SUMMARY
Recent genome-wide association studies have linked common variants in the human genome to Parkinson’s disease (PD) risk. Here we show that the consequences of variants at 2 such loci, PARK16 and LRRK2, are highly interrelated, both in terms of their broad impacts on human brain transcriptomes of unaffected carriers, and in terms of their associations with PD risk. Deficiency of the PARK16 locus gene RAB7L1 in primary rodent neurons, or of a RAB7L1 orthologue in Drosophila dopamine neurons, recapitulated degeneration observed with expression of a familial PD mutant form of LRRK2, whereas RAB7L1 overexpression rescued the LRRK2 mutant phenotypes. PD-associated defects in RAB7L1 or LRRK2 led to endolysosomal and Golgi apparatus sorting defects and deficiency of the VPS35 component of the retromer complex. Expression of wild-type VPS35, but not a familial PD-associated mutant form, rescued these defects. Taken together, these studies implicate retromer and lysosomal pathway alterations in PD risk.
INTRODUCTION
Parkinson’s disease (PD) is a common neurodegenerative disorder of aging, characterized by slowed movements and a distinctive tremor at rest (Lang and Lozano, 1998). Defining pathological features of the disease include neurodegeneration that is most prominent among midbrain dopamine neurons (DNs) in the Substantia Nigra (SN) and Lewy body protein aggregates that are composed in part of alpha-Synuclein (aSyn) protein. As the course of PD is thought to last decades, and as at the time of autopsy the vast majority of DNs are long lost, the molecular pursuit of initial etiological events has proven difficult.
In rare inherited familial forms of PD, specific causative mutations have been identified, and this has significantly advanced the field (Abeliovich and Beal, 2006; Hardy et al., 2006). For instance, autosomal dominantly inherited mutations in aSyn, including missense mutations and triplication of the locus, lead to a familial PD variant, implicating aSyn directly in the disease process. Another familial genetic cause of PD is the presence of autosomal dominantly inherited mutations in the Leucine rich-repeat kinase-2 (LRRK2) protein, which encodes a large multidomain protein with GTPase and kinase activities. Although the precise functions of aSyn and LRRK2 in neurons remain to be determined, both proteins have been broadly implicated in intraneuronal protein sorting. aSyn mutations have been reported to modify synaptic vesicle kinetics (Abeliovich et al., 2000) as well as trafficking to the Golgi apparatus in a variety of model systems (Cooper et al., 2006; Thayanidhi et al., 2010), whereas LRRK2 mutations are implicated in defective lysosomal protein degradation and macroautophagy, which is a cellular process that delivers cytosolic proteins and protein agregates to the lysosome (Dodson et al., 2012; Heo et al., 2010; MacLeod et al., 2006), and Golgi Apparatus integrity (Stafa et al., 2012). The recent identification of rare autosomal dominant familial PD mutations in VPS35 (Vilarino-Guell et al., 2011; Zimprich et al., 2011), which encodes a component of the retromer complex that guides protein sorting from the endosome-lysosome degradation pathway retrogradely to the Golgi Apparatus (Bonifacino and Hurley, 2008; Seaman, 2009; Seaman et al., 1998), suggests that defective protein sorting in vesicular compartments may play a role in PD.
Several genome-wide association studies (GWAS) have described common genetic variants (at single nucleotide polymorphisms; SNPs) that modify PD risk in nonfamilial ‘sporadic’ cases (Hamza et al., 2010; Simon-Sanchez et al., 2009; Lill et al., 2012). Strikingly, a subset of these common variants lie within genomic loci previously associated with familial disease, such as aSyn or LRRK2, supporting the notion that common pathogenic pathways underlie familial and sporadic forms of PD. However, mechanisms that underlie the impact of non-familial genetic loci on PD risk, or that relate the functions of such loci to familial PD genes, remain unclear.
Here we describe a series of human brain transcriptome, human genetic, and cell biological studies, that together implicate a PD-associated genetic and cellular pathway. RAB7L1 -- one of 5 genes within the PARK16 non-familial PD risk-associated locus -- functions together with LRRK2 to impact non-familial PD risk in the human population. this genetic interaction is apparent even in unaffected individuals who carry both risk alleles, as quantified in terms of a broad transcriptomic analysis of brain gene expression. Similarly, these genes together modify neuronal survival and neurite integrity in model systems. At a cellular level, defects in this PD-associated RAB7L1-LRRK2 pathway lead to abnormal lysosomal structures and defective retromer complex function, that normally links the endolysosomal protein degradation system with the Golgi apparatus (Bonifacino and Hurley, 2008; Seaman, 2009; Seaman et al., 1998). Consistent with a role for such cellular defects in disease pathology, mutations in a retromer complex component, VPS35, have recently been associated with rare forms of autosomal dominantly inherited familial PD (Vilarino-Guell et al., 2011; Zimprich et al., 2011).
RESULTS
LRRK2 and PARK16 PD risk variants impart a common brain transcriptome impact
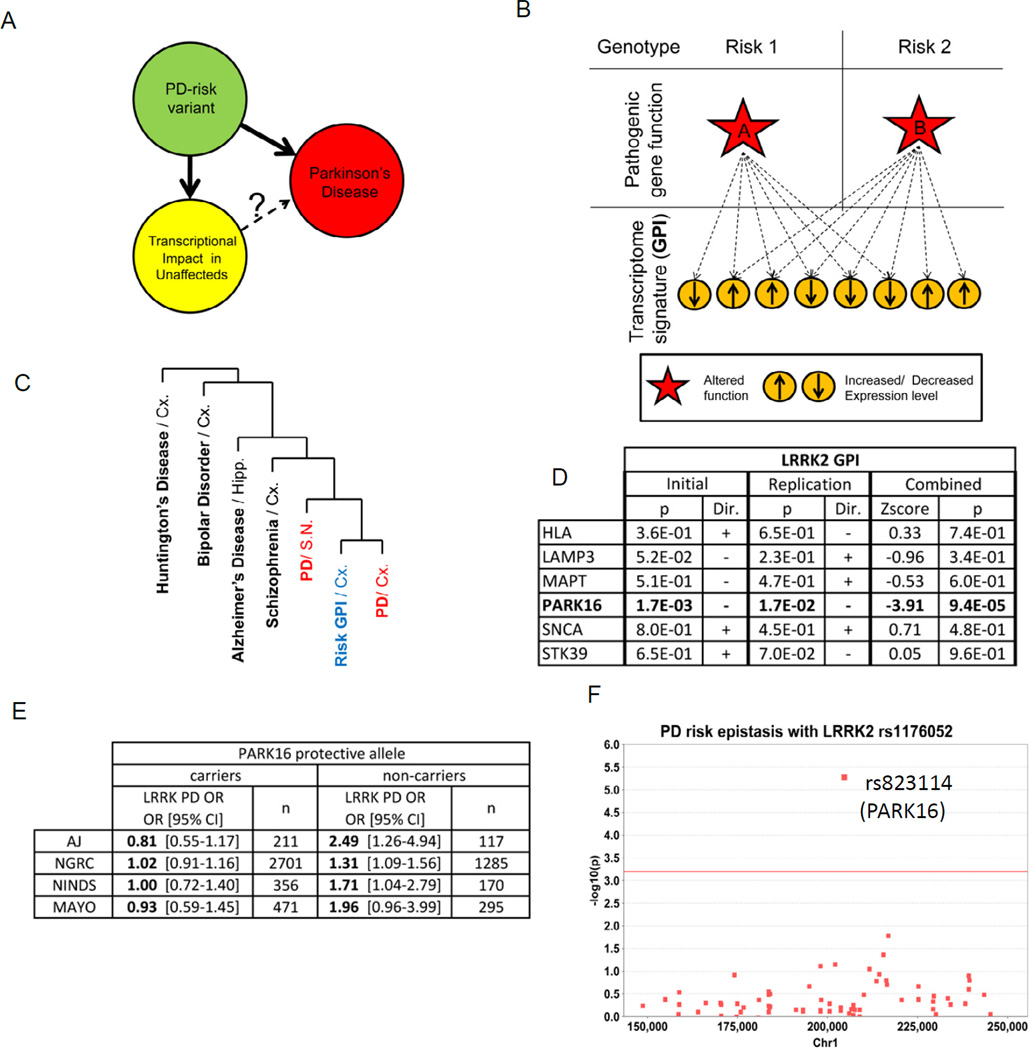
We sought an unbiased and systematic approach to assess the phenotypic impacts of common genetic variants associated with PD risk, particularly in brain tissue from yet unaffected carriers (Figure 1A), in order to circumvent the limitations of the analysis of diseased patient autopsy tissue. To this end, we compared the transcriptome-wide gene expression profiles of brain tissue samples from cohorts of unaffected individuals who share either a risk or a protective allele at any given PD risk SNP (Figure 1B). Such a Global Phenotypic Impact (GPI) quantifies the effect of disease risk variants onto the transcriptome-wide gene expression profile in brain. A key aspect of the GPI analysis herein is that we focus on tissue from unaffected individuals, in hope of avoiding secondary effects of disease pathology such as cell loss.
Figure 1. LRRK2 and PARK16 PD risk-associated variants function in a common genetic pathway.
(A)PD risk-associated variants exert functional effects in the CNS of unaffected individuals that is assessed in terms of a global transcriptome impact. Similar to the one observed in PD affected brain, it may reflect a pre-disease prodromal state. (B) Schematic of GPI analysis. PD risk-associated genotypes at 2 independent loci (upper panels) are hypothesized to differentially alter the function of a nearby gene (red star in middle panel). This secondarily impacts the brain transcriptome (lower panels), with significant overlap for different PD-risk genotype shows. (C)Hierarchical clustering dendrogram shows that the gene expression signatures across 7 PD-associated variant GPIs (“Risk GPI”; in unaffected cerebral cortex Broadmann Area 9 [BA9]) are most similar to the signatures seen in PD brain (BA9 or substantia nigra; SN; in red) rather than in other CNS diseases such as Alzheimer’s disease, Huntington’s disease, Bipolar Disorder or Schizophrenia. 352 gene transcript expression patterns - corresponding to the intersection of the PD risk variants GPIs (Figure S1A–C) - were interrogated. Clustering was performed using Pearson’s distance with complete linkage (see Methods). (D) Genetic interaction between PARK16 and LRRK2 alleles revealed by GPI analysis in 185 unaffected brain samples (GEO GSE15222 “Initial”) and in an independent cohort of 143 unaffected brain samples (GEO GSE15745, “Replication”), as established by the interaction factor between pairs of GPIs as indicated, in a linear regression model (see Methods). The p-value (“p”) associated with the interaction term as well as its orientation (“Dir.”) are presented. Results combined across both cohorts presented (“Combined”) with the resulting Z-scores and p-values for interaction. (E) The PARK16 genotype modifies LRRK2 associated risk in sporadic PD. A table presents the odds ratios for PD at the LRRK2 locus as a function of the PARK16 genotype in 4 independent GWAS cohorts: 1 of Ashkenazi Jews (“AJ”, n= 417) and 3 of Caucasians (“NGRC”, n= 4008; “NINDS”, n=537; “MAYO”, n=886). (F) Manhattan plot of the Chr1 region reported as a modifier of age of disease onset in familial PD with LRRK2 mutation (Latourelle et al., 2011). Epistasis was evaluated for 74 SNPs in 4 independent sporadic PD GWAS datasets. X-axis represents chromosomal location, Y-axis represents -log10 of the combined p-value for epistasis of each SNP with the PD risk SNP rs11176052 at the LRRK2 locus. The PARK16 locus PD-associated SNP rs823114 (arrow) exhibited the most significant association (p=4.6 E-6; red line represents the significance threshold after correction for multiple testing).
We assessed the transcriptome-wide GPI at 7 PD-associated loci (SNCA, LRRK2, MAPT, HLA-DRA, PARK16, LAMP3, STK39, Table S1) (Simon-Sanchez et al., 2009) in a publically available gene expression dataset from cerebral cortex autopsy brain tissue of 185 individuals not apparently affected by a neurodegenerative disease (GSE15222). The GPIs of the 7 loci revealed a high degree of overlap in terms of the identity of transcripts altered in expression level and the valence of such alterations: genes were coordinately altered in their expression by each of the 7 PD-associated loci (over 15-fold greater than expected by chance; p=1.5E-5 by resampling statistics; Figure S1A–B, Table S2). This observation of an overlapping GPI for these 7 PD-associated loci was moreover confirmed in an additional independent dataset of cerebral frontal cortex autopsy brain tissue of 143 individuals (p=1.6 E-3 by resampling statistics; derived from GEO GSE15745).
Function annotation was performed on the gene expression changes that underlie the common GPIs among PD risk variants. Strikingly, among the annotated gene sets most significantly reduced in expression are “mitochondria” functions (Figure S1C–D), consistent with the well-described association of defects in mitochondria with PD pathology (Zheng et al., 2010). Furthermore, the common overlapping transcriptomic signature of gene expression changes associated with these 7 PD risk variants revealed a pattern most similar to the transcriptome changes observed in the context of PD patient brain tissue (relative to unaffected brain tissue; Figure S1C), rather than to other CNS disorders such as Alzheimer’s disease or schizophrenia.
LRRK2 and PARK16 variants cooperatively determine PD risk
Among the 7 analyzed PD risk locus GPIs, those at the PARK16 and LRRK2 loci were found to be the most similar. Furthermore, variants at these two loci impacted the transcriptome in a non-additive manner, signifying a genetic interaction (as determined by analysis of carriers of both risk variants; Figure 1D). We thus investigated whether these loci similarly genetically interact in terms of their impact on PD risk: namely, whether harboring either a risk (or protective) allele at one of these loci would modify the association of the second locus with PD risk. In an initial study on an Ashkenazi Jewish (AJ) population, the effect of a risk-associated variant at the LRRK2 locus was in fact highly dependent on the presence of the risk variant at the PARK16 locus, and vice versa (Figure 1E). Such ‘epistasis’ between the LRRK2 and PARK16 loci regarding PD risk was replicated by reanalysis of 3 other independent GWAS, strongly supporting a common mechanism of action (Figure 1E). Although to our knowledge, prior studies have not reported genetic interactions with the common sporadic PD risk-associated variants at the LRRK2 locus, a GWAS of patients who harbor rare familial LRRK2 mutations identified a broad 15 Mb region of Chromosome 1 as harboring a genetic modifier of age of PD onset (Latourelle et al., 2011). We note that this region encompassed the PARK16 locus. Meta-analysis in 4 independent sporadic PD GWAS datasets (as above) of the 74 identified SNP variants within this Chromosome 1 region for epistasis with the common LRRK2 SNP variant regarding PD risk identified the PARK16-associated variant as by far the most significantly interacting variant (combined p-value for epistasis: 4.6E-6 ; Figure 1F, Table S3). Taken together, these data strongly support a genetic interaction between LRRK2 and PARK16 that initially impacts human CNS tissue physiology, as reflected by the transcriptome signature in unaffected carriers, and ultimately favors PD pathology in a small subset of individuals at risk.
Evidence of a LRRK2-RAB7L1 pathway
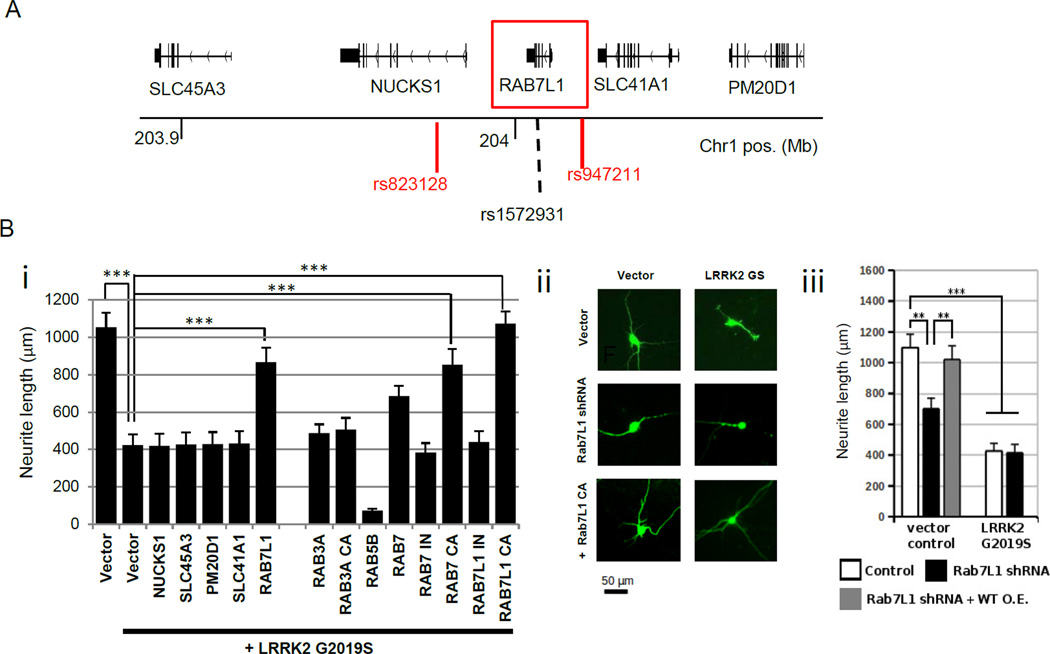
As 5 candidate genes are present within the PARK16 locus (SLC45A3, NUCKS, RAB7L1, SLC41A1, and PM20D1), we sought to experimentally screen each of these for a functional interaction with LRRK2 (Figure 2A). We took advantage of a previouslydescribed primary rat neuron in vitro culture model, in which transient expression of familial PD-associated LRRK2 G2019S or R1441C mutant alleles leads to a marked reduction in neurite process length (MacLeod et al., 2006). Overexpression of RAB7L1, but not other genes in the PARK16 locus, significantly suppressed the LRRK2 mutation-induced neurite length phenotype (Figure 2B). RAB7L1 did not modify neurite length in the context of over-expression of wild-type LRRK2 (Figure 2A). RAB7L1 is a small cytosolic GTPase, structurally distinct from RAB7 despite its name (also known as RAB29) (Shimizu et al., 1997). One of ~60 small GTPases in the human genome, RAB7L1 has previously been shown to localize primarily to the Golgi apparatus and implicated in vesicular sorting in the context of Salmonella or Hepatitis C infection (Berger et al., 2009; Spano et al., 2011). But the function of RAB7L1 in CNS neurons remains unknown. Orthologues of RAB7L1 in other organisms, including C. elegans Glo-1 and Drosophila melanogaster Lightoid, have been implicated in trafficking to lysosome-related organelles (Hermann et al., 2005) and in the regulation of neurite process length (Grill et al., 2007), reminiscent of LRRK2 mutant phenotypes (MacLeod et al., 2006). Thus this gene was of particular interest.
Figure 2. Overexpression of the PARK16 locus gene RAB7L1 specifically rescues a LRRK2 mutant phenotype.
(A)Schematics of the PARK16 locus on chromosome 1. (B)RAB7L1 modifies a LRRK2-associated neurite process length phenotype. Rat primary cortical neuron cultures transfected with a vector expressing G2019S mutant LRRK2 displayed reduced total neurite length relative to vector alone (cells are co-transfected with GFP for visualization by fluorescence microscopy). (i) (ii) Co-transfection of a wild-type or constitutively active (CA) RAB7L1 expression vector (1µg/well) along with LRRK2 G2019S (0.5µg/well) significantly rescued neurite length; other PARK16 genes—NUCKS1, SLC45A3, PM20D1, and SLC41A1 -- failed to rescue. CA or inactive (IN) RAB vectors were also tested as indicated (left panel; GFP-tagged at the N-terminus; 1µg/well). (iii) Knock-down of RAB7L1 by shRNA vector transfection led to a similar decrease in neurite length as with LRRK2 G2019S expression. n=20 neurons in 4 independent cultures per group. Mean total neurite lengths are displayed; error bars represent SEM, *: p<0.05, **: p<0.01, ***: p<0.001 for ANOVA followed by Tukey’s HSD post hoc analysis
Because GTPases such as RAB7L1 are typically only active in the GTP-bound state, we generated mutant forms that are constitutively active (CA; Q67L; this mutation is deficient in GTPase activity [data not shown]) or inactive (IN; T21N; a mutation within the presumptive GTP binding site; data not shown). As expected, overexpression of the CA RAB7L1, but not IN RAB7L1, significantly suppressed the LRRK2 mutation-induced neurite length phenotype. Of other Rab family members, expression of RAB3A or RAB5A failed to rescue the phenotype, whereas RAB7 CA was effective in suppressing the process length shortening induced by LRRK2 mutation (Figure 2B). In contrast to RAB7L1 overexpression, knockdown of RAB7L1 alone led to a significant reduction in neurite process length, similar to the effect of the LRRK2 G2019S mutant expression (Figure 2B, S2B).
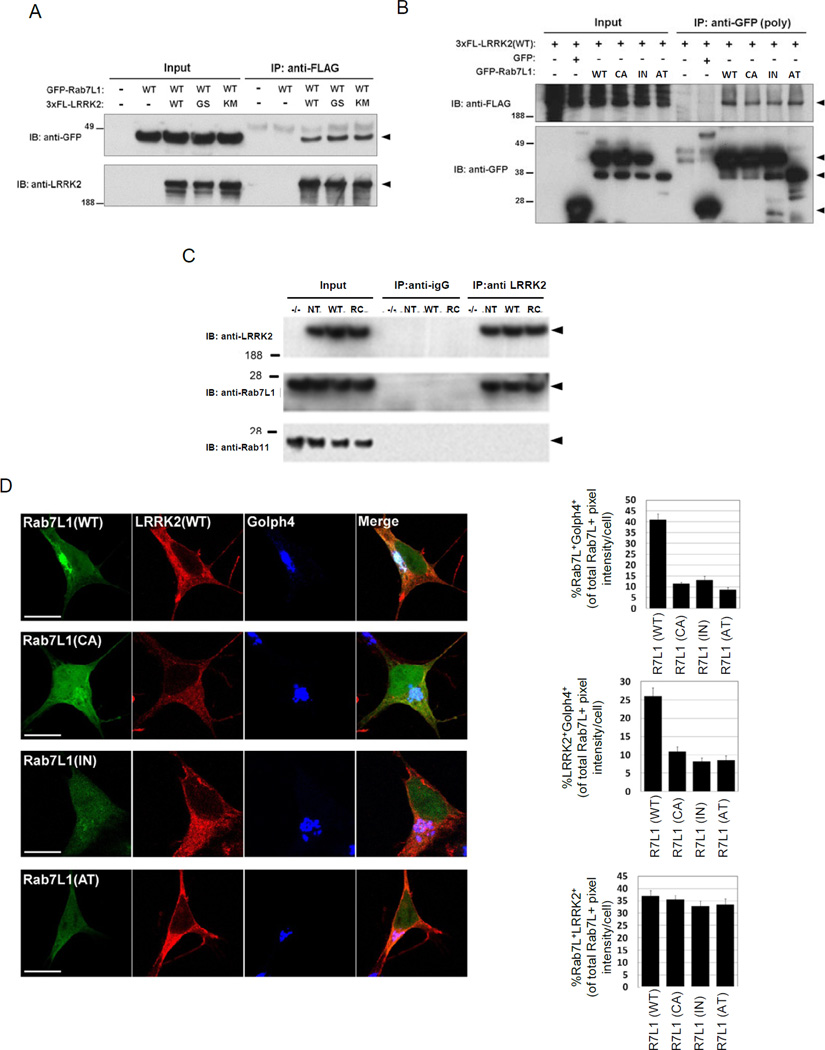
We next sought more direct evidence of a physical interaction between LRRK2 and RAB7L1 and thus performed co-immunoprecipitation studies. Epitope-tagged forms of LRRK2 and RAB7L1 (3xFlag-LRRK2 and GFP-RAB7L1) were co-transfected into HEK293T cells, and after 48 hrs, cell lysates were immunoprecipitated with an anti-Flag antibody and then probed for RAB7L1. Flag-immunoprecipitation of LRRK2 effectively co-precipitated RAB7L1 (Figure 3A). The interaction did not appear to be altered by the presence of the G2019S mutant, or using a kinase-dead variant K1906M of LRRK2 (MacLeod et al., 2006). Similarly, immunoprecipitation of RAB7L1 with an antibody to the GFP tag co-precipitated LRRK2 only in the presence of RAB7L1-GFP (Figure 3B). To probe for an interaction between LRRK2 and RAB7L1 in a more physiological context, we examined RAB7L1 protein in brain lysates from transgenic mice that harbor human wild-type LRRK2 or a familial PD mutant form of LRRK2, R1441C, within a large bacterial artificial chromosome (BAC) construct. Transgenic LRRK2 is broadly expressed throughout the CNS of these mice, although at relatively low levels (Figure S3A). Brain tissue lysates were immunoprecipitated for LRRK2 protein with a rabbit monoclonal antibody. Western blotting of the lysates for RAB7L1 demonstrated coimmunoprecipitation of RAB7L1 (Figure 3C).
Figure 3. Evidence of a RAB7L1-LRRK2 complex.
(A)Immunoprecipitation (IP) analysis of RAB7L1 from lysates of HEK293T cells transfected with plasmids encoding a GFP-RAB7L1 fusion protein (or vector alone) and a 3xflag (3FL) epitope-tagged LRRK2 construct (either wild type [WT], G2019S [GS], K1906M [KM], or empty vector). IP with an anti-flag antibody was followed with immunoblot (IB) analysis with an anti-GFP or an anti-LRRK2 antibody as indicated. Arrowheads indicate the expected protein sizes. (B) Co-immunoprecipitation of LRRK2 with RAB7L1 from lysates of HEK293T cells transfected with a plasmid encoding a 3xflag LRRK2 construct and a plasmid encoding a GFP-RAB7L1 fusion protein (either WT, CA, IN, or GFP only). (C) Immunoprecipitation using an anti-LRRK2 antibody from whole brain lysates of non-transgenic (NT), LRRK2 wild type transgenic (WT), LRRK2 R1441C (RC) transgenic, or LRRK2 knockout (−/−) mice. IB was subsequently performed for RAB7L1, RAB11 and LRRK2.
(D)Subcellular co-localization of RAB7L1 and LRRK2. Human neuroblastoma SH-SY5Y cells were transfected with GFP-tagged RAB7L1 vectors (in green; either WT, CA, or IN forms, as well as a RAB7L1 construct lacking exon 2 and 3 and corresponding to an alternatively spliced RAB7L1 transcript, “AT”) and a 3xflag-tagged LRRK2 vector (in red, left panel). Subcellular localization was determined by immunostaining with a marker for the Golgi apparatus (Golph4; in blue). The CA form leads to a reduced localization to the Golgi apparatus. Co-localization is evaluated by quantifying the fraction of RAB7L1/Golph4, RAB7L1/ LRRK2 and LRRK2/Golph4 staining overlap (Upper, lower and middle right panels respectively). Results represent mean ± SEM (n=15 per group)
In vitro fluorescence microscopy studies were consistent with the presence of RAB7L1 and LRRK2 in common subcellular compartments. GFP-tagged RAB7L1, transfected into SH-SY5Y cells, localized primarily to the Golgi apparatus (as identified with the Golph4 marker), as well as along tubular structures emerging from Golgi apparatus, consistent with prior reports (Spano et al., 2011). LRRK2 staining appeared more diffuse than RAB7L1, but there was significant overlap (Figure 3D). In contrast to the wild-type form, the RAB7L1 CA or IN mutant forms appeared more diffusely localized through the cytoplasm, as did a RAB7L1 alternative transcript (AT) deficient in the predicted GTP-binding region (Figure 3D; see below); accumulation of the IN and AT mutant proteins was significantly reduced (Figures 3D and S3B).
In vivo analysis of a LRRK2-RAB7L1 pathway in Drosophila dopamine neurons
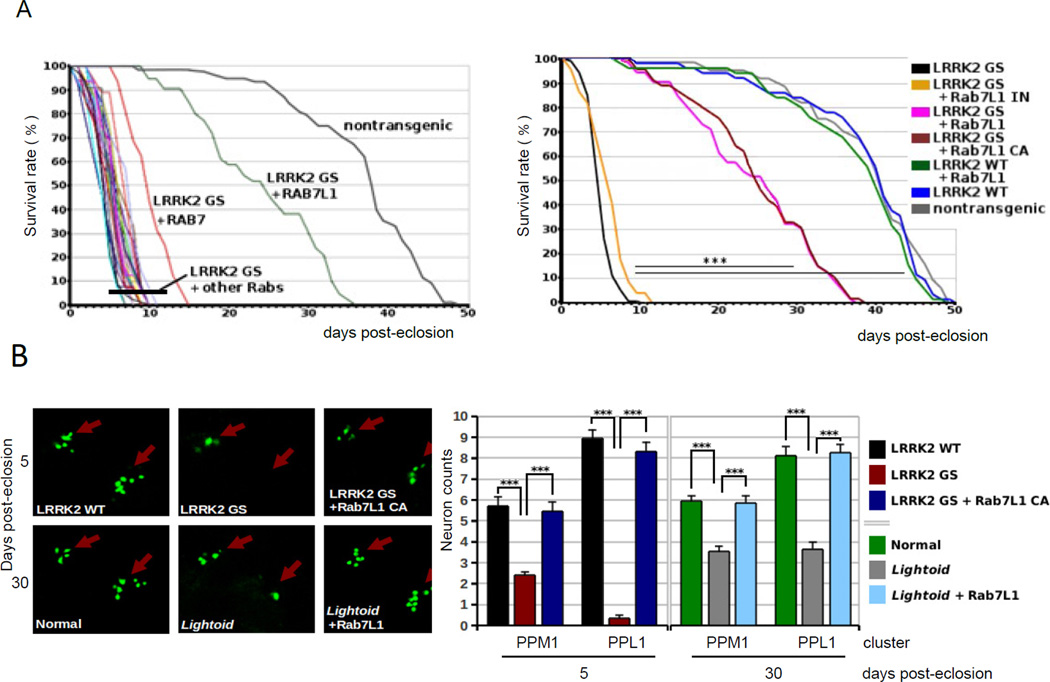
To pursue potential mechanisms of LRRK2 pathology in vivo, we established a Drosophila model. Although transgenic mouse models expressing mutant LRRK2 have been widely described (Andres-Mateos et al., 2009; Li et al., 2009; Piccoli et al., 2011; Tong et al., 2009), these do not show consistent neurodegenerative phenotypes. Dopamine neuron-selective expression of human familial PD-associated G2019S-mutant LRRK2 -- using either a tyrosine hydroxylase (TH) (Friggi-Grelin et al., 2003) or dopa decarboxylase (DDC) promoter-Gal4 driver (Fischer et al., 1988) -- induced premature mortality of young adult animals (Figure 4A and data not shown; nontransgenic mean lifespan 37.1 days +/− 1; G2019S mean lifespan 4.8 days +/− 0.2), akin to previous reports (Ng et al., 2009). In contrast, transgenic expression of wild-type human LRRK2 did not lead to a discernible phenotype. Furthermore, expression of the mutant G2019S LRRK2 transgene in several other cell types, including motor neurons, eye tissues, or muscles (using a variety of promoter-Gal4 driver constructs; data not shown), failed to lead to a discernible effect on survival or otherwise (data not shown).
Figure 4. RAB7L1 rescues lethality and dopamine neuron loss in a Drosophila model of LRRK2 G2019S neurodegeneration.
(A) Modifier screen for suppressors of an early adult lethality phenotype seen with expression of LRRK2 G2019S selectively in tyrosine hydroxylase (TH)-positive dopamine neurons. Left, a panel of 16 Drosophila RAB transgenes was screened (of 31 total; see Table S3). Adult survival (days post-eclosion) curves are presented for individual strains harboring different RABs along with the LRRK2 G2019S transgene. Non-transgenic survival curve is shown for comparison. n > 25 for all conditions. Right, Adult survival (days post-eclosion) of Drosophila is presented in the context of transgenic expression of LRRK2 (WT or G2019S), with or without RAB7L1 (WT, CA or IN), using a tyrosine hydroxylase promoter GAL4 driver for dopaminergic neuron expression. Non-transgenic survival is also shown for comparison. n > 25 for all conditions. (B) (i) Confocal microscopy of mushroom bodies of the CNS from transgenic Drosophila as in (B), with dopaminergic neuron nuclei visualized using an additional marker transgene, a nuclear localization sequence (NLS)-GFP fusion, also driven by TH-Gal4. (ii) Quantitation of surviving dopaminergic neurons in the PPM1 and PPL1 clusters of Drosophila CNS mushroom bodies. Means are displayed; error bars represent SEM; *** : p<0.001 by ANOVA followed by Tukey’s HSD post hoc analysis for (A) and (B).
We subsequently performed a targeted screen for potential genetic modifiers of the LRRK2 G2019S mutant phenotype, based on the hypothesis that LRRK2 may modify a specific intracellular trafficking process, and focused on RAB7L1. A series of 16 Drosophila Rab genes, (see Table S4; out of 33 identified in Drosophila), or CA or IN forms of these (Zhang et al., 2007), were investigated. Briefly, LRRK2 G2019S mutants were mated with a panel of previously described transgenic Drosophila strains that allow for overexpression of wild-type (WT) or constitutively active (CA), forms of the Rab genes (Zhang et al., 2007), using a standard balancer chromosome-based mating scheme. Co-expression of a majority of these Rab transgenes with LRRK2 within dopamine neurons produced no effect on the survival of animals co-expressing LRRK2 G2019S (Figure 4A; Table S4). In contrast, overexpression of wild-type and CA forms of the Drosophila RAB7L1 orthologue (termed lightoid) afforded a dramatic rescue of the LRRK2 G2019S premature mortality phenotype (mean lifespan 24.0 days +/− 1 for the CA; Figure 4A). Of note, among the other Rabs screened, only Rab7 led to a statistically significant -- albeit much weaker -- survival benefit (mean lifespan 14.3 days +/− 0.6). Rab1, which was previously found to rescue a defect in vesicular trafficking to the Golgi apparatus in α-Synuclein overexpression models of PD (Cooper et al., 2006), did not rescue the LRRK2 defect, suggesting distinct mechanisms.
Next, dopamine neuron survival at the dorsomedial posterior protocerebral (PPM1) and dorsolateral posterior protocerebral (PPL1) clusters of Drosophila CNS mushroom bodies was quantified in terms of the loss of expression of a dopamine neuron-specific nuclear localization signal (NLS)-GFP marker protein, using fluorescent confocal microscopy analysis of whole mounted tissue. Expression of LRRK2 G2019S, but not the WT form, led to the preferential loss of neurons in the dorsomedial cluster, reminiscent of the phenotype in other Drosophila models of PD (Feany and Bender, 2000). Co-expression of CA RAB7L1 rescued the LRRK2 G2019S dopamine neuron loss phenotype (Figure 4B). Deficiency of the RAB7L1 orthologue (in lightoid homozygous mutants) selectively in dopamine neurons by expression of an siRNA construct (Dietzl et al., 2007), led to a significant loss of dopamine neurons (Figure 4B).
PARK16 risk variants modify RAB7L1 splicing and expression
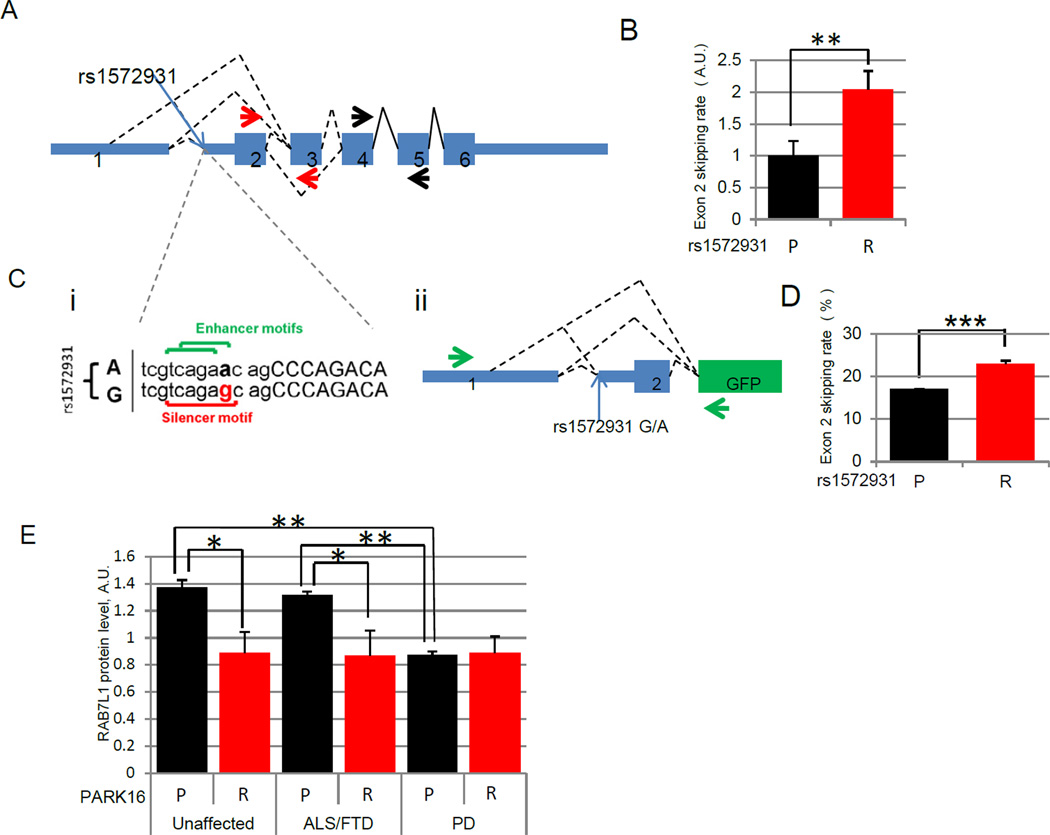
The combination of human brain transcriptomic, human genetic, and model system studies support a role for PARK16, and specifically the PARK16 locus gene RAB7L1, in a pathway with LRRK2. We next sought to query possible molecular mechanisms at play at the PARK16 locus that may be responsible for a link between common genetic variants, RAB7L1 function, and PD risk. A challenge to this is that typically many variants at a given chromosomal location are so closely associated (in ‘linkage dysequilibrium’) so as to make impossible the identification of which is truly ‘causal’ rather than just coincidental. On re-analysis of existing genome-wide splicing data from human lymphoblasts (Montgomery et al., 2010), the PD-associated PARK16 haplotype was found to be associated with alternative splicing of RAB7L1, characterized by the skipping of exons 2 and 3. We note that a common SNP variant within the PARK16 locus, rs1572931, that is in linkage dysequilibrium with SNP rs947211 (Hamza et al., 2010) and thus similarly linked to PD risk, falls precisely within regulatory sequences for splicing at the Intron1-exon2 boundary (Figure 5A). Akin to the lymphoblast transcriptome splicing data, our analysis of a set of human cortical brain samples revealed that the rs1572931 genotype is similarly associated with modified splicing of RAB7L1 in human forebrain (Figure 5B, S5A), where the protective PARK16 haplotype is associated with increased exon 2 inclusion in RAB7L1 mRNA. Based strictly on human gene expression data, we cannot directly assign a causal role for SNP rs1572931 in altered splicing of RAB7L1 (as other SNPs in linkage disequilibrium could be responsible for the observed association). We thus sought to evaluate the causal effect of rs1572931 using minigene reporter vectors that harbor either the risk-associated or protective allele at rs1572931, but are otherwise identical (Figure 5Cii, S5B). Upon transfection into SHSY5Y human neuroblastoma cells, the rs1572931 risk allele led to increased RAB7L1 exon 2 skipping relative to the protective allele (Figure 5 D, S5CDE).
Figure 5. PARK16 PD risk-associated variants modify RAB7L1 splicing and protein accumulation.
(A) Exonic structure of the human RAB7L1 gene.(B)Analysis of RAB7L1 alternative splicing in human cortical brain samples. The rs1572931 allele G, linked to the PD high-risk haplotype (R), is associated with an increase in the fraction of RAB7L1 transcripts that lack the exon 2 to exon 3 junction region (termed exon 2 skipping; presented relative to the extent of exon 2 skipping seen in carriers of the rs1572931 protective allele A; quantified by qrtPCR using primers as depicted by red and black arrows in (A) detecting respectively the amount of total and unskipped RAB7L1 mRNA ; n=15 and 57 for P and R respectively; details in Table S6). (C)i) Schematic of predicted splice site enhancer and silencer motifs upstream of RAB7L1 exon2 and affected by rs1572931 variants G (associated with increased PD risk, “R”) and A (protective, “P”, associated with decreased PD risk). (ii) Structure of a minigene construct to assess the effect of rs1572931 variants on RAB7L1 exon2 inclusion in vitro. Green arrows indicate the position of the primers used to assess exon 2 inclusion. (D) Impact of PARK16 variants on splicing in vitro. The rs1572931 allele G (associated with increased PD risk, R; relative to the allele A associated with decreased PD risk, P) leads to a relative decrease in RAB7L1 exon 2 inclusion in transfected human SH-SY5Y cells as assess by PCR gel quantification (pictures in Figure S5DE; n=6/group) (E)Impact of rs1572931 on RAB7L1protein level in human cortical brain samples. rs1572931 allele G is associated with a decrease in RAB7L1 protein level in non-PD post-mortem human cortical brain samples, as assessed by Western Blot from individuals homozygous for the risk allele (R, n=25) and from carriers of the protective allele (P, n=13). Mean levels are displayed ; errors bars are SEM ; *: p < 0.05, **: p < 0.01, ***: p < 0.001 by two-tailed t-test (B,D) or by linear regression analysis (E).
Exon skipping is predicted to lead to the formation of a truncated form of RAB7L1 protein that lacks the predicted GTP-binding domain in the amino-terminal region (Figure S5C). Overexpression of this truncated form leads to low level accumulation of a shortened protein product (Figure S3B), and reduced localization to the Golgi apparatus (Figure 3D); although the shortened product can bind with LRRK2 protein (Figure 3B), expression of this truncation mutant in primary neurons failed to rescue the reduced neurite length phenotype associated with G2019S mutant LRRK2 (Figure S5F), whereas expression of the wild-typeRAB7L1 effectively rescued the phenotype. Consistent with these in vitro findings, we observed a significant reduction in full-length RAB7L1 protein in cerebral cortex tissue from unaffected individuals who carry the PARK16 risk allele, when compared to non-carrier individuals (Figure 5E). We note that a similar reduction is seen in PD patient cerebral cortex tissue regardless of the PARK16 genotype. This appears specific to PD, as no such decrease is observed in tissue from patients suffering from other neurodegenerative disorders examined (frontotemporal dementia or amyotrophic lateral sclerosis) who do not carry the PARK16 risk allele (Figure 5E). Taken together, these findings argue in favor of a post-transcriptional (splicing) mechanism of action for the PARK16 PD risk variant’s impact on RAB7L1 levels. However, given the linkage disequilibrium structure of the region, we cannot exclude additional transcriptional regulatory effects (Gan-Or et al., 2012).
Lysosomal changes and retromer-associated sorting defects in LRRK2 and RAB7L1 mutant neurons
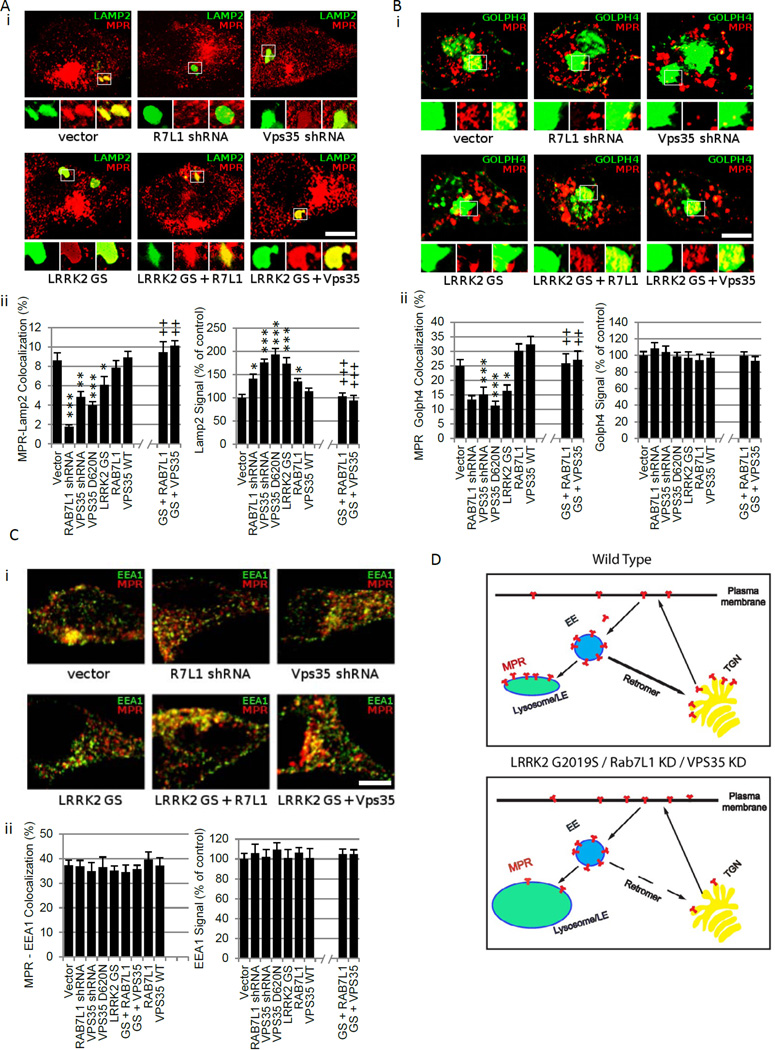
We sought to pursue a cellular role for the LRRK2-RAB7L1 pathway. Prior studies have broadly implicated both of these gene products in intracellular sorting (Sakaguchi-Nakashima et al., 2007; Spano et al., 2011). Expression of the LRRK2 G2019S clinical mutation in rat primary neurons induced lysosomal swelling, as quantified by immunostaining for the lyosomal marker LAMP2 or using the lysosomotropic dye Lysotracker, consistent with our prior work and other studies (Dodson et al., 2012; MacLeod et al., 2006; Stafa et al., 2012) (Figure 6A and data not shown). In addition to lysosomal enlargement, there was significant reduction in lysosomal accumulation of the cation-independent mannose 6-phosphate receptor (MPR) in terms of the fraction of LAMP2-positive structures stained with MPR. As MPR is required also for the recruitment of lysosomal hydrolases, its deficiency is predicted to lead to functional lysosomal deficits. Knockdown of RAB7L1 was similarly associated with swollen lysosomes and reduced lysosomal MPR, whereas overexpression of RAB7L1 suppressed the lysosomal phenotypes in the context of LRRK2 G2019S expression (Figure 6A).
Figure 6. RAB7L1 and LRRK2 modulate lysosome and Golgi apparatus sorting in a retromer-dependent manner.
(A–C) Analysis of MPR sorting in primary rat neuron cultures transfected with vectors encoding LRRK2 G2019S mutant (GS), RAB7L1, VPS35, or VPS35 D620N; or with shRNA plasmids for VPS35, RAB7L1 or vector only, co-transfected with GFP vector for visualization and immunostained for MPR as well as either the Golgi marker Golph4 (A, upper panel), the lysosome marker Lamp2 (B, upper panel) or with the early endosome marker EEA1(B, upper panel). MPR colocalization with either the Golph4 or LAMP2 marker was reduced with G2019S LRRK2, VPS35 D620N, or knockdown of either RAB7l1 or VPS35 (A, lower panel; B, lower panel). These manipulations also increased total LAMP2 staining (but not Golph4 staining). Scale bar represents 10um. Quantifications of the MPR co-localization and of total organelle marker analyses are presented in the lower panels. Error bars represent SEM. n>10 cells in 3 independent wells per group. *: p<0.05 , **: p<0.01 , ***: p<0.001 for comparisons with “vector” group, ++: p<0.01 , +++: p<0.001 for comparisons with “LRRK2 G2019S” group by ANOVA followed by Tukey’s HSD post hoc analysis. (D) Schematic of cell sorting phenotype associated with defects in the LRRK2-Rab7L1 pathway or knockdown of the VPS35 retromer component. MPR accumulation at Golph4-positive structures (trans-golgi network [TGN]) and at LAMP2-positive structures (lysosomes and late endosomes [LE]) is reduced, and lysosomes appear swollen.
MPR is typically recycled between the endolysosome compartment and the Golgi apparatus by the retromer complex (Arighi et al., 2004; Bonifacino and Hurley, 2008; Seaman, 2009; St. George-Hyslop et al., 2009). Given the primary apparent localization of RAB7L1 to the Golgi apparatus (Figure 3D;), as well as the enrichment of LRRK2 at this organelle (Figure 3D)(Stafa et al., 2012), we hypothesized that the lysosomal compartment defects described above may be secondary to altered retromer mediated trafficking machinery between these organelles (Bonifacino and Hurley, 2008; Seaman, 2004). Analysis of Golgi structures by immunostaining with the Golph4 marker in primary neurons transfected with either LRRK2 G2019S or shRNA for RAB7L1 did not reveal evidence of gross structural changes, but MPR co-localization at the Golph4-positive Golgi apparatus structures was significantly reduced (Figure 6B). Accumulation of MPR at early endosomes, assessed by co-staining with the marker early endosomal antigen-1 (EEA1; Figure 6C), did not appear altered, whereas accumulation at the cell surface appeared increased (data not shown). The total areas of Golph4, MPR, or EEA1 staining were unaffected by G2019S LRRK2 expression or RAB7L1 knockdown (Figure 6ABC).
The retromer complex is required for retrograde transport of selective cargo – including MPR – between lysosomes and the Golgi apparatus, through endosomal intermediates, in mammalian cells (Figure 6D) (Bonifacino and Hurley, 2008; St. George-Hyslop et al., 2009), and defects can lead to lysosomal swelling (Arighi et al., 2004). Furthermore, rare mutations in a retromer component, VPS35, were recently linked to rare familial forms of PD (Vilarino-Guell et al., 2011; Zimprich et al., 2011). Knockdown of VPS35 in primary neuron cultures led to reduced MPR co-localization with the Golgi apparatus and with late endosomes/lysosome markers (Figure 6AB), as previously described (Seaman, 2009). Similarly, expression of a familial PD-associated mutation in VPS35, D620N (Vilarino-Guell et al., 2011; Zimprich et al., 2011), phenocopied the MPR missorting phenotype of G2019S mutant LRRK2 expression or VPS35 knockdown (Figure 6AB), suggesting a dominant negative mode of action which is consistent with a predicted structural alteration of a retromer complex interaction motif (Vilarino-Guell et al., 2011; Zimprich et al., 2011). In contrast, overexpression of wild-type VPS35, which promotes trafficking through the retromer pathway, suppressed the altered MPR localization seen with G2019S mutant LRRK2 expression (Figure 6AB). Thus, although it is likely that the LRRK2-RAB7L1 pathway impacts intracellular sorting processes in addition to retromer complex function, suppression of retromer dysfunction is sufficient to rescue the deficits associated with defects in the LRRK2-RAB7L1 pathway.
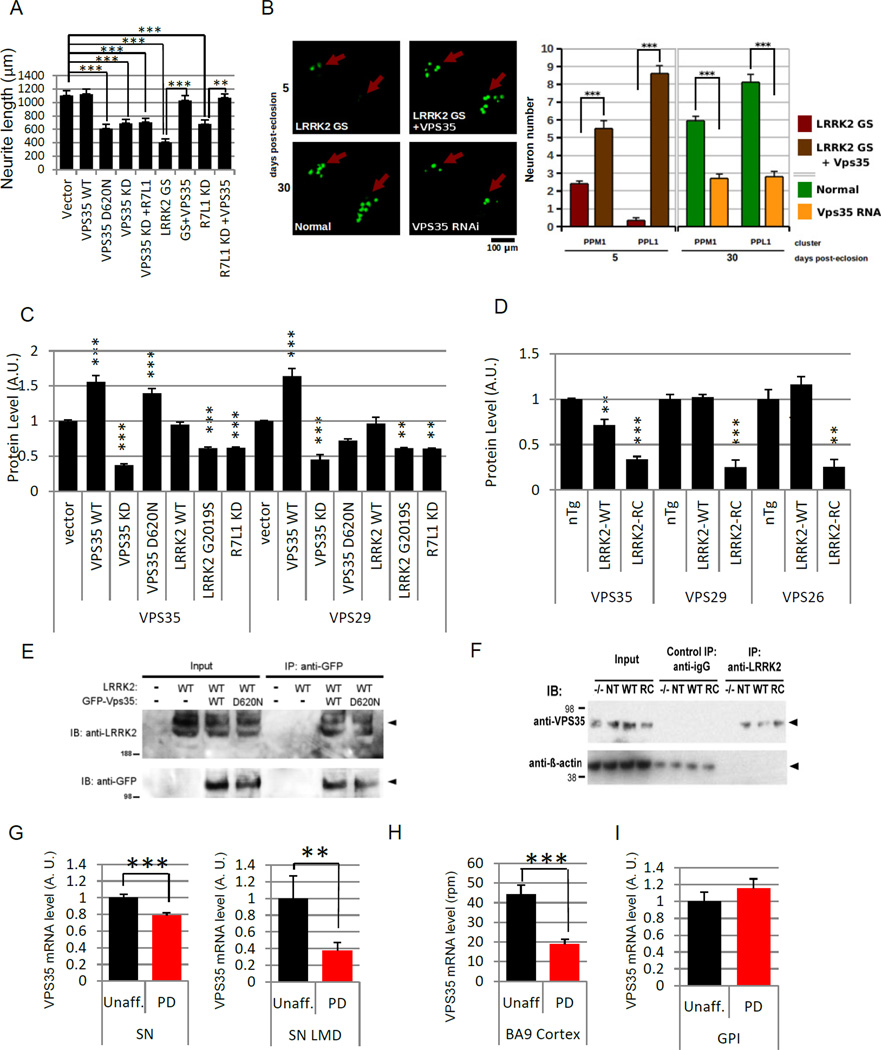
We further investigated the functional relationship of VPS35 with the LRRK2-RAB7L1 pathway in the context of neurite process maintenance. In rat primary neurons, overexpression of VPS35 alone did not directly modify neurite process length, but effectively suppressed the loss of neurite processes in the context of LRRK2 G2019S expression or RAB7L1 knockdown (Figure 7A). In contrast, knockdown of VPS35 with an shRNA vector, or expression of the VPS35 D620N mutant form, led to neurite process length reduction that phenocopied the effect of LRRK2 G2019S expression. In vivo analysis in the Drosophila CNS further supported a role for retromer dysfunction in the context of LRRK2-RAB7L1 pathway defects. Overexpression of Drosophila VPS35 in Drosophila CNS dopamine neurons rescued the LRRK2 G2019S dopamine neuron loss phenotype (Figure 7B), and similarly extended the lifespan of G2019S LRRK2 mutant-expressing flies. In contrast, knockdown of VPS35 selectively in Drosophila TH-positive dopamine neurons led to significant cell loss and a reduced lifespan (Figure 7B).
Figure 7. Evidence of retromer insufficiency in the context of LRRK2-RAB7L1 pathway defects.
(A) Transfection of rat primary cortical neuron cultures with a wild-type (WT) VPS35 expression vector rescued the reduced neurite length phenotype associated with LRRK2 G2019S (GS) mutant expression or with Rab7L1 (R7L1) knockdown. Overexpression of a familial PD mutant VPS35 D620N vector leads to reduced neurite length relative to vector alone. Knockdown of VPS35 by shRNA leads to similarly reduced neurite length relative to vector alone, which is not rescued by Rab7L1 overexpression (n=20 neurons in 4 cultures per group). (B) (i) Confocal microscopy of mushroom bodies of the CNS from transgenic Drosophila as in (B), with dopaminergic neuron nuclei visualized using a TH-Gal4-driven nuclear localization sequence (NLS)-GFP fusion. (ii) Quantitation of surviving dopaminergic neurons in the PPM1 and PPL1 clusters of Drosophila CNS mushroom bodies. (C) Relative quantification by western blot of VPS35 (left) or VPS29 (right) protein levels in lysates from mouse neuroblastoma (N2a) cells transfected with vectors encoding VPS35 WT, VPS35 shRNA, VPS35 D620N, LRRK2 WT, LRRK2 G2019S (GS), RAB7L1, RAB7L1 shRNA, or vector control (N=3 /group). (D) LRRK2 impacts the levels of retromer components in mouse brain. Relative quantification by Western blotting of VPS35 (left), VPS29 (middle) and VPS26 (right) levels in brain tissue samples from non-transgenic (“nTg”), LRRK2 wild-type (“LRRK2-WT”) and LRRK2 R1441C mutant (“LRRK2-RC”) BAC transgenic mice (N=3 /group). (E) Immunoprecipitation (IP) analysis of RAB7L1 from lysates of SH-SY5Y cells transfected with plasmids encoding a GFP-VPS35 fusion protein with VPS35 wild-type sequence (“WT”) or the familial PD mutant D620M (“D620N) or vector alone, along with a LRRK2 construct or an empty vector. IP with an anti-GFP antibody was followed with Western immunoblot analysis with an anti-LRRK2 or anti-GFP antibody as indicated. Arrowheads indicate the expected protein sizes. (F) IP using an anti-LRRK2 antibody from whole brain lysates of non-transgenic (NT), LRRK2 wild type transgenic (WT), LRRK2 R1441C (RC) transgenic, or LRRK2 knockout (−/−) mice as in Figure 3D. Immunoblot was subsequently performed for VPS35 and β-Actin. (G) VPS35 mRNA levels in substantia nigra tissue as determined by meta-analysis of 5 gene expression microarray datasets (Table S5) in 63 unaffected individuals and 81 PD patients samples (left panel) and in laser-microdissected substantia nigra dopaminergic neurons from 8 unaffected individuals and 10 PD patients samples (right panel, GEO GSE20141). Expression levels are normalized to mean of the unaffected group. (H) VPS35 mRNA in cerebral cortex tissue as determined by high-throughput sequencing of the 3’UTR ends of polyadenylated mRNA transcripts on a cohort of 17 unaffected and 17 PD cerebral cortical tissue samples. Levels are expressed as reads per million (rpm). (I) VPS35 mRNA levels in Globus Pallidus Interna (Gpi) samples (n=10/group GEO GSE20146). Expression levels are normalized to mean of the unaffected group. For all graphs means are displayed, error bars represent SEM; p<0.05(*) p<0.01(**) for ANOVA followed by Tukey’s HSD post hoc analysis (A,B,C) or by two-tailed t-test (G,H).
Reduction in retromer complex component levels in the context of LRRK2-RAB7L1 pathway defects
We next sought to pursue potential molecular mechanisms for the apparent defects in retromer pathway function in the context of LRRK2 G2019S mutation or RAB7L1 knockdown. In mouse N2A neuroblastoma cells, expression of LRRK2 G2019S or knockdown of RAB7L1 led to a significant reduction in the levels of accumulated VPS35 as well as VPS29, a second component of the retromer complex (Figure 7C). Levels of retromer complex components are dependent on the formation of the entire complex, which also includes VPS29, and thus loss of any complex component is predicted to impact levels of others (Kim et al., 2010). Analysis of transgenic mouse total brain tissue overexpressing the R1441C mutant form of LRRK2 also led to a significant reduction in the accumulation of VPS35 and VPS29, and VPS26 (Figure 7D).
Although the precise mechanism by which the LRRK2-RAB7L1 pathway impacts retromer complex function and levels remains to be determined, co-immunoprecipitation studies of LRRK2 with VPS35 support a direct interaction between these proteins: Lysates from SH-SY5Y cells co-expressing epitope-tagged V5-LRRK2 (or vector) and eGFP-VPS35 forms, were immunprecipitated for the eGFP tag. Subsequent Western blotting revealed co-purification of LRRK2 with eGFP-VPS35 (Figure 7E). Similarly, immunoprecipitation of LRRK2 from LRRK2 transgenic mouse brain tissue led to the co-precipitation of endogenous VPS35 (Figure 7F). It remains to be determined whether the interactions of LRRK2 with VPS35 and RAB7L1 are within a single complex or multiple complexes.
To relate those findings to sporadic PD, we analyzed VPS35 levels in PD or unaffected human brain tissue. We first conducted a meta-analysis of substantia nigra (SN) mRNA expression levels in 5 publically available microarray gene expression datasets from patients and controls (Table S5; totally 144 individuals, 63 unaffected individuals and 81 PD patients), and observed a highly significant decreased in VPS35 mRNA levels (Figure 7G). Such a decrease was also observed in gene expression data from laser-microdissected PD SN dopamine neurons, when compared to similar cells isolated from unaffected patients (Figure 7G), as well as in PD cerebral cortex tissue (Figure 7H – 7I). Taken together, these results support a role for retromer deficiency in the impact of PD-associated genetic risk variants on brain neurons.
DISCUSSION
Using a brain transcriptomic approach as a starting point, we provide evidence that the impacts of several distinct PD risk-associated common genetic variants are overlapping, even in unaffected PD-free carrier tissue. This points to a convergent pathway of action for such variants. Focusing subsequently on LRRK2 and the PARK16 locus gene RAB7L1, in vitro and in vivo studies support a functional interaction: these gene products bound together and functionally interacted in the regulation of neurite process length in vitro, as well as in the context of dopamine neuron survival in vivo. We emphasize that the impact of LRRK2 and PARK16 variants on brain gene expression was observed even in unaffected carriers of the PARK16 or LRRK2 locus risk variants; this suggests the existence of a pre-disease prodromal state in such carriers, that favors subsequent progression.
The most prominent neuronal sorting phenotypes we observed in the context of PD-associated LRRK2-RAB7L1 pathway changes were at lysosomes and the Golgi apparatus. We hypothesize that the proximal site of action for these proteins may be in defective retromer function at the Golgi apparatus, given the enrichment of both proteins at this structure. Trafficking of MPR to the Golgi apparatus -- a function of the retromer complex -- is defective, and associated with lysosomal swelling. Although the precise mechanism of retromer dysfunction is unclear, retromer pathway components including VPS35 appear reduced in the context of LRRK2 mutation or RAB7l1 knockdown. Recently described familial PD-associated clinical mutations in VPS35 phenocopy the deficits associated with LRRK2-RAB7L1 pathway dysfunction, whereas overexpression of VPS35 can rescue such deficits. We also note that RAB7 was identified in both our in vitro and in vivo screens of RAB proteins as suppressing the phenotype of LRRK2 mutant pathology, albeit less robustly than RAB7L1. RAB7 is the only RAB protein previously implicated in the regulation of retromer function (Rojas et al., 2008).
Prior studies have supported a role for LRRK2 in vesicular trafficking (Biskup et al., 2006; Dodson et al., 2012; Higashi et al., 2009; MacLeod et al., 2006; Stafa et al., 2012). However, cellular mechanisms of LRRK2 relevant in human brain -- and in the context of PD or PD risk variants -- have remained unclear. The studies herein are unusual in pursuing a PD genetic pathway using both human brain and model system analyses. We identify a genetic interaction between LRRK2 and RAB7L1 in the context of PD risk, and variants at the loci of these genes impact the brain transcriptome in an overlapping manner. Subsequent cell and animal model studies support a model where LRRK2 and RAB7L1 defects may modify intracellular sorting and retromer pathway function.
It is possible that PD-related defects in LRRK2 and RAB7L1 adversly impact aspects of vesicular trafficking in addition to retromer function. Nonetheless, inducing retromer function appears sufficient to rescue cellular defects and neuronal survival in these models, suggesting a therapeutic venue in PD patients. It is interesting to note that VPS35 deficits, as well as genetic variants at retromer complex receptor loci such as SORLA (Rogaeva et al., 2007), have also been associated with a second major neurodegenerative disorder, Alzheimer’s disease (Muhammad et al., 2008); this suggests a broader role for retromer dysfunction in neurodegeneration. We speculate that perhaps different cargos may be involved in the association of the retromer pathway with distinct pathological processes in Alzeimer’s and Parkinson’s. To this end, it is of interest to investigate the impact of such retromer dysfunction on aSyn and other proteins associated with PD pathology.
EXPERIMENTAL PROCEDURES
Drosophila methods
>Drosophila were cultured by standard methods on yeast-cornmeal-agar medium at 25°C. Wild-type and mutant G2019S LRRK2 transgenes were expressed specifically in catecholaminergic neurons, including dopamine neurons, using the Gal4-UAS system described (Fischer et al., 1988). Driver lines used include OK6 (motor neuron), Gmr (eye), G14 (muscle), TH (dopaminergic neuron), and DDC (dopaminergic neuron). A UAS-GFP::nuclear localization sequence (NLS) marker was used to visualize nucleii of cells in which trangenes were expressed (stock 4775) (w1118; P{UAS-GFP.nls}14), Drosophila Stock Center, Bloomington, IN). For the RAB screen, UAS-LRRK2 (G2019S) transgenic Drosophila, crossed with the TH-Gal4 driver, were screened against a UAS-Rab transgenic library (Zhang et al., 2007). Crossings were typically performed using standard balancer chromosome techniques. To generate strains in which the homozygous LRRK2 transgene and another (Driver or marker) transgene lay on the same chromosome (III), genetic recombination was using standard techniques. Adult Drosophila mushroom bodies were dissected as in (Wu and Luo, 2006) and imaged immediately, without fixation, using a Zeiss LSM510 Meta confocal fluorescent microscope. For mortality curves, transgenic Drosophila surviving through adult metamorphosis were counted daily, from the day of pupal eclosion onward.
Primary Neurons Culture
Sprague-Dawley rat or mouse P1 primary dissociated cortical cultures were prepared and transfected essentially as described (Xia et al., 1996) with the following modifications: cells were plated at high density, approximately 250,000 cells/cm2, in 24-well plates with 500ul medium/well. Culture medium used for plating cells was Neurobasal-A supplemented with 2% B-27 and 10% FBS. At 1 day after plating, medium was changed to reduced serum (1% FBS+ added antimitotic agents: 70 µM uridine and 25 µM 5-fluorodeoxyuridine) and replaced weekly thereafter; for transfections no DMSO was added to the transfection mixture, cells were not subjected to glycerol shock, and a total of 3 µg plasmid DNA was used per well. Cells were fixed in 4% PFA and immunostained with mouse α-RAB7L1 (Santa Cruz, 1:100), and rabbit monoclonal α-LRRK2 (Michal J. Fox Foundation MJFF4, 1:100), then with appropriate fluorescent secondaries (Jackson, 1:1000–2000). Neurite length and neurite puncta (defined as swellings greater than 2um in diameter) were counted for for at least 20 neurons per condition. Mean puncta number per neuron was normalized to total average neurite length versus wild-type LRRK2 transfected cells. Fluorescent microscopy was performed using a Nikon TE 2000-S microscope and a Zeiss LSM510 Meta confocal microscope. Images were analyzed using Image-Pro Plus (Mediacybernetics) software version 5.1.0.20.
Colocalization Analysis
Primary rat cortical neurons were cultured on glass coverslips, transfected, and fixed as previously described in this methods section. Cells were immunostained for MRP (Abcam #ab2733, 1:400), Golph4(Abcamab #28049, 1:500), Lamp2 (Sigma L0668, 1:500). Fluorescent microscopy was performed using a Zeiss LSM510 Meta confocal microscope. Images were analysed using NIH ImageJ software version 1.45.
Cell culture, transfection and cytochemistry
HEK293T and SH-SY5Y cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a 5% CO2 atmosphere. Transient expression was performed by transfecting the plasmids using Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions. The transfected SH-SY5Y cells grown on glass coverslips for 24hours were fixed with 4% paraformaldehyde in PBS for 30 minutes, washed three times with PBS and subjected to the observation of fluorescence. For immunostaining of golgi, fixed cells were blocked and permeabilized with PBS containing 0.1% TritonX-100 and 3% bovine serum albumin followed by incubation with anti-Golph4 polyclonal antibody (abcam) and Alexa Fluor 555 goat anti-rabbit IgG (Invitrogen). Staining of nuclei was performed by using SYTOX Orange nucleic acid stain (Invitrogen). Fluorescence was detected using Zeiss LSM 510 confocal microscope.
Immunohistochemistry and Signal Quantification
LRRK2 R1441C or Wt BAC transgenic mice (Li et al., 2009) (Jackson Laboratory) were sacrificed and perfused immediately with 4% PFA for 20min. Brains were cut by vibratome into sections 60 µm thick. Sections were blocked in 5% NDS overnight at 4 C, then incubated with primary antibodies overnight at 4 C. Antibodies used were sheep monoclonal α-TH (Pelfreeze, 1:500), mouse α-RAB7L1 (Santa Cruz, 1:100), and rabbit monoclonal α-LRRK2 (Michal J. Fox Foundation MJFF4, 1:100). Sections were incubated at room temperature for 2 hours with appropriate fluorescent secondaries (Jackson Laboratories, 1:1000). Microscopy was performed with a Zeiss LSM510 Meta confocal. Fluorescence signal intensity was quantified using NIH ImageJ.
Human autopsied brain samples
Cortical BA9 area brain samples were obtained from the New York Brain Bank and are detailed in Suppl. Table 5. Anonymous, de-identified tissue from the brain bank was used.
Quantitative real time RT-PCR
RT-qPCR was done as described in (Rhinn et al., 2008) RAB7L1 ratio was quantified using ΔΔCt method using primers pairs
RAB7L1_Ex2_fw (CAGCAAACACTACAAGTCCACG)
RAB7L1_Ex3_rv (CAGCTGAAGCCGCACTATCTCG) and
RAB7L1_Ex4_fw (GACAGCAAGCTCACACTACCCA)
RAB7L1_Ex5_rv (TCTGTCCAACCTGTGAAACCGT) for human brain samples.
Minigene Splicing assay
The human SH-SY5Y neuroblastoma cell line (ATCC) was cultured following ATCC’s instructions, plated at densities of 4.10e5 cells per well (48-well plates) in wells coated with 0.1% gelatin (Specialty Media, Millipore) 24 hours prior to transfections. Transfections were performed with Lipofectamine 2000 reagent (Invitrogen) following manufacturer's instructions. After transfection with plasmids encoding the reporter contruct, RNA was extracted using miRNeasy kit (Qiagen) and reverse transcribed using Superscript III reverse transcriptase (Invitrogen) following manufacturer’s instructions. The cDNA was amplified by PCR using the following primers:
GGAGGGCGTCTAGGGAATCGAG (Fw, complementary to exon1 of RAB7L1) and CTTCAGGGTCAGCTTGCCGTAG (Rev., complementary to GFP CDS) and Accuprime high-fidelity polymerase (Invitrogen) following manufacturer’s instruction with an hybridization at 55C and an elongation step of 1 min.
Supplementary Material
HIGHLIGHTS.
Genetic variants at PARK16 and LRRK2 interact to modify Parkinson’s disease risk.
Splicing of the PARK16 locus gene RAB7L1 is modified by genetic variants.
RAB7L1 and LRRK2 coordinately regulate protein sorting through the retromer pathway.
Expression of the retromer component VPS35 can suppress LRRK2 mutant pathology.
Acknowledgments
We thank the New York Brain Bank and Jean Paul Vonsattel for banked tissue samples. We are grateful to all the contributors who made their data publicly available at the Gene Expression Omnibus. We thank Karen Marder for contribution of the PD patient data (NS36630), and Oliver Hobert for reviewing the manuscript. The work was supported by grants from the Michael J. Fox Foundation (A.A.) and the NIH (NS064433 and NS060876 to A.A., NS060113 to L.N.C., and AG025161 to S.A.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: The authors declare no competing interest.
Authors contributions
A.A. designed studies and interpreted data. H.R. designed and performed the human genetics analyses, and performed the bioinformatics and the splicing experiments. D.A.M. performed the Drosophila and mouse experiments. D.A.M., T. K. and A.Z performed the cell culture experiments. B.D.M. helped designing and interpreting the Drosophila experiments, L.N.C. the human genetics, G.D.P. and S.A.S. the retromer and trafficking experiments. A.A. and H.R wrote the manuscript.
REFERENCES
- Abeliovich A, Beal M. Parkinsonism genes: culprits and clues. J Neurochem. 2006;99:1062–1072. doi: 10.1111/j.1471-4159.2006.04102.x. [DOI] [PubMed] [Google Scholar]
- Abeliovich A, Schmitz Y, Farinas I, Choi-Lundberg D, Ho WH, Castillo PE, Shinsky N, Verdugo JM, Armanini M, Ryan A, et al. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 2000;25:239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- Andres-Mateos E, Mejias R, Sasaki M, Li X, Lin BM, Biskup S, Zhang L, Banerjee R, Thomas B, Yang L, et al. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) J Neurosci. 2009;29:15846–15850. doi: 10.1523/JNEUROSCI.4357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165:123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, Kurkinen K, Yu SW, Savitt JM, Waldvogel HJ, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Hurley JH. Retromer. Curr Opin Cell Biol. 2008;20:427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dodson MW, Zhang T, Jiang C, Chen S, Guo M. Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum Mol Genet. 2012;21:1350–1363. doi: 10.1093/hmg/ddr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB, Bender WW. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. Journal of neurobiology. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- Gan-Or Z, Bar-Shira A, Dahary D, Mirelman A, Kedmi M, Gurevich T, Giladi N, Orr-Urtreger A. Association of sequence alterations in the putative promoter of RAB7L1 with a reduced parkinson disease risk. Arch Neurol. 2012;69:105–110. doi: 10.1001/archneurol.2011.924. [DOI] [PubMed] [Google Scholar]
- Grill B, Bienvenut WV, Brown HM, Ackley BD, Quadroni M, Jin Y. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron. 2007;55:587–601. doi: 10.1016/j.neuron.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, Yearout D, Kay DM, Doheny KF, Paschall J, Pugh E, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat Genet. 2010;42:781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Cai H, Cookson MR, Gwinn-Hardy K, Singleton A. Genetics of Parkinson's disease and parkinsonism. Ann Neurol. 2006;60:389–398. doi: 10.1002/ana.21022. [DOI] [PubMed] [Google Scholar]
- Heo HY, Kim KS, Seol W. Coordinate Regulation of Neurite Outgrowth by LRRK2 and Its Interactor, Rab5. Exp Neurobiol. 2010;19:97–105. doi: 10.5607/en.2010.19.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, Grant BD, Priess JR. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi S, Moore DJ, Yamamoto R, Minegishi M, Sato K, Togo T, Katsuse O, Uchikado H, Furukawa Y, Hino H, et al. Abnormal localization of leucine-rich repeat kinase 2 to the endosomal-lysosomal compartment in lewy body disease. J Neuropathol Exp Neurol. 2009;68:994–1005. doi: 10.1097/NEN.0b013e3181b44ed8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Lee Y, Lee HJ, Kim JS, Song BS, Huh JW, Lee SR, Kim SU, Kim SH, Hong Y, et al. Implication of mouse Vps26b-Vps29-Vps35 retromer complex in sortilin trafficking. Biochem Biophys Res Commun. 2010;403:167–171. doi: 10.1016/j.bbrc.2010.10.121. [DOI] [PubMed] [Google Scholar]
- Lang AE, Lozano AM. Parkinson's disease. First of two parts. N Engl J Med. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- Latourelle JC, Hendricks AE, Pankratz N, Wilk JB, Halter C, Nichols WC, Gusella JF, Destefano AL, Myers RH, Foroud T. Genomewide linkage study of modifiers of LRRK2-related Parkinson's disease. Mov Disord. 2011;26:2039–2044. doi: 10.1002/mds.23781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu W, Oo TF, Wang L, Tang Y, Jackson-Lewis V, Zhou C, Geghman K, Bogdanov M, Przedborski S, et al. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson's disease. Nat Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, Schjeide BM, Schjeide LM, Meissner E, Zauft U, Allen NC, et al. Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database. PLoS Genet. 2012;8:e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad A, Flores I, Zhang H, Yu R, Staniszewski A, Planel E, Herman M, Ho L, Kreber R, Honig LS, et al. Retromer deficiency observed in Alzheimer's disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc Natl Acad Sci U S A. 2008;105:7327–7332. doi: 10.1073/pnas.0802545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CH, Mok SZ, Koh C, Ouyang X, Fivaz ML, Tan EK, Dawson VL, Dawson TM, Yu F, Lim KL. Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. J Neurosci. 2009;29:11257–11262. doi: 10.1523/JNEUROSCI.2375-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli G, Condliffe SB, Bauer M, Giesert F, Boldt K, De Astis S, Meixner A, Sarioglu H, Vogt-Weisenhorn DM, Wurst W, et al. LRRK2 controls synaptic vesicle storage and mobilization within the recycling pool. J Neurosci. 2011;31:2225–2237. doi: 10.1523/JNEUROSCI.3730-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn H, Marchand-Leroux C, Croci N, Plotkine M, Scherman D, Escriou V. Housekeeping while brain's storming Validation of normalizing factors for gene expression studies in a murine model of traumatic brain injury. BMC Mol Biol. 2008;9:62. doi: 10.1186/1471-2199-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJ, Raposo G, van der Sluijs P, Bonifacino JS. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol. 2008;183:513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi-Nakashima A, Meir JY, Jin Y, Matsumoto K, Hisamoto N. LRK-1, a C. elegans PARK8-related kinase, regulates axonal-dendritic polarity of SV proteins. Current biology : CB. 2007;17:592–598. doi: 10.1016/j.cub.2007.01.074. [DOI] [PubMed] [Google Scholar]
- Seaman CF, SaMNJ The Role of Retromer in Neurodegenerative Disease. Intracellular Traffic and Neurodegenerative Disorders. 2009:125–140. [Google Scholar]
- Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu F, Katagiri T, Suzuki M, Watanabe TK, Okuno S, Kuga Y, Nagata M, Fujiwara T, Nakamura Y, Takahashi E. Cloning and chromosome assignment to 1q32 of a human cDNA (RAB7L1) encoding a small GTP-binding protein, a member of the RAS superfamily. Cytogenet Cell Genet. 1997;77:261–263. doi: 10.1159/000134591. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano S, Liu X, Galan JE. Proteolytic targeting of Rab29 by an effector protein distinguishes the intracellular compartments of human-adapted and broad-host Salmonella. Proc Natl Acad Sci U S A. 2011;108:18418–18423. doi: 10.1073/pnas.1111959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. George-Hyslop P, Mobley WC, Christen Y. Intracellular traffic and neurodegenerative disorders. Berlin; London: Springer; 2009. [Google Scholar]
- Stafa K, Trancikova A, Webber PJ, Glauser L, West AB, Moore DJ. GTPase activity and neuronal toxicity of Parkinson's disease-associated LRRK2 is regulated by ArfGAP1. PLoS Genet. 2012;8:e1002526. doi: 10.1371/journal.pgen.1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayanidhi N, Helm JR, Nycz DC, Bentley M, Liang Y, Hay JC. Alpha-synuclein delays endoplasmic reticulum (ER)-to-Golgi transport in mammalian cells by antagonizing ER/Golgi SNAREs. Mol Biol Cell. 2010;21:1850–1863. doi: 10.1091/mbc.E09-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Pisani A, Martella G, Karouani M, Yamaguchi H, Pothos EN, Shen J. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proc Natl Acad Sci U S A. 2009;106:14622–14627. doi: 10.1073/pnas.0906334106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarino-Guell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, et al. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP. Thirty-one flavors of Drosophila rab proteins. Genetics. 2007;176:1307–1322. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson's disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Benet-Pages A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.