Abstract
Bardet–Biedl syndrome (BBS) is a rare, primarily autosomal-recessive ciliopathy. The phenotype of this pleiotropic disease includes retinitis pigmentosa, postaxial polydactyly, truncal obesity, learning disabilities, hypogonadism and renal anomalies, among others. To date, mutations in 15 genes (BBS1–BBS14, SDCCAG8) have been described to cause BBS. The broad genetic locus heterogeneity renders mutation screening time-consuming and expensive. We applied a strategy of DNA pooling and subsequent massively parallel resequencing (MPR) to screen individuals affected with BBS from 105 families for mutations in 12 known BBS genes. DNA was pooled in 5 pools of 21 individuals each. All 132 coding exons of BBS1–BBS12 were amplified by conventional PCR. Subsequent MPR was performed on an Illumina Genome Analyzer II™ platform. Following mutation identification, the mutation carrier was assigned by CEL I endonuclease heteroduplex screening and confirmed by Sanger sequencing. In 29 out of 105 individuals (28%), both mutated alleles were identified in 10 different BBS genes. A total of 35 different disease-causing mutations were confirmed, of which 18 mutations were novel. In 12 additional families, a total of 12 different single heterozygous changes of uncertain pathogenicity were found. Thus, DNA pooling combined with MPR offers a valuable strategy for mutation analysis of large patient cohorts, especially in genetically heterogeneous diseases such as BBS.
Introduction
Bardet–Biedl syndrome (BBS; OMIM# 209900) is a clinically pleiotropic disorder caused by defects of primary cilia (Zaghloul and Katsanis 2009). The cardinal diagnostic criteria are retinitis pigmentosa, postaxial polydactyly, truncal obesity, learning disabilities, abnormalities of the urogenital tract and renal anomalies (Baker and Beales 2009; Bardet 1920; Biedl 1922). Moreover, a large spectrum of secondary features can occur, which have been updated in a recent review by Baker and Beales (2009).
The prevalence of BBS is low in the general population, ranging from 1:125,000 to 1:160,000 in Europe (Beales et al. 1997; Haim 1992; Klein and Ammann 1969), and 1:65,000 in Arab populations (Farag and Teebi 1988). A higher incidence is found in certain isolated populations, such as those in Newfoundland, Kuwait and the Faroe islands (Hjortshoj et al. 2010; Moore et al. 2005; Teebi 1994).
To date, mutations in 15 genes (BBS1–BBS12, MKS1, CEP290 and SDCCAG8) have been shown to cause BBS under an autosomal recessive mode of inheritance (Ansley et al. 2003; Badano et al. 2003a; Chiang et al. 2004, 2006; Katsanis et al. 2000; Leitch et al. 2008; Li et al. 2004; Mykytyn et al. 2001, 2002; Nishimura et al. 2001, 2005; Otto et al. 2010a; Slavotinek et al. 2000; Stoetzel et al. 2006, 2007; Young et al. 1999). Mutations in known BBS genes are found in about 75% of families, of which BBS1 and BBS10 each account for 20–25% (Beales et al. 2003; Stoetzel et al. 2006), BBS12 for about 5% (Stoetzel et al. 2007), and each of the other 12 genes for less than 5% (Chiang et al. 2006; Katsanis 2004) in Caucasians. Second-site phenotypic modification, whereby mutations at a second gene modulate the penetrance and/or expressivity of recessive mutations at a primary locus, has been suggested to play a role in certain cases (Badano et al. 2006; Katsanis et al. 2001; Khanna et al. 2009). Two common mutations have been described: p.M390R in BBS1 (Mykytyn et al. 2002) and p.C91fsX95 in BBS10 (Stoetzel et al. 2006). The BBS proteins can roughly be divided into two groups. BBS1, -2, -4, -5, -7, -8 and -9 form a complex called the BBSome, which cooperates with the GTPase Rab8 to promote ciliogenesis (Nachury et al. 2007). A second group is formed by the chaperonin-like proteins BBS6, -10 and -12, which represent a vertebrate-specific branch of the type II chaperonin superfamily (Billingsley et al. 2010; Stoetzel et al. 2007).
The broad genetic locus heterogeneity in BBS renders mutational analysis expensive and time-consuming. Genome-wide homozygosity mapping greatly reduces the number of genes to be sequenced by defining candidate regions of homozygosity by descent, but can only be applied in consanguineous families (Hildebrandt et al. 2009; Lander and Botstein 1987; Nishimura et al. 2005). Here, we report the use of a combined approach of DNA pooling and massively parallel resequencing (MPR) (Otto et al. 2010b) to screen individuals from 105 families from both inbred and outbred backgrounds for mutations in BBS1–BBS12. Mutations were assigned to their mutation carrier using CEL I endonuclease heteroduplex screening and confirmed by direct Sanger sequencing (Otto et al. 2008). In 29 out of 105 families (28%), two mutated alleles in 10 different BBS genes were identified. In these 29 families, a total of 35 different pathogenic mutations were identified, 18 of which were novel. Two families carried a novel change of uncertain pathogenicity in addition to two mutated alleles. In addition, 12 different single heterozygous mutations, of which 4 have previously been published, were found in 12 out of 105 (11%) families. Thus, DNA pooling combined with MPR offers a valuable strategy for mutation analysis of large patient cohorts of both inbred and outbred backgrounds, especially in heterogeneous diseases such as BBS.
Patients and methods
Patients and DNA pooling
DNA samples and clinical information were obtained after receiving informed consent from 132 individuals diagnosed with BBS from 105 different families. The diagnoses were ascertained according to previously established criteria (Baker and Beales 2009). This study was approved by the Internal Review Board of the University of Michigan, the Ethics Committee of the UCL Institute of Child Health and The Internal Review Boards of the Johns Hopkins University and Duke University. Previous mutation screening by linkage and homozygosity mapping in a subset of patients could not identify two mutated alleles in a known BBS gene (Harville et al. 2010; Katsanis et al. 2001; Stoetzel et al. 2006). Evidence for consanguinity was found in 16 families after total genome search for linkage was performed in 28/105 families. DNA of 105 families (one affected individual per family) was divided over 5 pools with 21 individuals each. Genomic DNA of 21 individuals was pooled with 2 μg per individual and diluted to 60 ng/μl. In addition, an equimolar DNA pool was generated by pooling 96 DNA samples derived from healthy individuals of Caucasian origin [Human Random Control DNA Panel-1 (HRC-1); European Collection of Cell Cultures, Salisbury, UK].
PCR amplification and massively parallel resequencing
DNA pools were used as templates to individually amplify all 132 exons of the genes BBS1-BBS12 by PCR (primer sequences and PCR conditions available upon request). For each pool, PCR products were combined, enzymatically modified and constructed into an Illumina sequencing library as previously described (Otto et al. 2010b). Each library was sequenced using a single lane on an Illumina Genome Analyzer II™ platform generating between about 9 and 26 million reads of 39 bases each. Reads were aligned to the hg18 genomic sequence of the 132 target exons ±100 bp adjacent intronic sequence (http://genome.ucsc.edu) using CLC Genomics Workbench software™ (CLC-bio, Aarhus, Denmark).
Mutation detection and carrier identification
For patient pools #2–4 and the healthy control pool, variant calls were obtained using the following filter parameters: coverage ≥300×, variant frequency ≥0.7%, and a minimum variant count of five reads (including duplicate reads). For patient pool #1 and 5, variant calls were obtained using the following filter parameters: coverage ≥300×, variant frequency ≥0.9%, and a minimum variant count of five reads (Supplementary Table 1). Variants present in dbSNP130, the “1,000 Genomes Project” (180 control individuals, http://www.1000genomes.org/page.php) or in the healthy control pool of 96 individuals (HRC-1) were excluded from further analysis. To prioritize for pathogenic mutations, only variants that were likely to truncate the protein (nonsense, frameshift or obligatory splice site mutations) or missense mutations predicted to be possibly damaging with a PolyPhen score of at least 1.4 were further analyzed. Polymorphism Phenotyping (Poly-Phen) is a software tool used to predict the effect of a non-synonymous SNP on protein structure and function (http://genetics.bwh.harvard.edu/pph/; Ramensky et al. 2002). These variants were amplified by PCR for each patient in the respective DNA pool and analyzed by either CEL I endonuclease heteroduplex screening (Otto et al. 2008) with subsequent Sanger sequencing or by Sanger sequencing alone to determine the mutation carrier. For all individuals in whom only one mutated allele was discovered, all exons of the respective gene were sequenced by direct Sanger sequencing.
Results
In an ethnically diverse cohort of individuals diagnosed with BBS, we screened for mutations in the genes BBS1–BBS12 using a combined approach of DNA pooling and MPR. DNA of 105 individuals was divided over five pools and amplified by PCR. All PCR products of each pool were subjected to MPR on a single lane of an Illumina Genome Analyzer II™ platform. This yielded on average 19,534,601 reads (ranging from 8,995,581 to 25,708,164) of 39 bases in length, of which 77% of all reads mapped back to one of the 132 exons of BBS1–BBS12 ±100 bp adjacent intronic sequence after alignment to the NCBI36/hg18 human reference sequence. The median coverage for coding nucleotides was 8,554 (mean 19,445) reads, resulting in an average coverage depth of 203 (median 452) reads per site, per single allele. For all five pools, an average of 96% of nucleotides met a 300-fold minimal coverage depth. This translates into an average count of 7 per allele, which is sufficient to call a heterozygous change in 1 out of 42 pooled alleles (21 patients) (Supplementary Table 1).
MPR mutation analysis of all PCR products from 21 individuals in a representative pool (pool #3) yielded a total of 51 non-synonymous variants from normal reference sequence (VRS) (Supplementary Table 1). Of these, 17 were known SNPs and 5 were present in a cohort of 96 Caucasian healthy control individuals and are thought to be as yet unannotated SNPs or sequencing and/or alignment artifacts. Of the remaining 29 changes, 17 changes were predicted to truncate the protein product or affect obligatory splice sites, or had a PolyPhen score higher than 1.4, and were thus assumed to affect the function of the encoded protein. These changes were followed up by CEL I endonuclease heteroduplex screening and subsequent Sanger sequencing, confirming a total of six different changes (Supplementary Table 1). For six cases in which CEL I endonuclease heteroduplex screening did not indicate a mutation carrier, direct Sanger sequencing of all patients in the pool was performed and confirmed one additional change. If initially only one mutated allele was identified in a patient, all exons of the gene involved were sequenced by direct Sanger sequencing in order to find a second mutated allele, which led to the identification of two additional mutations. An analogous approach was applied to the remaining four pools (Supplementary Table 1). In all five pools together, 49 different changes out of 143 possibly damaging changes were confirmed (Supplementary Table 1), of which 28 were novel findings. Of these, 18 mutations are considered to be pathogenic, while 10 changes are of uncertain pathogenicity. This increases the number of mutations known to be involved in BBS from 276 to 294 (6.5%).
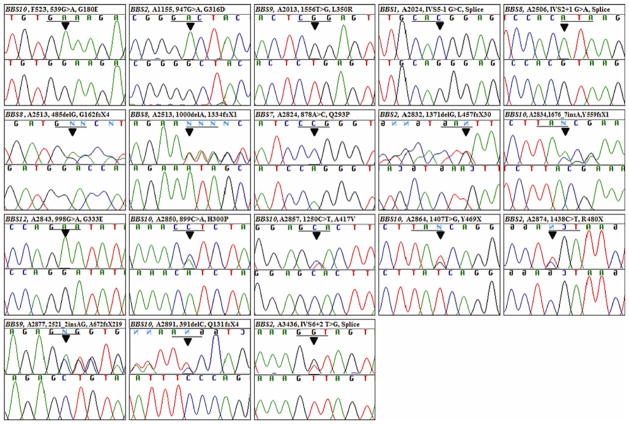
Both mutated alleles were identified in 29 out of 105 families (Table 1). The primary genes most frequently involved were BBS10 (9 families) and BBS2 (8 families). The other mutated genes were BBS1 (4 families), BBS4 (1 family), BBS5 (1 family), BBS7 (1 family), BBS8 (2 families), BBS9 (2 families) and BBS12 (1 family). No primary mutations were identified in BBS3 (ARL6), BBS6 (MKKS) or BBS11 (TRIM32). Of the 37 different mutations identified (Table 1), 35 mutations were assumed to be pathogenic, whereas two changes were present as third alleles and of unknown pathogenicity. Of the 35 pathogenic mutations, 18 were novel (Fig. 1). The novel mutations were present in eight different BBS genes (BBS1, -2, -6, -7, -8, -9, -10, and -12). They are distributed as follows: two nonsense mutations (p.R480X in BBS2; and p.Y469X in BBS10), six small insertions/deletions leading to a frame-shift (p.L457fsX30 in BBS2; p.G162fsX4 and p.I334fsX1 in BBS8; p.A672fsX219 in BBS9; and p.Q131fsX4 and p.Y559fsX1 in BBS10), three obligatory splice site mutations (c.IVS5-1G > C in BBS1; c.IVS6+2 in BBS2, and c.IVS2+1G > A in BBS8) and seven missense mutations (p.G316D in BBS2; p.Q293P in BBS7; p.L350R in BBS9; p.G180E, p.H300P and p.A417V in BBS10; and p.G333E in BBS12). Of these, 7 were found in the homozygous state and 11 in the compound heterozygous state (Table 1).
Table 1.
Genotypes and phenotypes of 39 individuals with BBS (29 families) with both mutated alleles detected in one of 12 known BBS genes (BBS1–BBS12)
| Family [Individual (alias)] | Cardinal features
|
Secondary signs | Origin | BBS genea | Nucleotide change (zygosity state)b | Amino acid change (segregation) | Count/coverage (% frequency) | Mutation assignment methodc | Mutation reference | PolyPhen PSIC score | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Retinitis pigmentosa | Postaxial polydactyly | Obesity | Hypogonadism | Renal anomalies | |||||||||||
| PB206 (A2024) | -II1 | ND | Y | Y | Y | Y | DD, MR | Turkey | 1 | c.IVS5-1 G>C (H) | Splice site | 891/24,085 (3.7%) | All pat seq | Present study | n/a |
| AR61 (A2826) | -04 | Y | N | Y | N | N | LD, UTIS | N. Eur. | 1 | c.223_224delCT (h) | p.L75fsX23 | 85/9,054 (0.9%) | CEL I | Beales et al. 2003 | n/a |
| 1 | c.1169T>G (h) | p.M390R | – | All exon seq | Mykytyn et al. 2002 | 2.703 | |||||||||
| -05 | Y | Y | Y | N | N | LD, SD | |||||||||
| 1 | c.616T>G (h) | p.L206V | – | All exon seq | Present study | 1.099 | |||||||||
| AR122 (A2831)d | -04 | Y | N | Y | Y | Y | MR | N. Eur. | 1 | c.436C>T (h) | p.R146X | 24/1,589 (1.5%) | CEL I | Beales et al. 2003 | n/a |
| 1 | c.1169T>G (h) | p.M390R | – | All exon seq | Mykytyn et al. 2002 | 2.703 | |||||||||
| AR603 (A2858) | -03 | Y | Y | Y | N | ND | LD, PP, SD | N. Eur. | 1 | c.436C>T (h) | p.R146X | 10/1,119 (0.9%) | All pat seq | Beales et al. 2003 | n/a |
| -05 | Y | Y | Y | ND | ND | ND | 1 | c.1169 T>G (h) | p.M390R | – | All exon seq | Mykytyn et al. 2002 | 2.703 | ||
| A1155 | -II1 | N | Y | ND | N | Y | ASD | N. Eur. | 2 | c.947G>A (H) | p.G316D | 235/6,551 (3.6%) | All pat seq | Present study | 2.158 |
| A1885 | -II1 | Y | N | ND | ND | Y | – | USA | 2 | c.823C>T (h) | p.R275X | 787/5,356 (14.7%) | All pat seq | Katsanis et al. 2001 | n/a |
| 2 | c.1895G>C (h) | p.R632P | 232/23,796 (1.0%) | All pat seq | Katsanis et al. 2001 | 2.698 | |||||||||
| A2296 | -II1 | N | Y | ND | ND | Y | ASD, IVS-AZ, SI | USA | 2 | c.823C>T (H) | p.R275X | 787/5,356 (14.7%) | All pat seq | Katsanis et al. 2001 | n/a |
| AR124 (A2832) | -03 | Y | Y | Y | N | Y | DD | N. Eur. | 2 | c.IVS3-1delG (h) | p.V158fsX43e | 6/526 (1.1%) | All pat seq | Katsanis et al. 2001 | n/a |
| -04 | Y | Y | Y | N | Y | DD | 2 | c.1371delG (h) | p.L457fsX30 | – | All exon seq | Present study | n/a | ||
| AR724 (A2866) | -05 | Y | Y | ND | N | Y | BCU, HMC, MD | N. Eur. | 2 | c.175C>T (h) | p.Q59Xf | 72/7,990 (0.9%) | CEL I | Katsanis et al. 2001 | n/a |
| 2 | c.72C>G (h) | p.Y24X | – | All exon seq | Katsanis et al. 2001 | n/a | |||||||||
| AR839 (A2874) | -04 | Y | Y | Y | N | N | CHD, MD, NSM, SD, SI | N. Eur. | 2 | c.1438C>T (h) | p.R480X | 13/358 (3.6%) | CEL I | Present study | n/a |
| 2 | c.1237C>T (h) | p.R413X | 97/5,450 (1.8%) | CEL I | Fauser 2003 | n/a | |||||||||
| AR850 (A2878) | -03 | Y | Y | Y | Y | Y | AST, DD, MC, SD, SYN | N. Eur. | 2 | c.72C>G (h) | p.Y24X | 198/27,120 (0.7%) | CEL I | Katsanis et al. 2001 | n/a |
| 2 | c.504delG (h) | p.L168fsX33 | 12/1,603 (0.7%) | CEL I | Muller et al. 2010 | n/a | |||||||||
| -04 | Y | Y | Y | Y | Y | MC, SD | |||||||||
| 6 | c.1294A>T (h) only in individual ‘-04’ | p.I432F | 137/8,059 (1.7%) | CEL I | Present study | 1.523 | |||||||||
| A3436 | -II1 | N | Y | Y | ND | Y | – | Turkey | 2 | c.940delA (h) | p.I314fsX11 (p) | 59/6,577 (0.9%) | All pat seq | Nishimura et al. 2001 | n/a |
| 2 | c.IVS6+2 (h) | Splice site (m) | 69/8,574 (0.8%) | All pat seq | Present study | n/a | |||||||||
| KK44 (A2884) | -08 | Y | Y | Y | Y | ND | – | Saudi Arabia | 4 | c.IVS3-2A>G (H) | Splice site | 84/4,953 (1.7%) | CEL I | Katsanis et al. 2002 | n/a |
| A2507 | Y | Y | Y | Y | N | AN, FL, MR, PS | Somalia | 5 | c.214 G>A (H) | p.G72S | 45/2,423 (1.9%) | CEL I | Hjortshoj et al. 2008 | 2.112 | |
| AR14 (A2824) | -04 | Y | Y | Y | Y | Y | MR | N. Eur. | 7 | c.878A>C (H) | p.Q293Pg | 261/5,626 (4.6%) | All pat seq | Present study | 2.167 |
| -08 | Y | Y | Y | Y | Y | MR, PN | |||||||||
| A2506 | Y | Y | Y? | N | N | AZ, MR, NE | ND | 8 | c.IVS2+1G>A (H) | Splice site | 393/9,334 (4.2%) | CEL I | Present study | n/a | |
| A2513 | Y | Y | Y | Y | Y | FL, GASTS, MR | Hispanic | 8 | c.485delG (h) | p.G162fsX4 | 47/1,946 (2.4%) | CEL I | Present study | n/a | |
| 8 | c.1000delA (h) | p.I334fsX1 | – | All exon seq | Present study | n/a | |||||||||
| PB233 (A2013) | -II1 | Y | Y | Y | Y | ND | MR | Turkey | 9 | c.1556T>G (H) | p.L350R | 163/4,646 (3.5%) | All pat seq | Present study | 1.885 |
| -II2 | ND | Y | Y | ND | ND | ND | |||||||||
| AR847 (A2877) | -03 | Y | ND | Y | N | Y | AST, DD, MY, SD | N. Eur. | 9 | c.2390_2393delAACA (h) | p.K628fsX22h | – | All exon seq | Nishimura et al. 2005 | n/a |
| -04 | Y | Y | Y | N | Y | DD, LD, Ri, SD | 9 | c.2521_2522insAG (h) | p.A672fsX219 | 14/930 (1.5%) | All pat seq | Present study | n/a | ||
| F523 | -II1 | ND | ND | ND | ND | Y | SM, TCP | N. Eur. | 10 | c.539G>A (H) | p.G180E | 603/15,920 (3.8%) | All pat seq | Present study | 2.172 |
| AR151 (A2834) | -04 | Y | Y | Y | N | ND | LD, SD | N. Eur. | 10 | c.271_272insT (h) | p.C91fsX5 | 33/1,310 (2.5%) | CEL I | Stoetzel et al. 2006 | n/a |
| 10 | c.1676_1677insA (h) | p.Y559fsX1 | 5/604 (0.8%) | CEL I | Present study | n/a | |||||||||
| AR232 (A2839) | -03 | Y | Y | Y | Y | N | SD | N. Eur. | 10 | c.1677delC (h) | p.Y559fsX1 (m)i | 8/673 (1.2%) | CEL I | Stoetzel et al. 2006 | n/a |
| -04 | Y | Y | Y | Y | N | SD | 10 | c.271_272insT (h) | p.C91fsX5 (p) | 33/1,310 (2.5%) | CEL I | Stoetzel et al. 2006 | n/a | ||
| AR371 (A2850) | -03 | Y | Y | Y | N | ND | AMRR, Scol, SD | N. Eur. | 10 | c.271_272insT (h) | p.C91fsX5j | 64/1,396 (4.6%) | CEL I | Stoetzel et al. 2006 | n/a |
| 10 | c.899A>C (h) | p.H300P | 2,056/119,460 (1.7%) | CEL I | Present study | 2.575 | |||||||||
| AR515 (A2857) | -03 | Y | Y | Y | Y | N | CTEV, SD, SS | N. Eur. | 10 | c.271_272insT (h) | p.C91fsX5 | 64/1,396 (4.6%) | CEL I | Stoetzel et al. 2006 | n/a |
| 10 | c.1250C>T (h) | p.A417V | – | All exon seq | Present study | 1.413 | |||||||||
| AR707 (A2864) | -08 | Y | Y | Y | N | Y | HMC, MR | N. Eur. | 10 | c.271_272insT (h) | p.C91fsX5j | 198/5,633 (3.5%) | CEL I | Stoetzel et al. 2006 | n/a |
| -09 | Y | Y | Y | N | Y | ND | 10 | c.1407T>G (h) | p.Y469X (p) | 451/23,019 (2.0%) | CEL I | Present study | n/a | ||
| AR880 (A2881) | -04 | Y | Y | Y | N | N | Cons, SD, UTIS | N. Eur. | 10 | c.271_272insT (H) | p.C91fsX5 | 64/1,396 (4.6%) | CEL I | Stoetzel et al. 2006 | n/a |
| F1 (A2891) | -04 | Y | Y | Y | Y | Y | BD, COM, DD, HT, SYN | N. Eur. | 10 | c.271_272insT (h) | p.C91fsX5 | 198/5,633 (3.5%) | All pat seq | Stoetzel et al. 2006 | n/a |
| -05 | Y | Y | Y | Y | Y | BD, COM, DD, IM, SYN | 10 | c.391delC (h) | p.Q131fsX4 | 1,036/78,357 (1.3%) | All pat seq | Present study | n/a | ||
| A3185 | -II1 | Y | Y | Y | Y | Y | BE, SD | USA | 10 | c.687delA (h) | p.P229fsX8 | 750/55,490 (1.4%) | All pat seq | Stoetzel et al. 2006 | n/a |
| 10 | c.271_272insT (h) | p.C91fsX5 | 203/16,449 (1.2%) | All pat seq | Stoetzel et al. 2006 | n/a | |||||||||
| AR248 (A2843) | -05 | Y | Y | Y | Y | Y | COM, SD | N. Eur. | 12 | c.998G>A (H) | p.G333E | 193/14,768 (1.3%) | All pat seq | Present study | 2.274 |
AMRR amenorrhea, AN anosmia, ASD atrial septal defect, AST astigmatism, AZ asthma, BCU bicornuate uterus, BD brachydactyly, BE bifid epiglottis, CHD congenital heart disease, COM chronic otitis media, Cons constipation, CTEV congenital talipes equinovarus, DD developmental delay, FL fatty liver, GASTS gall stones, h heterozygous change, H homozygous change, HAP high arched palate, HMC hydrometrocolpos, HT hypotonia, IM intestinal malrotation, IVC-AZ inferior vena cava interruption with azygos continuation, LD learning disabilities, MC macrocephaly, MD motor delay, MR mental retardation, MY (severe) myopia, n/a not available, ND no data, NE nasal encephalocele, N. Eur. Northern Europe, NSM narrow small mouth, PN peripheral neuropathy, PP precocious puberty, PS psychosis, Ri rickets, Scol scoliosis, SD speech delay, SDys sensory dysesthesia, SI situs inversus, SM splenomegaly, SS short stature, SYN syndactyly, TCP thrombocytopenia, UTIS recurrent urinary tract infections
Accession numbers: BBS1, NM_024649; BBS2, NM_031885; BBS3, NM_177976; BBS4, NM_033028; BBS5, NM_152384; BBS6, NM_018848; BBS7, NM_176824; BBS8, NM_198309.2; BBS9, NM_198428; BBS10, NM_024685; BBS11, NM_012210; BBS12, NM_152618
Mutation numbering is based on the cDNA position in reference sequences indicated in Table 1 with +1 corresponding to the A of the ATG translation initiation codon. All changes were absent from the healthy control pool (96 HRC-1 individuals) and the “1,000 genomes project” (180 individuals)
The mutation carrier assignment was performed by heteroduplex based CEL I endonuclease screening (CEL I) or by direct Sanger sequencing of the respective 21 DNA samples (All pat seq). If initially only one mutated allele was found, all exons of the respective gene were sequenced (All exon seq)
Family AR122 (A2831) has previously been published for linkage to the BBS1 locus by Katsanis et al. 1999
Family AR124 (A2832) has previously been published for this heterozygous change in BBS2 (p.V158fsX43) by Katsanis et al. 2001
Family AR724 (A2866) has previously been published for this heterozygous change in BBS2 (p.Q59X) by Katsanis et al. 2001
Family AR14 (A2824) has previously been published for a heterozygous change in BBS12 (p.G540D) present in individual ‘-04’ (but not ‘-08’) by Stoetzel et al. 2007
The mutation p.K628fsX22 has previously been published by Nishimura et al. 2005 as p.K626fsX647
Family AR232 (A2839) has previously been published for this heterozygous change in BBS10 (p.Y559fsX1) by Stoetzel et al. 2006
Family AR371 (A2850) and AR707 (A2864) have previously been published for this heterozygous change in BBS10 (p.C91fsX5) by Stoetzel et al. 2006
Fig. 1.
Sequence chromatograms of 18 different novel mutations identified in individuals with BBS. Gene name, patient identifier, nucleotide change, and inferred amino acid alteration are given above sequence traces. The mutation position is indicated with an arrow. Wild type sequence chromatograms are shown below mutated sequences. Reading frames are underlined
A number of mutations were present in more than one family. Families AR14 (Northern Europe) and AR634 (Northern Europe) shared the same novel change in BBS7 (p.Q293P). In families AR724 (Northern Europe) and AR850 (Northern Europe), the same heterozygous change in BBS2 (p.Y24X) was present. Likewise, another heterozygous nonsense mutation (p.R275X) in BBS2 was shared between families A1885 (USA) and A2296 (USA). Affected individuals from both families AR122 (Northern Europe) and AR603 (Northern Europe) carried the same compound heterozygous mutations in BBS1 (p.R146X and p.M390R) (Beales et al. 2003). The previously described recurrent mutation in BBS1, p.M390R (Mykytyn et al. 2002) was present in the heterozygous state in two additional families, AR61 and AR786. A recurrent mutation in BBS10, p.C91fsX95 (Stoetzel et al. 2006) was present in eight families. This change was present in the homozygous state in one family and as a heterozygous change in seven families, in which case the second mutated allele in BBS10 was consistently found. Two affected individuals from family AR61 carried three heterozygous changes in BBS1, two of which have previously been published (p.L75fsX23, p.M390R) (Beales et al. 2003; Mykytyn et al. 2002), while one is novel (p.L206V) and of uncertain pathogenicity. In family AR850, a missense change in BBS6 (p.I432F) with a moderate PolyPhen score (1.523) was present in addition to compound heterozygous mutations in BBS2 (p.Y24X, p.L168fsX33) in one affected individual (AR850-04), but not in the second affected sibling (AR850-03) (Table 1).
In addition to these families in which at least two mutated alleles in one BBS gene were found, 12 different single heterozygous changes of uncertain pathogenicity have been identified in 12 different families (Table 2). These cover the genes BBS1, -2, -4, -5, -6, -7, -9, -10 and -12, and are all missense mutations, four of which have previously been published. In family A2499, two novel heterozygous missense mutations were present in two different genes: BBS1 (p.P245L) and BBS9 (p.L781Q). The affected individual from family A3227 carried one novel heterozygous missense change in BBS10 (p.R530S) and one in BBS12 (p.V503M).
Table 2.
Genotypes and phenotypes of 14 individuals with BBS (12 families) with only one mutated allele detected in a known BBS gene (BBS1–BBS12)
| Family [Individual (alias)] | Cardinal features
|
Secondary signs | Origin | BBS genea | Nucleotide change (zygosity state)b | Amino acid change (segregation) | Count/coverage (% frequency) | Mutation assignmentc | Mutation reference | PolyPhen PSIC score | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Retinitis pigmentosa | Postaxial polydactyly | Obesity | Hypogonadism | Renal anomalies | |||||||||||
| A2499 | Y | Y | N | N | Y | – | ND | 1 | c.734 C>T (h) | p.P245L | 6/382 (1.6%) | All pat seq | Present study | 2.328 | |
| 9 | c.2849 T>A (h) | p.L781Q | 4/425 (0.9%) | CEL I | Present study | 1.885 | |||||||||
| A2517 | Y | Y | N | N | Y | DD, HAP, OB | ND | 1 | c.871 C>T (h) | p.Q291X | 1,607/54,862 (2.9%) | CEL I | Beales et al. 2003 | n/a | |
| AR786 (A2868) | -03 | Y | N | Y | ND | N | MC, MD, MR, SD | N. Eur. | 1 | c.1169 T>G (h) | p.M390R | 2/22 (9.1%) | All pat seq | Mykytyn et al. 2002 | 2.703 |
| -04 | Y | Y | ND | ND | N | MR, SD, SDys | c.1169 T>G (H) | ||||||||
| PB236 (A2010) | -II1 | Y | Y | Y | ND | Y | ATX, BD, STBI | Turkey | 2 | c.1891G>A (h) | p.A631T | 10/494 (2.0%) | All pat seq | Present study | 1.522 |
| AR348 (A2848) | -03 | Y | Y | Y | ND | ND | – | N. Eur. | 4 | c.218A>G (h) | p.Q73R | 74/3,159 (2.3%) | CEL I | Present study | 1.574 |
| AR364 (A2849) | -02 | Y | Y | Y | Y | Y | DD, HNP | N. Eur. | 5 | c.551A>G (h) | p.N184S | 41/1,353 (3.0%) | All pat seq | Li et al. 2004 | 2.211 |
| AR755 (A2867)d | -03 | Y | Y | Y | ND | N | BD, DD, DF, SD | N. Eur. | 5 | c.551A>G (h) | p.N184S | 41/1,353 (3.0%) | All pat seq | Li et al. 2004 | 2.211 |
| A786 | -II1 | N | Y | Y | Y | Y | ASD, MR, MY, Sz | Macedonia | 6 | c.724G>T (h) | p.A242S | 216/12,651 (1.7%) | All pat seq | Stone et al. 2000 | 1.280 |
| AR800 (A2870) | -03 | Y | Y | Y | Y | ND | MR, SS, STBI | N. Eur. | 6 | c.724G>T (h) | p.A242S | 1,878/55,174 (3.4%) | CEL I | Stone et al. 2000 | 1.280 |
| AR634 (A2862) | -II1 | Y | N | Y | Y? | ND | DD, MR, SD | N. Eur. | 7 | c.878A>C (h) | p.Q293P | 261/5,626 (4.6%) | All pat seq | Present study | 2.167 |
| -II2 | Y | N | Y | Y | N | MC, MD, SS | |||||||||
| A3260 | -II1 | N | Y | ND | N | Y | PSp, SI | USA | 9 | c.2983C>T (h) | p.R826C | 1,575/25,567 (6.2%) | All pat seq | Present study | 2.257 |
| A3227 | -II1 | N | N | Y | N | Y | BD, MR, PHP, SS | Egypt | 10 | c.1590A>C (h) | p.R530S | 102/11,606 (0.9%) | All pat seq | Present study | 1.686 |
| 12 | c.1507G>A (h) | p.V503M | 509/37,380 (1.4%) | All pat seq | Present study | 1.430 | |||||||||
ASD atrial septum defect, ATX ataxia, BD brachydactyly, DD developmental delay, DF dysmorphic features, HAP high arched palate, HNP herniated nucleus pulposus, MC macrocephaly, MD motor delay, MR mental retardation, MY (severe) myopia, OB overbite, PHP pseudohypoparathyroidism, PSp polysplenia, SD speech delay, SDys sensory dysesthesia, SI situs inversus, SS short stature, STBI strabismus, Sz seizures, ND no data available, N. Eur. Northern Europe
Accession numbers: BBS1, NM_024649; BBS2, NM_031885; BBS3, NM_177976; BBS4, NM_033028; BBS5, NM_152384; BBS6, NM_018848; BBS7, NM_176824; BBS8, NM_198309.2; BBS9, NM_198428; BBS10, NM_024685; BBS11, NM_012210; BBS12, NM_152618
Mutation numbering is based on the cDNA position in reference sequences indicated in Table 1 with +1 corresponding to the A of the ATG translation initiation codon. All changes were absent in the healthy control pool (96 HRC-1 individuals) and the “1,000 genomes project” (180 individuals)
The mutation carrier assignment was performed by heteroduplex based CEL I endonuclease screening (“CEL I”) or by direct Sanger sequencing of the respective 21 DNA samples (“All pat seq”). If initially only one mutated allele was found, all exons of the respective gene were sequenced (“All exon seq”)
Unpublished mutation analysis of family AR755 (A2867) showed the presence of a homozygous change in BBS1 (p.M390R)
In all cases where DNA from relatives was available, mutations segregated as expected. All changes stated were absent from 96 Caucasian healthy control individuals and were not present in 180 control individuals from the “1,000 Genomes Project” (http://www.1000genomes.org/page.php).
Discussion
Conventional mutation analysis is tedious, and expensive to perform in diseases that exhibit broad genetic locus heterogeneity such as BBS. Most previous large scale mutation analysis studies focused solely on a subset of the known BBS loci (Billingsley et al. 2010; Hjortshoj et al. 2010; Muller et al. 2010). Here, we present the application of a DNA pooling and massively parallel resequencing strategy that we developed recently (Otto et al. 2010b) to identify mutations in patients with BBS. We estimate the cost of this MPR mutation analysis to be in the range of $50–60 per patient. We discovered the disease-causing mutations in 29 out of 105 families (28%). In total, we identified 49 different mutations, 28 of which are novel. Of these, we consider 18 mutations to be pathogenic, while 10 changes are of uncertain pathogenicity. We thereby increase the number of known causative BBS mutations from 276 to 294 (6.5%).
In this study, we were able to identify both disease causing alleles in 28% of our patient cohort. In contrast, Muller et al. 2010 identified both disease causing alleles in 67% of their patient cohort. This discrepancy is most likely due to the fact that the majority of our cohort has previously been screened for homozygous mutations by linkage and homozygosity mapping (Harville et al. 2010; Katsanis et al. 2001; Stoetzel et al. 2006). Furthermore, this may reflect potential differences in the sensitivity or specificity of sequencing technology: our method might miss mutations, if they are positioned in an area of low coverage (<300 reads) or if they are insertions or deletions larger than 2 bp in length. We did not search for copy-number variants (CNV’s) to detect large heterozygous deletions. Mutations such as complete exon deletions, promoter mutations, or intronic exon splice enhancers (ESE) are not detected by our method.
In one family (AR850), we found a third mutated allele in addition to the primary BBS locus which could represent a form of oligogenic inheritance (Badano et al. 2003b). However, this potential modifier allele is a missense mutation of unknown pathogenicity and functional studies are required in order to draw conclusions about its potential modifying effects. In family AR850, the p.I432F allele in BBS6 was only present in one of the two affected siblings who exhibit differences in secondary symptoms (Table 1). It has recently been shown that variants predicted in silico to be neutral, can act as modifiers to exacerbate phenotypes across the ciliopathy spectrum (Badano et al. 2003b; Khanna et al. 2009; Zaghloul et al. 2010). However, we did not investigate the presence of these variants in our patient cohort. Due to the relatively high false positive error rate in next generation sequencing, we prioritized our analysis to detect mutations that are predicted to functionally damage the encoded proteins.
Our samples (pool #1–5) were sequenced successively over a period of 6 months, during which a clear improvement of next-generation sequencing was observed, with an error rate reduction due to higher coverage. The number of false positive variants in the pool sequenced first (pool #1) [29/43 (67%)] was remarkably higher than in the pool sequenced 6 months later (pool #5) [1/12 (8%)] (Supplementary Table 1). This considerable decrease in false positive variants in pool #5 allowed us to assign variants to their mutation carrier by direct Sanger sequencing alone, without tedious CEL I endonuclease heteroduplex screening. In addition, the number of variants that we discovered upon sequencing of the whole gene if a first mutation was found in a family (“false negative variants”) decreased over the course of our study, with 3 false-negatives in pool # 1 [3/43 (7%)] and none in pool #5 [0/12 (0%)]. Future application of paired-end technology would reduce the number of false-positive and negative calls even further, which would be of major importance for clinical utility. Other technological improvements to next-generation sequencing such as increasing read length and read number will also substantially cut down on the number of false-positive and negative variants detected. This will enable comprehensive, robust, rapid and cost effective mutation analysis for many patients across genetically heterogeneous diseases.
Supplementary Material
Acknowledgments
The authors wish to thank the physicians and families for participating in this study. We would like to thank Dr. E. Héon (Toronto) and Dr. J.L. Duncan (San Francisco) for their contribution of DNA and clinical data from patients. We also thank Robert Lyons and the University of Michigan DNA Sequencing Core for excellent Illumina sequencing. F.H. is an Investigator of the Howard Hughes Medical Institute, a Doris Duke Distinguished Clinical Scientist, and a Frederick G. L. Huetwell Professor. N.K. is a Distinguished George W. Brumley professor. This research was supported by grants from the National Institutes of Health to F.H. (DK1069274, DK1068306, DK064614) and N.K. (HD04260, DK072301, DK075972). P.L.B. is funded by Wellcome Trust.
Abbreviations
- BBS
Bardet–Biedl Syndrome
- bp
Base pairs
- GTP
Guanosine triphosphate
- HRC-1
Human random control DNA panel-1
- MPR
Massively parallel resequencing
- nt
Nucleotides
- PCR
Polymerase chain reaction
- VRS
Variants from reference sequence
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-010-0902-8) contains supplementary material, which is available to authorized users.
Contributor Information
Sabine Janssen, Departments of Pediatrics and of Human Genetics, University of Michigan, Ann Arbor, MI, USA.
Gokul Ramaswami, Departments of Pediatrics and of Human Genetics, University of Michigan, Ann Arbor, MI, USA.
Erica E. Davis, Center for Human Disease Modeling, Duke University Medical Center, Durham, NC, USA. Department of Pediatrics, Duke University Medical Center, Durham, NC, USA
Toby Hurd, Departments of Pediatrics and of Human Genetics, University of Michigan, Ann Arbor, MI, USA.
Rannar Airik, Departments of Pediatrics and of Human Genetics, University of Michigan, Ann Arbor, MI, USA.
Jennifer M. Kasanuki, Departments of Pediatrics and of Human Genetics, University of Michigan, Ann Arbor, MI, USA
Lauren Van Der Kraak, Department of Genetics and Genome Biology, The Hospital for Sick Children, Toronto, Ontario, Canada.
Susan J. Allen, Departments of Pediatrics and of Human Genetics, University of Michigan, Ann Arbor, MI, USA
Philip L. Beales, Molecular Medicine Unit, UCL Institute of Child Health, London, UK
Nicholas Katsanis, Center for Human Disease Modeling, Duke University Medical Center, Durham, NC, USA. Department of Pediatrics, Duke University Medical Center, Durham, NC, USA. Cell biology, Duke University Medical Center, Durham, NC, USA.
Edgar A. Otto, Departments of Pediatrics and of Human Genetics, University of Michigan, Ann Arbor, MI, USA
Friedhelm Hildebrandt, Email: fhilde@umich.edu, Departments of Pediatrics and of Human Genetics, University of Michigan, Ann Arbor, MI, USA. Howard Hughes Medical Institute, Chevy Chase, MD, USA. University of Michigan Health System, 8220C MSRB III, 1150 West Medical Center Drive, Ann Arbor, MI 48109-5646, USA.
References
- Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, Mah AK, Johnsen RC, Cavender JC, Lewis RA, Leroux MR, Beales PL, Katsanis N. Basal body dysfunction is a likely cause of pleiotropic Bardet–Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Badano JL, Ansley SJ, Leitch CC, Lewis RA, Lupski JR, Katsanis N. Identification of a novel Bardet–Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am J Hum Genet. 2003a;72:650–658. doi: 10.1086/368204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Kim JC, Hoskins BE, Lewis RA, Ansley SJ, Cutler DJ, Castellan C, Beales PL, Leroux MR, Katsanis N. Heterozygous mutations in BBS1, BBS2 and BBS6 have a potential epistatic effect on Bardet–Biedl patients with two mutations at a second BBS locus. Hum Mol Genet. 2003b;12:1651–1659. doi: 10.1093/hmg/ddg188. [DOI] [PubMed] [Google Scholar]
- Badano JL, Leitch CC, Ansley SJ, May-Simera H, Lawson S, Lewis RA, Beales PL, Dietz HC, Fisher S, Katsanis N. Dissection of epistasis in oligogenic Bardet–Biedl syndrome. Nature. 2006;439:326–330. doi: 10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- Baker K, Beales PL. Making sense of cilia in disease: the human ciliopathies. Am J Med Genet C Semin Med Genet. 2009;151C:281–295. doi: 10.1002/ajmg.c.30231. [DOI] [PubMed] [Google Scholar]
- Bardet G. On congenital obesity syndrome with polydactyly and retinitis pigmentosa (a contribution to the study of clinical forms of hypophyseal obesity) Obes Res. 1920;3:387–399. doi: 10.1002/j.1550-8528.1995.tb00165.x. [DOI] [PubMed] [Google Scholar]
- Beales PL, Warner AM, Hitman GA, Thakker R, Flinter FA. Bardet–Biedl syndrome: a molecular and phenotypic study of 18 families. J Med Genet. 1997;34:92–98. doi: 10.1136/jmg.34.2.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales PL, Badano JL, Ross AJ, Ansley SJ, Hoskins BE, Kirsten B, Mein CA, Froguel P, Scambler PJ, Lewis RA, Lupski JR, Katsanis N. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet–Biedl syndrome. Am J Hum Genet. 2003;72:1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedl A. A pair of siblings with adiposo-genital dystrophy. Obes Res. 1922;3:404. doi: 10.1002/j.1550-8528.1995.tb00167.x. [DOI] [PubMed] [Google Scholar]
- Billingsley G, Bin J, Fieggen KJ, Duncan JL, Gerth C, Ogata K, Wodak SS, Traboulsi EI, Fishman GA, Paterson A, Chitayat D, Knueppel T, Millan JM, Mitchell GA, Deveault C, Heon E. Mutations in chaperonin-like BBS genes are a major contributor to disease development in a multiethnic Bardet–Biedl syndrome patient population. J Med Genet. 2010;47:453–463. doi: 10.1136/jmg.2009.073205. [DOI] [PubMed] [Google Scholar]
- Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, Sheffield VC. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet–Biedl syndrome (BBS3) Am J Hum Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AP, Beck JS, Yen HJ, Tayeh MK, Scheetz TE, Swiderski RE, Nishimura DY, Braun TA, Kim KY, Huang J, Elbedour K, Carmi R, Slusarski DC, Casavant TL, Stone EM, Sheffield VC. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet–Biedl syndrome gene (BBS11) Proc Natl Acad Sci USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag TI, Teebi AS. Bardet–Biedl and Laurence–Moon syndromes in a mixed Arab population. Clin Genet. 1988;33:78–82. doi: 10.1111/j.1399-0004.1988.tb03414.x. [DOI] [PubMed] [Google Scholar]
- Fauser S, Munz M, Besch D. Further support for digenic inheritance in Bardet-Biedl syndrome. J Med Genet. 2003;40:e104. doi: 10.1136/jmg.40.8.e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim M. Prevalence of retinitis pigmentosa and allied disorders in Denmark. II. Systemic involvement and age at onset. Acta Ophthalmol (Copenh) 1992;70:417–426. doi: 10.1111/j.1755-3768.1992.tb02109.x. [DOI] [PubMed] [Google Scholar]
- Harville HM, Held S, Diaz-Font A, Davis EE, Diplas BH, Lewis RA, Borochowitz ZU, Zhou W, Chaki M, MacDonald J, Kayserili H, Beales PL, Katsanis N, Otto E, Hildebrandt F. Identification of 11 novel mutations in eight BBS genes by high-resolution homozygosity mapping. J Med Genet. 2010;47:262–267. doi: 10.1136/jmg.2009.071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Heeringa SF, Rüschendorf F, Attanasio M, Nürnberg G, Becker C, Seelow D, Huebner N, Chernin G, Vlangos CN, Zhou W, O’Toole JF, Hoskins BE, Wolf MTF, Hinkes BG, Chaib H, Ashraf S, Allen SJ, Vega-Warner V, Wise E, Harville HM, Lyons RH, Washburn J, MacDonald J, Nürnberg P, Otto EA. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PloS Genet. 2009;5:31000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjortshøj TD, Grønskov K, Philp AR, Nishimura DY, Adeyemo A, Rotimi CN, Sheffield VC, Rosenberg T, Brøndum-Nielsen K. Novel mutations in BBS5 highlight the importance of this gene in non-Caucasian Bardet–Biedl syndrome patients. Am J Med Genet. 2008;146:517–520. doi: 10.1002/ajmg.a.32136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjortshoj TD, Gronskov K, Philp AR, Nishimura DY, Riise R, Sheffield VC, Rosenberg T, Brondum-Nielsen K. Bardet–Biedl syndrome in Denmark—report of 13 novel sequence variations in six genes. Hum Mutat. 2010;31:429–436. doi: 10.1002/humu.21204. [DOI] [PubMed] [Google Scholar]
- Katsanis N. The oligogenic properties of Bardet–Biedl syndrome. Hum Mol Genet. 2004;13(Spec1):R65–R71. doi: 10.1093/hmg/ddh092. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Lewis RA, Stockton DW, Mai PM, Baird L, Beales PL, Leppert M, Lupski JR. Delineation of the critical interval of Bardet-Biedl syndrome 1 (BBS1) to a small region of 11q13, through linkage and haplotype analysis of 91 pedigrees. Am J Hum Genet. 1999;65:1672–1679. doi: 10.1086/302684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, Parfrey PS, Ansley SJ, Davidson WS, Lupski JR. Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet–Biedl syndrome. Nat Genet. 2000;26:67–70. doi: 10.1038/79201. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR. Triallelic inheritance in Bardet–Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Eichers ER, Ansley SJ, Lewis RA, Kayserili H, Hoskins BE, Scambler PJ, Beales PL, Lupski JR. BBS4 is a minor contributor to Bardet–Biedl syndrome and may also participate in triallelic inheritance. Am J Hum Genet. 2002;71:22–29. doi: 10.1086/341031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna H, Davis EE, Murga-Zamalloa CA, Estrada-Cuzcano A, Lopez I, den Hollander AI, Zonneveld MN, Othman MI, Waseem N, Chakarova CF, Maubaret C, Diaz-Font A, Macdonald I, Muzny DM, Wheeler DA, Morgan M, Lewis LR, Logan CV, Tan PL, Beer MA, Inglehearn CF, Lewis RA, Jacobson SG, Bergmann C, Beales PL, Attie-Bitach T, Johnson CA, Otto EA, Bhattacharya SS, Hildebrandt F, Gibbs RA, Koenekoop RK, Swaroop A, Katsanis N. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D, Ammann F. The syndrome of Laurence–Moon–Bardet–Biedl and allied diseases in Switzerland. Clinical, genetic and epidemiological studies. J Neurol Sci. 1969;9:479–513. doi: 10.1016/0022-510x(69)90091-4. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- Leitch CC, Zaghloul NA, Davis EE, Stoetzel C, Diaz-Font A, Rix S, Alfadhel M, Lewis RA, Eyaid W, Banin E, Dollfus H, Beales PL, Badano JL, Katsanis N. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet–Biedl syndrome. Nat Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Moore SJ, Green JS, Fan Y, Bhogal AK, Dicks E, Fernandez BA, Stefanelli M, Murphy C, Cramer BC, Dean JC, Beales PL, Katsanis N, Bassett AS, Davidson WS, Parfrey PS. Clinical and genetic epidemiology of Bardet–Biedl syndrome in Newfoundland: a 22-year prospective, population-based, cohort study. Am J Med Genet A. 2005;132:352–360. doi: 10.1002/ajmg.a.30406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Stoetzel C, Vincent MC, Leitch CC, Laurier V, Danse JM, Helle S, Marion V, Bennouna-Greene V, Vicaire S, Megarbane A, Kaplan J, Drouin-Garraud V, Hamdani M, Sigaudy S, Francannet C, Roume J, Bitoun P, Goldenberg A, Philip N, Odent S, Green J, Cossee M, Davis EE, Katsanis N, Bonneau D, Verloes A, Poch O, Mandel JL, Dollfus H. Identification of 28 novel mutations in the Bardet–Biedl syndrome genes: the burden of private mutations in an extensively heterogeneous disease. Hum Genet. 2010;127:583–593. doi: 10.1007/s00439-010-0804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, Riise R, Baldi A, Raas-Rothschild A, Gorman SW, Duhl DM, Jacobson SG, Casavant T, Stone EM, Sheffield VC. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, Beck JS, Braun T, Streb LM, Cornier AS, Cox GF, Fulton AB, Carmi R, Luleci G, Chandrasekharappa SC, Collins FS, Jacobson SG, Heckenlively JR, Weleber RG, Stone EM, Sheffield VC. Identification of the gene (BBS1) most commonly involved in Bardet–Biedl syndrome, a complex human obesity syndrome. Nat Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Searby CC, Carmi R, Elbedour K, Van Maldergem L, Fulton AB, Lam BL, Powell BR, Swiderski RE, Bugge KE, Haider NB, Kwitek-Black AE, Ying L, Duhl DM, Gorman SW, Heon E, Iannaccone A, Bonneau D, Biesecker LG, Jacobson SG, Stone EM, Sheffield VC. Positional cloning of a novel gene on chromosome 16q causing Bardet–Biedl syndrome (BBS2) Hum Mol Genet. 2001;10:865–874. doi: 10.1093/hmg/10.8.865. [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Swiderski RE, Searby CC, Berg EM, Ferguson AL, Hennekam R, Merin S, Weleber RG, Biesecker LG, Stone EM, Sheffield VC. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet–Biedl syndrome gene. Am J Hum Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Ramaswami G, Janssen S, Chaki M, Allen SJ, Zhou W, Airik R, Hurd TW, Ghosh AK, Wolf MT, Hoppe B, Neuhaus TJ, Bockenhauer D, Milford DV, Soliman NA, Antignac C, Saunier S, Johnson CA, Hildebrandt F the GPN Study Group. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J Med Genet. 2010b doi: 10.1136/jmg.2010.082552. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Helou J, Allen SJ, O’Toole JF, Wise EL, Ashraf S, Attanasio M, Zhou W, Wolf MT, Hildebrandt F. Mutation analysis in nephronophthisis using a combined approach of homozygosity mapping, CEL I endonuclease cleavage, and direct sequencing. Hum Mutat. 2008;29:418–426. doi: 10.1002/humu.20669. [DOI] [PubMed] [Google Scholar]
- Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, Patil SB, Levy S, Ghosh AK, Murga-Zamalloa CA, van Reeuwijk J, Letteboer SJ, Sang L, Giles RH, Liu Q, Coene KL, Estrada-Cuzcano A, Collin RW, McLaughlin HM, Held S, Kasanuki JM, Ramaswami G, Conte J, Lopez I, Washburn J, Macdonald J, Hu J, Yamashita Y, Maher ER, Guay-Woodford LM, Neumann HP, Obermüller N, Koenekoop RK, Bergmann C, Bei X, Lewis RA, Katsanis N, Lopes V, Williams DS, Lyons RH, Dang CV, Brito DA, Dias MB, Zhang X, Cavalcoli JD, Nürnberg G, Nürnberg P, Pierce EA, Jackson PK, Antignac C, Saunier S, Roepman R, Dollfus H, Khanna H, Hildebrandt F. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;420:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek AM, Stone EM, Mykytyn K, Heckenlively JR, Green JS, Heon E, Musarella MA, Parfrey PS, Sheffield VC, Biesecker LG. Mutations in MKKS cause Bardet–Biedl syndrome. Nat Genet. 2000;26:15–16. doi: 10.1038/79116. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Laurier V, Davis EE, Muller J, Rix S, Badano JL, Leitch CC, Salem N, Chouery E, Corbani S, Jalk N, Vicaire S, Sarda P, Hamel C, Lacombe D, Holder M, Odent S, Holder S, Brooks AS, Elcioglu NH, Silva ED, Rossillion B, Sigaudy S, de Ravel TJ, Lewis RA, Leheup B, Verloes A, Amati-Bonneau P, Megarbane A, Poch O, Bonneau D, Beales PL, Mandel JL, Katsanis N, Dollfus H. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet. 2006;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Muller J, Laurier V, Davis EE, Zaghloul NA, Vicaire S, Jacquelin C, Plewniak F, Leitch CC, Sarda P, Hamel C, de Ravel TJ, Lewis RA, Friederich E, Thibault C, Danse JM, Verloes A, Bonneau D, Katsanis N, Poch O, Mandel JL, Dollfus H. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet–Biedl syndrome. Am J Hum Genet. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone DL, Slavotinek A, Bouffard GG, Banerjee-Basu S, Baxevanis AD, Barr M, Biesecker LG. Mutation of a gene encoding a putative chaperonin causes McKusick-Kaufman syndrome. Nat Genet. 2000;25:79–82. doi: 10.1038/75637. [DOI] [PubMed] [Google Scholar]
- Teebi AS. Autosomal recessive disorders among Arabs: an overview from Kuwait. J Med Genet. 1994;31:224–233. doi: 10.1136/jmg.31.3.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TL, Penney L, Woods MO, Parfrey PS, Green JS, Hefferton D, Davidson WS. A fifth locus for Bardet–Biedl syndrome maps to chromosome 2q31. Am J Hum Genet. 1999;64:900–904. doi: 10.1086/302301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul NA, Katsanis N. Mechanistic insights into Bardet–Biedl syndrome, a model ciliopathy. J Clin Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul NA, Liu Y, Gerdes JM, Gascue C, Oh EC, Leitch CC, Bromberg Y, Binkley J, Leibel RL, Sidow A, Badano JL, Katsanis N. Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet–Biedl syndrome. Proc Natl Acad Sci USA. 2010;107:10602–10607. doi: 10.1073/pnas.1000219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.