Abstract
Background
Common variable immunodeficiency (CVID) is a heterogeneous immune defect characterized by hypogammaglobulinemia, failure of specific antibody production, susceptibility to infections, and an array of comorbidities.
Objective
To address the underlying immunopathogenesis of CVID and comorbidities, we conducted the first genome-wide association and gene copy number variation (CNV) study in patients with CVID.
Methods
Three hundred sixty-three patients with CVID from 4 study sites were genotyped with 610,000 single nucleotide polymorphisms (SNPs). Patients were divided into a discovery cohort of 179 cases in comparison with 1,917 control subjects and a replication cohort of 109 cases and 1,114 control subjects.
Results
Our analyses detected strong association with the MHC region and association with a disintegrin and metalloproteinase (ADAM) genes (P combined = 1.96 × 10−7) replicated in the independent cohort. CNV analysis defined 16 disease-associated deletions and duplications, including duplication of origin recognition complex 4L (ORC4L) that was unique to 15 cases (P = 8.66 × 10−16), as well as numerous unique rare intraexonic deletions and duplications suggesting multiple novel genetic causes of CVID. Furthermore, the 1,000 most significant SNPs were strongly predictive of the CVID phenotype by using a Support Vector Machine algorithm with positive and negative predictive values of 1.0 and 0.957, respectively.
Conclusion
Our integrative genome-wide analysis of SNP genotypes and CNVs has uncovered multiple novel susceptibility loci for CVID, both common and rare, which is consistent with the highly heterogeneous nature of CVID. These results provide new mechanistic insights into immunopathogenesis based on these unique genetic variations and might allow for improved diagnosis of CVID based on accurate prediction of the CVID clinical phenotypes by using our Support Vector Machine model.
Keywords: Primary immunodeficiency, common variable immunodeficiency, antibody deficiency, genetics, genome-wide association, copy number variation, immunodiagnostics
Common variable immunodeficiency (CVID) is manifested by insufficient quantity and quality of immunoglobulin, leading to susceptibility to bacterial infections.1 CVID is a primary immunodeficiency disease because it is believed to result from intrinsic deficits affecting immunologic functions. CVID is heterogeneous, can present early or later in life, and is associated with specific comorbidities.2 Efforts to subcategorize CVID to predict outcomes and comorbid conditions both clinically and based on immunologic phenotypes are ongoing.3 B cell–activating factor of the TNF family receptor,4 transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI),5–7 and certain HLA haplotypes8,9 have been identified as potential gene candidates for susceptibility to CVID. Inducible costimulator, 10,11 CD81,12 CD19,13,14 and CD2015 harbor disease-causing mutations that presently explain only a small percentage of cases.16 The heterogeneous presentations of patients with CVID complicate clinical management, and searches for novel genetic predictors of disease or comorbidity have been of limited success.
Genome-wide single nucleotide polymorphism (SNP) arrays have enabled high-throughput genotyping of genomic DNAwith tagging of the whole genome based on linkage disequilibrium. Given both intensity and genotype content, copy number variations (CNVs) can also be detected with the same genome-wide association study (GWAS) platforms and are predicted to be responsible for generation of rare phenotypes. We genotyped 363 CVID cases and 3031 healthy control subjects on the Illumina Infinium Human Hap610K BeadChip (Illumina, San Diego, Calif). We sought to associate SNPs and CNVs with CVID, as well as with the characteristic clinical subphenotypes of this syndrome, to address disease heterogeneity. Our hypothesis was that although complex, CVID is sufficiently genetically distinct from the healthy population to allow genetic prediction of this uncommon but clinically relevant disease.
METHODS
Patients
The diagnosis of CVID was established in concordance with existing diagnostic criteria.16,17 All patients were enrolled in institutionally approved research protocols to enable genetic analysis and collection of clinical data. Subsets of the patients reported here have been previously included in published studies.2,18,19
Illumina Infinium assay for SNP genotyping and CNV discovery
We performed high-throughput, genome-wide SNP genotyping with the InfiniumII HumanHap610 BeadChip technology at the Center for Applied Genomics at the Children’s Hospital of Philadelphia. The genotype data content together with the intensity data provided by the genotyping array provides high confidence for CNV calls. Importantly, the simultaneous analysis of intensity and genotype data in the same experimental setting establishes a highly accurate definition for normal diploid states and any deviation from the norm. To call CNVs, we used the PennCNV algorithm,20 which combines multiple sources of information, including log R ratio and B-allele frequency at each SNP marker, along with SNP spacing and population frequency of the B allele to generate CNV calls. Rare recurrent CNVs were the focus of our study.
CNV quality control
We calculated quality control measures on our HumanHap610 GWAS data based on statistical distributions to exclude poor-quality DNA samples and false-positive CNVs. The first threshold is the percentage of attempted SNPs that were successfully genotyped. Only samples with call rates of greater than 98% were included. The genome-wide intensity signal must have as little noise as possible. Only samples with an SD of normalized intensity (log R ratio) of less than 0.35 were included. All samples must have white ethnicity based on principle components analysis, and all other samples were excluded. Furthermore, case and control matching was ensured by calculating a genomic inflation factor between groups. Wave artifacts roughly correlating with guanine-cytosine content resulting from hybridization bias of low full-length DNA quantity are known to interfere with accurate inference of CNVs. Only samples in which the guanine-cytosine-corrected wave factor of the log R ratio −0.02 <×< 0.02 were accepted. If the count of CNV calls made by using PennCNV exceeds 100, it is suggestive of poor DNA quality, and those samples were excluded. Thus only samples with CNV call counts of less than 100 were included. Any duplicate samples (eg, monozygotic twins or repeats on the same patient) were identified, and as a result, 1 sample was excluded.
Statistical analysis of CNVs
CNV frequency between cases and control subjects was evaluated at each SNP by using the Fisher exact test. We only considered loci that were nominally significant between cases and control subjects (P < .05) for which patients had the same variations that were observed in multiple cohorts or were not observed in any of the control subjects and were validated with an independent method. We report statistical local minimums to narrow the association in reference to a region of nominal significance, including SNPs residing within 1 Mb of each other. Resulting nominally significant CNVs were excluded if they met any of the following criteria: (1) residing on telomere or centromere proximal cytobands; (2) arising in a “peninsula” of common CNVs arising from variation in boundary truncation of CNV calling; (3) being genomic regions with extremes in GC content, which produces hybridization bias; or (4) contributing to multiple CNVs. Three lines of evidence establish statistical significance: an independent replication P value of less than .05, permutation of observations, and no loci observed with control-enriched significance. We used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) to assess the significance of functional annotation clustering of independently associated results into InterPro categories.
RESULTS
Our CVID case cohort was composed of 223 patients from Mount Sinai School of Medicine, 76 patients from the University of Oxford, 37 patients from the Children’s Hospital of Philadelphia, and 27 patients from the University of South Florida. The diagnosis in each case was validated against the European Society for Immunodeficiencies/Pan-American Group for Immunodeficiency diagnostic criteria.16
We first evaluated the quality and suitability of the data for a case-control study. Seven samples had call rates of less than 98% and were excluded. Based on principle components analysis with EIGENSTRAT software,21 the cases were stratified into 3 clusters: 288 cases were of confirmed European ancestry, whereas 30 samples were significant outliers from white race and were omitted from this analysis. A random split of 179 cases was matched with 1917 white control subjects. We identified an additional 1114 control samples that clustered well with the balance of 109 white cases used for replication. We excluded subjects with observed cryptic relatedness of genotypes. For CNV calling, samples with high noise (SD of log R ratio >0.35) in intensity signal were additionally removed.
SNP association analysis
We first performed a GWAS using the SNP genotype data. Genotype frequencies were compared between cases and control subjects by using a χ2 test statistic applied in PLINK22 for SNPs with an at least 90% call rate and 1% minor allele frequency. The discovery cohort of 179 cases and 1917 control subjects yielded a low genomic inflation factor of 1.02783 (Fig 1, A). The most significant association was to the MHC, with the most significant SNP, rs3117426, having a P value of 8.62 × 10−10 (Table I). The second best association was to a locus on 8p21.2 harboring a disintegrin and metalloproteinase 28 (ADAM28), ADAM7, ADAMDEC1, and STC1. However, this locus did not meet genome-wide significance criteria (P = 6.24 × 10−6 for rs4872262) and results, which is in keeping with modest power from the relatively small sample sizes.
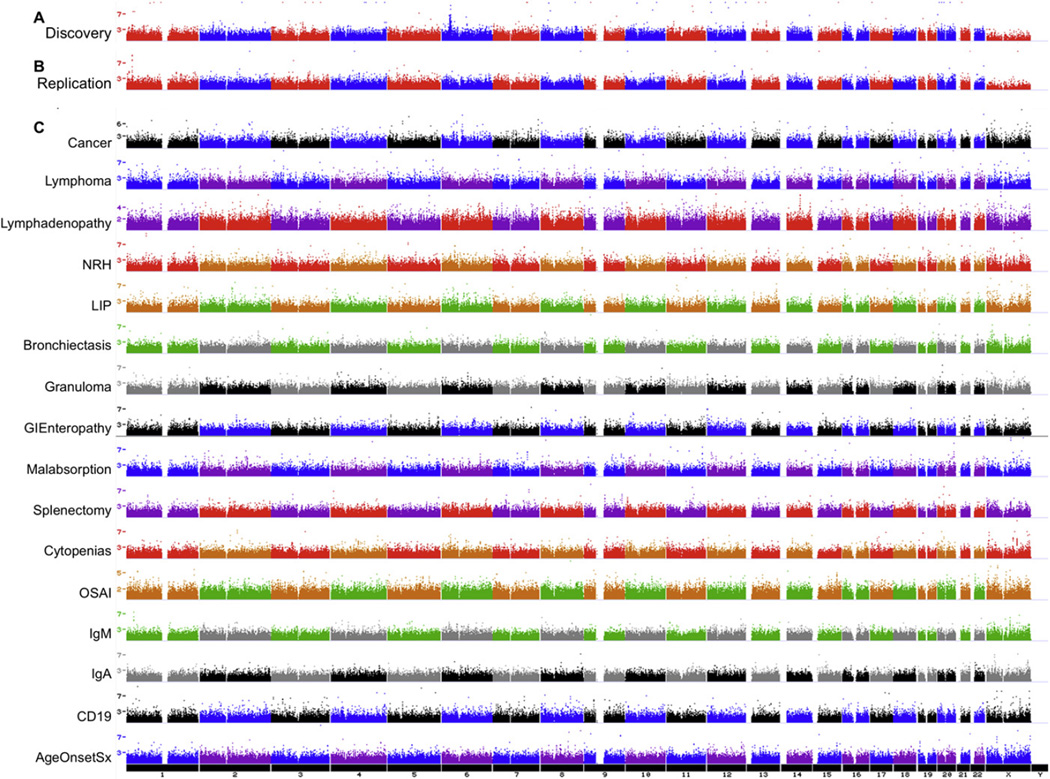
FIG 1.
Case-control allele frequency significance. SNP-based case-control significance is shown in negative log base 10 genome-wide analysis for (A) discovery- and (B) replication-inclusive CVID cohorts. (C) Subsequently, CVID cases with specific disease subphenotypes were compared with CVID cases without the subphenotype. The single-SNP tests are shown as single points. Multiple neighboring SNPs of similar significance boost confidence in the association, as shown in the strong peak on 6p22.1-p21.32 of the discovery case-control cohort. Conversely, the median significance P value is kept low by minimizing population stratification, which minimizes the genomic inflation factor.
TABLE I.
Most significant associated regions based on genotype association
| Region genomic span |
Gene | Count SNPs top 1000 |
Distance from exon |
P value |
SNP | A1 | F_A | F_U | OR |
P value, replication |
SNP, replication |
F_A, replication |
F_U, replication |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr6:27236785-32521295 | MHC | 110 | 0 | 8.62 × 10−10 | rs3117426 | T | 0.3268 | 0.1907 | 2.061 | .0004 | rs2156875 | 0.5734 | 0.4475 |
| chr8:23746576-24681608 | ADAM28, ADAM7, ADAMDEC1, STC1 | 4 | 0 | 6.24 × 10−6 | rs4872262 | A | 0.0391 | 0.0107 | 3.765 | .0314 | rs4872262 | 0.0321 | 0.0135 |
| chr7:4270929-4287047 | SDK1 | 5 | 0 | 4.70 × 10−5 | rs895710 | T | 0.2905 | 0.4003 | 0.6135 | .0235 | rs4720301 | 0.3991 | 0.4793 |
| chr1:20369369-20490791 | FLJ32784, UBXN10 | 7 | 0 | 9.09 × 10−5 | rs7514144 | C | 0.2654 | 0.1808 | 1.636 | 2.25E-8 | rs6426636 | 0.4679 | 0.2857 |
| chr1:59987116-59993733 | FGGY | 2 | 2458 | 1.52 × 10−4 | rs11207520 | G | 0.3994 | 0.3026 | 1.533 | .0015 | rs11207520 | 0.2569 | 0.1706 |
| chr2:65518304-65520560 | FLJ16124 | 2 | 784 | 5.20 × 10−4 | rs1194849 | G | 0.3659 | 0.4614 | 0.6737 | .0119 | rs1876518 | 0.3853 | 0.4744 |
Known immunologic function genes are shown in boldface and underlined. Additional details are shown in Tables E7 and E8 in this article’s Online Repository at www.jacionline.org.
A1, Allele 1, which is the minor allele, F_A, frequency of allele 1 in affected subjects; F_U, frequency of allele 1 in unaffected subjects.
In an attempt to replicate the 8p21.2 locus, we searched for direction of effects in our independent CVID cohort of 109 cases and 1114 control subjects (genomic inflation factor, 1.04789; Fig 1, B). Interestingly, 4 SNPs displayed significance (P < .05) and allele frequency in the same direction as the discovery cohort, including rs11207520, rs1194849, rs17790790, and rs4872262 (Table I). When all SNPs contributing to the discovery region were queried in replication rather than just the most significant SNP, a total of 8 SNPs displayed significance (P <.05) and allele frequencies in the same direction as discovery (Table I).
We next addressed the heterogeneity of the CVID phenotype by assessing the various CVID subphenotypes. On the basis of recent phenotypic characterization,18 we established 16 distinct clinical subgroups within the CVID cohort, including cancer, lymphoma, lymphadenopathy, nodular regenerative hyperplasia of the liver, lymphoid interstitial pneumonitis (LIP), bronchiectasis, biopsyproved granuloma, gastrointestinal enteropathy, malabsorption, splenectomy, cytopenias, organ-specific autoimmunity, low IgM level (<50 mg/dL), low IgA level (<10 mg/dL), low B cell number (CD19+ cells <1%), and young age at symptom onset (<10 years, see Fig E1 and Table E1 in this article’s Online Repository at www.jacionline.org).
Differences in genotype frequencies were assigned significance, and regions with multiple significant SNPs were then scored. Significant SNP associations were made for all subsets (Table II and Fig 1, C). The most significant association was observed with nodular regenerative hyperplasia of the liver on AK096081-AK124028 (P = 2.29 × 10−10). Lymphoma was associated with KIAA0834, PFTK1, and HAVCR1, with corresponding P values ranging from 1.69 × 10−8 to 3.62 × 10−8. An association was also observed between LIP and FGF14 and ZNF81, with corresponding P values of 5.76 × 10−8 and 7.70 × 10−8, respectively. Organ-specific autoimmunity also associated with SNX31 (P = 6.89 × 10−8), and low IgM levels (<50 mg/dL) in the case subjects were associated with LDLRAP1 (P = 6.02 × 10−8).
TABLE II.
Most significant associated regions based on subphenotype genotype association
| Significant region | P value | SNP | A1 | F_A | F_U | OR | Count SNPs |
Gene | Distance | Distance from exon |
Subphenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|
| chr1:68153970-68161019 | 2.29 × 10−10 | rs1926283 | C | 0.2308 | 0.01807 | 16.3 | 3 | AK096081, AK124028 | 0 | 33408 | NRH |
| chr20:55901032-55985359 | 3.21 × 10−9 | rs8124301 | T | 0.4231 | 0.07631 | 8.877 | 5 | C20orf85 | 174030 | 174030 | Malabsorption |
| chr7:90245135-90245280 | 1.69 × 10−8 | rs975004 | G | 0.3333 | 0.03953 | 12.15 | 2 | KIAA0834, PFTK1 | 0 | 12544 | Lymphoma |
| chr5:156402979-156407976 | 3.62 × 10−8 | rs10038271 | T | 0.7222 | 0.1877 | 11.25 | 2 | HAVCR1 | 0 | 651 | Lymphoma |
| chrX:22422160-22428888 | 5.71 × 10−8 | rs5925651 | A | 0.3462 | 0.1386 | 3.291 | 3 | ZNF645 | 219665 | 219665 | Bronchiectasis |
| chr13:101641990-101673539 | 5.76 × 10−8 | rs1336698 | G | 0.65 | 0.168 | 9.197 | 6 | FGF14 | 0 | 69139 | LIP |
| chr1:25735669-25753653 | 6.02 × 10−8 | rs2065970 | G | 0.06553 | 0.2411 | 4.529 | 5 | LDLRAP1 | 0 | 0 | Low IgM |
| chr8:101728102-101728344 | 6.89 × 10−8 | rs7815950 | G | 0.2133 | 0.05615 | 4.559 | 2 | SNX31 | 0 | 2334 | OSAI |
| chr8:13834782-13842376 | 7.62 × 10−8 | rs2682665 | C | 0.1818 | 0.01394 | 15.71 | 3 | SGCZ | 149368 | 149368 | Low B cells |
| chrX:152295068-152303019 | 9.84 × 10−8 | rs5987017 | A | 0.2143 | 0.04773 | 5.442 | 4 | ZNF275 | 24829 | 24829 | Young age |
| chr12:2531014-2538733 | 1.21 × 10−7 | rs4765961 | C | 0.4737 | 0.142 | 5.439 | 3 | CACNA1C | 0 | 0 | GI Enteropathy |
| chrX:30733771-30736604 | 1.44 × 10−7 | rs11095197 | T | 0.1469 | 0.3431 | 3.034 | 2 | MAP3K7IP3 | 18876 | 18876 | Low IgA |
| chrX:53035117-53039801 | 1.90 × 10−7 | rs10127016 | G | 0.3833 | 0.125 | 4.351 | 2 | TMEM29 | 0 | 1409 | Lymphadenopathy |
| chr14: 23168471- 23169215 | 4.57 × 10−7 | rs222723 | C | 0.6154 | 0.1968 | 6.531 | 2 | DHRS2 | 0 | 0 | Cancer |
Known immunologic function genes are shown in boldface and underlined. Additional details are shown in Table E9 in this article’s Online Repository at www.jacionline.org.
GI, Gastrointestinal; NRH, nodular regenerative hyperplasia of the liver; OR, odds ratio; OSAI, organ-specific autoimmunity.
Finally, patients with CVID manifesting enteropathy and their resulting subphenotype significance were additionally compared with patients with inflammatory bowel disease from a previously reported cohort23 to assess whether specific loci contributed to a common cause. The SNP rs12889533 was significant in both the CVID cases with enteropathy and the inflammatory bowel disease cohort, with allele frequency difference in the same direction (see Table E2 in this article’s Online Repository at www.jacionline.org).
CNV analysis
We next performed a CNV analysis, in which rare (<1%) CNVs were called by using PennCNV.20 We analyzed deletion and duplication CNV frequencies of 311 cases and 2766 control subjects (see Fig E2 in this article’s Online Repository at www.jacionline.org). We discovered 5 deletions and 11 duplications to be recurrent and significantly enriched in the CVID cases compared with the control subjects (Tables III and IV). In addition, 15 regions were exclusive to CVID cases. The most noteworthy duplication resided at 2q23.1 and affected multiple exons of origin recognition complex 4L (ORC4L) that were exclusive to 15 cases, and were not identified in control subjects (P = 8.66 × 10−16, see Fig E3 in this article’s Online Repository at www.jacionline.org).
TABLE III.
Most significant CVID-associated regions based on CNV association: deletions
| CNV deletion | Count SNPs |
Distance from exon |
P value, deletion |
Cases, deletion |
Control subjects, deletion |
OR | Gene | MSSM | Oxford | USF | CHOP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| chr11:85365857-85381622 | 5 | 0 | .01 | 2 | 0 | Infinity | PICALM, PICALM variant protein | 0 | 2 | 0 | 0 |
| chr20:57735790-57741780 | 6 | 5902 | .026 | 3 | 4 | 6.73 | PHACTR3 | 2 | 1 | 0 | 0 |
| chr22:17396663-18417315 | 270 | 0 | .029 | 2 | 1 | 17.90 | ARVCF, COMT, TBX1, others | 2 | 0 | 0 | 0 |
| chr4:10256682-10264316 | 4 | 2915 | .029 | 2 | 1 | 17.90 | CLNK | 0 | 1 | 1 | 0 |
| chr10:46003146-46042543 | 8 | 0 | .029 | 2 | 1 | 17.90 | DKFZp566K0524, PTPN20A, PTPN20B | 2 | 0 | 0 | 0 |
Known immunologic function genes are shown in boldface and underlined. Additional details are available in Table E10 in this article’s Online Repository at www.jacionline.org.
CHOP, Children’s Hospital of Philadelphia; MSSM, Mount Sinai School of Medicine; OR, odds ratio; USF, University of South Florida.
TABLE IV.
Most significant CVID-associated regions based on CNV association: duplications
| CNV duplication | Count SNPs |
Distance from exon |
P value, duplication |
Cases, duplication |
Control subjects, duplication |
OR | Gene | MSSM | Oxford | USF | CHOP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| chr2:148396730-148433180 | 6 | 0 | 8.66 × 10−16 | 15 | 0 | Infinity | ACVR2A, ORC4L | 10 | 5 | 0 | 0 |
| chr15:35053039-35063531 | 6 | 23146 | 5.72 × 10−13 | 19 | 10 | 17.93 | MEIS2 | 17 | 2 | 0 | 0 |
| chr7:91789778-91801963 | 4 | 0 | 1.20 × 10−9 | 12 | 4 | 27.71 | ANKIB1 | 9 | 3 | 0 | 0 |
| chr2:163316298-163316595 | 3 | 17100 | 1.00 × 10−7 | 7 | 0 | Infinity | KCNH7 | 6 | 1 | 0 | 0 |
| chr7:87180702-87191931 | 4 | 202 | 7.41 × 10−7 | 7 | 1 | 63.67 | RPIB9 | 5 | 2 | 0 | 0 |
| chr19:9256584-9277749 | 4 | 0 | 0.001 | 3 | 0 | Infinity | ZNF699 | 2 | 0 | 1 | 0 |
| chr4:39190766-39201960 | 6 | 0 | 0.004 | 3 | 1 | 26.93 | UGDH | 1 | 2 | 0 | 0 |
| chr7:18903915-18905725 | 4 | 23171 | 0.009 | 3 | 2 | 13.46 | HDAC9 | 1 | 2 | 0 | 0 |
| chr5:170531269-170536993 | 4 | 374 | 0.01 | 2 | 0 | Infinity | DKFZp666P032, RANBP17 | 1 | 1 | 0 | 0 |
| chr7:34685030-34686172 | 4 | 4517 | 0.01 | 2 | 0 | Infinity | AAA1, NPSR1 | 1 | 1 | 0 | 0 |
| chr1:170400944-170403742 | 4 | 26274 | 0.01 | 2 | 0 | Infinity | DNM3 | 0 | 1 | 1 | 0 |
Known immunologic function genes are shown in boldface and underlined. Additional details are available in Table E10 in this article’s Online Repository at www.jacionline.org.
CHOP, Children’s Hospital of Philadelphia; MSSM, Mount Sinai School of Medicine; OR, odds ratio; USF, University of South Florida.
We additionally observed large CNVs captured by 10 or more SNPs that were exclusive to CVID cases totaling 84 deletions and 98 duplications. (see Table E3 in this article’s Online Repository at www.jacionline.org). No significant overrepresentation was observed in the control subjects in this frequency range. Of clinical relevance, 2 patients were newly identified with deletions in the 22q11 region, suggesting an alternative presentation of the well-characterized congenital immunodeficiency associated with this microdeletion. Interestingly, 10 regions of homozygous deletion were observed exclusively in CVID cases (see Table E4 in this article’s Online Repository at www.jacionline.org).
We next examined the overall CNV burden in cases compared with control subjects for both exonic CNVs and for large (100 kb) and rare (<1%) CNVs. We found deletions to be significantly enriched in the CVID cases when considering CNV observations on all loci genome wide (see Table E5 in this article’s Online Repository at www.jacionline.org). All but 5 of the 182 CNVs exclusive to the CVID cohort were intraexonic. To assess the reliability of our CNV detection method, we reviewed the B-allele frequency (genotype) and log R ratio (intensity) values of our Illumina data and experimentally validated all the significant CNVs by using an independent method, the Affymetrix Cytogenetics Whole-Genome 2.7M Array (Affymetrix, Santa Clara, Calif), which provides high resolution with 400,103 SNP and 2,387,595 CN probes (see Fig E4 in this article’s Online Repository at www.jacionline.org).
CVID disease prediction
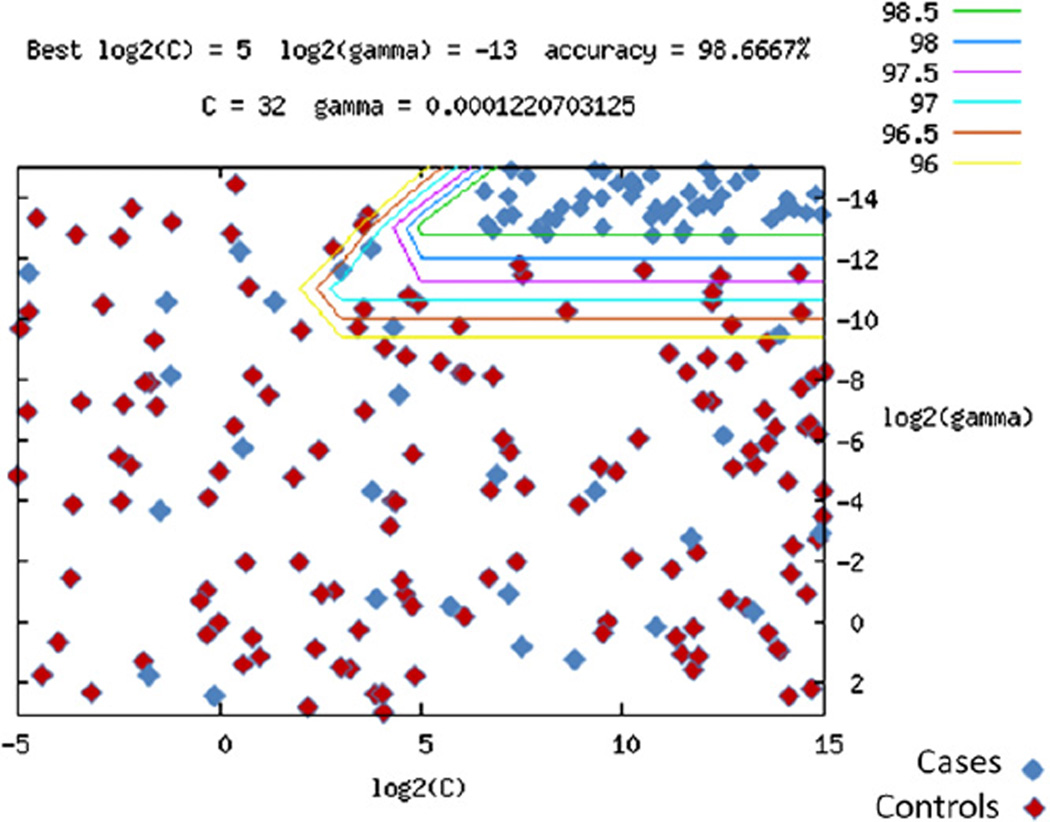
We used a Support Vector Machine (SVM) algorithm to determine how well we could predict the CVID phenotype in a pool of “unknown” samples (see the Methods section in this article’s Online Repository at www.jacionline.org). The SVM algorithm was trained on the discovery cohort of 179 CVID cases and 1917 control subjects. We identified 1000 SNPs from association analysis that accurately separated the CVID cases from the control subjects with 98.7% accuracy overall. When applied on the independent cohort of 109 cases and 1114 control subjects, the positive and negative predictive values amounted to 1.0 and 0.957, respectively (Fig 2), suggesting that these markers were robust in distinguishing the genetic architecture of CVID cases from that of healthy control subjects.
FIG 2.
SVM prediction of CVID status. Plotted are log base 2 SVM scaling values of cost C (the penalty parameter of SVMs) and γ (1/number of variables) from LIMSVM software. Data were coded as +1 for affected and −1 for unaffected subjects. Genotype was scaled as follows: AA, 0; AB, 0.5; and BB, 1. Lines represent modeling with increasing resolution, with the green line representing the final model solution. The model was generated by using the discovery cohort and control subjects, whereas the subjects represented (blue, CVID cases; red, control subjects) were from the separate replication cohort.
DISCUSSION
CVID was described more than 50 years ago, but aside from a small number of recessively inherited genes in a few families and the more prevalent but poorly understood contribution of mutations in TNFRSF13B,5,6,7,24 other causes have remained obscure. CVID has thus been hypothesized to represent a diverse collection of genetic lesions resulting in a similar immunologic phenotype. The MHC region has been associated with a myriad of complex diseases,25 including immune-related conditions26 and CVID.8,9 MHC was robustly associated with CVID in our study, further validating the prior work that was not performed at the genome-wide level.
The SNP genotype association analysis presented here revealed a suggestive association outside of the MHC region with a locus encompassing ADAM28, ADAM7, ADAMDEC1, and STC1 (P = 6.24 × 10−6) that we also replicated in an independent case-control cohort (P value range = 2.25 × 10−8 to .0314). The ADAM family proteins are zinc metalloproteases involved in diverse biologic processes, including immune responses. Interestingly, the metallopeptidase MMP27 was also nominally associated with CVID (P = 1.69 × 10−4). Proteins of the matrix metalloproteinase family are involved in the breakdown of extracellular matrix to promote routine physiological processes but might also facilitate disease pathogenesis. Related genes have demonstrated immunologic function involved in the regulation of cytokine release, TH2 immune responses, and specific inflammatory processes. ADAM28 is also known as the lymphocyte metalloprotease MDC-L, which is expressed on the lymphocyte cell surface.27 As such, it has been defined as a ligand for α4β1 integrin, which enables the adhesion of other leukocytes expressing this integrin. STC1, which is located in the same region, is a gene that encodes stanniocalcin 1 protein, which is involved in regulation of calcium, including in antioxidant pathways of marcophages. 28 In this light calcium regulation within immune cells has been previously identified as aberrant in certain samples of patients with CVID.29 UBXN10 might be a compelling candidate potentially involved in immunopathogenesis of primary antibody failure in that it encodes an ubiquitin-like protein, which, in addition to phosphorylation, has been shown to regulate nuclear factor κB activity.30 Sphingosine-dependent protein kinase 1 (SDK1) is important in the survival of alveolar macrophages.31 DEPDC6 is a negative regulator of mammalian target of rapamycin signaling pathways, and RNA expression levels were found to be significantly different between those mice resistant to H5N1 influenza virus and those that were susceptible.32 No significant associations in the region of TACI were observed, and subjects with TACI mutations were not separately identified. We reviewed the reported amino acid changes and looked up their corresponding nonsynonymous SNP IDs (see Table E6 in this article’s Online Repository at www.jacionline.org). No imputation reference files, including these SNPs, were available to infer genotypes.
We additionally performed an SNP genotype association to the particular features of CVID to define potential common mechanistic links among the specific clinical and immunologic variants to potentially enable the prediction of CVID clinical phenotypes. Significant individual associations were made with all CVID variables studied (Table II). In this regard PFTK1 is a member of the CDC2-related protein kinase family found to be constitutively expressed at high levels in B-cell lymphomas33 and also found to be associated with lymphoma in the subjects with this complication. Interestingly, SNPs in HAVCR1, which is also associated in the same subjects (P= 3.62 × 10−8), are genes that play a role in TH cell development and the regulation of asthma and allergic diseases. 34 FGF14, which is associated with LIP and low IgA levels, is a member of the fibroblast growth factor family, which has crucial roles in embryonic development, cell growth, morphogenesis, tissue repair, tumor growth, and invasion.35 SNX31, which is associated with organ-specific autoimmunity, is a sorting nexin that might be involved in protein trafficking; SNX family proteins are subunits required for CD28-mediated T-cell costimulation.36
Although these SNP association results are not powered for definitive conclusions, the CNV association analysis uncovered several novel genes that were significantly associated with CVID. In fact, 84 CNV deletions and 98 duplications were identified in 1 or more patients with CVID but were not found in any of the 3031 control subjects. Most were intraexonic and thus likely to disrupt the genes involved. Some of the genes potentially affected by the identified CNV were also discovered in the GWAS part of this study. Many others have direct or potential relevance to the immune system, and many were unique to individual patients, thus underscoring the great mechanistic diversity that is likely to underlie this collection of disorders, which is also reflected in the variability in clinical presentation and disease natural history.1 Among those, we noted a highly significant number of subjects with duplications in ORC4L, a gene previously associated with B-cell lymphoproliferative disorders.37 This gene is essential for initiation of DNA replication and potentially in rapidly proliferating immune cells.
In addition to the potential of these findings to provide mechanistic insight into how CVID and its subphenotypes arise, we used SVM to exploit the overall genetic uniqueness of patients with CVID compared with healthy control subjects. These results suggest that the genetic signatures of the disease might also allow for accurate prediction of CVID based on genetic profiling. This has the potential to greatly improve the clinical management of the CVID and address a major unmet need in the field of primary immunodeficiency. By using current diagnostic criteria, diagnostic delay of multiple years can occur38 which can in part be because of the evolving immunoglobulin phenotype.38 This can lead to repeated infection and irreparable organ damage during the period of diagnostic delay. The ability to determine increased likelihood for CVID at the time of initial clinical suspicion could allow for improved intervention and reduced morbidity.
This study represents the first genome-wide population-based study of CVID. The use of the relatively large cohorts of this rare disorder assembled here was essential both to discover and confirm the findings and demonstrates the potential of genome-wide association in complicated polygenic rare diseases. This type of unbiased study has discovered many novel loci that might underlie the development of CVID. It can provide clues to the pathogenesis of the heterogeneous clinical complications and subtypes of CVID, providing a solid foundation for further studies to understand the mechanism, interplay, and clinical manifestations of CVID, with the immediate potential of improving clinical management through enhanced diagnosis. Finally, the great diversity of genetic findings identified with regard to unique CNVs substantiates the hypothesis that CVIDs are a collection of diverse mechanisms leading to complex phenotypes.
Supplementary Material
Clinical implications.
We define CVID as a genetically heterogeneous condition through genome-wide analysis. Patients with CVID can be distinguished from healthy subjects by using multiparameter predictive algorithms that could prove useful in facilitating diagnosis.
Acknowledgments
Children’s Hospital of Philadelphia support was from the Children’s Hospital of Philadelphia Institutional Development Award to the Center for Applied Genomics, which funded all genotyping (to H.H.); a Research Development Award from the Cotswold Foundation (to H.H.); the Jeffrey Modell foundation (to J.S.O.); and National Institutes of Health (NIH) grant AI-079731 (to J.S.O.). Oxford support was from the NIHR Oxford Biomedical Research Centre, Baxter Healthcare (general support to the department not specific to this project), Talecris (general support to the department not specific to this project), and the Jeffrey Modell Foundation for unrestricted gifts; the Primary Immunodeficiency Association for the Centre of Excellence award; and the European Commission for EU 7th FP EURO-PADnet number 201549. University of South Florida support was from NIH grant 5R03AI083904 (to E.E.P.). Mount Sinai support was from NIH grants, AI-101093, AI-467320, AI-48693, NIAID Contract 03-22, and the David S Gottesman Immunology Chair (all to C.C.-R.).
J. S. Orange has consultant arrangements with Talecris Biotherapeutics, Baxter Health, CSL Behring, and IBT Reference Labs; is a speaker for Baxter Health; receives research support from the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID); is an Elected Officer of the American Academy of Allergy, Asthma & Immunology (AAAAI); and is on the advisory board for the Immune Deficiency Foundation. K. E. Sullivan receives research support from the NIH and is a consultant for the Immune Deficiency Foundation. J. W. Sleasman receives research support from the NIH, the Florida Department of Health, and the National Oceanic Atmospheric Administration. E. E. Perez has consultant arrangements with Baxter and CSL Behring.
We thank the clinical immunology and laboratory immunology staff in Oxford and Li Zhang, who prepared DNA at Mount Sinai Medical Center.
Abbreviations used
- ADAM
A disintegrin and metalloproteinase
- CNV
Copy number variation
- CVID
Common variable immunodeficiency
- GWAS
Genome-wide association study
- LIP
Lymphoid interstitial pneumonitis
- ORC
Origin recognition complex
- SNP
Single nucleotide polymorphism
- SVM
Support Vector Machine
- TACI
Transmembrane activator and calcium modulator and cyclophilin ligand interactor
Footnotes
Disclosure of potential conflict of interest: The rest of the authors have declared that they have no conflict of interest.
REFERENCES
- 1.Chapel H, Cunningham-Rundles C. Update in understanding common variable immunodeficiency disorders (CVIDs) and the management of patients with these conditions. Br J Haematol. 2009;145:709–727. doi: 10.1111/j.1365-2141.2009.07669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunningham-Rundles C, Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin Immunol. 1999;92:34–48. doi: 10.1006/clim.1999.4725. [DOI] [PubMed] [Google Scholar]
- 3.Wehr C, Kivioja T, Schmitt C, Ferry B, Witte T, Eren E, et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood. 2008;111:77–85. doi: 10.1182/blood-2007-06-091744. [DOI] [PubMed] [Google Scholar]
- 4.Bacchelli C, Buckridge S, Thrasher AJ, Gaspar HB. Translational mini-review series on immunodeficiency: molecular defects in common variable immunodeficiency. Clin Exp Immunol. 2007;149:401–409. doi: 10.1111/j.1365-2249.2007.03461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzer U, Chapel HM, Webster AD, Pan-Hammarström Q, Schmitt-Graeff A, Schlesier M, et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat Genet. 2005;37:820–828. doi: 10.1038/ng1600. [DOI] [PubMed] [Google Scholar]
- 6.Castigli E, Wilson SA, Garibyan L, Rachid R, Bonilla F, Schneider L, et al. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat Genet. 2005;37:829–834. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- 7.Pan-Hammarström Q, Salzer U, Du L, Björkander J, Cunningham-Rundles C, Nelson DL, et al. Reexamining the role of TACI coding variants in common variable immunodeficiency and selective IgA deficiency. Nat Genet. 2007;39:429–430. doi: 10.1038/ng0407-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volanakis JE, Zhu ZB, Schaffer FM, Macon KJ, Palermos J, Barger BO, et al. Major histocompatibility complex class III genes and susceptibility to immunoglobulin A deficiency and common variable immunodeficiency. J Clin Invest. 1992;89:1914–1922. doi: 10.1172/JCI115797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olerup O, Smith CI, Björkander J, Hammarström L. Shared HLA class II-associated genetic susceptibility and resistance, related to the HLA-DQB1 gene, in IgA deficiency and common variable immunodeficiency. Proc Natl Acad Sci U S A. 1992;89:10653–10657. doi: 10.1073/pnas.89.22.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimbacher B, Hutloff A, Schlesier M, Glocker E, Warnatz K, Dräger R, et al. Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nat Immunol. 2003;4:261–268. doi: 10.1038/ni902. [DOI] [PubMed] [Google Scholar]
- 11.Salzer U, Maul-Pavicic A, Cunningham-Rundles C, Urschel S, Belohradsky BH, Litzman J, et al. ICOS deficiency in patients with common variable immunodeficiency. Clin Immunol. 2004;113:234–240. doi: 10.1016/j.clim.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 12.van Zelm MC, Smet J, Adams B, Mascart F, Schandene L, Janssen F, et al. CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J Clin Invest. 2010;120:1265–1274. doi: 10.1172/JCI39748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Zelm MC, Reisli I, van den Burg M, Castano D, van Noesel CJ, van Tol MJ, et al. An antibody-deficiency syndrome due to mutations in the CD19 gene. N Engl J Med. 2006;354:1901–1912. doi: 10.1056/NEJMoa051568. [DOI] [PubMed] [Google Scholar]
- 14.Kanegane H, Agematsu K, Futatani T, Sira MM, Suga K, Sekiguchi T, et al. Novel mutations in a Japanese patient with CD19 deficiency. Genes Immun. 2007;8:663–670. doi: 10.1038/sj.gene.6364431. [DOI] [PubMed] [Google Scholar]
- 15.Kuijpers TW, Bende RJ, Baars PA, Grummels A, Derks IA, Dolman KM, et al. CD20 deficiency in humans results in impaired T cell-independent antibody responses. J Clin Invest. 2010;120:214–222. doi: 10.1172/JCI40231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conley ME, Notarangelo LD, Etzioni A. Diagnostic criteria for primary immunodeficiencies. Representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies) Clin Immunol. 1999;93:190–197. doi: 10.1006/clim.1999.4799. [DOI] [PubMed] [Google Scholar]
- 17.Bonilla FA, Bernstein IL, Khan DA, Ballas ZK, Chinen J, Frank MM, et al. Practice parameter for the diagnosis and management of primary immunodeficiency. Ann Allergy Asthma Immunol. 2005;94(suppl 1):S1–S63. doi: 10.1016/s1081-1206(10)61142-8. [DOI] [PubMed] [Google Scholar]
- 18.Chapel H, Lucas M, Lee M, Bjorkander J, Webster D, Grimbacher B, et al. Common variable immunodeficiency disorders: division into distinct clinical phenotypes. Blood. 2008;112:277–286. doi: 10.1182/blood-2007-11-124545. [DOI] [PubMed] [Google Scholar]
- 19.Yong PL, Orange JS, Sullivan KE. Pediatric common variable immunodeficiency: immunologic and phenotypic associations with switched memory B cells. Pediatr Allergy Immunol. 2010;21:852–858. doi: 10.1111/j.1399-3038.2010.01004.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Radigan L, Salzer U, Behrens TW, Grimbacher B, Diaz G, et al. Transmembrane activator and calcium-modulating cyclophilin ligand interactor mutations in common variable immunodeficiency: clinical and immunologic outcomes in heterozygotes. J Allergy Clin Immunol. 2007;120:1178–1185. doi: 10.1016/j.jaci.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiina T, Inoko H, Kulski JK. An update of the HLA genomic region, loci information and disease associations. Tissue Antigens. 2004;64:631–649. doi: 10.1111/j.1399-0039.2004.00327.x. [DOI] [PubMed] [Google Scholar]
- 26.International MHC and Autoimmunity Genetics Network. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci U S A. 2009;106:18680–18685. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bridges LC, Tani PH, Hanson KR, Roberts CM, Judkins MB, Bowditch RD. The lymphocyte metalloprotease MDC-L (ADAM 28) is a ligand for the integrin α4β1. J Biol Chem. 2002;277:3784–3792. doi: 10.1074/jbc.M109538200. [DOI] [PubMed] [Google Scholar]
- 28.Sheikh-Hamad D. Mammalian stanniocalcin-1 activates mitochondrial antioxidant pathways: new paradigms for regulation of macrophages and endothelium. Am J Physiol Renal Physiol. 2010;298:F248–F254. doi: 10.1152/ajprenal.00260.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foerster C, Voelxen M, Rakhmanov M, Keller B, Gutenberger S, Goldacker S, et al. B cell receptor-mediated calcium signaling is impaired in B lymphocytes of type Ia patients with common variable immunodeficiency. J Immunol. 2010;184:7305–7313. doi: 10.4049/jimmunol.1000434. [DOI] [PubMed] [Google Scholar]
- 30.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Ann Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 31.Monick MM, Mallampalli RK, Bradford M, McCoy D, Gross TJ, Flaherty DM, et al. Cooperative prosurvival activity by ERK and Akt in human alveolar macrophages is dependent on high levels of acid ceramidase activity. J Immunol. 2004;173:123–135. doi: 10.4049/jimmunol.173.1.123. [DOI] [PubMed] [Google Scholar]
- 32.Boon AC, deBeauchamp J, Hollmann A, Luke J, Kotb M, Rowe S, et al. Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J Virol. 2009;83:10417–10426. doi: 10.1128/JVI.00514-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saltis M, Davidson W. CDC2-related kinase and metabolic regulation in B cell malignancies. J Immunol. 2007;178:49. [Google Scholar]
- 34.Umetsu SE, Lee WL, McIntire JJ, Downey L, Sanjanwala B, Akbari O, et al. TIM-1 induces T cell activation and inhibits the development of peripheral tolerance. Nat Immunol. 2005;6:447–454. doi: 10.1038/ni1186. [DOI] [PubMed] [Google Scholar]
- 35.Broadley KN, Aquino AM, Woodward SC, Buckley-Sturrock A, Sato Y, Rifkin DB, et al. Monospecific antibodies implicate basic fibroblast growth factor in normal wound repair. Lab Invest. 1989;61:571–575. [PubMed] [Google Scholar]
- 36.Badour K, McGavin MK, Zhang J, Freeman S, Vieira C, Filipp D, et al. Interaction of the Wiskott-Aldrich syndrome protein with sorting nexin 9 is required for CD28 endocytosis and cosignaling in T cells. Proc Natl Acad Sci U S A. 2007;104:1593–1598. doi: 10.1073/pnas.0610543104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radojkovic M, Ristic S, Divac A, Tomic B, Nestorovic A, Radojkovic D. Novel ORC4L gene mutation in B-cell lymphoproliferative disorders. Am J Med Sci. 2009;338:527–529. doi: 10.1097/MAJ.0b013e3181b7f17c. [DOI] [PubMed] [Google Scholar]
- 38.Thickett KM, Kumararatne DS, Banerjee AK, Dudley R, Stableforth DE. Common variable immune deficiency: respiratory manifestations, pulmonary function and high-resolution CT scan findings. QJM. 2002;95:655–662. doi: 10.1093/qjmed/95.10.655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.