macroglobulinemia neoplastic cells lack immunoglobulin heavy chain locus translocations but have frequent 6q deletions. Blood. 2002;100:2996–3001. doi: 10.1182/blood.V100.8.2996. [DOI] [PubMed] [Google Scholar]
- 3.Schop RF, Van Wier SA, Xu R, et al. 6q deletion discriminates Waldenstrom macroglobulinemia from IgM monoclonal gammopathy of undetermined significance. Cancer Genet Cytogenet. 2006;169:150–153. doi: 10.1016/j.cancergencyto.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Terre C, Nguyen-Khac F, Barin C, et al. Trisomy 4, a new chromosomal abnormality in Waldenstrom's macroglobulinemia: a study of 39 cases. Leukemia. 2006;20:1634–1636. doi: 10.1038/sj.leu.2404314. [DOI] [PubMed] [Google Scholar]
- 5.Schop RF, Jalal SM, Van Wier SA, et al. Deletions of 17p13.1 and 13q14 are uncommon in Waldenstrom macroglobulinemia clonal cells and mostly seen at the time of disease progression. Cancer Genet Cytogenet. 2002;132:55–60. doi: 10.1016/s0165-4608(01)00526-x. [DOI] [PubMed] [Google Scholar]
- 6.Chng WJ, Schop RF, Price-Troska T, et al. Gene-expression profiling of Waldenstrom macroglobulinemia reveals a phenotype more similar to chronic lymphocytic leukemia than multiple myeloma. Blood. 2006;108:2755–2763. doi: 10.1182/blood-2006-02-005488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutierrez NC, Ocio EM, de Las Rivas J, et al. Gene expression profiling of B lymphocytes and plasma cells from Waldenstrom's macroglobulinemia: comparison with expression patterns of the same cell counterparts from chronic lymphocytic leukemia, multiple myeloma and normal individuals. Leukemia. 2007;21:541–549. doi: 10.1038/sj.leu.2404520. [DOI] [PubMed] [Google Scholar]
- 8.Hatjiharissi E, Ngo H, Leontovich AA, et al. M. Proteomic analysis of walden-strom macroglobulinemia. Cancer Res. 2007;67:3777–3784. doi: 10.1158/0008-5472.CAN-06-3089. [DOI] [PubMed] [Google Scholar]
- 9.Amiel A, Mulchanov I, Elis A, et al. Deletion of 6q27 in chronic lymphocytic leukemia and multiple myeloma detected by fluorescence in situ hybridization. Cancer Genet Cytogenet. 1999;112:53–56. doi: 10.1016/s0165-4608(98)00255-6. [DOI] [PubMed] [Google Scholar]
- 10.Cigudosa JC, Parsa NZ, Louie DC, et al. Cytogenetic analysis of 363 consecutively ascertained diffuse large B-cell lymphomas. Genes Chromosomes Cancer. 1999;25:123–133. [PubMed] [Google Scholar]
- 11.Tilly H, Rossi A, Stamatoullas A, et al. Prognostic value of chromosomal abnormalities in follicular lymphoma. Blood. 1994;84:1043–1049. [PubMed] [Google Scholar]
- 12.Offit K, Louie DC, Parsa NZ, et al. Clinical and morphologic features of B-cell small lymphocytic lymphoma with del(6)(q21q23) Blood. 1994;83:2611–2618. [PubMed] [Google Scholar]
- 13.Honma K, Tsuzuki S, Nakagawa M, et al. TNFAIP3 is the target gene of chromosome band 6q23.3-q24.1 loss in ocular adnexal marginal zone B cell lymphoma. Genes Chromosomes Cancer. 2008;47:1–7. doi: 10.1002/gcc.20499. [DOI] [PubMed] [Google Scholar]
- 14.Pasqualucci L, Compagno M, Houldsworth J, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thelander EF, Ichimura K, Corcoran M, et al. Characterization of 6q deletions in mature B cell lymphomas and childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2008;49:477–487. doi: 10.1080/10428190701817282. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira BI, Garcia JF, Suela J, et al. Comparative genome profiling across subtypes of low-grade B-cell lymphoma identifies type-specific and common aberrations that target genes with a role in B-cell neoplasia. Haematologica. 2008;93:670–679. doi: 10.3324/haematol.12221. [DOI] [PubMed] [Google Scholar]
- 17.Streubel B, Simonitsch-Klupp I, Mullauer L, et al. Variable frequencies of MALT lymphoma-associated genetic aberrations in MALT lymphomas of different sites. Leukemia. 2004;18:1722–1726. doi: 10.1038/sj.leu.2403501. [DOI] [PubMed] [Google Scholar]
- 18.Dierlamm J, Pittaluga S, Wlodarska I, et al. Marginal zone B-cell lymphomas of different sites share similar cytogenetic and morphologic features. Blood. 1996;87:299–307. [PubMed] [Google Scholar]
- 19.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]