Abstract
ATP-binding cassette transporter A1 (ABCA1) transporter regulates cholesterol efflux and is an essential mediator of high-density lipoprotein (HDL) formation. In amyloid precursor protein (APP) transgenic mice, Abca1 deficiency increased amyloid deposition in the brain paralleled by decreased levels of Apolipoprotein E (ApoE). The APOEε4 allele is the major genetic risk factor of sporadic Alzheimer's disease (AD). Here, we reveal the effect of Abca1 deficiency on phenotype in mice expressing human ApoE3 or ApoE4. We used APP/E3 and APP/E4 mice generated by crossing APP/PS1ΔE9 transgenic mice to human APOE3- and APOE4-targeted replacement mice and examined Abca1 gene dose effect on amyloid deposition and cognition. The results from two behavior tests demonstrate that lack of one copy of Abca1 significantly exacerbates memory deficits in APP/E4/Abca1−/+ but not in APP/E3/Abca1−/+ mice. The data for amyloid plaques and insoluble amyloid-β (Aβ) also show that Abca1 hemizygosity increases Aβ deposition only in APP/E4/Abca1−/+ but not in APP/E3/Abca1−/+ mice. Our in vivo microdialysis assays indicate that Abca1 deficiency significantly decreases Aβ clearance in ApoE4-expressing mice, while the effect of Abca1 on Aβ clearance in ApoE3-expressing mice was insignificant. In addition, we demonstrate that plasma HDL and Aβ42 levels in APP/E4/Abca1−/+ mice are significantly decreased, and there is a negative correlation between plasma HDL and amyloid plaques in brain, suggesting that plasma lipoproteins may be involved in Aβ clearance. Overall, our results prove that the presence of functional Abca1 significantly influences the phenotype of APP mice expressing human ApoE4 and further substantiate therapeutic approaches in AD based on ABCA1–APOE regulatory axis.
Introduction
Alzheimer's disease (AD) is a late-onset dementia characterized by the presence of senile plaques made of amyloid-β (Aβ), neurofibrillary tangles, and cognitive decline. Although the inheritance of ε4 allele of Apolipoprotein E (APOE) is the major genetic risk factor for late-onset AD (Saunders et al., 1993), the mechanisms underlying this association remain elusive (Kim et al., 2009). It is important to note that, while the incidence of AD among APOE4 allele carriers is substantially increased, not everybody with this allele develops the disease (Corder et al., 1993). It is conceivable that additional genetic factors influence the risk and precipitate the development of dementia.
ATP-binding cassette transporter A1 (ABCA1) is a lipid pump that regulates cholesterol and phospholipids efflux from cells to lipid-poor apolipoprotein A-I (ApoA-I) and ApoE in a process essential for the formation of high-density lipoproteins (HDLs) (Brooks-Wilson et al., 1999). We and others have shown that Abca1 deficiency increased amyloid deposition in different AD model mice in parallel with reduced ApoE and ApoA-I levels (Hirsch-Reinshagen et al., 2005; Koldamova et al., 2005b; Wahrle et al., 2005). A recent report from our group also demonstrated that amyloid precursor protein (APP) mice with one functional copy of Abca1 have significant memory deficits that correlated with the levels of soluble Aβ oligomers (Lefterov et al., 2009). In contrast, it has been shown that transgenic overexpression of ABCA1 (Wahrle et al., 2008) or treatments with liver X receptor (LXR) and retinoid X receptor (RXR) agonists (Koldamova et al., 2005a; Fitz et al., 2010; Cramer et al., 2012) ameliorate AD phenotype in APP transgenic mice. So far, the role of Abca1 has been studied only in mice expressing mouse ApoE. Considering the risk for developing AD in association with APOE4 carrier status, the phenotypic characterization of AD animal model on human ApoE3 or ApoE4 background would be helpful to further understand the role of ABCA1 in the pathogenesis of the disease. APOE-targeted replacement mice expressing human ApoE isoforms under the control of mouse promoter have been used to study the effect of human ApoE on AD-like phenotype (Sullivan et al., 1997). Two recent studies characterized the phenotype of PDAPP mice crossed to APOE-targeted replacement mice and demonstrated that APOE4-expressing mice have more amyloid and less APOE than the other two isoforms (Bales et al., 2009; Castellano et al., 2011). Moreover, it has been shown that, in APOE4 mice, Aβ clearance is significantly delayed (Castellano et al., 2011).
In this study, we used APP/PS1ΔE9 transgenic mice crossed to human APOE3- and APOE4-targeted replacement mice (APP/E3 and APP/E4, respectively). To examine the effect of Abca1 deficiency, APP/E3 and APP/E4 were crossed to Abca1 knock-out mice. Here, we compare amyloid deposition and cognitive decline in APP/E3 and APP/E4 mice expressing wild-type Abca1 to Abca1 hemizygous mice (APP/E3/Abca1−/+ and APP/E4/Abca1−/+). Unexpectedly, our results demonstrate that Abca1 deficiency affected amyloid load and memory deficits only in APP/E4 but not in APP/E3 mice. Importantly, our data also suggest that the level of ApoE and HDL in plasma may affect Aβ clearance.
Materials and Methods
Materials
All chemicals and plastics were purchased through Thermo Fisher Scientific, unless noted otherwise.
Animals
Mouse strains.
The study fully conformed to the guidelines outlined in the Guide for the Care and Use of Laboratory Animals from the United States Department of Health and Human Services and was approved by the University of Pittsburgh Institutional Animal Care and Use Committee. APP/PS1ΔE9 mice [B6.Cg-Tg (APPswe, PSEN1ΔE9)85Dbo/J] were purchased from The Jackson Laboratory on C57BL/6J background (Lefterov et al., 2010). APP/PS1ΔE9 mice express mutant familial variants of human amyloid precursor protein (APP) with Swedish mutation (APPsw), and human PS1 (presenilin 1) with deletion in exon 9 (PS1ΔE9). ABCA1+/− heterozygous mice (DBA/1-Abca1tm1Jdm/J) were purchased from The Jackson Laboratory on C57BL/6 × DBA/1 background and crossbred for 10 generations to pure C57BL/6 background in our laboratory. Human ApoE4 (B6.129P2-Apoetm3(APOE*4)Mae N8) and ApoE3 (B6.129P2-Apoetm3(APOE*3)Mae N8) were purchased from Taconic on C57BL/6 background.
Breeding.
APP/PS1ΔE9 mice were bred to human ApoE4+/+ and ApoE3+/+ targeted replacement mice to generate APP/PS1ΔE9/ApoE4+/+ (referred to as APP/E4) and APP/PS1ΔE9/ApoE3+/+ (referred to as APP/E3) mice expressing only human ApoE isoforms and wild-type Abca1. Separately, Abca1−/− mice were bred to human ApoE4+/+ and ApoE3+/+ targeted replacement mice to generate ApoE4+/+/Abca1+/+ (E4) and ApoE4+/+/Abca1−/+ (E4/Abca1−/+); as well as ApoE3+/+/Abca1+/+ (E3) and ApoE3+/+/Abca1−/+ (E3/Abca1−/+) (in all these lines, the mouse ApoE is entirely replaced). E4/Abca1−/+ mice were crossbred to APP/E4 to generate APP/PS1ΔE9/ApoE4+/+/Abca1−/+ (referred to as APP/E4/Abca1−/+). E3/Abca1−/+ mice were crossbred to APP/E3 to generate APP/PS1ΔE9/ApoE3+/+/Abca1−/+ (referred to as APP/E3/Abca1−/+). All mice were littermates and were fed normal mouse chow diet. Male and female mice from each genotype were used for this study. Radial arm water maze tests were performed at 6–8 months or age (mean age, 7.5 months) with mice perfused soon after for evaluation of amyloid phenotype. We used E4, E4/Abca1−/+; as well as E3 and E3/Abca1−/+ as nontransgenic controls for the behavior experiments. Contextual fear conditioning was performed on a separate group of ApoE3 and ApoE4 transgenic mice with an average age of 4.9 months. For microdialysis experiments, we used 4.5-month-old APP transgenic mice: APP/E3, APP/E3/Abca1−/+, APP/E4, and APP/E4/Abca1−/+.
Radial arm water maze
Two day radial arm water maze (RWM) was used to assess spatial navigational learning as before (Alamed et al., 2006) with minor modifications. The RWM consisted of six arms (20 cm wide, 40 cm long, 8 cm high walls above the water) and a central area (30 cm in diameter), filled with water (temperature, 21 ± 1°C) to a level 1 cm above the hidden platform (10 cm in diameter) (Stoelting). All animals are handled for 2 min for 2 consecutive days before any behavior testing.
Acquisition phase.
We measured the ability of mice to form a representation of the spatial relationship between a safe, but hidden platform and visual cues surrounding the maze. Acquisition testing was performed over 2 consecutive days with mice trained in groups of five or six. Each day, a mouse received two 6 trial blocks and a final 3 trial block (total of 15 trials per day) with a 30 min rest period between blocks. During day 1 of training, a visible platform (flag projecting 6 cm from the platform) was used during trials 1, 3, 5, 7, 9, and 11 to define the rule of a safe platform. All other trials consisted of animals finding the location of a hidden platform. Animals were allowed 60 s to find the platform and 20 s to rest on it. Mice that failed to find the platform were led there by the experimenter and allowed to rest there for 20 s. All animals in a group completed the trial before proceeding, providing a 5 min intertrial interval. The start location was changed for each trial and the platform location was changed between groups.
Open pool task with visible platform.
Following acquisition phase, visible platform training was performed to measure swim speed, motivation, and visual acuity. Briefly, all arms were removed from the water maze, distal visual cues were removed from around the maze, and the platform was marked with a flag projecting 6 cm above the surface of the water. Mice were placed in the maze and allowed 60 s to locate the platform and 20 s to rest on it. If they did not find the platform within 60 s, they were led there by the experimenter and remained there for 20 s. Once all animals in a group completed a trial, the position of the platform was moved while the start position remained constant throughout training. Animals were trained in groups of five or six, and training consisted of two 6 trial blocks and a final 3 trial block (15 total trials) with a 30 min rest period between blocks.
Performance was recorded with an automated tracking system (AnyMaze; Stoelting) during all phases of training. During the acquisition phase, total number of incorrect arm entries and time errors were combined for the overall performance of an animal during a trial. An incorrect arm entry was defined as the entry of 50% of the animal's body into an arm that did not contain the hidden platform. A time error was defined as the failure of an animal to enter an arm after 15 s elapsed. For the 15 daily trials, performance during 3 consecutive trials was averaged into a block (total of 5 blocks per day). During the open pool task of training, speed and latency to the platform were used to compare the performance between genotypes.
Contextual fear conditioning
Contextual fear conditioning (Stoelting) was performed on a separate group of ApoE3 and ApoE4 transgenic mice with an average age of 4.9 months as before (Puzzo et al., 2008; Kornecook et al., 2010) with slight modifications. Briefly, mice were placed in a conditioning chamber for 2 min before the onset of a tone [conditioned stimulus (CS), duration of 30 s, 85 dB sound at 2800 Hz]. In the last 2 s of the CS, mice were given a 2 s, 0.7 mA footshock through the bars on the floor of the chamber, and this cycle was repeated twice. Finally, the mice were allowed to remain in the chamber for 30 s before being returned to their housing cages. Contextual fear was evaluated 24 h after training by measuring freezing behavior for 5 min in the original chamber before mice were returned to their housing cages. Freezing behavior, defined as the absence of movement except for that needed for breathing, was scored using AnyMaze software (Stoelting). Cued fear learning was assessed 24 h after contextual testing by placing mice in a novel context (gray walls were replaced with black-and-white stripped walls) for 2 min, after which they were exposed to the CS for 3 min, and freezing behavior was measured. All chambers were cleaned between animals with 70% ethanol. Data are represented as percentage freezing during all stages of testing.
In vivo microdialysis
In vivo microdialysis to assess brain interstitial fluid (ISF) Aβ40 and Aβ42 in the hippocampus of awake, freely moving mice was performed as previously described (Cirrito et al., 2003; Fitz et al., 2010). Briefly, mice were anesthetized with Avertin (250 mg/kg, i.p.), head shaved, and placed into a stereotaxic frame (Stoelting). A bore hole (0.75 mm) was made above the left hippocampus (coordinates: −3.1 mm from bregma, 2.4 lateral), as well as the right aspect of the skull for placement of an anchoring screw. MD-2250 guide cannulas (Bioanalytical Systems) were stereotactically lowered into the hippocampus (12° angle, −0.6 mm relative to dura mater) and anchored to the bone screw using binary dental cement.
After washing of the MD-2200 microdialysis probes (Bioanalytical Systems) and FEP tubing with 0.15% BSA in aCSF perfusion buffer (in mm: 1.3 CaCl2, 1.2 MgSO4, 3 KCL, 0.4 KH2PO4, 25 NaHCO3, and 122 NaCl, pH 7.35), the probes were manually inserted through the guide cannula into the hippocampus. To measure Aβ40 and Aβ42, microdialysis probes had a constant flow rate of 1.0 μl/min and samples were collected every 75 min on ice, 12 h after probe implantation. To determine ISF Aβ40 and Aβ42 half-life, 1 h baseline samples were taken from hours 12–15 after probe implantation, and at the beginning of hour 16, animals were injected with 10 mg/kg (aS)-N-[(1S)-2-[[(7S)-6,7-dihydro-5-methyl-6-oxo-5H-dibenz[b,d]azepin-7-yl]amino]-1-methyl-2-oxoethyl]-3,5-difluoro-α-hydroxybenzeneacetamide (LY411575), a γ-secretase inhibitor (synthesized at Chemical Synthesis Core Facility, Mayo Clinic Jacksonville, Jacksonville, FL), diluted in corn oil. Five additional 75 min samples were collected for each animal. The levels of Aβ40 and Aβ42 in each sample were determined by ELISA as described below. At the conclusion of the microdialysis experiment, animals were perfused as described below, and 30 μm brain sections were stained with cresyl violet to confirm probe placement.
Animal tissue processing
Mice were anesthetized with Avertin (250 mg/kg of body weight, i.p.) and perfused transcardially (Lefterov et al., 2010). Blood was drawn from the heart before the mice were perfused transcardially with 25 ml of cold 0.1 m PBS, pH 7.4. Brains were rapidly removed and divided into hemispheres, with one of the hemispheres being dissected into the cortex and hippocampus. These parts were snap-frozen on dry ice, while the other hemisphere was drop fixed in 4% phosphate-buffered paraformaldehyde at 4°C for 48 h before storage in 30% sucrose.
Histology and immunohistochemistry
All procedures were as reported previously (Lefterov et al., 2010). Histoprep-embedded hemibrains were cut in the coronal plane at 30 μm sections and stored in a glycol-based cryoprotectant at −20°C until staining. Sections were selected 700 μm apart, starting from a randomly chosen section ∼150 μm caudal to the first appearance of the CA3 and dentate gyrus. Sections mounted on slides were washed in PBS for 10 min and stained with 1,4-bis(3-carboxy-4-hydroxyphenylethenyl)-benzene (X-34) (100 μm) for 10 min. Following the staining, sections were washed in water, incubated in 0.2% NaOH in 80% unbuffered ethanol for 2 min, washed again in water, and soaked in PBS for 10 min.
Another set of adjacent sections were immunostained with 6E10 antibody (Koldamova et al., 2005b) (SIG-39340; Covance). Briefly, free-floating sections were blocked for endogenous peroxidases, avidin–biotin quenched, antigen retrieval was performed with 70% formic acid, and tissue was blocked with 3% normal goat serum. Sections were then incubated in 6E10 biotin-labeled antibody (1:1000) at room temperature for 2 h before being developed with Vectastain ABC Elite kit and a DAB substrate (Vector Laboratories). A final group of adjacent sections were immunostained with polyclonal rabbit anti-GFAP antibody (DK-2600; Dako) for detection of activated microglia as in the study by Lefterov et al. (2009). Briefly, free-floating sections were washed with PBS and blocked with 3% normal goat serum. Sections were then incubated in anti-GFAP antibody (1:1000) at room temperature for 3 h before incubation with anti-rabbit 594-labeled secondary antibody (DI-1594; Vector Laboratories) for 1 h (1:250). Sections were then mounted on slides. Microscopic examination was performed using a Nikon Eclipse 80i microscope (20× magnification). For quantitative analysis, staining in the cortex and hippocampus was defined as the percentage area covered by X-34, GFAP, or 6E10 positivity using MetaMorph 7.0 software (Molecular Devices).
Western blotting and ELISA
The frozen cortices and hippocampi were homogenized separately (Fitz et al., 2010; Lefterov et al., 2010) in tissue homogenization buffer (250 mm sucrose, 20 mm Tris base, 1 mm EDTA, 1 mm EGTA, 1 ml per 100 mg tissue) and protease inhibitors (10 μg/ml leupeptin, 10 μg/ml aprotinin, and 10 μg/ml AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride]) using a glass Dounce. To prepare soluble brain extracts, the initial homogenate was spun at 100,000 × g for 1 h, and the supernatant was used for determination of soluble Aβ as well as soluble ApoE in the brain. To extract insoluble Aβ, the remaining pellet was resuspended with 70% formic acid, and samples were sonicated for 30 s, and then spun at 100,000 × g for 1 h as before (Fitz et al., 2010; Lefterov et al., 2010).
Protein concentration.
The Bradford assay was used to determine the protein concentration of all samples. Briefly, diluted sample are mixed 1:1 with 40% Bradford reagent dye (Bio-Rad) and absorbance at 595 nm was read on a microplate reader. Concentration is determined by comparison to bovine serum albumin standard curve using linear regression analysis.
Aβ ELISA.
Aβ ELISA was performed essentially as in the studies by Fitz et al. (2010) and Lefterov et al. (2010). Briefly, we used 6E10 as the capture antibody (reactive with very high specificity to amino acid residues 1–16 of human Aβ with no cross-reactivity to mouse Aβ), and anti-Aβ40 (G2-10 mAb) (specific to amino acids 31–40 of Aβ40 and has no cross-reactivity with other Aβ peptides) and anti-Aβ42 (G2-13 mAb) (specific for Aβ1–42 and not reactive to Aβ1–38, Aβ1–39, or Aβ1–40 but does have ∼10% reactivity to Aβ1–43 and Aβ1–44) monoclonal antibodies conjugated to horseradish peroxidase (Genetics Company) were used as the detection antibodies. The final values of Aβ were based on Aβ40 and Aβ42 peptide standards (Bachem Biosciences) and are normalized to the total protein concentration in the sample, assayed by Bradford assay, and Aβ values are expressed as picomoles per milligram. Aβ concentration in ISF and plasma was determined using the same ELISA protocol. With this protocol, our interassay and intraassay CV% is <10, and the standard curve demonstrates good linearity (r2 ≥ 0.98) over a wide range of Aβ concentrations from 800 to 6.25 fmol. To reduce interassay variation, all samples are run in a single group when possible and read in duplicate.
Western blotting analysis.
For Western blotting (WB) analysis, extracts containing 50–100 μg of total protein were mixed with Tris/glycine or NUPAGE denaturing loading buffer, loaded, and electrophoresed on 10% Tris/glycine gels or NuPAGE Bis-Tris gels (Invitrogen). Gels were transferred to nitrocellulose membranes, incubated with the respective primary antibodies followed by secondary antibodies conjugated to horseradish peroxidase, and processed for visualization by enhanced chemiluminescence Plus-ECL (PerkinElmer). β-Actin was used as an internal standard. The relative intensities of the bands were quantified by densitometry (ImageQuant, version 5.2; GE Healthcare).
RIPA extraction.
To detect Abca1, full-length APP (APPfl) and C-terminal fragments result of β-secretase cleavages (CTFβ) protein extracts were prepared by 1:1 dilution of the initial homogenate with 2× RIPA buffer in the presence of protease inhibitors, and WBs were performed as before (Koldamova et al., 2005a; Lefterov et al., 2010). Abca1 was detected using monoclonal antibody, ab7360 (Abcam), and APPfl and CTFβ with 6E10 antibody. β-Actin was used as a loading control for all WBs and detected with monoclonal antibody, A5441 (Sigma-Aldrich). Tau phosphorylation was examined on the RIPA extracts from cortices and hippocampi using PHF phosphospecific monoclonal (pSer202/Thr205) antibody (generously provided by Dr. Peter Davies, Albert Einstein College of Medicine, Bronx, NY). The bands were scanned and normalized to total tau using tau-5 antibody from Abcam.
ApoE and ApoA-I.
ELISA for ApoE was performed on soluble brain extract and plasma samples. We used sandwich ELISA assay with goat anti-human ApoE antibody (AB947; Calbiochem) as the capture antibody and biotin-labeled goat anti-ApoE (K74180B; Meridian Life Sciences) as the detection antibody followed by HRP-labeled streptavidin (Fitzgerald). The ELISA was developed using tetramethylbenzidine (TMB) as substrate (KPL), and the absorbance was read at 650 nm wavelength. To detect ApoA-I, we used solid-phase ELISA assay as before (Koldamova et al., 2001). Briefly, plasma or soluble brain extracts samples were diluted in bicarbonate buffer and loaded directly on polysorb ELISA plate. After overnight incubation, the plate was incubated with blocking solution (0.4% Block Ace; AbD Serotec) for 4 h at room temperature. As detection antibody we used mouse-specific anti-ApoA-I polyclonal antibody (600-101-196; Rockland) followed by HRP-labeled anti-goat antibody (sc-2020; Santa Cruz Biotechnology). As a substrate, we used TMB, and the absorbance was read at 650 nm wavelength. As a standard for ApoA-I ELISA, we used recombinant mouse ApoA-I generously provided by Dr. M. Phillips (Children's Hospital of Philadelphia, Philadelphia, PA). Alternatively, ApoA-I level in serum or soluble brain fraction was determined by WB using anti-ApoA-I polyclonal antibody 600-101-196 (see above).
Total cholesterol
Aliquots of cleared soluble brain extract were used to determine the total cholesterol levels in the brain using Amplex Red Cholesterol Assay Kit from Invitrogen according to the manufacturer's protocol and as before (Fitz et al., 2010). The fluorescence of the sample was measured in a clear 96-well microplate using a Spectra Max 300 microplate reader (Molecular Devices) with 540 nm excitation and 590 nm emission. The cholesterol amount of the sample was determined from a standard curve using GraphPad Prism, version 4.0, and normalized to total protein in the extract.
HDL and LDL concentration
At time of collection, blood samples were centrifuged at 16,000 rpm for 5 min, and the plasma was collected and snap-frozen on dry ice. Aliquots of 50 μl of plasma were assayed for HDL and LDL/very-low-density lipoprotein (VLDL) levels using the Abcam HDL and LDL/VLDL Cholesterol Assay Kit (ab65390) according to the manufacturer's protocol. The OD of the sample was measured in a clear 96-well microplate at 570 nm wavelength using a Spectra Max 300 microplate reader (Molecular Devices). The cholesterol amount of the sample was determined from a standard curve using GraphPad Prism, version 4.0, and expressed in milligrams per milliliter.
Statistical analysis
All results are reported as means ± SEM. Statistical significance of differences between mean scores during acquisition phase of training in the RWM were assessed with two-way repeated-measures ANOVA (general linear model/repeated-measures ANOVA) and Tukey's post hoc analysis for multiple comparisons using Genotype and Trial Block Number as sources of variation. The rest of the data (contextual fear conditioning, immunohistochemistry, ELISA, etc.) were analyzed by two-way ANOVA followed by Bonferroni's post hoc test with Abca1 status and ApoE isoform as sources of variation. Additional comparisons between two groups were made by t test. Aβ half-life was determined by nonlinear regression analysis using an equation of one-phase exponential decay [Y = span*exp(−K*X) + plateau with a rate constant K; the half-life is 0.69/K]. Correlation was assessed using nonparametric analysis to determine Spearman's rank coefficient (ρ). All statistical analyses were performed in GraphPad Prism, version 4.0, or SPSS, version 19, IBM, and differences were considered significant where p < 0.05.
Results
Abca1 effect on cognitive performance depends on human ApoE isoform
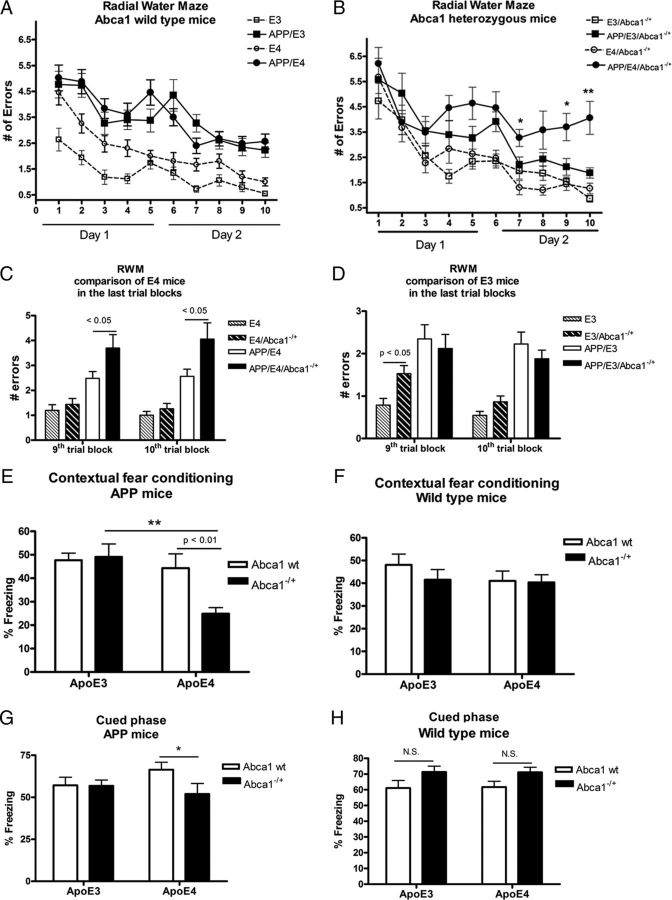
To examine the effects of Abca1 on AD-like phenotype, we used APP/PS1ΔE9 mice expressing human ApoE3 (referred to as APP/E3) and ApoE4 (APP/E4) isoforms. First, we determined cognitive deficits in mice aged 7.5 months using the RWM paradigm. As shown in Figure 1A, among mice expressing wild-type Abca1 there was a significant difference between APP transgenic mice and their respective nontransgenic controls (compare APP/E3 to E3 mice and APP/E4 to E4 mice). However, we did not find a statistical difference between APP/E3 and APP/E4 mice. Next, we crossed APP/E3 and APP/E4 mice to Abca1 knock-out mice to generate mice lacking one copy of Abca1, namely APP/E3/Abca1−/+ and APP/E4/Abca1−/+ mice. As seen in Figure 1B, APP/E4/Abca1−/+ mice performed significantly worse than APP/E3/Abca1−/+ particularly during the last trial blocks (p < 0.05 for trial blocks 10, 9, and 7). This result suggests that the lack of one copy of Abca1 significantly impairs the ability of APP/E4 mice to acquire spatial memory. We also directly compared the performance of all ApoE4 and all ApoE3 mice in the last two trial blocks and found that Abca1 deficiency significantly aggravated memory deficits in APP/E4/Abca1−/+ (Fig. 1C) but not in APP/E3/Abca1−/+ mice (Fig. 1D). The differences in behavior could not be attributed to swim speed or motivation as there was no significant difference between genotypes in performance during the open pool visual platform testing (data not shown).
Figure 1.
Abca1 gene dose affects cognitive decline in APP/E4 but not in APP/E3 mice. A–D, RWM was used to assess memory deficits in APP/E3 and APP/E4, as well as in APP/E3/Abca1−/+ and APP/E4/Abca1−/+ mice, between 6 and 8 months of age (mean age, 7.5). Nontransgenic E3, E4, E3/Abca1−/+, and E4/Abca1−/+ mice were used as controls. A, There was no significant difference in cognitive performance between APP/E3 and APP/E4 mice. Analysis by two-way repeated-measures ANOVA demonstrates a significant effect on genotype (p < 0.0001) but no interaction. There was a significant difference between APP transgenic mice and their respective nontransgenic controls. N = 16–25 mice per group. B, RWM revealed a significant difference in cognitive decline between APP/E3/Abca1−/+ and APP/E4/Abca1−/+ mice. Analysis by two-way repeated-measures ANOVA showed an interaction (p < 0.05) and significant effect on genotype (p < 0.0001). There was a significant difference between APP transgenic mice and their respective nontransgenic controls. *p < 0.05 and **p < 0.01 when APP/E4/Abca1−/+ were compared with APP/E3/Abca1−/+ by t test. N = 10–17 mice per group. C, The graph shows the performance of all ApoE4 mice on the last two trial blocks. N = 10–22 mice per group. D shows the performance of all ApoE3 mice on the last two trial blocks. N = 11–25 mice per group. In C and D, the two groups were compared by t test. E, F, Contextual fear conditioning paradigm was used to assess memory deficits in younger APP/E3, APP/E4, APP/E3/Abca1−/+, and APP/E4/Abca1−/+ mice (mean age, 4.9 months). Nontransgenic E3, E4, E3/Abca1−/+, and E4/Abca1−/+ age- and gender-matched mice were used as wild-type controls. E, Results for APP mice. Analysis by two-way ANOVA revealed an interaction between Abca1 and ApoE genotype (p < 0.05) and a main effect of ApoE (p < 0.01) but not of Abca1 genotype (p = 0.06). p < 0.01, by Bonferroni's posttest; **p < 0.01 by t test. N = 7–13 per group. F, Results from contextual fear conditioning for wild-type mice. Analysis by two-way ANOVA showed no interaction and no main effects of Abca1 and ApoE. No significant difference between APP mice and their non-APP controls except for APP/E4/Abca1−/+, which differed significantly from E4/Abca1−/+ (APP/E4/Abca1−/+ percentage freezing, 24.89 ± 2.56, vs E4/Abca1−/+ percentage freezing, 40.31 ± 3.40; p < 0.01 by t test). N = 15–20 mice per group. G, Cued phase of fear conditioning test was performed on the same APP/E3, APP/E4, APP/E3/Abca1−/+, and APP/E4/Abca1−/+ mice as in E. Analysis by two-way ANOVA showed no interaction and no main effects of Abca1 and ApoE. **p < 0.01 by t test; N = 7–13 per group. H, Cued phase of fear conditioning test was performed on the same wild-type controls as in F. N.S., Not significant. N = 15–20 mice per group. Error bars indicate SEM.
Memory deficits in mice of the same genotypes were also examined at an earlier age (mean age, 4.9 months) using a contextual fear conditioning paradigm (Kornecook et al., 2010). We specifically chose younger mice, which are at the onset of amyloid pathology and only rarely have amyloid plaques. The results for APP-expressing mice are shown in Figure 1E and for wild-type controls in Figure 1F. As visible from Figure 1E, there is no difference between APP/E3, APP/E4, and APP/E3/Abca1−/+ mice. Furthermore, these mice did not differ from their nontransgenic controls (compare the results for APP mice shown in Fig. 1E with the results for wild mice in Fig. 1F). This suggests that amyloid pathology particularly in APP/E3, APP/E4, and APP/E3/Abca1−/+ is not fully advanced to cause memory deficits. However, APP/E4/Abca1−/+ mice differed significantly from APPE4 and APP/E3/Abca1−/+ mice. APP/E4/Abca1−/+ mice also differed significantly from their nontransgenic controls (APP/E4/Abca1−/+ percentage freezing, 24.89 ± 2.56, vs E4/Abca1−/+ percentage freezing, 40.31 ± 3.40; p < 0.01). The results from cued phase of this test are shown in Figure 1, G and H; APP/E4/Abca1−/+ were significantly impaired compared with APP/E4 mice. The conclusion from this study is that Abca1 deficiency accelerates the appearance of memory deficits in APP/E4 mice but not in APP/E3 mice.
Overall, the results from the behavior experiments suggest that, in terms of cognition, APP/E4 mice are more vulnerable to Abca1 deficiency than APP/E3 mice.
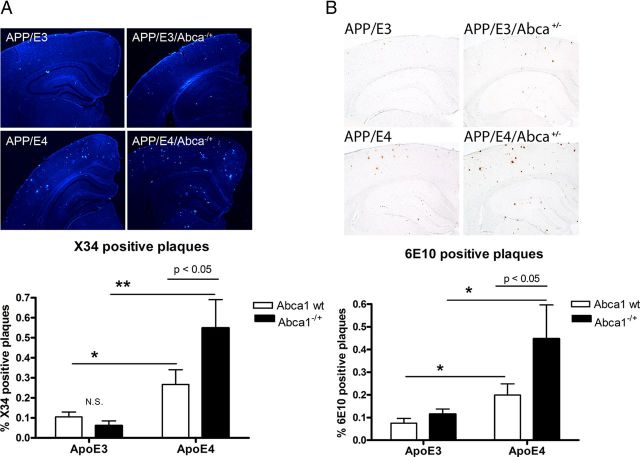
Amyloid load is increased by Abca1 hemizygosity in APP/E4 mice but not in APP/E3 mice
To visualize compact fibrillar amyloid plaques, brain sections were stained with X-34, and the results are presented in Figure 2A. Analysis by two-way ANOVA demonstrated that there is an interaction between Abca1 and ApoE genotypes (p < 0.05). As visible in Figure 2A, there was no significant difference between amyloid plaque levels in APP/E3 and APP/E3/Abca1−/+, while there was more than twofold increase when APP/E4 were compared with APP/E4/Abca1−/+ (posttest, p < 0.05). Remarkably, the compact amyloid plaques in APP/E4/Abca1−/+ compared with APP/E3/Abca1−/+mice were increased ninefold.
Figure 2.
Amyloid plaques are increased by Abca1 hemizygosity in APP/E4 mice but not in APP/E3 mice. Amyloid plaques were compared in APP/E3 and APP/E4 as well as in APP/E3/Abca1−/+ and APP/E4/Abca1−/+ mice between 6 and 8 months of age (mean age, 7.5). A, Brain sections were stained with X-34 to visualize compact fibrillar amyloid plaques. Representative pictures for X-34 staining are shown above the graphs (20× magnification). X-34-positive amyloid load analyzed by two-way ANOVA and Bonferroni's posttest. There is an interaction between Abca1 and ApoE genotypes (p < 0.05) and a significant effect of ApoE genotype (p < 0.001), but no effect of Abca1 genotype. Note that the level of compact amyloid plaques in APP/E4/Abca1−/+ is increased by ninefold compared with the level in APP/E3/Abca1−/+ mice (Bonferroni's posttest, p < 0.05). *p < 0.05 and **p < 0.01, two-group comparisons by t test. N = 8–12 mice per group. B, Brain sections were stained with anti-Aβ antibody, 6E10, to visualize diffuse and compact amyloid plaques. Representative pictures for 6E10 staining are shown above the graph (20× magnification). Analysis is by two-way ANOVA and Bonferroni's posttest. There is no interaction between Abca1 and ApoE genotypes, and there is a significant effect of ApoE (p < 0.01), and Abca1 (p < 0.05) genotypes; p < 0.05, by Bonferroni's posttest. Note that APP/E4 have more than twofold increase of plaque load (*p < 0.05 by t test), and APP/E4/Abca1−/+ mice have more than threefold increase in amyloid load versus APP/E3/Abca1−/+ (*p < 0.05 by t test). N = 8–13 mice per group. Error bars indicate SEM.
To visualize all, including diffuse and compact amyloid plaques, brain sections were stained with 6E10 anti-Aβ antibody (Fig. 2B). Analysis by two-way ANOVA showed that there was no interaction between Abca1 and ApoE genotypes, but there were significant main effects of ApoE (p < 0.01) and Abca1 (p < 0.05) genotypes. Similarly to X-34 results, we found a significant difference between APP/E4 and APP/E4/Abca1−/+ mice (Bonferroni's posttest, p < 0.05), but no significant difference between APP/E3 and APP/E3/Abca1−/+ mice. Furthermore, as visible in Figure 2B, APP/E4/Abca1−/+ mice had threefold more amyloid load over APP/E3/Abca1−/+ mice, which was statistically significant. These results demonstrate that Abca1 deficiency exacerbates amyloid phenotype in mice expressing human ApoE4 mice but not in mice expressing human ApoE3.
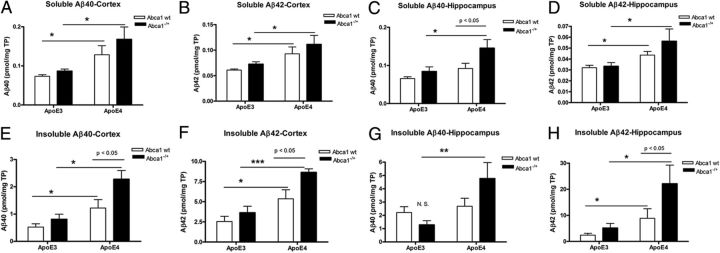
Soluble Aβ level is increased in the brain of APP/E4 and APP/E4/Abca1−/+ mice
Next, we compared the level of soluble Aβ in the brains. Soluble Aβ was extracted separately from cortices and hippocampi, followed by extraction of the insoluble Aβ. Aβ level was measured by ELISA; the results for cortex are shown in Figure 3, A and B, and for hippocampus in Figure 3, C and D. Analysis by two-way ANOVA did not show an interaction between ApoE and Abca1 genotypes, and there was a main effect of ApoE (p < 0.001) but not of Abca1 genotype. We found a significant difference in Aβ40 and Aβ42 levels in the cortex (Fig. 3A,B), and in Aβ42 level in the hippocampus of APP/E4 mice (Fig. 3D) compared with APP/E3 mice confirming previous studies (Bales et al., 2009; Castellano et al., 2011). There was a significant difference between soluble Aβ40 and Aβ42 levels in the cortex and hippocampus of APP/E4/Abca1−/+ when compared directly with APP/E3/Abca1−/+ mice (Fig. 3A–D; p < 0.5 by t test).
Figure 3.
Lack of one Abca1 gene copy increases soluble and insoluble Aβ levels in APP/E4 but not in APP/E3 mice. Soluble proteins were extracted from the cortices (A, B, E, F) and hippocampi (C, D, G, H) of 6- to 8-month-old APP/E3, APP/E4, APP/E3/Abca1−/+, and APP/E4/Abca1−/+ mice. Aβ40 and Aβ42 were measured by ELISA as explained in Materials and Methods. A, Soluble Aβ40 in cortex. N = 6–9 mice per group. B, Soluble Aβ42 level in cortex. N = 9–13 mice per group. For A and B, analysis by two-way ANOVA did not show an interaction between ApoE and Abca1. There was a main effect of ApoE (p < 0.001) but not of Abca1. *p < 0.05 by t test. C, Soluble Aβ40 level in hippocampus. N = 7–13 mice per group. Two-way ANOVA: no interaction between ApoE and Abca1, main effect of ApoE (p < 0.001) and Abca1 genotypes (p < 0.01). p < 0.05 by Bonferroni's posttest; *p < 0.5 by t test. D, Soluble Aβ42 level in hippocampus. N = 7–14 mice per group. Two-way ANOVA: no interaction between ApoE and Abca1, main effect of ApoE (p < 0.001) but not of Abca1. *p < 0.5 by t test. E, Insoluble Aβ40 in cortex. Two-way ANOVA: no interaction between ApoE and Abca1; main effects of Abca1 (p < 0.01) and ApoE (p < 0.0001). p < 0.05 by Bonferroni's posttest. *p < 0.05; **p < 0.01 by t test. N = 6–14 mice per group. F, Insoluble Aβ42 in cortex. Two-way ANOVA: no interaction between ApoE and Abca1; main effects of Abca1 (p < 0.05) and ApoE (p < 0.001). p < 0.05 by Bonferroni's posttest. *p < 0.5, ***p < 0.001 by t test. N = 6–14 mice per group. G, Insoluble Aβ40 in hippocampus. Two-way ANOVA: interaction between ApoE and Abca1 (p < 0.05); main effect of ApoE (p < 0.01) but not of Abca1. **p < 0.01 by t test. N = 7–12 mice per group. H, Insoluble Aβ42 in hippocampus. Two-way ANOVA: no interaction between ApoE and Abca1; main effects of Abca1 (p < 0.05) and ApoE (p < 0.001). p < 0.05 by Bonferroni's posttest. *p < 0.05, by t test. N = 6–14 mice per group. Error bars indicate SEM.
Abca1 deficiency increases insoluble Aβ level in APP/E4 but not in APP/E3 mice
Next, we compared the level of insoluble Aβ in the cortex and hippocampus. As shown in Figure 3, E and F, insoluble Aβ40 and Aβ42 levels in the cortex of APP/E4/Abca1−/+ mice were increased more than twofold compared with APP/E3/Abca1−/+ mice. For Figure 3, E and F, analysis by two-way ANOVA demonstrated no interaction between Abca1 and ApoE, but there were significant main effects of both genotypes. Importantly, there was a significant difference between Aβ40 level in APP/E4 and APP/E4/Abca1−/+ mice (posttest, p < 0.05) but not between APP/E3 and APP/E3/Abca1−/+ mice.
Figure 3G demonstrates that insoluble Aβ40 in the hippocampus of APP/E4/Abca1−/+ mice was increased more than threefold compared with APP/E3/Abca1−/+ mice (p < 0.05), but there was no difference when these mice were compared with APP/E4. Furthermore, analysis for Aβ40 showed that there was an interaction between Abca1 and ApoE genotypes. Finally, as seen in Figure 3H, insoluble Aβ42 in the hippocampus of APP/E4/Abca1−/+ was significantly different from Aβ42 level of APP/E4 (posttest, p < 0.05). Similarly, insoluble Aβ42 in the hippocampus of APP/E4/Abca1−/+ mice was increased more than fourfold compared with APP/E3/Abca1−/+ mice. Two-way ANOVA for Aβ42 demonstrated no interaction between Abca1 and ApoE but significant main effects of both genotypes. The conclusion from these experiments is that the deficiency of Abca1 significantly increases insoluble Aβ level in APP/E4 mice but not in APP/E3 confirming the immunohistochemistry results (Fig. 2).
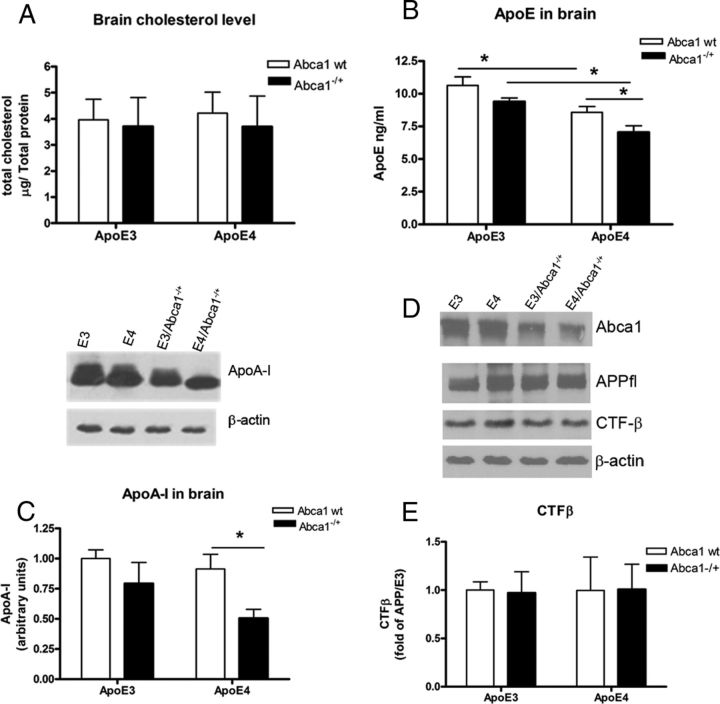
Soluble ApoE and ApoA-I levels are decreased in the brain of APP/E4/Abca1−/+ mice
Next, we examined the levels of total cholesterol and apolipoproteins in the soluble brain fraction. There was no difference in cholesterol concentration between mice of any genotype, confirming previous results that Abca1 deficiency does not affect brain cholesterol level (Wahrle et al., 2004). For ApoE level (Fig. 4B), analysis by two-way ANOVA demonstrated no interaction between ApoE and Abca1 genotypes, but there were significant main effects of Abca1 (p < 0.01) and ApoE (p < 0.001) genotypes. Furthermore, there was a statistical significance when APP/E3 were compared with APP/E4 mice, confirming previous report (Bales et al., 2009), as well as between APP/E3/Abca1−/+ and APP/E4/Abca1−/+ mice. Finally, Abca1 deficiency caused a differential decrease of ApoE level in APP/E4 (compare APP/E4 with APP/E4/Abca1−/+, p < 0.05), but not in APP/E3 mice. As visible from Figure 4C, ApoA-I level was not significantly different between APP/E3 and APP/E4 mice. Analysis by two-way ANOVA demonstrated that there is a main effect of Abca1 (p < 0.05) but not of ApoE genotype, which was anticipated. Furthermore, the result in Figure 4C demonstrates that there is a significant difference between ApoA-I level of APP/E4 and APP/E4/Abca1−/+ mice (p < 0.05). To examine whether there was an effect on APP processing, we measured CTF-β, a result of β-secretase cleavage of APP, and did not find a difference between the genotypes (Fig. 4, see the graph in E and the picture shown in D). This result confirms previous studies that demonstrated neither Abca1 nor ApoE isoform affect APP processing (Koldamova et al., 2005b; Castellano et al., 2011).
Figure 4.
ApoE and ApoA-I levels in soluble brain fraction are decreased in APP/E4/Abca1−/+ but not in APP/E3/Abca1−/+ mice. Soluble brain exacts were used from the cortices and hippocampi of 6- to 8-month-old APP/E3, APP/E4, APP/E3/Abca1−/+, and APP/E4/Abca1−/+ mice. A, Cholesterol concentration was measured in the soluble brain fraction as explained in the text. Two-way ANOVA demonstrated no interaction and no effects of genotypes. N = 6–9 mice per group. B, ApoE was detected in the soluble fraction of cortex and hippocampus by ELISA as explained in Materials and Methods. Analysis is by two-way ANOVA. There is no interaction between ApoE and Abca1; there are main effects of Abca1 (p < 0.01) and ApoE (p < 0.001). *p < 0.5 by t test. N = 6–10 mice per group. C, ApoA-I was detected in the soluble fraction of cortex and hippocampus by WB. The intensity of the bands were quantified and analyzed by two-way ANOVA. There is no interaction between ApoE and Abca1, and there is a main effect of Abca1 (p < 0.05) but not of ApoE. *p < 0.5 by t test. N = 6–8 mice per group. D, Western blotting for Abca1, full-length APP (APPfl), and C-terminal fragments result of β-secretase cleavage (CTFβ). β-Actin is shown as a loading control. E represents the quantification of CTFβ. Note that there is no difference between the genotypes. N = 6–8 mice per group. Error bars indicate SEM.
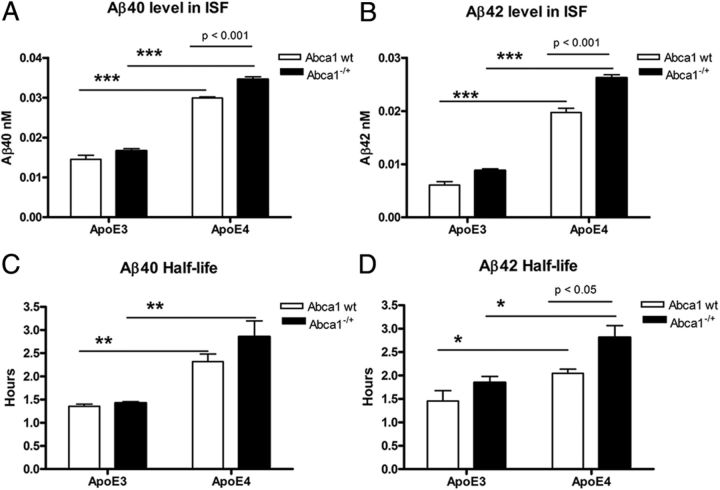
Aβ clearance from CNS is delayed in APP/E4 mice and worsens by Abca1 deficiency
To examine Aβ level in ISF, we performed in vivo microdialysis in freely moving mice (Fig. 5A,B). For Aβ40 and Aβ42, there was no interaction between Abca1 and ApoE, but there were significant main effects of both genotypes. Most importantly, Abca1 deficiency caused statistically significant increase of ISF Aβ level in APP/E4/Abca1−/+ mice compared with APP/E4 (Fig. 5A,B; p < 0.001 by posttest). In contrast, there was no difference in ISF Aβ concentration between APP/E3 and APP/E3/Abca1−/+ mice. As visible from Figure 5, A and B, Aβ40 level in ISF was increased twofold and Aβ42 was increased threefold when ApoE3-expressing mice were compared with their respective ApoE4 counterparts (compare APP/E3/Abca1−/+ with APP/E4/Abca1−/+, and APP/E3 with APP/E4).
Figure 5.
Aβ clearance from the brain is decreased in APP/E4 and APP/E4/Abca1−/+ mice. In vivo microdialysis was performed in the hippocampus of 4.5-month-old awake freely moving mice, and Aβ40 and Aβ42 concentration was determined by ELISA. Analysis is by two-way ANOVA followed by Bonferroni's posttest. A, Aβ40 level in ISF. There is no interaction between ApoE and Abca1. There is a main effect of Abca1 (p < 0.001) and ApoE (p < 0.001). p < 0.001 by Bonferroni's posttest. ***p < 0.001 by t test. N = 4–5 mice per group. B, Aβ42 level in ISF. There is an interaction between ApoE and Abca1 (p < 0.01); main effects of Abca1 (p < 0.001) and ApoE (p < 0.001). p < 0.001 by Bonferroni's posttest. ***p < 0.001 by t test. N = 4–5 mice per group. C, D, Aβ40 and Aβ42 half-life is increased in APP/E4 and APP/E4/Abca1−/+ mice. To determine ISF Aβ half-life, 1 h baseline samples were taken from hours 12–15 after probe implantation, and at the beginning of hour 16, animals were injected with γ-secretase inhibitor LY411575 (10 mg/kg). Aβ half-life was calculated by nonlinear regression analysis (one-phase exponential decay) as described in Materials and Methods. C, Aβ40 half-life is increased in APP/E4 and APP/E4/Abca1−/+ mice. There is no interaction between ApoE and Abca1; main effect of ApoE (p < 0.001) but not of Abca1. **p < 0.01 by t test. N = 4–5 mice per group. D, Aβ42 half-life is increased in APP/E4 and APP/E4/Abca1−/+ mice. There is no interaction between ApoE and Abca1; main effects of ApoE (p < 0.01) and Abca1 (p < 0.01). p < 0.05 by Bonferroni's posttest; *p < 0.05 by t test. N = 4–5 mice per group. Error bars indicate SEM.
To measure the time of Aβ elimination from the brain, we used a γ-secretase inhibitor to block the production of Aβ (Fig. 5C,D) (Cirrito et al., 2003). In general, the results shown in Figure 5, C and D, demonstrate that Aβ40 and Aβ42 half-lives are increased in APP/E4 and APP/E4/Abca1−/+ mice versus their respective ApoE3 isoforms. When we compared APP/E4 with APP/E4/Abca1−/+ mice, Aβ40 clearance showed a trend toward decrease, but the difference was insignificant (Fig. 5C). Two-way ANOVA analysis demonstrated effect only of ApoE but not of Abca1 genotype. In contrast, Aβ42 half-life was significantly affected by ApoE (p < 0.01) and Abca1 (p < 0.01) genotypes. We also found that Aβ42 clearance was significantly decreased in APP/E4/Abca1−/+ mice versus APP/E4 (Fig. 5D; p < 0.05 by Bonferroni). Importantly, the lack of one copy of Abca1 did not affect Aβ40 and Aβ42 half-lives in APP/E3 mice. The conclusion from these studies is that ApoE4 delays the clearance of Aβ from the brain and Abca1 deficiency exacerbates this effect.
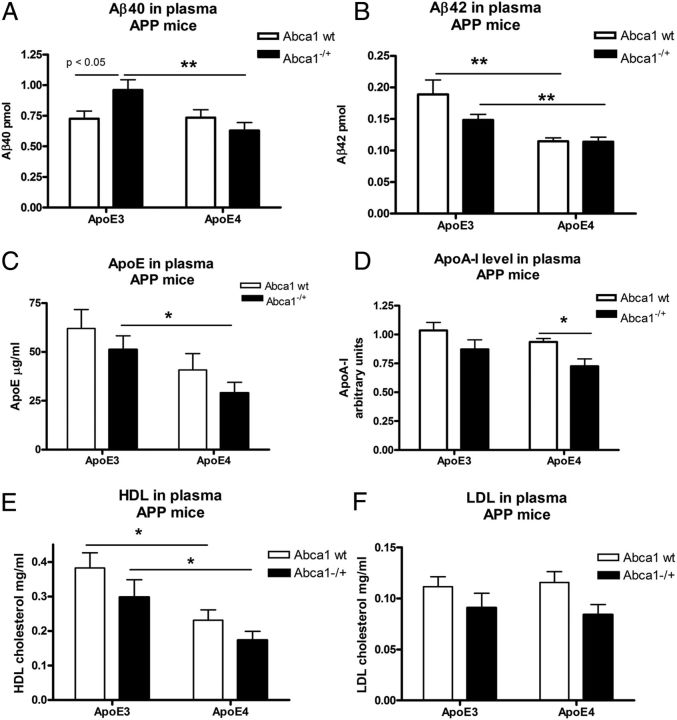
Plasma levels of Aβ42 and HDL are decreased in APP/E4 and APP/E4/Abca1−/+ mice
Next, we compared Aβ levels in the plasma of APP/E3, APP/E4, APP/E3/Abca1−/+, and APP/E4/Abca1−/+ mice. We reasoned that, if human ApoE isoforms differentially affect Aβ clearance from the brain, this will reflect on peripheral Aβ concentration. Figure 6, A and B, shows that APP/E3/Abca1−/+ mice had 50% more Aβ40 in plasma, and 30% more Aβ42 than APP/E4/Abca1−/+ mice. However, we did not find a difference between Aβ40 and Aβ42 level when APP/E4 mice were compared with APP/E4/Abca1−/+. Importantly, the results from two-way ANOVA demonstrated that there is a significant main effect of Abca1 on Aβ42 level in plasma but not on Aβ40.
Figure 6.
Plasma Aβ42, ApoE, and HDL levels are decreased in APP/E4 and APP/E4/Abca1−/+ mice. Amyloid β, ApoE, ApoA-I, HDL, and LDL levels were measured in plasma of APP/E3, APP/E4, APP/E3/Abca1−/+, and APP/E4/Abca1−/+ mice aged between 6 and 8 months. For all panels, analysis is by two-way ANOVA followed by Bonferroni's posttest. Two groups were compared by t test. A shows ELISA results for Aβ40. There is an interaction between ApoE and Abca1 (p < 0.05). There is a significant main effect of ApoE (p < 0.05) but not of Abca1 (p < 0.05 by Bonferroni's posttest). **p < 0.01 by t test. B shows ELISA results for Aβ42. There is no interaction between ApoE and Abca1. There is a main effect of Abca1 (p < 0.01) but not of ApoE. **p < 0.01 by t test. For A and B, N = 8–14 mice per group. C shows ELISA results for ApoE level in plasma. There is no interaction between ApoE and Abca1. There is a main effect of ApoE genotype (p < 0.05) but not of Abca1. *p < 0.05 by t test. N = 7–12 mice per group. D shows ApoA-I level in plasma. ApoA-I level was measured by ELISA and WB as described in Materials and Methods, and the mean values expressed as a fold of ApoA-I level in APP/E3 mice. For each mouse, mean value from both measurements is shown. There is no interaction between ApoE and Abca1. There is a main effect of Abca1 (p < 0.01) but not of ApoE. *p < 0.05 by t test. N = 8–15 mice per group. E, F, HDL and LDL levels in plasma were measured using commercially available kit as described in Materials and Methods. E, For HDL, two-way ANOVA demonstrated no interaction between Abca1 and ApoE. There was a significant main effect of Abca1 (p < 0.01) but not of ApoE. *p < 0.05 by t test. N = 8–14. F, For LDL, two-way ANOVA revealed no interaction between Abca1 and ApoE. There was a significant main effect of ApoE (p < 0.01) but not of Abca1. N = 8–14 mice per group. Error bars indicate SEM.
We also measured ApoE and ApoA-I levels in plasma. For ApoE, two-way ANOVA showed a main effect of ApoE but not of Abca1 genotype (Fig. 6C). Furthermore, APP/E4/Abca1−/+ mice had a statistically significant decrease of ApoE level compared with APP/E3/Abca1−/+ mice. Previously, we and others have shown that ApoA-I and HDLs are important for Aβ aggregation and cognitive decline in experimental mice (Lefterov et al., 2010; Lewis et al., 2010). Interestingly, Abca1 genotype had a significant main effect on ApoA-I levels and its deficiency affected significantly ApoA-I concentration in APP/E4 mice (Fig. 6D, compare APP/E4 with APP/E4/Abca1−/+). There was a trend toward decrease of ApoA-I in APP/E3/Abca1−/+, but the difference was insignificant.
Next, we measured the level of HDL in the plasma of all four genotypes. The results presented in Figure 6E demonstrate a statistically significant difference between ApoE3 and ApoE4 genotypes, which was apparent in mice expressing one or two copies of Abca1 (Fig. 6E, compare APP/E3 with APP/E4 and APP/E3/Abca1−/+ with APP/E4/Abca1−/+). Abca1 deficiency caused a trend toward decreased HDL level in both APP/E3 and APP/E4 mice, but the difference was not statistically significant due to the variability between mice. We measured LDLs, and the results presented in Figure 6F show that there was no statistical difference between genotypes.
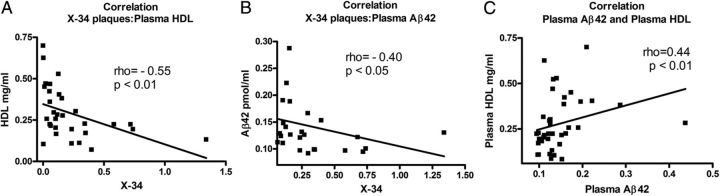
Higher plasma HDL level correlates to a lower amyloid load in the brain
To assess the relationship between plasma HDL and X-34-positive plaques in the brain, we performed a correlation analysis. Figure 7A shows that there is a negative correlation between the two variables (p < 0.01). This result demonstrates that mice with a higher level of HDL in the plasma display less amyloid plaques in their brains. In contrast, we did not find a correlation between ApoE level in brain or plasma and X-34-positive plaques (data not shown). Furthermore, as seen in Figure 7B, there is a negative correlation between Aβ42 level in plasma and X-34-positive plaques. In contrast, we did not find a correlation between Aβ40 level in plasma and X-34 plaques (data not shown). Finally, we examined the relationship between the levels of HDL and Aβ42 in plasma. As visible from Figure 7C, there was a positive correlation between HDL and Aβ42 (p < 0.01). Similar positive correlation was found between ApoE concentration and Aβ42 in plasma (ρ = 0.37, p < 0.05; data not shown). These experiments suggest that ApoE and HDL in periphery may have a role in Aβ clearance from the brain.
Figure 7.
HDL level in plasma negatively correlates to amyloid plaques in the brain. For A–C, Spearman's correlation analysis was performed on groups of APP/E3, APP/E3/Abca1−/+, APP/E4, and APP/E4/Abca1−/+ mice. A, There is a negative correlation between plasma HDL and X-34-positive amyloid plaques in the brain. ρ = −0.55; p < 0.01; N = 31. B, There is a negative correlation between X-34-positive amyloid plaques in brain and Aβ42 level in plasma. ρ = −0.40; p < 0.05; N = 38. C, There is a positive correlation between plasma levels of Aβ42 and HDL. ρ = 0.44; p < 0.01; N = 39.
Discussion
In this study, we examined Abca1 gene dose effect on the phenotype of APP transgenic mice expressing human ApoE3 or ApoE4 isoforms. Surprisingly, our results demonstrate that the lack of one copy of Abca1 significantly aggravates memory deficits and amyloid pathology in APP/E4 but not in APP/E3 mice. An important finding of the study was that Aβ clearance from the brain was decreased by Abca1 deficiency in APP/E4/Abca1−/+ but not in APP/E3/Abca1−/+ mice. In contrast, Abca1 deficiency did not affect cognition, amyloid phenotype, and Aβ clearance in APP mice expressing ApoE3. Interestingly, the correlation between plasma HDL and brain amyloid load (Fig. 7) suggests that there may be a causative connection between peripheral lipoproteins and Aβ load in the CNS.
Previous studies demonstrated that ABCA1 affects amyloid deposition and memory deficits in experimental animals (Hirsch-Reinshagen et al., 2005; Koldamova et al., 2005b; Wahrle et al., 2005; Lefterov et al., 2009). Furthermore, treatment with LXR and RXR ligands, which increases Abca1 expression significantly, ameliorates amyloid pathology and improves cognitive function (Koldamova et al., 2005a; Jiang et al., 2008; Donkin et al., 2010; Fitz et al., 2010; Cramer et al., 2012). Last, transgenic mice overexpressing Abca1 in brain have less amyloid plaques (Wahrle et al., 2008). There are several genome-wide association studies that reported a genetic link between single-nucleotide polymorphisms spanning ABCA1 and late-onset AD (Wollmer et al., 2003; Katzov et al., 2004; Sundar et al., 2007; Reynolds et al., 2009). However, other studies did not replicate the published association (Li et al., 2004; Shibata et al., 2006; Wahrle et al., 2007) and some did observe a possible interaction between ABCA1 and APOE in Hispanics (Shibata et al., 2006). The effect of ABCA1 is attributed to its role in modifying the lipidation and stability of ApoE and ApoA-I, and one consequence of its deficiency is the decreased level of these apolipoproteins in the brain (Hirsch-Reinshagen et al., 2005; Koldamova et al., 2005b; Wahrle et al., 2005; Lefterov et al., 2009). It is speculated that the lack of Abca1 can affect Aβ aggregation or clearance. Previously, we reported that APP23 mice lacking one copy of Abca1 experienced memory deficits in correlation to the increased level of soluble Aβ oligomers (Lefterov et al., 2009). Here, we show that the level of ApoE and ApoA-I in the brain is significantly affected by Abca1 deficiency only in APP/E4 mice but not in APP/E3 mice (Fig. 4B,C), thus supporting this hypothesis. Furthermore, the decreased level or lipidation of ApoE as a result of Abca1 deficiency can affect Aβ clearance, which is apparent in APP/E4 but not APP/E3 mice (Fig. 5).
The exact mechanism by which APOE genotype affects the risk for AD remains unresolved. It has been consistently shown that patients carrying APOE4 allele, compared with those with the other two isoforms, have more amyloid plaques, and this has been replicated in experimental AD mice (Fryer et al., 2005; Bales et al., 2009; Castellano et al., 2011). In agreement with prior studies (Bales et al., 2009; Castellano et al., 2011), our results demonstrate that APP mice expressing ApoE4 isoform have increased amyloid deposition compared with those expressing ApoE3. Furthermore, our microdialysis experiments (Fig. 5) show that, in comparison with ApoE3, expression of ApoE4 delays Aβ clearance, confirming a conclusion from a recent study (Castellano et al., 2011). It is unclear how ApoE affects Aβ clearance from the brain. One possibility is that binding of Aβ to ApoE decreases Aβ aggregation and keeps Aβ in a soluble form, thus facilitating its efflux out of CNS. Another possibility is that ApoE is important for Aβ clearance by microglia or astrocytes inside the brain (Jiang et al., 2008) or mediates Aβ removal from the brain through the blood–brain barrier (BBB). There are numerous reports demonstrating that ApoE binds Aβ (Wisniewski and Frangione, 1992; Strittmatter et al., 1993; LaDu et al., 1994), but it is still unclear how this binding affects Aβ aggregation (for review, see Kim et al., 2009; Holtzman et al., 2012). One frequently discussed possibility for these discrepancies is the use of nonphysiological lipid-free ApoE preparations in most in vitro studies (Kim et al., 2009). Recent in vitro studies have demonstrated that ApoE lipidation is essential for the clearance of the ApoE–Aβ complex by receptor-mediated endocytosis (Jiang et al., 2008; Lee et al., 2012).
However, in vivo data have consistently demonstrated that the absence of ApoE decreases compact amyloid plaques in the brain (Bales et al., 1997; Holtzman et al., 2000; Fryer et al., 2003). Supporting these studies are Aβ clearance experiments, demonstrating that ApoE delays Aβ efflux from the brain (Bell et al., 2007; Deane et al., 2008). Two recent reports demonstrated that amyloid pathology is significantly decreased by eliminating one copy of ApoE in mice expressing human ApoE4 and ApoE3 (Kim et al., 2011; Bien-Ly et al., 2012). Therefore, considering all data generated with APP/ApoE knock-out and hemizygous mice, it seems reasonable to conclude that the presence of ApoE in the brain retains Aβ in CNS and delays its clearance. In this regard, it seems difficult to reconcile the phenotype of APP/Abca1 knock-out mice, which have less ApoE but more amyloid when compared with APP/ApoE knock-out mice.
A very distinctive characteristic of APP/Abca1 knock-out mice not seen in APP/ApoE knock-out mice is decreased total plasma cholesterol, including both HDL and LDL cholesterol. In an effort to explain the differential effect of ABCA1 on the phenotype of ApoE3- and ApoE4-expressing mice, we examined the level of Aβ and apolipoproteins in plasma and analyzed the relationship with brain amyloid. First, we found that the level of Aβ42 in plasma of APP/E4 and APP/E4/Abca1−/+ is lower than in APP/E3 and APP/E4/Abca1 mice (Fig. 6B). Thus, changes of plasma Aβ42 is in opposite direction of the brain Aβ42 level (compare Figs. 6B, 5B). However, since human APP is expressed in the periphery of these mice another possibility should also be considered, namely that there is different secretion of peripheral Aβ42 among the genotypes. Second, we demonstrate that there is a negative correlation between plasma HDL and amyloid plaques. It is possible that HDL and apolipoproteins in plasma have a role in Aβ clearance because they can affect Aβ balance on both sides of BBB. In this regard, our data are in concert with conclusions from other studies that showed proteins in plasma such as soluble LRP (lipoprotein receptor-related protein) can affect Aβ clearance (Sagare et al., 2007; Sehgal et al., 2012).
The question raised by the current study is why Abca1 deficiency affects more significantly APP/E4 than APP/E3 mice? In humans with ABCA1 deficiency, cholesterol efflux is compromised, and as a result, HDL concentration is decreased (Frikke-Schmidt et al., 2004; Koldamova et al., 2010). Data from mice and humans have demonstrated that the concentration of ApoE4 in plasma and brain is lower than the concentrations of ApoE3 (Riddell et al., 2008; Bales et al., 2009; Gupta et al., 2011). A recent cross-sectional study demonstrated that total ApoE and ApoE4 levels were significantly lower in patients with AD, and they further decrease as the Aβ load, assessed by PET, increases (Gupta et al., 2011). Here, we demonstrate that the level of plasma HDL is decreased significantly in APP/E4 compared with APP/E3 mice and in APP/E4/Abca1−/+ compared with APP/E3/Abca1−/+ mice. Moreover, when HDL levels in APP/E4/Abca1−/+ mice were contrasted to those in APP/E3 mice, the difference was more than twofold (Fig. 6E). This result demonstrates that lack of one copy of Abca1 doubles the effect of ApoE4 genotype on HDL concentration. Potentially, in humans, this could render patients expressing APOE4 allele more vulnerable to the consequences of ABCA1 deficiency than ApoE3 carriers.
It has to be recognized that there are differences in lipoprotein metabolism between mice and humans. For example, HDLs are the main carrier of cholesterol in mouse plasma with levels higher than LDL cholesterol (Pendse et al., 2009). In contrast, in humans, LDLs are the predominant cholesterol-transporting particles with much higher levels of cholesterol compared with HDL (Pendse et al., 2009). The consequence is that, in the human population, LDLs or other circulating lipoproteins might be equally important for Aβ clearance as are HDLs. Most studies agree that the total cholesterol (HDL plus non-HDL) is decreased in AD patients (Reitz et al., 2004, 2010; Blasko et al., 2011). A recent study demonstrated that the decreased total plasma cholesterol correlated positively to decreased plasma Aβ in a group of patients who converted to AD (Blasko et al., 2011).
In conclusion, our results clearly demonstrate that Abca1 deficiency affects significantly the phenotype of APP mice expressing human ApoE4 and has little effect on mice expressing ApoE3 isoform. Our data also indicate that APOE4 genotype is more susceptible to gene–gene interactions with negative consequences for the animal. Finally, the results presented here suggest that future therapies targeting ABCA1/ApoE expression such as LXR/RXR agonists could have a favorable effect on the outcome in ApoEε4 carriers.
Footnotes
This work was supported in part by NIH–NIA Grants R01AG027973 and R01AG037481 (R.K.), Institute for the Study of Aging grant (I.L.), and Alzheimer's Art Quilt Initiative Grant (N.F.F.). N.F.F. is supported by NIA F32 fellowship (Grant F32AG034031). We are grateful to Sai Pratyusha Kancherla, Hiral Jayantibhai Patel, and Xiaozhuo Lu for their excellent technical assistance. The content of this report is solely responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging, National Institutes of Health, or the private foundations.
References
- Alamed J, Wilcock DM, Diamond DM, Gordon MN, Morgan D. Two-day radial-arm water maze learning and memory task; robust resolution of amyloid-related memory deficits in transgenic mice. Nat Protoc. 2006;1:1671–1679. doi: 10.1038/nprot.2006.275. [DOI] [PubMed] [Google Scholar]
- Bales KR, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, Hyslop P, Johnstone EM, Little SP, Cummins DJ, Piccardo P, Ghetti B, Paul SM. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat Genet. 1997;17:263–264. doi: 10.1038/ng1197-263. [DOI] [PubMed] [Google Scholar]
- Bales KR, Liu F, Wu S, Lin S, Koger D, DeLong C, Hansen JC, Sullivan PM, Paul SM. Human APOE isoform-dependent effects on brain β-amyloid levels in PDAPP transgenic mice. J Neurosci. 2009;29:6771–6779. doi: 10.1523/JNEUROSCI.0887-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RD, Sagare AP, Friedman AE, Bedi GS, Holtzman DM, Deane R, Zlokovic BV. Transport pathways for clearance of human Alzheimer's amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J Cereb Blood Flow Metab. 2007;27:909–918. doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien-Ly N, Gillespie AK, Walker D, Yoon SY, Huang Y. Reducing human apolipoprotein E levels attenuates age-dependent Ab accumulation in mutant human amyloid precursor protein transgenic mice. J Neurosci. 2012;32:4803–4811. doi: 10.1523/JNEUROSCI.0033-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasko I, Kemmler G, Jungwirth S, Wichart I, Weissgram S, Jellinger K, Tragl KH, Fischer P. Prospective study on association between plasma amyloid β-42 and atherosclerotic risk factors. J Neural Transm. 2011;118:663–672. doi: 10.1007/s00702-011-0599-4. [DOI] [PubMed] [Google Scholar]
- Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, Loubser O, Ouelette BF, Fichter K, Ashbourne-Excoffon KJ, Sensen CW, Scherer S, Mott S, Denis M, Martindale D, Frohlich J, et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, Goate AM, Bales KR, Paul SM, Bateman RJ, Holtzman DM. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci Transl Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirrito JR, May PC, O'Dell MA, Taylor JW, Parsadanian M, Cramer JW, Audia JE, Nissen JS, Bales KR, Paul SM, DeMattos RB, Holtzman DM. In vivo assessment of brain interstitial fluid with microdialysis reveals plaque-associated changes in amyloid-β metabolism and half-life. J Neurosci. 2003;23:8844–8853. doi: 10.1523/JNEUROSCI.23-26-08844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Cramer PE, Cirrito JR, Wesson DW, Lee CY, Karlo JC, Zinn AE, Casali BT, Restivo JL, Goebel WD, James MJ, Brunden KR, Wilson DA, Landreth GE. ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models. Science. 2012;335:1503–1506. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane R, Sagare A, Hamm K, Parisi M, Lane S, Finn MB, Holtzman DM, Zlokovic BV. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest. 2008;118:4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkin JJ, Stukas S, Hirsch-Reinshagen V, Namjoshi D, Wilkinson A, May S, Chan J, Fan J, Collins J, Wellington CL. ATP-binding cassette transporter A1 mediates the beneficial effects of the liver X receptor agonist GW3965 on object recognition memory and amyloid burden in amyloid precursor protein/presenilin 1 mice. J Biol Chem. 2010;285:34144–34154. doi: 10.1074/jbc.M110.108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz NF, Cronican A, Pham T, Fogg A, Fauq AH, Chapman R, Lefterov I, Koldamova R. Liver X receptor agonist treatment ameliorates amyloid pathology and memory deficits caused by high-fat diet in APP23 mice. J Neurosci. 2010;30:6862–6872. doi: 10.1523/JNEUROSCI.1051-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frikke-Schmidt R, Nordestgaard BG, Jensen GB, Tybjaerg-Hansen A. Genetic variation in ABC transporter A1 contributes to HDL cholesterol in the general population. J Clin Invest. 2004;114:1343–1353. doi: 10.1172/JCI20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JD, Taylor JW, DeMattos RB, Bales KR, Paul SM, Parsadanian M, Holtzman DM. Apolipoprotein E markedly facilitates age-dependent cerebral amyloid angiopathy and spontaneous hemorrhage in amyloid precursor protein transgenic mice. J Neurosci. 2003;23:7889–7896. doi: 10.1523/JNEUROSCI.23-21-07889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JD, Simmons K, Parsadanian M, Bales KR, Paul SM, Sullivan PM, Holtzman DM. Human apolipoprotein E4 alters the amyloid-β 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J Neurosci. 2005;25:2803–2810. doi: 10.1523/JNEUROSCI.5170-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AIBL Research Group. Gupta VB, Laws SM, Villemagne VL, Ames D, Bush AI, Ellis KA, Lui JK, Masters C, Rowe CC, Szoeke C, Taddei K, Martins RN. Plasma apolipoprotein E and Alzheimer disease risk: the AIBL study of aging. Neurology. 2011;76:1091–1098. doi: 10.1212/WNL.0b013e318211c352. [DOI] [PubMed] [Google Scholar]
- Hirsch-Reinshagen V, Maia LF, Burgess BL, Blain JF, Naus KE, McIsaac SA, Parkinson PF, Chan JY, Tansley GH, Hayden MR, Poirier J, Van Nostrand W, Wellington CL. The absence of ABCA1 decreases soluble ApoE levels but does not diminish amyloid deposition in two murine models of Alzheimer disease. J Biol Chem. 2005;280:43243–43256. doi: 10.1074/jbc.M508781200. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Fagan AM, Mackey B, Tenkova T, Sartorius L, Paul SM, Bales K, Ashe KH, Irizarry MC, Hyman BT. Apolipoprotein E facilitates neuritic and cerebrovascular plaque formation in an Alzheimer's disease model. Ann Neurol. 2000;47:739–747. [PubMed] [Google Scholar]
- Holtzman DM, Herz J, Bu G. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006312. doi: 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Lee CY, Mandrekar S, Wilkinson B, Cramer P, Zelcer N, Mann K, Lamb B, Willson TM, Collins JL, Richardson JC, Smith JD, Comery TA, Riddell D, Holtzman DM, Tontonoz P, Landreth GE. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58:681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzov H, Chalmers K, Palmgren J, Andreasen N, Johansson B, Cairns NJ, Gatz M, Wilcock GK, Love S, Pedersen NL, Brookes AJ, Blennow K, Kehoe PG, Prince JA. Genetic variants of ABCA1 modify Alzheimer disease risk and quantitative traits related to beta-amyloid metabolism. Hum Mutat. 2004;23:358–367. doi: 10.1002/humu.20012. [DOI] [PubMed] [Google Scholar]
- Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer's disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jiang H, Park S, Eltorai AE, Stewart FR, Yoon H, Basak JM, Finn MB, Holtzman DM. Haploinsufficiency of human APOE reduces amyloid deposition in a mouse model of amyloid-β amyloidosis. J Neurosci. 2011;31:18007–18012. doi: 10.1523/JNEUROSCI.3773-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koldamova RP, Lefterov IM, Lefterova MI, Lazo JS. Apolipoprotein A-I directly interacts with amyloid precursor protein and inhibits A beta aggregation and toxicity. Biochemistry. 2001;40:3553–3560. doi: 10.1021/bi002186k. [DOI] [PubMed] [Google Scholar]
- Koldamova RP, Lefterov IM, Staufenbiel M, Wolfe D, Huang S, Glorioso JC, Walter M, Roth MG, Lazo JS. The liver X receptor ligand T0901317 decreases amyloid β production in vitro and in a mouse model of Alzheimer's disease. J Biol Chem. 2005a;280:4079–4088. doi: 10.1074/jbc.M411420200. [DOI] [PubMed] [Google Scholar]
- Koldamova R, Staufenbiel M, Lefterov I. Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. J Biol Chem. 2005b;280:43224–43235. doi: 10.1074/jbc.M504513200. [DOI] [PubMed] [Google Scholar]
- Koldamova R, Fitz NF, Lefterov I. The role of ATP-binding cassette transporter A1 in Alzheimer's disease and neurodegeneration. Biochim Biophys Acta. 2010;1801:824–830. doi: 10.1016/j.bbalip.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornecook TJ, McKinney AP, Ferguson MT, Dodart JC. Isoform-specific effects of apolipoprotein E on cognitive performance in targeted-replacement mice overexpressing human APP. Genes Brain Behav. 2010;9:182–192. doi: 10.1111/j.1601-183X.2009.00545.x. [DOI] [PubMed] [Google Scholar]
- LaDu MJ, Falduto MT, Manelli AM, Reardon CA, Getz GS, Frail DE. Isoform-specific binding of apolipoprotein E to beta-amyloid. J Biol Chem. 1994;269:23403–23406. [PubMed] [Google Scholar]
- Lee CY, Tse W, Smith JD, Landreth GE. Apolipoprotein E promotes beta-amyloid trafficking and degradation by modulating microglial cholesterol levels. J Biol Chem. 2012;287:2032–2044. doi: 10.1074/jbc.M111.295451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterov I, Fitz NF, Cronican A, Lefterov P, Staufenbiel M, Koldamova R. Memory deficits in APP23/Abca1+/− mice correlate with the level of Abeta oligomers. ASN Neuro. 2009;1:pii:e00006. doi: 10.1042/AN20090015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterov I, Fitz NF, Cronican AA, Fogg A, Lefterov P, Kodali R, Wetzel R, Koldamova R. Apolipoprotein A-I deficiency increases cerebral amyloid angiopathy and cognitive deficits in APP/PS1DeltaE9 mice. J Biol Chem. 2010;285:36945–36957. doi: 10.1074/jbc.M110.127738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis TL, Cao D, Lu H, Mans RA, Su YR, Jungbauer L, Linton MF, Fazio S, LaDu MJ, Li L. Overexpression of human apolipoprotein A-I preserves cognitive function and attenuates neuroinflammation and cerebral amyloid angiopathy in a mouse model of Alzheimer disease. J Biol Chem. 2010;285:36958–36968. doi: 10.1074/jbc.M110.127829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tacey K, Doil L, van Luchene R, Garcia V, Rowland C, Schrodi S, Leong D, Lau K, Catanese J, Sninsky J, Nowotny P, Holmans P, Hardy J, Powell J, Lovestone S, Thal L, Owen M, Williams J, Goate A, et al. Association of ABCA1 with late-onset Alzheimer's disease is not observed in a case-control study. Neurosci Lett. 2004;366:268–271. doi: 10.1016/j.neulet.2004.05.047. [DOI] [PubMed] [Google Scholar]
- Pendse AA, Arbones-Mainar JM, Johnson LA, Altenburg MK, Maeda N. Apolipoprotein E knock-out and knock-in mice: atherosclerosis, metabolic syndrome, and beyond. J Lipid Res. 2009;(50 Suppl):S178–S182. doi: 10.1194/jlr.R800070-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzzo D, Privitera L, Leznik E, Fà M, Staniszewski A, Palmeri A, Arancio O. Picomolar amyloid-β positively modulates synaptic plasticity and memory in hippocampus. J Neurosci. 2008;28:14537–14545. doi: 10.1523/JNEUROSCI.2692-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C, Tang MX, Luchsinger J, Mayeux R. Relation of plasma lipids to Alzheimer disease and vascular dementia. Arch Neurol. 2004;61:705–714. doi: 10.1001/archneur.61.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C, Tang MX, Schupf N, Manly JJ, Mayeux R, Luchsinger JA. Association of higher levels of high-density lipoprotein cholesterol in elderly individuals and lower risk of late-onset Alzheimer disease. Arch Neurol. 2010;67:1491–1497. doi: 10.1001/archneurol.2010.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CA, Hong MG, Eriksson UK, Blennow K, Bennet AM, Johansson B, Malmberg B, Berg S, Wiklund F, Gatz M, Pedersen NL, Prince JA. A survey of ABCA1 sequence variation confirms association with dementia. Hum Mutat. 2009;30:1348–1354. doi: 10.1002/humu.21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell DR, Zhou H, Atchison K, Warwick HK, Atkinson PJ, Jefferson J, Xu L, Aschmies S, Kirksey Y, Hu Y, Wagner E, Parratt A, Xu J, Li Z, Zaleska MM, Jacobsen JS, Pangalos MN, Reinhart PH. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci. 2008;28:11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagare A, Deane R, Bell RD, Johnson B, Hamm K, Pendu R, Marky A, Lenting PJ, Wu Z, Zarcone T, Goate A, Mayo K, Perlmutter D, Coma M, Zhong Z, Zlokovic BV. Clearance of amyloid-beta by circulating lipoprotein receptors. Nat Med. 2007;13:1029–1031. doi: 10.1038/nm1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Sehgal N, Gupta A, Valli RK, Joshi SD, Mills JT, Hamel E, Khanna P, Jain SC, Thakur SS, Ravindranath V. Withania somnifera reverses Alzheimer's disease pathology by enhancing low-density lipoprotein receptor-related protein in liver. Proc Natl Acad Sci U S A. 2012;109:3510–3515. doi: 10.1073/pnas.1112209109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Kawarai T, Lee JH, Lee HS, Shibata E, Sato C, Liang Y, Duara R, Mayeux RP, St George-Hyslop PH, Rogaeva E. Association studies of cholesterol metabolism genes (CH25H, ABCA1 and CH24H) in Alzheimer's disease. Neurosci Lett. 2006;391:142–146. doi: 10.1016/j.neulet.2005.08.048. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PM, Mezdour H, Aratani Y, Knouff C, Najib J, Reddick RL, Quarfordt SH, Maeda N. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J Biol Chem. 1997;272:17972–17980. doi: 10.1074/jbc.272.29.17972. [DOI] [PubMed] [Google Scholar]
- Sundar PD, Feingold E, Minster RL, DeKosky ST, Kamboh MI. Gender-specific association of ATP-binding cassette transporter 1 (ABCA1) polymorphisms with the risk of late-onset Alzheimer's disease. Neurobiol Aging. 2007;28:856–862. doi: 10.1016/j.neurobiolaging.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J Biol Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, Holtzman DM. Deletion of Abca1 increases Aβ deposition in the PDAPP transgenic mouse model of Alzheimer disease. J Biol Chem. 2005;280:43236–43242. doi: 10.1074/jbc.M508780200. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Shah AR, Fagan AM, Smemo S, Kauwe JS, Grupe A, Hinrichs A, Mayo K, Jiang H, Thal LJ, Goate AM, Holtzman DM. Apolipoprotein E levels in cerebrospinal fluid and the effects of ABCA1 polymorphisms. Mol Neurodegener. 2007;2:7. doi: 10.1186/1750-1326-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, Jain S, Hirsch-Reinshagen V, Wellington CL, Bales KR, Paul SM, Holtzman DM. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J Clin Invest. 2008;118:671–682. doi: 10.1172/JCI33622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski T, Frangione B. Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci Lett. 1992;135:235–238. doi: 10.1016/0304-3940(92)90444-c. [DOI] [PubMed] [Google Scholar]
- Wollmer MA, Streffer JR, Lütjohann D, Tsolaki M, Iakovidou V, Hegi T, Pasch T, Jung HH, Bergmann K, Nitsch RM, Hock C, Papassotiropoulos A. ABCA1 modulates CSF cholesterol levels and influences the age at onset of Alzheimer's disease. Neurobiol Aging. 2003;24:421–426. doi: 10.1016/s0197-4580(02)00094-5. [DOI] [PubMed] [Google Scholar]