Abstract
Protein oxidative modifications, also known as protein oxidation, are a major class of protein posttranslational modifications. They are caused by reactions between protein amino acid residues and reactive oxygen species (ROS) or reactive nitrogen species (RNS) and can be classified into two categories: irreversible modifications and reversible modifications. Protein oxidation has been often associated with functional decline of the target proteins, which are thought to contribute to normal aging and age-related pathogenesis. However, it has now been recognized that protein oxidative modifications can also play beneficial roles in disease and health. This review summarizes and highlights certain positive roles of protein oxidative modifications that have been documented in the literature. Covered oxidatively modified protein adducts include carbonylation, 3-nitrotyrosine, s-sulfenation, s-nitrosylation, s-glutathionylation, and disulfide formation. All of which have been widely analyzed in numerous experimental systems associated with redox stress conditions. The authors believe that selected protein targets, when modified in a reversible manner in prophylactic approaches such as preconditioning or ischemic tolerance, may provide potential promise in maintaining health and fighting disease.
Keywords: carbonylation, cysteine, glutathionylation, ischemic tolerance, nitrosylation, nitrotyrosine, sulfenation, sulfenic acid, postconditioning, preconditioning, oxidative modifications, reactive oxygen species
1. Introduction
In order to cope with environmental challenges, cells rely on a variety of posttranslational modification mechanisms to expand protein function [1-4]. Of all the documented posttranslational modifications, oxidative modification of the side chains of various amino acid residues forms a major category of protein posttranslational modifications [5-7]. Protein oxidative modifications can be generally classified into two categories: irreversible oxidation and reversible oxidation [8-10]; both of which can be selectively induced by reactive oxygen species (ROS) and reactive nitrogen species (RNS) [11, 12].
Earlier studies of protein oxidation nearly exclusively focused on the detrimental effects of protein oxidation in aging and diseases [5, 9, 13-17]. It has now been firmly recognized, however, that protein oxidation can also play a positive role in many cellular functions. This gradual realization of the beneficial roles of protein oxidation may be attributed to accumulating evidence that ROS and RNS are indispensible for cell survival [18-22] and regeneration [23], and in many cases, they are required for recovery of cellular functions by creating positive stress conditions whereby cell survival mechanisms are reprogrammed to extend life span [24-27] or to withstand severe, or lethal challenges [28-31].
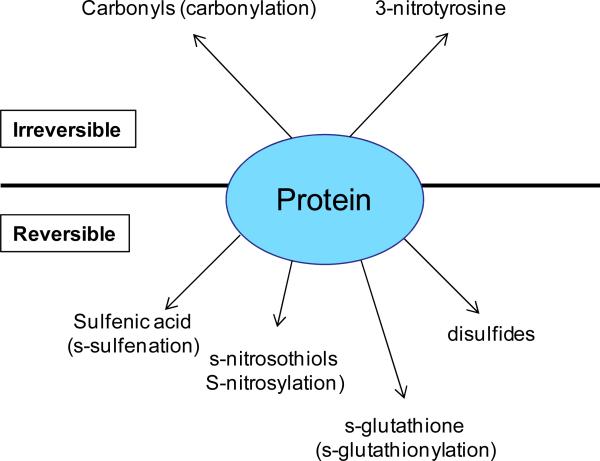
In this article, we review both irreversible and reversible oxidative modifications that are beneficial in health and disease. Modification adducts to be discussed include protein carbonyls, 3-nitrotyrosine, and cysteine oxidation products (Fig. 1). As protein cysteine residue is the one that often undergoes reversible redox modifications by ROS or RNS [32-34], we have inclined to devote more space on cysteine modifications including s-sulfenation, s-nitrosylation, s-glutathionylation, and disulfide formation that are all reversible [35-38]. It should be noted that protein oxidative modifications that have deleterious effects in health and disease are beyond the scope of this review and will only be sporadically mentioned.
Fig. 1.
Irreversible and reversible protein oxidation products discussed in this review. Irreversible oxidation includes protein carbonyls and 3-nitrotyrosine while reversible oxidation includes cysteine modification products such as sulfenic acid, nitrosothiols, and s-glutathione.
2. Cellular sources of oxidants
There are many systems inside a cell that can generate ROS. Mitochondria are recognized as the major site for ROS production [39-41]; and both complexes I and III have been established to be the specific sites for mitochondrial ROS generation [42-45]. Besides mitochondria, many enzymes are also capable of producing ROS. These include, but not limited to, NADPH oxidase [46, 47], xanthine oxidase [48, 49], α-ketoglutarate dehydrogenase complex [50-52], d-amino acid oxidases [53-55], and dihydrolipoamide dehydrogenase [56-62]. On the other hand, nitric oxide production in vivo is mainly achieved by nitric oxide synthases [63-65] though under certain conditions deoxygenated myoglobin [66] or xanthine oxidoreductase [67] or cytochrome c oxidase [68] can be involved in NO production; and in vitro nitric oxide donors are also frequently used either in experimental systems [69-71] or for therapeutic purpose [72-74]. It should be noted that in the presence of superoxide anion, nitric oxide can rapidly react with superoxide anion to yield peroxynitrite [75-77], a reactive species that is highly reactive toward redox-sensitive amino acid residues including tyrosine and cysteine [78, 79].
3. Irreversible protein oxidative modifications
First, we would like to discuss briefly the possible beneficial role of irreversible modifications. These types of modifications include mainly protein carbonylation and tyrosine nitration [11, 80-84]. Both modifications are often associated with oxidative damage and have been used as biomarkers for assessment of oxidative stress in aging and diseases [13, 15-17, 85]. While both carbonylation and nitration can have detrimental effects on the target proteins, evidence has also emerged that such modifications can also play positive roles in cellular function under stress conditions.
3.1. Protein carbonyls
Protein carbonyls formed on several amino acids residues, including arginine, histidine, lysine, proline, threonine and cysteine, are the most widely used biomarker for measurement of protein oxidation and oxidative stress in aging and diseases [5, 8, 11-14, 86-90]. As the modification occurs on multiple amino acid residues on selected protein targets [15-17, 91], its magnitude is much greater than any other modifications that occur only on a specific amino acid residue [11, 12], and thus is more readily detectable. Many studies have employed protein carbonylation to evaluate the detrimental effects of protein oxidation and oxidative stress [13-16, 87-90]; evidence for positive effects of this modification, however, has started to accumulate. For example, protein carbonylation has been shown to be involved in signal transduction [92-95] and is known to be involved in ischemic preconditioning eliciting protection against reperfusion-induced tissue injuries [96, 97].
3.2. Protein nitrotyrosine
Nitrotyrosine, usually 3-nitrotyrosine, is formed between reactive nitrogen species and a protein's tyrosine residue [78, 98, 99]. This modification is a highly selective process as not all proteins or all tyrosine residues on a target protein can get nitrated [100]. Formation of nitrotyrosine is often thought to be accompanied with acute or chronic inflammation disease [101-104], whereby level of nitric oxide is elevated [102, 104-106]. While numerous studies have investigated the deleterious effects of 3-nitrotyrosines [107, 108], concurrent with development of methods for detection and quantitation [109, 110], this modification has been detected under normal physiological conditions such as healthy pregnancy [111, 112], indicating that formation of 3-nitrotyrosine has physiological function.
4. Reversible protein oxidative modifications: protein cysteine modifications
4.1. Chemistry of protein cysteine residues
At neutral pH under physiological conditions, free cysteine residues have a pKa value that is around 8.5, which makes oxidative modifications impossible [113]. To be susceptible to oxidation, the pKa value of a cysteine residue needs to be lower than the physiological pH value (pH 7.4), a condition under which, the cysteine –SH group becomes thiolated (thiolate anion) [113-115]. It is those thiolated cysteine residues that are redox reactive [35, 116]. This thiolation process, decreasing the pKa value to 7.2 or lower, can be achieved via many factors such as hydrogen bonding [117, 118], the effect of adjacent basic amino acid residues [117], the microenvironment of the target cysteine residues [117], and substrate binding [119]. For example, albumin cysteine 34 has a very low pKa value of 5 [120]. Hence under physiological condition, it exists as thiolate anion and is very reactive towards oxidants, thiols, metals, and disulfides [121-123].
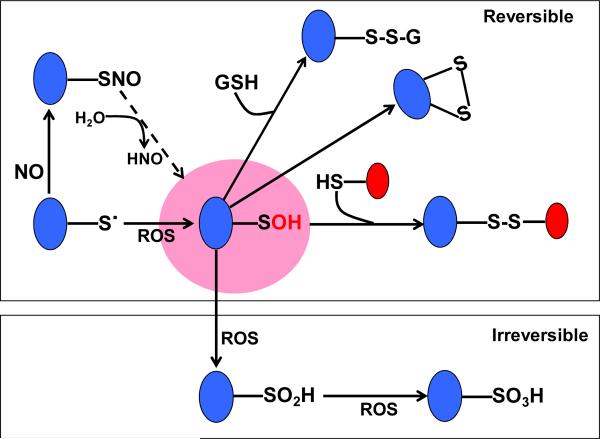
As described above, thiols with low pKa values are more reactive because they are usually deprotonated or thiolated at physiological pH [124-126]. Therefore, oxidation of protein cysteines that are redox reactive is also a highly selective process [127, 128]. As shown in Fig. 2, cysteine oxidation usually starts with the formation of sulfenic acid, from which a variety of oxidation products can be furtherly formed and many of them are reversible and well defined chemically. These cysteine oxidation products include disulfide formation (S-S-), S-glutathionylation (protein-SSG), S-nitrosylation (-SNO), sulfenic acid formation (-SOH, or S-sulfenation) and have all been demonstrated in redox regulation of protein functions by ROS and RNS [35, 129, 130]. Importantly, all of which have been implicated to play beneficial roles in disease and health because they may protect the target proteins from further oxidation that will otherwise permanently damage the target proteins [131-133]. Another mechanism is that these modifications also play a role in redox signaling cascades that boost cellular defense systems to better counteract stress insults [134-136].
Fig. 2.
Chemistry of cysteine oxidative modifications. Sulfenic acid is truly an intermediate product during cysteine oxidation. Given appropriate conditions, s-nitrosothiols can also be discomposed to yield sulfenic acids with concurrent production of nitroxyl [137]. Sulfenic acid can be further oxidized to form disulfide bonds, s-glutathionylation. Irreversible oxidation products sulfinic and sulfonic acids are also shown.
4.2. Protein sulfenic acid formation (S-sulfenation)
This sulfur-hydroxylation product (P-SOH) possesses powerful redox chemistry and has been demonstrated to play a key redox regulatory role in a growing number of proteins [34, 138-141]. Its formation is mainly induced by ROS such as hydrogen peroxide, alkyl hydroperoxides, and RNS such as peroxynitrite [38, 129, 137, 142, 143]. Although being a simple chemical modification, sulfenic acid formation can have a dramatic effect on protein function [130, 137, 144]. It was for a long time regarded as an intermediate, unstable cysteine oxidation product, which may still be true for many proteins [137, 145, 146]. Growing evidence, however, has demonstrated that stable-SOH indeed exists, making trapping, labeling, detecting, and quantitating possible for further evaluation of the formed –SOH [142, 147-150]. A beneficial effect of protein SOH formation has been elegantly demonstrated in studies whereby s-sulfenation of aldose reductase protects the heart against ischemic/reperfusion injury [151-153]. Specifically, these studies found that cyse-298's sulfenation of aldose reductase by peroxynitrite shows great protection against cardiac ischemic injury; and administration of peroxynitrite scavengers not only eliminates cys-298's sulfenation, but also abolishes cardiac protection against ischemic injury. In unrelated studies, Michalek et al. demonstrated that protein sulfenation is indispensible for T-cell growth and proliferation as arrest of sulfenic acids greatly impairs T cell maturation [154]. Another example of a beneficial role of P-SOH is that of the sulfenation of nitrile hydratase; sulfenic acid formation on this enzyme's Cys114 residue is absolutely essential for the enzyme's catalytic activity [155].
4.3. Protein s-nitrosylation
Protein s-nitrosylation can be induced by nitric oxide, nitroxyl, and peroxynitrite [156, 157]. This modification has been regarded as functionally equivalent to protein phosphorylation and dephosphorylation [158-160]. Besides occurring on cysteine residues other than on tyrosine, serine, or threonine residues, s- nitrosylation is also potentially different from phosphorylation in that nitrosylation may not involve a delicate network consisting of kinases or enzymes that catalyze, respectively, nitrosylation and denitrosylation, though the existence of denitrosylases, including Cu,Zn-superoxide dismutase and bilirubin, has been reported [161-165]. Nonetheless, s-nitrosylation has been demonstrated to be a key modification of cysteine residues under a variety of physiological and pathophysiological conditions [157, 166, 167]. In particular, in connection with nitric oxide-based redox regulation of protein function, s-nitrosylation has been found to be involved in protective mechanisms in many disorders [157, 168-170]. For example, Sheng et al. have demonstrated that chemically-enhanced s-nitrosylation can improve recovery from subarachnoid hemorrhage [171], and Penna et al. have demonstrated that protein s-nitrosylation is favorably produced during cardiac postconditioning [172].
4.4. Protein s-glutathionylation
Protein cysteine residues can also undergo s-glutathionylation under oxidative stress conditions [173-175]. Glutathione (GSH) is the major cellular antioxidant, yet, it can also modify proteins via mixed disulfide formation (P-S-S-G), leading to functional changes of the target proteins [176]. This reversible oxidation of critical cysteine residues on proteins has been found to be involved in oxidative signal transduction, control of gene expression, cell proliferation, apoptosis, and cellular responses to protecting key regulatory molecules from oxidative insults [173, 176-178]. Similar to s-sulfenation and s-nitrosylation, protein-S-S-G is also often associated with a detrimental effect on the target protein's function [179-182], but can also protect the target protein from irreversible and permanent damage [183-186]. Therefore, protein glutathionylation has increasingly gained great attention as a possible means of redox regulation of protein functions in response to oxidative stress under physiological and pathophysiological conditions [185, 187]. For example, actin glutathionylation regulates actin dynamics in polymorphonuclear neutrophils [188], manipulation of uncoupling protein 2's glutathionylation may provide a strategy for cancer treatment [189], and glutathionylation of adenine nuclear translocase induced by preconditioning can prevent mitochondrial membrane permeabilization and apoptosis [190].
4.5. Protein disulfides
This is different from protein s-glutathionylation, where a mixed disulfide between GSH and a protein-linked cysteine residue is formed [191-193]. Native disulfide bond formation is usually involved in correct protein folding and is catalyzed by disulfide isomerase in the endoplasmic reticulum and the mitochondrial intermembrane space [194-197], and should be considered different from those formed under oxidative stress or pathophysiological conditions. Hence herein, disulfide formation is strictly meant to reflect inter- or intra- protein disulfide formation that is caused by ROS or RNS [198-205]. Disulfide bonds formed between free cysteine residues upon oxidative stress have been reported to play a beneficial role in cellular defense systems against a variety of stress challenges [191, 206-210]. For example, intra-protein disulfide formation in Cdc25c upon hydrogen peroxide exposure regulates the stability of the protein [211], and in the brain type creatine kinase, disulfide formation between two cysteine residues (cys74 and cys254) can serve as a cellular defense mechanism [212]. Additionally and importantly, it is well established that formation of disulfide linkage within Keap1 in response to cellular stimuli by electrophiles and oxidants [213-217] is essential for activation of the NF-E2-related factor 2 (Nrf2) that then upregulates the expression of phase II antioxidant enzymes under a variety of physiological and pathophysiological conditions [218-224].
5. Protein oxidative modifications and ischemic tolerance
Posttranslational protein oxidative modifications, in particular cysteine modifications, have been implicated in ischemic tolerance or preconditioning [168, 225-231]. Ischemic tolerance constitutes a positive stress that reporgrams cellular defense systems to prevent subsequent lethal injuries [232-237]. The phenomenon of preconditioning seems to be universal as all tissues in mamalian systems as well as all organisms can be preconditioned. In particular, the heart and the brain can be preconditioned by a variety of mechanisms to prevent further injuries caused by ischemia reperfusion [232, 238]. Therefore, preconditioning has both prophylactic and therapeutic value. Despite intensive studies, the mechanisms of preconditioning has not been well understood. Nonetheless, ROS are known to be the key molecules involved in preconditioning development [239-243] as antioxidants administered during induction of preconditioning can block the preconditioning effect [28, 30]. Moreover, a moderately-elevated level of ROS, in particular, H2O2, has been shown to be neuroprotective [221, 244-246]. Nevertheless, how ROS work in preconditioning induction and tissue protection remains elusive. As ROS can impart their effects by modifying proteins, identification of endogenous protein targets of ROS may elucidate mechanisms of protection induced by ischemic tolerance. It is thus conceivable that identification of oxidatively modified protein targets, especially those that can undergo reversible oxidative modifications, may provide insights into novel therapeutic strategies for ischemic tolerance. It is also worth mentioning that a concept of postconditioning, whereby the reperfusion procedure can be disrupted and intervened to elicit protection against lethal injury, has been recently established [247-250]. We think that postconditioning can also be placed under the notion of ischemic tolerance. In fact, preconditioning and postconditioning may share similar pathways or mechanisms [250-254].
So why could protein oxidation, in particular, reversible oxidation-induced by ischemic tolerance be involved in protection against subsequent ischemic injury? As it is the reperfusion stage that often incurs the injury due to a sudden burst in ROS production concurrent with resupply of oxygen [255-259], oxidized proteins with altered protein function could slow down the rate of ROS production during reperfusion and hence could attenuate ischemic injury [230, 260]. In addition, as already pointed out earlier in this review, oxidized proteins induced by ischemic tolerance could also be involved in eliciting cellular defense systems to protect against further severe ischemia reperfusion injury [261, 262].
6. Summary and Perspectives
While studies on the detrimental or deleterious effects of protein oxidative modifications are, and will still be, dominating the field of protein oxidation, investigation of the beneficial roles of protein oxidation appears to be gaining increasing interest [135, 263]. For beneficial purposes, efforts should be focused on proteomic identification of reversibly oxidized proteins that may exhibit protective effects. Further, studies on a comprehensive understanding of the mechanisms or pathways that regulate the reversible nature of the corresponding modifications should be undertaken. This should be particularly true for reversible cysteine oxidation, which not only reflects changes in cellular redox state, but can also protect the target proteins from further damage. Additionally, reversible cysteine oxidation is also involved in redox signaling cascades [264-267] that can elicit positive stress responses to prevent unpredicted disastrous events such as stroke and heart attack. Therefore, equal efforts will also be needed to identify those protein targets that undergo reversible cysteine modifications in preventative or protective approaches as such identified protein targets may provide therapeutic values in fighting diseases, in particular, ischemia associated cerebral and cardiovascular diseases.
Acknowledgments
The authors wish to apologize to those whose work could not be cited due to space limitations. LJY was supported in part by the National Institutes of Health (Grant: AG022550) and by the University of North Texas Health Science Center (UNTHSC-UAEM seed grant: RI6044).
Footnotes
Conflict of interest: None declared.
References
- 1.Glazer AN, Delange RJ, Sigman DS. Chemical Modification of proteins: Selected methods and analytical procedures. Elsevier Biomedical Press; Amsterdam: 1975. [Google Scholar]
- 2.Johnson BC. Posttranslational covalent modifications of proteins. Academic Press; New York: 1983. [Google Scholar]
- 3.Graves DJ, Martin BL, Wang JH. Co- and post-translational modification of proteins: Chemical principles and biological effects. Oxford University Press; Oxford: 1994. [Google Scholar]
- 4.Walsh CT. Posttranslational modification of proteins: Expanding nature's inventory. Robert and Company Publishers; Eaglewood, Colorado: 2006. [Google Scholar]
- 5.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 6.Shacter E. Quantification and significance of protein oxidation in biological samples. Drug Metab Rev. 2000;32:307–326. doi: 10.1081/dmr-100102336. [DOI] [PubMed] [Google Scholar]
- 7.Shacter E. Protein oxidative damage. Methods Enzymol. 2000;319:428–436. doi: 10.1016/s0076-6879(00)19040-8. [DOI] [PubMed] [Google Scholar]
- 8.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 9.Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz AG, Ahn BW, Shaltiel S, Stadtman ER. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 10.Fedorova M, Kuleva N, Hoffmann R. Reversible and irreversible modifications of skeletal muscle proteins in a rat model of acute oxidative stress. Biochim Biophys Acta. 2009;1792:1185–1193. doi: 10.1016/j.bbadis.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Yan LJ. Analysis of oxidative modification of proteins. Curr Protoc Protein Sci. 2009 doi: 10.1002/0471140864.ps1404s55. Chapter 14:Unit14 4. [DOI] [PubMed] [Google Scholar]
- 12.Yan LJ, Forster MJ. Chemical probes for analysis of carbonylated proteins: A review. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:1308–1315. doi: 10.1016/j.jchromb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stadtman ER. Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 14.Stadtman ER. Protein oxidation and aging. Free Radic Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 15.Yan LJ, Levine RL, Sohal RS. Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl. Acad. Sci. USA. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan LJ, Sohal RS. Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc Natl Acad Sci USA. 1998;95:12896–12901. doi: 10.1073/pnas.95.22.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan LJ, Christians ES, Liu L, Xiao X, Sohal RS, Benjamin IJ. Mouse heat shock transcription factor 1 deficiency alters cardiac redox homeostasis and increases mitochondrial oxidative damage. EMBO J. 2002;21:5164–5172. doi: 10.1093/emboj/cdf528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groeger G, Quiney C, Cotter TG. Hydrogen peroxide as a cell-survival signaling molecule. Antioxid Redox Signal. 2009;11:2655–2671. doi: 10.1089/ars.2009.2728. [DOI] [PubMed] [Google Scholar]
- 19.Knoefler D, Thamsen M, Koniczek M, Niemuth NJ, Diederich AK, Jakob U. Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Mol Cell. 2012;47:767–776. doi: 10.1016/j.molcel.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groeger G, Doonan F, Cotter TG, Donovan M. Reactive oxygen species regulate prosurvival ERK1/2 signaling and bFGF expression in gliosis within the retina. Invest Ophthalmol Vis Sci. 2012;53:6645–6654. doi: 10.1167/iovs.12-10525. [DOI] [PubMed] [Google Scholar]
- 21.Bevilacqua E, Gomes SZ, Lorenzon AR, Hoshida MS, Amarante-Paffaro AM. NADPH oxidase as an important source of reactive oxygen species at the mouse maternal-fetal interface: putative biological roles. Reprod Biomed Online. 2012;25:31–43. doi: 10.1016/j.rbmo.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Watson J. Oxidants, antioxidants and the current incurability of metastatic cancers. Open Biol. 2013;3:120144. doi: 10.1098/rsob.120144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, Gallop JL, Dorey K, Amaya E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol. 2013 doi: 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis). Exp Gerontol. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 25.Ristow M, Schmeisser S. Extending life span by increasing oxidative stress. Free Radic Biol Med. 2011;51:327–336. doi: 10.1016/j.freeradbiomed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko YS, Ota A, Nakashima A, Mori K, Nagatsu I, Nagatsu T. Regulation of oxidative stress in long-lived lipopolysaccharide-activated microglia. Clin Exp Pharmacol Physiol. 2012;39:599–607. doi: 10.1111/j.1440-1681.2012.05716.x. [DOI] [PubMed] [Google Scholar]
- 27.Mesquita A, Weinberger M, Silva A, Sampaio-Marques B, Almeida B, Leao C, Costa V, Rodrigues F, Burhans WC, Ludovico P. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc Natl Acad Sci U S A. 2010;107:15123–15128. doi: 10.1073/pnas.1004432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mori T, Muramatsu H, Matsui T, McKee A, Asano T. Possible role of the superoxide anion in the development of neuronal tolerance following ischaemic preconditioning in rats. Neuropathol Appl Neurobiol. 2000;26:31–40. doi: 10.1046/j.1365-2990.2000.00215.x. [DOI] [PubMed] [Google Scholar]
- 29.Das DK, Maulik N, Sato M, Ray PS. Reactive oxygen species function as second messenger during ischemic preconditioning of heart. Mol Cell Biochem. 1999;196:59–67. [PubMed] [Google Scholar]
- 30.Puisieux F, Deplanque D, Bulckaen H, Maboudou P, Gele P, Lhermitte M, Lebuffe G, Bordet R. Brain ischemic preconditioning is abolished by antioxidant drugs but does not up-regulate superoxide dismutase and glutathion peroxidase. Brain Res. 2004;1027:30–37. doi: 10.1016/j.brainres.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 31.Bhagatte Y, Lodwick D, Storey N. Mitochondrial ROS production and subsequent ERK phosphorylation are necessary for temperature preconditioning of isolated ventricular myocytes. Cell Death Dis. 2012;3:e345. doi: 10.1038/cddis.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beinert H. A tribute to sulfur. Eur J Biochem. 2000;267:5657–5664. doi: 10.1046/j.1432-1327.2000.01637.x. [DOI] [PubMed] [Google Scholar]
- 33.Leonard SE, Carroll KS. Chemical ‘omics’ approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Thamsen M, Jakob U. The redoxome: Proteomic analysis of cellular redox networks. Curr Opin Chem Biol. 2011;15:113–119. doi: 10.1016/j.cbpa.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, Stamler JS, Rhee SG, van der Vliet A. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic Biol Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddie KG, Carroll KS. Expanding the functional diversity of proteins through cysteine oxidation. Curr Opin Chem Biol. 2008;12:746–754. doi: 10.1016/j.cbpa.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 37.Jacob C, Knight I, Winyard PG. Aspects of the biological redox chemistry of cysteine: from simple redox responses to sophisticated signalling pathways. Biol Chem. 2006;387:1385–1397. doi: 10.1515/BC.2006.174. [DOI] [PubMed] [Google Scholar]
- 38.Poole LB, Nelson KJ. Discovering mechanisms of signaling-mediated cysteine oxidation. Curr Opin Chem Biol. 2008;12:18–24. doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ames BN, Shigenaga MK. Oxidants are a major contributor to aging. Ann N Y Acad Sci. 1992;663:85–96. doi: 10.1111/j.1749-6632.1992.tb38652.x. [DOI] [PubMed] [Google Scholar]
- 40.Starkov AA. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann N Y Acad Sci. 2008;1147:37–52. doi: 10.1196/annals.1427.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenaz G. Mitochondria and reactive oxygen species. Which role in physiology and pathology? Adv Exp Med Biol. 2012;942:93–136. doi: 10.1007/978-94-007-2869-1_5. [DOI] [PubMed] [Google Scholar]
- 42.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 44.Miwa S, St-Pierre J, Partridge L, Brand MD. Superoxide and hydrogen peroxide production by Drosophila mitochondria. Free Radic Biol Med. 2003;35:938–948. doi: 10.1016/s0891-5849(03)00464-7. [DOI] [PubMed] [Google Scholar]
- 45.Drose S, Brandt U. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv Exp Med Biol. 2012;748:145–169. doi: 10.1007/978-1-4614-3573-0_6. [DOI] [PubMed] [Google Scholar]
- 46.Manea A. NADPH oxidase-derived reactive oxygen species: involvement in vascular physiology and pathology. Cell Tissue Res. 2010;342:325–339. doi: 10.1007/s00441-010-1060-y. [DOI] [PubMed] [Google Scholar]
- 47.Bylund J, Brown KL, Movitz C, Dahlgren C, Karlsson A. Intracellular generation of superoxide by the phagocyte NADPH oxidase: how, where, and what for? Free Radic Biol Med. 2010;49:1834–1845. doi: 10.1016/j.freeradbiomed.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 48.Harrison R. Physiological roles of xanthine oxidoreductase. Drug Metab Rev. 2004;36:363–375. doi: 10.1081/dmr-120037569. [DOI] [PubMed] [Google Scholar]
- 49.Agarwal A, Banerjee A, Banerjee UC. Xanthine oxidoreductase: a journey from purine metabolism to cardiovascular excitation-contraction coupling. Crit Rev Biotechnol. 2011;31:264–280. doi: 10.3109/07388551.2010.527823. [DOI] [PubMed] [Google Scholar]
- 50.Starkov AA, Fiskum G, Chinopoulos C, Lorenzo BJ, Browne SE, Patel MS, Beal MF. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J Neurosci. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ambrus A, Tretter L, Adam-Vizi V. Inhibition of the alpha-ketoglutarate dehydrogenase-mediated reactive oxygen species generation by lipoic acid. J Neurochem. 2009;109(Suppl 1):222–229. doi: 10.1111/j.1471-4159.2009.05942.x. [DOI] [PubMed] [Google Scholar]
- 52.Ambrus A, Torocsik B, Tretter L, Ozohanics O, Adam-Vizi V. Stimulation of reactive oxygen species generation by disease-causing mutations of lipoamide dehydrogenase. Hum Mol Genet. 2011;20:2984–2995. doi: 10.1093/hmg/ddr202. [DOI] [PubMed] [Google Scholar]
- 53.Pollegioni L, Molla G. New biotech applications from evolved D-amino acid oxidases. Trends Biotechnol. 2011;29:276–283. doi: 10.1016/j.tibtech.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 54.Fang J, Sawa T, Akaike T, Maeda H. Tumor-targeted delivery of polyethylene glycol-conjugated D-amino acid oxidase for antitumor therapy via enzymatic generation of hydrogen peroxide. Cancer Res. 2002;62:3138–3143. [PubMed] [Google Scholar]
- 55.Haskew-Layton RE, Payappilly JB, Smirnova NA, Ma TC, Chan KK, Murphy TH, Guo H, Langley B, Sultana R, Butterfield DA, Santagata S, Alldred MJ, Gazaryan IG, Bell GW, Ginsberg SD, Ratan RR. Controlled enzymatic production of astrocytic hydrogen peroxide protects neurons from oxidative stress via an Nrf2-independent pathway. Proc Natl Acad Sci U S A. 2010;107:17385–17390. doi: 10.1073/pnas.1003996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bando Y, Aki K. Mechanisms of generation of oxygen radicals and reductive mobilization of ferritin iron by lipoamide dehydrogenase. J Biochem (Tokyo) 1991;109:450–454. doi: 10.1093/oxfordjournals.jbchem.a123402. [DOI] [PubMed] [Google Scholar]
- 57.Sreider CM, Grinblat L, Stoppani AO. Catalysis of nitrofuran redox-cycling and superoxide anion production by heart lipoamide dehydrogenase. Biochem Pharmacol. 1990;40:1849–1857. doi: 10.1016/0006-2952(90)90366-s. [DOI] [PubMed] [Google Scholar]
- 58.Gazaryan IG, Krasnikov BF, Ashby GA, Thorneley RN, Kristal BS, Brown AM. Zinc is a potent inhibitor of thiol oxidoreductase activity and stimulates reactive oxygen species production by lipoamide dehydrogenase. J Biol Chem. 2002;277:10064–10072. doi: 10.1074/jbc.M108264200. [DOI] [PubMed] [Google Scholar]
- 59.Tahara EB, Barros MH, Oliveira GA, Netto LE, Kowaltowski AJ. Dihydrolipoyl dehydrogenase as a source of reactive oxygen species inhibited by caloric restriction and involved in Saccharomyces cerevisiae aging. Faseb J. 2007;21:274–283. doi: 10.1096/fj.06-6686com. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Q, Zou P, Zhan H, Zhang M, Zhang L, Ge RS, Huang Y. Dihydrolipoamide dehydrogenase and cAMP are associated with cadmium-mediated Leydig cell damage. Toxicol Lett. 2011;205:183–189. doi: 10.1016/j.toxlet.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 61.Kareyeva AV, Grivennikova VG, Cecchini G, Vinogradov AD. Molecular identification of the enzyme responsible for the mitochondrial NADH-supported ammonium-dependent hydrogen peroxide production. FEBS Lett. 2011;585:385–389. doi: 10.1016/j.febslet.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kareyeva AV, Grivennikova VG, Vinogradov AD. Mitochondrial hydrogen peroxide production as determined by the pyridine nucleotide pool and its redox state. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbabio.2012.03.033. [DOI] [PubMed] [Google Scholar]
- 63.Jin RC, Loscalzo J. Vascular Nitric Oxide: Formation and Function. J Blood Med. 2010;2010:147–162. doi: 10.2147/JBM.S7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tennyson AG, Lippard SJ. Generation, translocation, and action of nitric oxide in living systems. Chem Biol. 2011;18:1211–1220. doi: 10.1016/j.chembiol.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Feng C. Mechanism of Nitric Oxide Synthase Regulation: Electron Transfer and Interdomain Interactions. Coord Chem Rev. 2012;256:393–411. doi: 10.1016/j.ccr.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hendgen-Cotta UB, Merx MW, Shiva S, Schmitz J, Becher S, Klare JP, Steinhoff HJ, Goedecke A, Schrader J, Gladwin MT, Kelm M, Rassaf T. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2008;105:10256–10261. doi: 10.1073/pnas.0801336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Samouilov A, Liu X, Zweier JL. Characterization of the magnitude and kinetics of xanthine oxidase-catalyzed nitrate reduction: evaluation of its role in nitrite and nitric oxide generation in anoxic tissues. Biochemistry. 2003;42:1150–1159. doi: 10.1021/bi026385a. [DOI] [PubMed] [Google Scholar]
- 68.Sarti P, Forte E, Mastronicola D, Giuffre A, Arese M. Cytochrome c oxidase and nitric oxide in action: molecular mechanisms and pathophysiological implications. Biochim Biophys Acta. 2012;1817:610–619. doi: 10.1016/j.bbabio.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 69.Yan LJ, Yang SH, Shu H, Prokai L, Forster MJ. Histochemical staining and quantification of dihydrolipoamide dehydrogenase diaphorase activity using blue native PAGE. Electrophoresis. 2007;28:1036–1045. doi: 10.1002/elps.200600574. [DOI] [PubMed] [Google Scholar]
- 70.Han P, Zhou X, Huang B, Zhang X, Chen C. On-gel fluorescent visualization and the site identification of S-nitrosylated proteins. Anal Biochem. 2008;377:150–155. doi: 10.1016/j.ab.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 71.Huang B, Chen C. Detection of protein S-nitrosation using irreversible biotinylation procedures (IBP). Free Radic Biol Med. 2010;49:447–456. doi: 10.1016/j.freeradbiomed.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 72.Marsh N, Marsh A. A short history of nitroglycerine and nitric oxide in pharmacology and physiology. Clin Exp Pharmacol Physiol. 2000;27:313–319. doi: 10.1046/j.1440-1681.2000.03240.x. [DOI] [PubMed] [Google Scholar]
- 73.Serafim RA, Primi MC, Trossini GH, Ferreira EI. Nitric oxide: state of the art in drug design. Curr Med Chem. 2012;19:386–405. doi: 10.2174/092986712803414321. [DOI] [PubMed] [Google Scholar]
- 74.Nossaman VE, Nossaman BD, Kadowitz PJ. Nitrates and nitrites in the treatment of ischemic cardiac disease. Cardiol Rev. 2010;18:190–197. doi: 10.1097/CRD.0b013e3181c8e14a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goldstein S, Merenyi G. The chemistry of peroxynitrite: implications for biological activity. Methods Enzymol. 2008;436:49–61. doi: 10.1016/S0076-6879(08)36004-2. [DOI] [PubMed] [Google Scholar]
- 76.Frein D, Schildknecht S, Bachschmid M, Ullrich V. Redox regulation: a new challenge for pharmacology. Biochem Pharmacol. 2005;70:811–823. doi: 10.1016/j.bcp.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 77.Poderoso JJ. The formation of peroxynitrite in the applied physiology of mitochondrial nitric oxide. Arch Biochem Biophys. 2009;484:214–220. doi: 10.1016/j.abb.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 78.Castro L, Demicheli V, Tortora V, Radi R. Mitochondrial protein tyrosine nitration. Free Radic Res. 2011;45:37–52. doi: 10.3109/10715762.2010.516254. [DOI] [PubMed] [Google Scholar]
- 79.Ullrich V, Kissner R. Redox signaling: bioinorganic chemistry at its best. J Inorg Biochem. 2006;100:2079–2086. doi: 10.1016/j.jinorgbio.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 80.Dalle-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A. Protein carbonylation, cellular dysfunction, and disease progression. J Cell Mol Med. 2006;10:389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wong PS, Eiserich JP, Reddy S, Lopez CL, Cross CE, van der Vliet A. Inactivation of glutathione S-transferases by nitric oxide-derived oxidants: exploring a role for tyrosine nitration. Arch Biochem Biophys. 2001;394:216–228. doi: 10.1006/abbi.2001.2532. [DOI] [PubMed] [Google Scholar]
- 82.Rao RS, Moller IM. Pattern of occurrence and occupancy of carbonylation sites in proteins. Proteomics. 2011;11:4166–4173. doi: 10.1002/pmic.201100223. [DOI] [PubMed] [Google Scholar]
- 83.Peluffo G, Radi R. Biochemistry of protein tyrosine nitration in cardiovascular pathology. Cardiovasc Res. 2007;75:291–302. doi: 10.1016/j.cardiores.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 84.Szabo C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- 85.Prokai L, Yan LJ, Vera-Serrano JL, Stevens SM, Forster MJ. Mass spectrometry-based survey of age-associated protein carbonylation in rat brain mitochondria. J. Mass Spectrom. 2007;42:1583–1589. doi: 10.1002/jms.1345. [DOI] [PubMed] [Google Scholar]
- 86.Madian AG, Regnier FE. Proteomic Identification of Carbonylated Proteins and Their Oxidation Sites. J Proteome Res. 2010;9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan LJ, Sohal RS. Prevention of flight activity prolongs the life span of the housefly, Musca domestica, and attenuates the age-associated oxidative damamge to specific mitochondrial proteins. Free Radic Biol Med. 2000;29:1143–1150. doi: 10.1016/s0891-5849(00)00423-8. [DOI] [PubMed] [Google Scholar]
- 88.Yan LJ, Orr WC, Sohal RS. Identification of oxidized proteins based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunochemical detection, isoelectric focusing, and microsequencing. Anal Biochem. 1998;263:67–71. doi: 10.1006/abio.1998.2799. [DOI] [PubMed] [Google Scholar]
- 89.Yan LJ, Sohal RS. Gel electrophoretic quantitation of protein carbonyls derivatized with tritiated sodium borohydride. Anal Biochem. 1998;265:176–182. doi: 10.1006/abio.1998.2868. [DOI] [PubMed] [Google Scholar]
- 90.Wehr NB, Levine RL. Quantification of protein carbonylation. Methods Mol Biol. 2013;965:265–281. doi: 10.1007/978-1-62703-239-1_18. [DOI] [PubMed] [Google Scholar]
- 91.Guidi F, Magherini F, Gamberi T, Bini L, Puglia M, Marzocchini R, Ranaldi F, Modesti PA, Gulisano M, Modesti A. Plasma protein carbonylation and physical exercise. Mol Biosyst. 2011;7:640–650. doi: 10.1039/c0mb00106f. [DOI] [PubMed] [Google Scholar]
- 92.Wong CM, Cheema AK, Zhang L, Suzuki YJ. Protein carbonylation as a novel mechanism in redox signaling. Circ Res. 2008;102:310–318. doi: 10.1161/CIRCRESAHA.107.159814. [DOI] [PubMed] [Google Scholar]
- 93.Wong CM, Marcocci L, Liu L, Suzuki YJ. Cell signaling by protein carbonylation and decarbonylation. Antioxid Redox Signal. 2010;12:393–404. doi: 10.1089/ars.2009.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Curtis JM, Hahn WS, Long EK, Burrill JS, Arriaga EA, Bernlohr DA. Protein carbonylation and metabolic control systems. Trends Endocrinol Metab. 2012;23:399–406. doi: 10.1016/j.tem.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wong CM, Bansal G, Marcocci L, Suzuki YJ. Proposed role of primary protein carbonylation in cell signaling. Redox Rep. 2012;17:90–94. doi: 10.1179/1351000212Y.0000000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Serviddio G, Di Venosa N, Federici A, D'Agostino D, Rollo T, Prigigallo F, Altomare E, Fiore T, Vendemiale G. Brief hypoxia before normoxic reperfusion (postconditioning) protects the heart against ischemia-reperfusion injury by preventing mitochondria peroxyde production and glutathione depletion. Faseb J. 2005;19:354–361. doi: 10.1096/fj.04-2338com. [DOI] [PubMed] [Google Scholar]
- 97.Oksala NK, Paimela H, Alhava E, Atalay M. Heat shock preconditioning induces protein carbonylation and alters antioxidant protection in superficially injured guinea pig gastric mucosa in vitro. Dig Dis Sci. 2007;52:1897–1905. doi: 10.1007/s10620-006-9214-1. [DOI] [PubMed] [Google Scholar]
- 98.Ferrer-Sueta G, Radi R. Chemical biology of peroxynitrite: kinetics, diffusion, and radicals. ACS Chem Biol. 2009;4:161–177. doi: 10.1021/cb800279q. [DOI] [PubMed] [Google Scholar]
- 99.Feeney MB, Schoneich C. Tyrosine modifications in aging. Antioxid Redox Signal. 2012;17:1571–1579. doi: 10.1089/ars.2012.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Radi R. Protein Tyrosine Nitration: Biochemical Mechanisms and Structural Basis of Functional Effects. Acc Chem Res. 2012 doi: 10.1021/ar300234c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Robbins RA, Hadeli K, Nelson D, Sato E, Hoyt JC. Nitric oxide, peroxynitrite, and lower respiratory tract inflammation. Immunopharmacology. 2000;48:217–221. doi: 10.1016/s0162-3109(00)00220-4. [DOI] [PubMed] [Google Scholar]
- 102.Salvemini D, Doyle TM, Cuzzocrea S. Superoxide, peroxynitrite and oxidative/nitrative stress in inflammation. Biochem Soc Trans. 2006;34:965–970. doi: 10.1042/BST0340965. [DOI] [PubMed] [Google Scholar]
- 103.Ohmori H, Kanayama N. Immunogenicity of an inflammation-associated product, tyrosine nitrated self-proteins. Autoimmun Rev. 2005;4:224–229. doi: 10.1016/j.autrev.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 104.Sugiura H, Ichinose M. Nitrative stress in inflammatory lung diseases. Nitric Oxide. 2011;25:138–144. doi: 10.1016/j.niox.2011.03.079. [DOI] [PubMed] [Google Scholar]
- 105.Cheyuo C, Wu R, Zhou M, Jacob A, Coppa G, Wang P. Ghrelin suppresses inflammation and neuronal nitric oxide synthase in focal cerebral ischemia via the vagus nerve. Shock. 2011;35:258–265. doi: 10.1097/SHK.0b013e3181f48a37. [DOI] [PubMed] [Google Scholar]
- 106.Codoner-Franch P, Tavarez-Alonso S, Murria-Estal R, Megias-Vericat J, Tortajada-Girbes M, Alonso-Iglesias E. Nitric oxide production is increased in severely obese children and related to markers of oxidative stress and inflammation. Atherosclerosis. 2011;215:475–480. doi: 10.1016/j.atherosclerosis.2010.12.035. [DOI] [PubMed] [Google Scholar]
- 107.Rubbo H, Radi R. Protein and lipid nitration: role in redox signaling and injury. Biochim Biophys Acta. 2008;1780:1318–1324. doi: 10.1016/j.bbagen.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 108.Souza JM, Peluffo G, Radi R. Protein tyrosine nitration--functional alteration or just a biomarker? Free Radic Biol Med. 2008;45:357–366. doi: 10.1016/j.freeradbiomed.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 109.Weber D, Kneschke N, Grimm S, Bergheim I, Breusing N, Grune T. Rapid and sensitive determination of protein-nitrotyrosine by ELISA: Application to human plasma. Free Radic Res. 2012;46:276–285. doi: 10.3109/10715762.2011.652627. [DOI] [PubMed] [Google Scholar]
- 110.Dekker F, Abello N, Wisastra R, Bischoff R. Enrichment and detection of tyrosine-nitrated proteins. Curr Protoc Protein Sci. 2012 doi: 10.1002/0471140864.ps1413s69. Chapter 14:Unit 14 13. [DOI] [PubMed] [Google Scholar]
- 111.Webster RP, Roberts VH, Myatt L. Protein nitration in placenta - functional significance. Placenta. 2008;29:985–994. doi: 10.1016/j.placenta.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Horvath EM, Magenheim R, Kugler E, Vacz G, Szigethy A, Levardi F, Kollai M, Szabo C, Lacza Z. Nitrative stress and poly(ADP-ribose) polymerase activation in healthy and gestational diabetic pregnancies. Diabetologia. 2009;52:1935–1943. doi: 10.1007/s00125-009-1435-3. [DOI] [PubMed] [Google Scholar]
- 113.Roos G, Foloppe N, Messens J. Understanding the pK(a) of Redox Cysteines: The Key Role of Hydrogen Bonding. Antioxid Redox Signal. 2013;18:94–127. doi: 10.1089/ars.2012.4521. [DOI] [PubMed] [Google Scholar]
- 114.Chiu J, Dawes IW. Redox control of cell proliferation. Trends Cell Biol. 2012;22:592–601. doi: 10.1016/j.tcb.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 115.Broniowska KA, Hogg N. The chemical biology of S-nitrosothiols. Antioxid Redox Signal. 2012;17:969–980. doi: 10.1089/ars.2012.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rhee SG, Chang TS, Bae YS, Lee SR, Kang SW. Cellular regulation by hydrogen peroxide. J Am Soc Nephrol. 2003;14:S211–S215. doi: 10.1097/01.asn.0000077404.45564.7e. [DOI] [PubMed] [Google Scholar]
- 117.Rhee SG, Bae YS, Lee SR, Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE. 2000;2000:pe1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 118.Lim JC, Gruschus JM, Kim G, Berlett BS, Tjandra N, Levine RL. A low pKa cysteine at the active site of mouse methionine sulfoxide reductase A. J Biol Chem. 2012;287:25596–25601. doi: 10.1074/jbc.M112.369116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Antoine M, Gand A, Boschi-Muller S, Branlant G. Characterization of the amino acids from Neisseria meningitidis MsrA involved in the chemical catalysis of the methionine sulfoxide reduction step. J Biol Chem. 2006;281:39062–39070. doi: 10.1074/jbc.M608844200. [DOI] [PubMed] [Google Scholar]
- 120.Sengupta S, Chen H, Togawa T, DiBello PM, Majors AK, Budy B, Ketterer ME, Jacobsen DW. Albumin thiolate anion is an intermediate in the formation of albumin-S-S-homocysteine. J Biol Chem. 2001;276:30111–30117. doi: 10.1074/jbc.M104324200. [DOI] [PubMed] [Google Scholar]
- 121.Turell L, Botti H, Carballal S, Ferrer-Sueta G, Souza JM, Duran R, Freeman BA, Radi R, Alvarez B. Reactivity of sulfenic acid in human serum albumin. Biochemistry. 2008;47:358–367. doi: 10.1021/bi701520y. [DOI] [PubMed] [Google Scholar]
- 122.Turell L, Botti H, Carballal S, Radi R, Alvarez B. Sulfenic acid--a key intermediate in albumin thiol oxidation. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3384–3392. doi: 10.1016/j.jchromb.2009.03.035. [DOI] [PubMed] [Google Scholar]
- 123.Alvarez B, Carballal S, Turell L, Radi R. Formation and reactions of sulfenic acid in human serum albumin. Methods Enzymol. 2010;473:117–136. doi: 10.1016/S0076-6879(10)73005-6. [DOI] [PubMed] [Google Scholar]
- 124.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 125.Riederer BM. Oxidation proteomics: The role of thiol modifications. Current Proteomics. 2009;6:51–62. [Google Scholar]
- 126.Murphy MP. Mitochondrial thiols in antioxidant protection and redox signaling: distinct roles for glutathionylation and other thiol modifications. Antioxid Redox Signal. 2012;16:476–495. doi: 10.1089/ars.2011.4289. [DOI] [PubMed] [Google Scholar]
- 127.Go YM, Halvey PJ, Hansen JM, Reed M, Pohl J, Jones DP. Reactive aldehyde modification of thioredoxin-1 activates early steps of inflammation and cell adhesion. Am J Pathol. 2007;171:1670–1681. doi: 10.2353/ajpath.2007.070218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Carvalho CM, Chew EH, Hashemy SI, Lu J, Holmgren A. Inhibition of the human thioredoxin system. A molecular mechanism of mercury toxicity. J Biol Chem. 2008;283:11913–11923. doi: 10.1074/jbc.M710133200. [DOI] [PubMed] [Google Scholar]
- 129.Salsbury FR, Jr., Knutson ST, Poole LB, Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17:299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jacob C, Battaglia E, Burkholz T, Peng D, Bagrel D, Montenarh M. Control of oxidative posttranslational cysteine modifications: from intricate chemistry to widespread biological and medical applications. Chem Res Toxicol. 2012;25:588–604. doi: 10.1021/tx200342b. [DOI] [PubMed] [Google Scholar]
- 131.Biswas S, Chida AS, Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochem Pharmacol. 2006;71:551–564. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 132.Requejo R, Hurd TR, Costa NJ, Murphy MP. Cysteine residues exposed on protein surfaces are the dominant intramitochondrial thiol and may protect against oxidative damage. Febs J. 2010;277:1465–1480. doi: 10.1111/j.1742-4658.2010.07576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen YY, Chu HM, Pan KT, Teng CH, Wang DL, Wang AH, Khoo KH, Meng TC. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J Biol Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lou YW, Chen YY, Hsu SF, Chen RK, Lee CL, Khoo KH, Tonks NK, Meng TC. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. Febs J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 135.Madian AG, Hindupur J, Hulleman JD, Diaz-Maldonado N, Mishra VR, Guigard E, Kay CM, Rochet JC, Regnier FE. Effect of single amino acid substitution on oxidative modifications of the Parkinson's disease-related protein, DJ-1. Mol Cell Proteomics. 2012;11:M111 010892. doi: 10.1074/mcp.M111.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang Y, Yang J, Yi J. Redox sensing by proteins: oxidative modifications on cysteines and the consequent events. Antioxid Redox Signal. 2012;16:649–657. doi: 10.1089/ars.2011.4313. [DOI] [PubMed] [Google Scholar]
- 137.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 138.van Montfort RL, Congreve M, Tisi D, Carr R, Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 139.Mansuy D, Dansette PM. Sulfenic acids as reactive intermediates in xenobiotic metabolism. Arch Biochem Biophys. 2011;507:174–185. doi: 10.1016/j.abb.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 140.Roos G, Messens J. Protein sulfenic acid formation: from cellular damage to redox regulation. Free Radic Biol Med. 2011;51:314–326. doi: 10.1016/j.freeradbiomed.2011.04.031. [DOI] [PubMed] [Google Scholar]
- 141.Kettenhofen NJ, Wood MJ. Formation, reactivity, and detection of protein sulfenic acids. Chem Res Toxicol. 2010;23:1633–1646. doi: 10.1021/tx100237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Saurin AT, Neubert H, Brennan JP, Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Natl Acad Sci U S A. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Charles RL, Schroder E, May G, Free P, Gaffney PR, Wait R, Begum S, Heads RJ, Eaton P. Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics. 2007;6:1473–1484. doi: 10.1074/mcp.M700065-MCP200. [DOI] [PubMed] [Google Scholar]
- 144.Yan LJ, Sumien N, Thangthaeng N, Forster MJ. Reversible inactivation of dihydrolipoamide dehydrogenase by mitochondrial hydrogen peroxide. Free Radic Res. 2013;47:123–133. doi: 10.3109/10715762.2012.752078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Poole LB. Formation and functions of protein sulfenic acids. Curr Protoc Toxicol. 2004 doi: 10.1002/0471140856.tx1701s18. Chapter 17:Unit17 1. [DOI] [PubMed] [Google Scholar]
- 146.Depuydt M, Leonard SE, Vertommen D, Denoncin K, Morsomme P, Wahni K, Messens J, Carroll KS, Collet JF. A periplasmic reducing system protects single cysteine residues from oxidation. Science. 2009;326:1109–1111. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- 147.Takanishi CL, Ma LH, Wood MJ. A genetically encoded probe for cysteine sulfenic acid protein modification in vivo. Biochemistry. 2007;46:14725–14732. doi: 10.1021/bi701625s. [DOI] [PubMed] [Google Scholar]
- 148.Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW, King SB. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug Chem. 2007;18:2004–2017. doi: 10.1021/bc700257a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Seo YH, Carroll KS. Quantification of protein sulfenic acid modifications using isotope-coded dimedone and iododimedone. Angew Chem Int Ed Engl. 2011;50:1342–1345. doi: 10.1002/anie.201007175. [DOI] [PubMed] [Google Scholar]
- 150.Seo YH, Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Natl Acad Sci U S A. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kaiserova K, Srivastava S, Hoetker JD, Awe SO, Tang XL, Cai J, Bhatnagar A. Redox activation of aldose reductase in the ischemic heart. J Biol Chem. 2006;281:15110–15120. doi: 10.1074/jbc.M600837200. [DOI] [PubMed] [Google Scholar]
- 152.Kaiserova K, Tang XL, Srivastava S, Bhatnagar A. Role of nitric oxide in regulating aldose reductase activation in the ischemic heart. J Biol Chem. 2008;283:9101–9112. doi: 10.1074/jbc.M709671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wetzelberger K, Baba SP, Thirunavukkarasu M, Ho YS, Maulik N, Barski OA, Conklin DJ, Bhatnagar A. Postischemic deactivation of cardiac aldose reductase: role of glutathione S-transferase P and glutaredoxin in regeneration of reduced thiols from sulfenic acids. J Biol Chem. 2010;285:26135–26148. doi: 10.1074/jbc.M110.146423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Michalek RD, Nelson KJ, Holbrook BC, Yi JS, Stridiron D, Daniel LW, Fetrow JS, King SB, Poole LB, Grayson JM. The requirement of reversible cysteine sulfenic acid formation for T cell activation and function. J Immunol. 2007;179:6456–6467. doi: 10.4049/jimmunol.179.10.6456. [DOI] [PubMed] [Google Scholar]
- 155.Murakami T, Nojiri M, Nakayama H, Odaka M, Yohda M, Dohmae N, Takio K, Nagamune T, Endo I. Post-translational modification is essential for catalytic activity of nitrile hydratase. Protein Sci. 2000;9:1024–1030. doi: 10.1110/ps.9.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 157.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang Y, Yun BW, Kwon E, Hong JK, Yoon J, Loake GJ. S-nitrosylation: an emerging redox-based post-translational modification in plants. J Exp Bot. 2006;57:1777–1784. doi: 10.1093/jxb/erj211. [DOI] [PubMed] [Google Scholar]
- 159.Akhtar MW, Sunico CR, Nakamura T, Lipton SA. Redox Regulation of Protein Function via Cysteine S-Nitrosylation and Its Relevance to Neurodegenerative Diseases. Int J Cell Biol. 2012;2012:463756. doi: 10.1155/2012/463756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sun J, Murphy E. Protein S-nitrosylation and cardioprotection. Circ Res. 2010;106:285–296. doi: 10.1161/CIRCRESAHA.109.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Okado-Matsumoto A, Fridovich I. Putative denitrosylase activity of Cu,Zn-superoxide dismutase. Free Radic Biol Med. 2007;43:830–836. doi: 10.1016/j.freeradbiomed.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 162.Duan S, Chen C. S-nitrosylation/denitrosylation and apoptosis of immune cells. Cell Mol Immunol. 2007;4:353–358. [PubMed] [Google Scholar]
- 163.Foster MW, Yang Z, Gooden DM, Thompson JW, Ball CH, Turner ME, Hou Y, Pi J, Moseley MA, Que LG. Proteomic characterization of the cellular response to nitrosative stress mediated by s-nitrosoglutathione reductase inhibition. J Proteome Res. 2012;11:2480–2491. doi: 10.1021/pr201180m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Beigi F, Gonzalez DR, Minhas KM, Sun QA, Foster MW, Khan SA, Treuer AV, Dulce RA, Harrison RW, Saraiva RM, Premer C, Schulman IH, Stamler JS, Hare JM. Dynamic denitrosylation via S-nitrosoglutathione reductase regulates cardiovascular function. Proc Natl Acad Sci U S A. 2012;109:4314–4319. doi: 10.1073/pnas.1113319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Barone E, Trombino S, Cassano R, Sgambato A, De Paola B, Di Stasio E, Picci N, Preziosi P, Mancuso C. Characterization of the S-denitrosylating activity of bilirubin. J Cell Mol Med. 2009;13:2365–2375. doi: 10.1111/j.1582-4934.2008.00680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yan LJ, Liu L, Forster MJ. Reversible inactivation of dihydrolipoamide dehydrogenase by Angeli's salt. Acta Biophysica Sinica (Sheng Wu Wu Li Hsueh Bao) 2012;28:341–350. [PMC free article] [PubMed] [Google Scholar]
- 167.Haldar SM, Stamler JS. S-nitrosylation: integrator of cardiovascular performance and oxygen delivery. J Clin Invest. 2013;123:101–110. doi: 10.1172/JCI62854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 169.Lin J, Steenbergen C, Murphy E, Sun J. Estrogen receptor-beta activation results in S-nitrosylation of proteins involved in cardioprotection. Circulation. 2009;120:245–254. doi: 10.1161/CIRCULATIONAHA.109.868729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Murphy E, Kohr M, Sun J, Nguyen T, Steenbergen C. S-nitrosylation: a radical way to protect the heart. J Mol Cell Cardiol. 2012;52:568–577. doi: 10.1016/j.yjmcc.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sheng H, Reynolds JD, Auten RL, Demchenko IT, Piantadosi CA, Stamler JS, Warner DS. Pharmacologically augmented S-nitrosylated hemoglobin improves recovery from murine subarachnoid hemorrhage. Stroke. 2011;42:471–476. doi: 10.1161/STROKEAHA.110.600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Penna C, Perrelli MG, Tullio F, Moro F, Parisella ML, Merlino A, Pagliaro P. Post-ischemic early acidosis in cardiac postconditioning modifies the activity of antioxidant enzymes, reduces nitration, and favors protein S-nitrosylation. Pflugers Arch. 2011;462:219–233. doi: 10.1007/s00424-011-0970-1. [DOI] [PubMed] [Google Scholar]
- 173.Xiong Y, Uys JD, Tew KD, Townsend DM. S-glutathionylation: from molecular mechanisms to health outcomes. Antioxid Redox Signal. 2011;15:233–270. doi: 10.1089/ars.2010.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pastore A, Piemonte F. S-Glutathionylation signaling in cell biology: progress and prospects. Eur J Pharm Sci. 2012;46:279–292. doi: 10.1016/j.ejps.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 175.Pimentel D, Haeussler DJ, Matsui R, Burgoyne JR, Cohen RA, Bachschmid MM. Regulation of cell physiology and pathology by protein S-glutathionylation: lessons learned from the cardiovascular system. Antioxid Redox Signal. 2012;16:524–542. doi: 10.1089/ars.2011.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zweier JL, Chen CA, Druhan LJ. S-glutathionylation reshapes our understanding of endothelial nitric oxide synthase uncoupling and nitric oxide/reactive oxygen species-mediated signaling. Antioxid Redox Signal. 2011;14:1769–1775. doi: 10.1089/ars.2011.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 178.Dalle-Donne I, Colombo G, Gagliano N, Colombo R, Giustarini D, Rossi R, Milzani A. S-glutathiolation in life and death decisions of the cell. Free Radic Res. 2011;45:3–15. doi: 10.3109/10715762.2010.515217. [DOI] [PubMed] [Google Scholar]
- 179.Velu CS, Niture SK, Doneanu CE, Pattabiraman N, Srivenugopal KS. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry. 2007;46:7765–7780. doi: 10.1021/bi700425y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xie Y, Kole S, Precht P, Pazin MJ, Bernier M. S-glutathionylation impairs signal transducer and activator of transcription 3 activation and signaling. Endocrinology. 2009;150:1122–1131. doi: 10.1210/en.2008-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kambe T, Song T, Takata T, Hatano N, Miyamoto Y, Nozaki N, Naito Y, Tokumitsu H, Watanabe Y. Inactivation of Ca2+/calmodulin-dependent protein kinase I by S-glutathionylation of the active-site cysteine residue. FEBS Lett. 2010;584:2478–2484. doi: 10.1016/j.febslet.2010.04.059. [DOI] [PubMed] [Google Scholar]
- 182.Kim YJ, Kim D, Illuzzi JL, Delaplane S, Su D, Bernier M, Gross ML, Georgiadis MM, Wilson DM., 3rd S-glutathionylation of cysteine 99 in the APE1 protein impairs abasic endonuclease activity. J Mol Biol. 2011;414:313–326. doi: 10.1016/j.jmb.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Coan C, Ji JY, Hideg K, Mehlhorn RJ. Protein sulfhydryls are protected from irreversible oxidation by conversion to mixed disulfides. Arch Biochem Biophys. 1992;295:369–378. doi: 10.1016/0003-9861(92)90530-a. [DOI] [PubMed] [Google Scholar]
- 184.Chae HZ, Oubrahim H, Park JW, Rhee SG, Chock PB. Protein glutathionylation in the regulation of peroxiredoxins: a family of thiol-specific peroxidases that function as antioxidants, molecular chaperones, and signal modulators. Antioxid Redox Signal. 2012;16:506–523. doi: 10.1089/ars.2011.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cooper AJ, Pinto JT, Callery PS. Reversible and irreversible protein glutathionylation: biological and clinical aspects. Expert Opin Drug Metab Toxicol. 2011;7:891–910. doi: 10.1517/17425255.2011.577738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.McLain AL, Szweda PA, Szweda LI. alpha-Ketoglutarate dehydrogenase: a mitochondrial redox sensor. Free Radic Res. 2011;45:29–36. doi: 10.3109/10715762.2010.534163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rasmussen HH, Hamilton EJ, Liu CC, Figtree GA. Reversible oxidative modification: implications for cardiovascular physiology and pathophysiology. Trends Cardiovasc Med. 2010;20:85–90. doi: 10.1016/j.tcm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 188.Sakai J, Li J, Subramanian KK, Mondal S, Bajrami B, Hattori H, Jia Y, Dickinson BC, Zhong J, Ye K, Chang CJ, Ho YS, Zhou J, Luo HR. Reactive oxygen species-induced actin glutathionylation controls actin dynamics in neutrophils. Immunity. 2012;37:1037–1049. doi: 10.1016/j.immuni.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pfefferie A, Mailloux RJ, Adjeitey CN, Harper ME. Glutathionylation of UCP2 sensitizes drug resistant leukemia cells to chemotherapeutics. Biochim Biophys Acta. 2013;1833:80–89. doi: 10.1016/j.bbamcr.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 190.Queiroga CS, Almeida AS, Martel C, Brenner C, Alves PM, Vieira HL. Glutathionylation of adenine nucleotide translocase induced by carbon monoxide prevents mitochondrial membrane permeabilization and apoptosis. J Biol Chem. 2010;285:17077–17088. doi: 10.1074/jbc.M109.065052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.O'Brian CA, Chu F. Post-translational disulfide modifications in cell signaling--role of inter-protein, intra-protein, S-glutathionyl, and S-cysteaminyl disulfide modifications in signal transmission. Free Radic Res. 2005;39:471–480. doi: 10.1080/10715760500073931. [DOI] [PubMed] [Google Scholar]
- 192.Huang KP, Huang FL. Glutathionylation of proteins by glutathione disulfide S-oxide. Biochem Pharmacol. 2002;64:1049–1056. doi: 10.1016/s0006-2952(02)01175-9. [DOI] [PubMed] [Google Scholar]
- 193.Cotgreave IA. Analytical developments in the assay of intraand extracellular GSH homeostasis: specific protein S-glutathionylation, cellular GSH and mixed disulphide compartmentalisation and interstitial GSH redox balance. Biofactors. 2003;17:269–277. doi: 10.1002/biof.5520170126. [DOI] [PubMed] [Google Scholar]
- 194.Xiao R, Wilkinson B, Solovyov A, Winther JR, Holmgren A, Lundstrom-Ljung J, Gilbert HF. The contributions of protein disulfide isomerase and its homologues to oxidative protein folding in the yeast endoplasmic reticulum. J Biol Chem. 2004;279:49780–49786. doi: 10.1074/jbc.M409210200. [DOI] [PubMed] [Google Scholar]
- 195.Narayan M. Disulfide bonds: protein folding and subcellular protein trafficking. Febs J. 2012;279:2272–2282. doi: 10.1111/j.1742-4658.2012.08636.x. [DOI] [PubMed] [Google Scholar]
- 196.Benham AM. The protein disulfide isomerase family: key players in health and disease. Antioxid Redox Signal. 2012;16:781–789. doi: 10.1089/ars.2011.4439. [DOI] [PubMed] [Google Scholar]
- 197.Depuydt M, Messens J, Collet JF. How proteins form disulfide bonds. Antioxid Redox Signal. 2011;15:49–66. doi: 10.1089/ars.2010.3575. [DOI] [PubMed] [Google Scholar]
- 198.Sommer A, Traut RR. Diagonal polyacrylamide-dodecyl sulfate gel electrophoresis for the identification of ribosomal proteins crosslinked with methyl-4-mercaptobutyrimidate. Proc Natl Acad Sci U S A. 1974;71:3946–3950. doi: 10.1073/pnas.71.10.3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 2004;279:21749–21758. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- 200.Danciu TE, Whitman M. Oxidative stress drives disulfide bond formation between basic helix-loop-helix transcription factors. J Cell Biochem. 2010;109:417–424. doi: 10.1002/jcb.22415. [DOI] [PubMed] [Google Scholar]
- 201.McDonagh B, Sheehan D. Effect of oxidative stress on protein thiols in the blue mussel Mytilus edulis: proteomic identification of target proteins. Proteomics. 2007;7:3395–3403. doi: 10.1002/pmic.200700241. [DOI] [PubMed] [Google Scholar]
- 202.McDonagh B, Sheehan D. Effects of oxidative stress on protein thiols and disulphides in Mytilus edulis revealed by proteomics: actin and protein disulphide isomerase are redox targets. Mar Environ Res. 2008;66:193–195. doi: 10.1016/j.marenvres.2008.02.069. [DOI] [PubMed] [Google Scholar]
- 203.An BC, Lee SS, Lee EM, Wi SG, Park W, Chung BY. Global analysis of disulfide bond proteins in Pseudomonas aeruginosa exposed to hydrogen peroxide and gamma rays. Int J Radiat Biol. 2010;86:400–408. doi: 10.3109/09553000903567953. [DOI] [PubMed] [Google Scholar]
- 204.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.McDonagh B. Diagonal electrophoresis for the detection of protein disulfides. Methods Mol Biol. 2012:309–315. doi: 10.1007/978-1-61779-821-4_26. [DOI] [PubMed] [Google Scholar]
- 206.Ghezzi P, Bonetto V, Fratelli M. Thiol-disulfide balance: from the concept of oxidative stress to that of redox regulation. Antioxid Redox Signal. 2005;7:964–972. doi: 10.1089/ars.2005.7.964. [DOI] [PubMed] [Google Scholar]
- 207.Shimizu Y, Hendershot LM. Oxidative folding: cellular strategies for dealing with the resultant equimolar production of reactive oxygen species. Antioxid Redox Signal. 2009;11:2317–2331. doi: 10.1089/ars.2009.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 209.Li W, Zhang J, An W. The conserved CXXC motif of hepatic stimulator substance is essential for its role in mitochondrial protection in H2O2-induced cell apoptosis. FEBS Lett. 2010;584:3929–3935. doi: 10.1016/j.febslet.2010.08.043. [DOI] [PubMed] [Google Scholar]
- 210.Wei PC, Hsieh YH, Su MI, Jiang XJ, Hsu PH, Lo WT, Weng JY, Jeng YM, Wang JM, Chen PL, Chang YC, Lee KF, Tsai MD, Shew JY, Lee WH. Loss of the Oxidative Stress Sensor NPGPx Compromises GRP78 Chaperone Activity and Induces Systemic Disease. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Savitsky PA, Finkel T. Redox regulation of Cdc25C. J Biol Chem. 2002;277:20535–20540. doi: 10.1074/jbc.M201589200. [DOI] [PubMed] [Google Scholar]
- 212.Li XH, Chen Z, Gao YS, Yan YB, Zhang F, Meng FG, Zhou HM. Generation of the oxidized form protects human brain type creatine kinase against cystine-induced inactivation. Int J Biol Macromol. 2011;48:239–242. doi: 10.1016/j.ijbiomac.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 213.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 216.Dinkova-Kostova AT, Talalay P. Direct and indirect antioxidant properties of inducers of cytoprotective proteins. Mol Nutr Food Res. 2008;52(Suppl 1):S128–S138. doi: 10.1002/mnfr.200700195. [DOI] [PubMed] [Google Scholar]
- 217.Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal. 2011;13:1699–1712. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yan W, Wang HD, Hu ZG, Wang QF, Yin HX. Activation of Nrf2-ARE pathway in brain after traumatic brain injury. Neurosci Lett. 2008;431:150–154. doi: 10.1016/j.neulet.2007.11.060. [DOI] [PubMed] [Google Scholar]
- 219.Abbas K, Breton J, Planson AG, Bouton C, Bignon J, Seguin C, Riquier S, Toledano MB, Drapier JC. Nitric oxide activates an Nrf2/sulfiredoxin antioxidant pathway in macrophages. Free Radic Biol Med. 2011;51:107–114. doi: 10.1016/j.freeradbiomed.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 220.Baird L, Dinkova-Kostova AT. The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol. 2011;85:241–272. doi: 10.1007/s00204-011-0674-5. [DOI] [PubMed] [Google Scholar]
- 221.Bell KF, Al-Mubarak B, Fowler JH, Baxter PS, Gupta K, Tsujita T, Chowdhry S, Patani R, Chandran S, Horsburgh K, Hayes JD, Hardingham GE. Mild oxidative stress activates Nrf2 in astrocytes, which contributes to neuroprotective ischemic preconditioning. Proc Natl Acad Sci U S A. 2011;108:E1–2. doi: 10.1073/pnas.1015229108. author reply E3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bell KFS, Fowler JH, Al-Mubarak B, Horsburgh K, Hardingham GE. Activation of Nrf2-Regulated Glutathione Pathway Genes by Ischemic Preconditioning 10.1155/2011/689524. Oxidative Medicine and Cellular Longevity. 2011;2011 doi: 10.1155/2011/689524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Li MH, Cha YN, Surh YJ. Peroxynitrite induces HO-1 expression via PI3K/Akt-dependent activation of NF-E2-related factor 2 in PC12 cells. Free Radic Biol Med. 2006;41:1079–1091. doi: 10.1016/j.freeradbiomed.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 224.Zheng H, Whitman SA, Wu W, Wondrak GT, Wong PK, Fang D, Zhang DD. Therapeutic potential of Nrf2 activators in streptozotocin-induced diabetic nephropathy. Diabetes. 2011;60:3055–3066. doi: 10.2337/db11-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Eaton P, Bell RM, Cave AC, Shattock MJ. Ischemic preconditioning: a potential role for protein S-thiolation? Antioxid Redox Signal. 2005;7:882–888. doi: 10.1089/ars.2005.7.882. [DOI] [PubMed] [Google Scholar]
- 226.Blanco M, Lizasoain I, Sobrino T, Vivancos J, Castillo J. Ischemic preconditioning: A novel target for neuroprotective therapy. Cerebrovasc Dis. 2006;21(Suppl 2):38–47. doi: 10.1159/000091702. [DOI] [PubMed] [Google Scholar]
- 227.Sun J, Steenbergen C, Murphy E. S-nitrosylation: NO-related redox signaling to protect against oxidative stress. Antioxid Redox Signal. 2006;8:1693–1705. doi: 10.1089/ars.2006.8.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sun J. Protein s-nitrosylation: A role of nitric oxide signaling in cardiac ischemic preconditioning. Acta Physiologica Sinica. 2007;59:544–552. [PubMed] [Google Scholar]
- 229.Shiva S, Sack MN, Greer JJ, Duranski M, Ringwood LA, Burwell L, Wang X, MacArthur PH, Shoja A, Raghavachari N, Calvert JW, Brookes PS, Lefer DJ, Gladwin MT. Nitrite augments tolerance to ischemia/reperfusion injury via the modulation of mitochondrial electron transfer. J Exp Med. 2007;204:2089–2102. doi: 10.1084/jem.20070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kohr MJ, Sun J, Aponte A, Wang G, Gucek M, Murphy E, Steenbergen C. Simultaneous measurement of protein oxidation and S-nitrosylation during preconditioning and ischemia/reperfusion injury with resin-assisted capture. Circ Res. 2011;108:418–426. doi: 10.1161/CIRCRESAHA.110.232173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hill BG, Darley-Usmar VM. S-nitrosation and thiol switching in the mitochondrion: a new paradigm for cardioprotection in ischaemic preconditioning. Biochem J. 2008;412:e11–e13. doi: 10.1042/BJ20080716. [DOI] [PubMed] [Google Scholar]
- 232.Kitagawa K, Matsumoto M, Tagaya M, Hata R, Ueda H, Niinobe M, Handa N, Fukunaga R, Kimura K, Mikoshiba K, et al. ‘Ischemic tolerance’ phenomenon found in the brain. Brain Res. 1990;528:21–24. doi: 10.1016/0006-8993(90)90189-i. [DOI] [PubMed] [Google Scholar]
- 233.Yang CW, Ahn HJ, Han HJ, Kim WY, Li C, Shin MJ, Kim SK, Park JH, Kim YS, Moon IS, Bang BK. Pharmacological preconditioning with low-dose cyclosporine or FK506 reduces subsequent ischemia/reperfusion injury in rat kidney. Transplantation. 2001;72:1753–1759. doi: 10.1097/00007890-200112150-00008. [DOI] [PubMed] [Google Scholar]
- 234.Ravingerova T. Intrinsic defensive mechanisms in the heart: a potential novel approach to cardiac protection against ischemic injury. Gen Physiol Biophys. 2007;26:3–13. [PubMed] [Google Scholar]
- 235.Stetler RA, Zhang F, Liu C, Chen J. Ischemic tolerance as an active and intrinsic neuroprotective mechanism. Handb Clin Neurol. 2009;92:171–195. doi: 10.1016/S0072-9752(08)01909-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kitagawa K. Ischemic tolerance in the brain: endogenous adaptive machinery against ischemic stress. J Neurosci Res. 2012;90:1043–1054. doi: 10.1002/jnr.23005. [DOI] [PubMed] [Google Scholar]
- 237.Della-Morte D, Guadagni F, Palmirotta R, Ferroni P, Testa G, Cacciatore F, Abete P, Rengo F, Perez-Pinzon MA, Sacco RL, Rundek T. Genetics and genomics of ischemic tolerance: focus on cardiac and cerebral ischemic preconditioning. Pharmacogenomics. 2012;13:1741–1757. doi: 10.2217/pgs.12.157. [DOI] [PubMed] [Google Scholar]
- 238.Yang X, Cohen MV, Downey JM. Mechanism of cardioprotection by early ischemic preconditioning. Cardiovasc Drugs Ther. 2010;24:225–234. doi: 10.1007/s10557-010-6236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Vanden Hoek TL, Becker LB, Shao Z, Li C, Schumacker PT. Reactive oxygen species released from mitochondria during brief hypoxia induce preconditioning in cardiomyocytes. J Biol Chem. 1998;273:18092–18098. doi: 10.1074/jbc.273.29.18092. [DOI] [PubMed] [Google Scholar]
- 240.da Silva MM, Sartori A, Belisle E, Kowaltowski AJ. Ischemic preconditioning inhibits mitochondrial respiration, increases H2O2 release, and enhances K+ transport. Am J Physiol Heart Circ Physiol. 2003;285:H154–H162. doi: 10.1152/ajpheart.00955.2002. [DOI] [PubMed] [Google Scholar]
- 241.Kunz A, Park L, Abe T, Gallo EF, Anrather J, Zhou P, Iadecola C. Neurovascular protection by ischemic tolerance: role of nitric oxide and reactive oxygen species. J Neurosci. 2007;27:7083–7093. doi: 10.1523/JNEUROSCI.1645-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Liu Y, Yang XM, Iliodromitis EK, Kremastinos DT, Dost T, Cohen MV, Downey JM. Redox signaling at reperfusion is required for protection from ischemic preconditioning but not from a direct PKC activator. Basic Res Cardiol. 2008;103:54–59. doi: 10.1007/s00395-007-0683-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Penna C, Mancardi D, Rastaldo R, Pagliaro P. Cardioprotection: a radical view Free radicals in pre and postconditioning. Biochim Biophys Acta. 2009;1787:781–793. doi: 10.1016/j.bbabio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 244.Geracitano R, Tozzi A, Berretta N, Florenzano F, Guatteo E, Viscomi MT, Chiolo B, Molinari M, Bernardi G, Mercuri NB. Protective role of hydrogen peroxide in oxygen-deprived dopaminergic neurones of the rat substantia nigra. J Physiol. 2005;568:97–110. doi: 10.1113/jphysiol.2005.092510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Nistico R, Piccirilli S, Cucchiaroni ML, Armogida M, Guatteo E, Giampa C, Fusco FR, Bernardi G, Nistico G, Mercuri NB. Neuroprotective effect of hydrogen peroxide on an in vitro model of brain ischaemia. Br J Pharmacol. 2008;153:1022–1029. doi: 10.1038/sj.bjp.0707587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Chadwick W, Zhou Y, Park SS, Wang L, Mitchell N, Stone MD, Becker KG, Martin B, Maudsley S. Minimal peroxide exposure of neuronal cells induces multifaceted adaptive responses. PLoS One. 2010;5:e14352. doi: 10.1371/journal.pone.0014352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhao H, Sapolsky RM, Steinberg GK. Interrupting reperfusion as a stroke therapy: ischemic postconditioning reduces infarct size after focal ischemia in rats. J Cereb Blood Flow Metab. 2006;26:1114–1121. doi: 10.1038/sj.jcbfm.9600348. [DOI] [PubMed] [Google Scholar]
- 248.Zhao H. The protective effect of ischemic postconditioning against ischemic injury: from the heart to the brain. J Neuroimmune Pharmacol. 2007;2:313–318. doi: 10.1007/s11481-007-9089-8. [DOI] [PubMed] [Google Scholar]
- 249.Zhao H. Ischemic postconditioning as a novel avenue to protect against brain injury after stroke. J Cereb Blood Flow Metab. 2009;29:873–885. doi: 10.1038/jcbfm.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhao H, Ren C, Chen X, Shen J. From rapid to delayed and remote postconditioning: the evolving concept of ischemic postconditioning in brain ischemia. Curr Drug Targets. 2012;13:173–187. doi: 10.2174/138945012799201621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hausenloy DJ, Yellon DM. Survival kinases in ischemic preconditioning and postconditioning. Cardiovasc Res. 2006;70:240–253. doi: 10.1016/j.cardiores.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 252.Lim SY, Davidson SM, Hausenloy DJ, Yellon DM. Preconditioning and postconditioning: the essential role of the mitochondrial permeability transition pore. Cardiovasc Res. 2007;75:530–535. doi: 10.1016/j.cardiores.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Hausenloy DJ, Tsang A, Yellon DM. The reperfusion injury salvage kinase pathway: a common target for both ischemic preconditioning and postconditioning. Trends Cardiovasc Med. 2005;15:69–75. doi: 10.1016/j.tcm.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 254.Pignataro G, Scorziello A, Di Renzo G, Annunziato L. Post-ischemic brain damage: effect of ischemic preconditioning and postconditioning and identification of potential candidates for stroke therapy. Febs J. 2009;276:46–57. doi: 10.1111/j.1742-4658.2008.06769.x. [DOI] [PubMed] [Google Scholar]
- 255.Zhao ZQ, Vinten-Johansen J. Postconditioning: reduction of reperfusion-induced injury. Cardiovasc Res. 2006;70:200–211. doi: 10.1016/j.cardiores.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 256.Rodriguez-Sinovas A, Abdallah Y, Piper HM, Garcia-Dorado D. Reperfusion injury as a therapeutic challenge in patients with acute myocardial infarction. Heart Fail Rev. 2007;12:207–216. doi: 10.1007/s10741-007-9039-9. [DOI] [PubMed] [Google Scholar]
- 257.Gursoy-Ozdemir Y, Yemisci M, Dalkara T. Microvascular protection is essential for successful neuroprotection in stroke. J Neurochem. 2012;123(Suppl 2):2–11. doi: 10.1111/j.1471-4159.2012.07938.x. [DOI] [PubMed] [Google Scholar]
- 258.Hausenloy DJ, Yellon DM. Preconditioning and postconditioning: united at reperfusion. Pharmacol Ther. 2007;116:173–191. doi: 10.1016/j.pharmthera.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 259.Hausenloy DJ, Wynne AM, Yellon DM. Ischemic preconditioning targets the reperfusion phase. Basic Res Cardiol. 2007;102:445–452. doi: 10.1007/s00395-007-0656-1. [DOI] [PubMed] [Google Scholar]
- 260.Burwell LS, Nadtochiy SM, Brookes PS. Cardioprotection by metabolic shut-down and gradual wake-up. J Mol Cell Cardiol. 2009;46:804–810. doi: 10.1016/j.yjmcc.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Muller BA, Dhalla NS. Mechanisms of the beneficial actions of ischemic preconditioning on subcellular remodeling in ischemic-reperfused heart. Curr Cardiol Rev. 2010;6:255–264. doi: 10.2174/157340310793566118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Murillo D, Kamga C, Mo L, Shiva S. Nitrite as a mediator of ischemic preconditioning and cytoprotection. Nitric Oxide. 2011;25:70–80. doi: 10.1016/j.niox.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wall SB, Oh JY, Diers AR, Landar A. Oxidative modification of proteins: an emerging mechanism of cell signaling. Front Physiol. 2012;3:369. doi: 10.3389/fphys.2012.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Chung HS, Wang SB, Venkatraman V, Murray CI, Van Eyk JE. Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system. Circ Res. 2013;112:382–392. doi: 10.1161/CIRCRESAHA.112.268680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Pagliaro P, Moro F, Tullio F, Perrelli MG, Penna C. Cardioprotective pathways during reperfusion: focus on redox signaling and other modalities of cell signaling. Antioxid Redox Signal. 2011;14:833–850. doi: 10.1089/ars.2010.3245. [DOI] [PubMed] [Google Scholar]