Abstract
Background
Pseudomonas fluorescens is a common inhabitant of soil and the rhizosphere environment. In addition to potential applications in biocontrol and bioremediation, P. fluorescens is of interest as a model for studying bacterial survival and fitness in soil. A previous study using in vivo expression technology (IVET) identified 22 genes in P. fluorescens Pf0-1 which are up-regulated during growth in Massachusetts loam soil, a subset of which are important for fitness in soil. Despite this and other information on adaptation to soil, downstream applications such as biocontrol or bioremediation in diverse soils remain underdeveloped. We undertook an IVET screen to identify Pf0-1 genes induced during growth in arid Nevada desert soil, to expand our understanding of growth in soil environments, and examine whether Pf0-1 uses general or soil type-specific mechanisms for success in soil environments.
Results
Twenty six genes were identified. Consistent with previous studies, these genes cluster in metabolism, information storage/processing, regulation, and ‘hypothetical’, but there was no overlap with Pf0-1 genes induced during growth in loam soil. Mutation of both a putative glutamine synthetase gene (Pfl01_2143) and a gene predicted to specify a component of a type VI secretion system (Pfl01_5595) resulted in a decline in arid soil persistence. When examined in sterile loam soil, mutation of Pfl01_5595 had no discernible impact. In contrast, the Pfl01_2143 mutant was not impaired in persistence in sterile soil, but showed a significant reduction in competitive fitness.
Conclusions
These data support the conclusion that numerous genes are specifically important for survival and fitness in natural environments, and will only be identified using in vivo approaches. Furthermore, we suggest that a subset of soil-induced genes is generally important in different soils, while others may contribute to success in specific types of soil. The importance of glutamine synthetase highlights a critical role for nitrogen metabolism in soil fitness. The implication of Type 6 secretion underscores the importance of microbial interactions in natural environments. Understanding the general and soil-specific genes will greatly improve the persistence of designed biocontrol and bioremediation strains within the target environment.
Background
Pseudomonas spp are frequently found among the numerous bacterial genera in soil and water environments. Pseudomonads are often closely associated with animals and plants, but are also found living free in bulk soil. Apart from their probable ecological importance, several P. fluorescens strains are of interest as potential biological control agents. A considerable body of research has shown that secondary metabolites are critical for biocontrol, both in vitro and in greenhouse experiments [1-7]. Unfortunately, greenhouse success has not consistently translated to success in field applications. Determining mechanisms by which pseudomonads persist and compete in soil would be of use in improving biocontrol strategies as well as in deepening the understanding of microbial success within natural environments.
A substantial body of work has given insight into bacterial fitness in laboratory culture systems, and to a lesser extent genetic experiments have been used to decipher environment-specific aspects of fitness which may not be apparent during growth in laboratory media [8-11]. IVET experiments have shown that a wide range of genes are induced during growth of Pseudomonas sp. on plant surfaces and in soil, and these genes fall into several broad functional categories. For example, genes important for utilization of various carbon sources, basic metabolism, transport, regulation, and antisense to known genes were identified as upregulated in Burkholderia multivorans[8] and P. fluorescens Pf0-1 [11,12] during growth in soil. These studies have provided insight into genetic circuits which promote fitness, and point the way to targets which may be manipulated to improve our ability to successfully apply exogenous bacteria to soil environments. Applications which could benefit from this knowledge include biological control of plant pathogens, and bioremediation. Despite progress, our knowledge on how microbes survive and potentially adapt to new soil environments still limits further applications of the use of microbes.
Studies aimed at deciphering genetics of survival and persistence in natural environments have generally focused on the known environment of the bacterium in question. Experiments on P. fluorescens isolates have identified genes induced in strain SBW25 on sugar beet, the plant from which SBW25 was originally isolated, and in Pf0-1 in the soil from which it was isolated. In P. fluorescens Pf0-1, an antisense gene termed cosA was shown to be important for optimal colonization of loam soil [13] and proper regulation of the gene ppk, specifying polyphosphate kinase, was demonstrated to be necessary for competitive fitness [14]. In P. fluorescens SBW25, genes controlling production of a cellulosic polymer were implicated as important for colonization of plant surfaces [12]. While informative, these experiments do not ask what is required to colonize and persist in new environments, an ability which is critical for expanding the ecological niche of the organism and for application to new environments in biocontrol. To address this question, we used a comparative approach based on IVET technology to identify the genetic basis of adaptation of P. fluorescens Pf0-1 to growth in soils. This type of approach is analogous to those used in determining the least number of genes required for growth in Staphylococcus aureus or sporulation in the Bacilli and Clostridia [15,16]. Those studies entailed comprehensive genetic searches for factors required for growth or sporulation in a target organism and a comparative analysis to a distantly related bacterium. We examined the complement of P. fluorescens genes expressed in arid soil, and tested a subset of these for their effect on colonization of both arid and agricultural loam soil. Our experiments suggest that nitrogen homeostasis is a key factor in adaptation to any soil.
Methods
Bacterial strains, plasmids, culture conditions, and primers
Bacterial strains and plasmids used in this study are described in Table 1. Wild type P. fluorescens Pf0-1 and Pf0-1Δdap were grown in LB [17] or Pseudomonas Minimal Medium (PMM) [18] at 27°C with shaking at 250 rpm. Escherichia coli strains were grown in LB medium at 37°C shaking at 250 rpm. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 10 μg/ml; tetracycline, 10 μg/ml or 25 μg/ml (for E. coli or P. fluorescens, respectively); carbenicillin, 100 μg/ml; and streptomycin, 20 μg/ml. In addition, agar media used to grow Pf0-1Δdap, or its derivatives, was amended with 10 μg/ml of diaminopimelic acid (DAP) (Sigma). Media used to detect β-galactosidase activity contained 35 μg/ml of X-Gal (5-bromo-4-chloro-3-indoyl-β-D-galactopyranoside). Oligonucleotide primers used in this study were synthesized by IDT (Coralville, IA) and are listed in Table 2.
Table 1.
Bacterial strains and plasmids
| Strains, plasmids or primers | Genotype or description | Reference or source |
|---|---|---|
|
E. coli |
|
|
| DH5αλ pir |
φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoR λpir |
[19] |
|
P. fluorescens |
|
|
| Pf0-1 |
Wild type, Ampr |
[20] |
| PfΔdap |
Pf0-1 ΔdapB |
This Study |
| Pf0-1::pKNOCK Fr2 |
Pf0-1 with partial deletion of Fr2 |
This Study |
| Pf0-1::pKNOCK Fr4 |
Pf0-1 with partial deletion of Fr4 |
This Study |
| Pf0-1::pKNOCK Fr9 |
Pf0-1 with partial deletion of Fr9 |
This Study |
| Pf0-1::pKNOCK Fr10 |
Pf0-1 with partial deletion of Fr10 |
This Study |
| Pf0-1::pKNOCK Fr2pJB |
Pf0-1 with partial deletion of Fr2 carrying pJB866 |
This Study |
| Pf0-1::pKNOCK Fr2pJB Fr2 |
Pf0-1 with partial deletion of Fr2 Complemented by pJB866::Fr2 |
This Study |
| Pf0-1::pKNOCK Fr10pJB |
Pf0-1 with partial deletion of Fr10 carrying pJB866 |
This Study |
| Pf0-1::pKNOCK Fr10pJB Fr10 |
Pf0-1 with partial deletion of Fr10 Complemented by pJB866::Fr10 |
This Study |
|
Plasmids |
|
|
| pGEM-T Easy |
Apr; cloning vector for PCR products |
Promega |
| pIVETdap |
dapB’ cloned in SpeI site of pUIC3 |
[11] |
| pJB866 |
Broad-host-range vector, Tcr |
[21] |
| pRK2013 |
Helper plasmid, IncP Tra+ Mob+ ColE1, Kmr |
[21] |
| pKNOCK-Km |
pBSL63 derivative carrying RP4 oriT and R6K γ-ori, Kmr |
[22] |
| pSR47s | Kmr; sacB-containing suicide vector (requiring R6K replication origin) | [23] |
Table 2.
Oligonucleotide primer sequences
| Primers | Description | Reference or source |
|---|---|---|
| Pbla |
5′-CAGGGTTATTGTCTCATGAGCG-3′ |
[12] |
| Pdap |
5′-CCGCCTCTACCAGCGTCTTGCC-3 |
[12] |
| DapB1 |
5′- GCATGAGAGCTCACCCTTTCCGTCAAAGTGC -3′ |
This Study |
| DapB2 |
5′- AAACCAGCGGCCGCTATACGTCGCATGCCGACTCC -3′ |
This Study |
| DapB3 |
5′- CGTATAGCGGCCGCTGGTTTGTACGACATGCAGG -3 |
This Study |
| DapB4 |
5′- TTACATGTCGACTTGCTCGCTACCAGCGG -3′ |
This Study |
| fFr2 |
5′- GTAACTGTTGGCCTGGAA -3′ |
This Study |
| rFr2 |
5′- GCCAAACGCGATCACA -3′ |
This Study |
| fFr4 |
5′- CCGCGTTATTCGCAGA -3′ |
This Study |
| rFr4 |
5′- TGTAATCATCCGGCCAGA -3′ |
This Study |
| fFr9 |
5′- GAGCCGACTGCACGAA -3′ |
This Study |
| rFr9 |
5′- TGGTCATGAGTTCGCTGA -3′ |
This Study |
| fFr10 |
5′- CGCACGTTCAGGCTGA -3′ |
This Study |
| rFr10 |
5′- CCAACAGCCACGAGCA -3′ |
This Study |
| fFr2com |
5′- ATTGCGGCCGCTCAGGCTTCGGTCAGATACC-3′ |
This Study |
| rFr2com |
5′- CGCACTAGTCGATGAAATTCGCAGCCATTGA -3′ |
This Study |
| fFr10com |
5′- GCGCAATTCTTACTCTTTGTCCAGCATGCCA -3′ |
This Study |
| rFr10com | 5′- ATTGCGGCCGCTATGAGCACTAGCGCAGCACA -3′ | This Study |
DNA manipulation and sequencing
Recombinant DNA techniques were carried out as described [17] or according to supplier instructions. Restriction enzymes and DNA modifying enzymes were purchased from Invitrogen (Carlsbad, CA), New England Biolabs (Ipswich, MA), and Promega (Madison, WI). Plasmid DNA was extracted using a QIAprep Spin Miniprep kit (Qiagen, Valencia, CA). DNA fragments were recovered from agarose gel slices using a QIAquick Gel Extraction kit (Qiagen). DNA was amplified by PCR using VentR DNA polymerase (NEB). PCRs to amplify DNA for cloning were all carried out using purified genomic DNA for the template (Wizard DNA Isolation Kit, Promega). Screening of mutants was carried out by colony PCR. When required, PCR products were cloned with pGEM-T Easy (Promega). DNA sequences were determined by the Nevada Genomic Center at the University of Nevada, Reno.
Construction of an in-frame dapB deletion in Pf0-1
The primer pairs DapB1/DapB2 and DapB3/DapB4 were used to PCR amplify upstream and downstream regions flanking dapB. The 5′ ends of DapB2 and DapB3 contained complementing linker sequences of 5′-AAACCAGCGGCCGCTATACG-3′ and 5′-CGTATAGCGGCCGCTGGTTT-3′ that were used to anneal both PCR products together. Annealed fragments were ligated into the plasmid pSR47s using the SalI and SacI sites, and used to transform E. coli DH5αλpir, resulting in pJGΔ101. The plasmid pJGΔ101 was transferred into Pf0-1 by conjugation to construct the dapB deletion by allele exchange, as we have described previously [11]. Deletion of dapB was confirmed by PCR, and by auxotrophy for DAP.
Construction of an IVET library
A Pf0-1 genomic library was constructed in the pIVETdap vector [11]. Pf0-1 genomic DNA was extracted from a culture grown in PMM for 18 h, using the Wizard® Genomic DNA Purification Kit (Promega; Madison, WI). The genomic DNA was partially digested with four units of Sau3A1 (New England Biolabs, Beverly, MA) for 18 minutes. The partially digested DNA was resolved by electrophoresis, and 1 to 3 kb fragments were isolated and purified from agarose fragments using a Qiaquick gel extraction kit (Qiagen, Valencia, CA). Fragments were ligated to dephosphorylated pIVETdap (Promega Calf Intestinal Alkaline Phosphatase) linearized with BglII, yielding the pIVETdap genomic library. Library DNA was used to transform E. coli DH5αλpir, and clones were selected in the presence of nalidixic acid and tetracycline. A pool of 9375 clones from several independent ligations was kept at -80°C.
Selection of soil-induced promoters
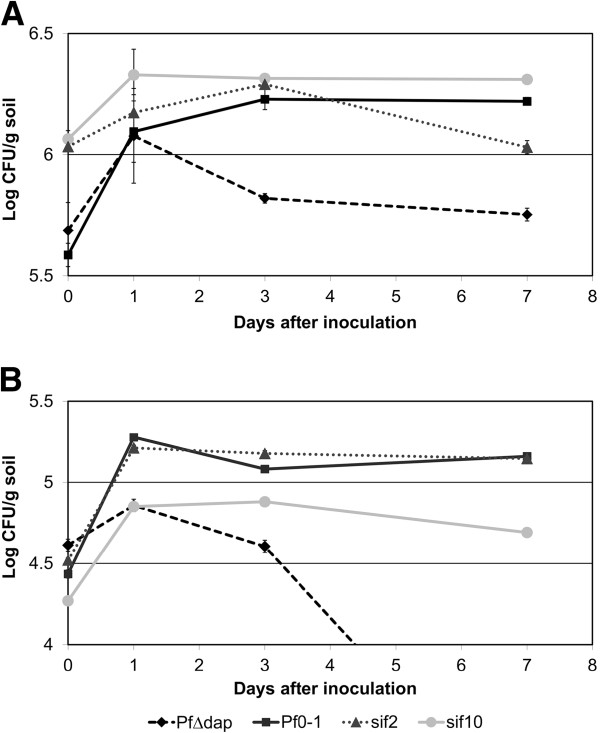
The Pf0-1 genomic library fused to a promoterless dapB in the plasmid pIVETdap (see above) was transferred by conjugation to Pf0-1ΔdapB. A pool of recombinant bacteria carrying pIVET fusion clones was diluted and adjusted with sterile double distilled water to 0.01 OD550. One mL of the bacterial suspension (approximately 5×105 CFU), was used to inoculate 5 g of arid Nevada desert soil (0.91% organic matter, 89.0% sand, 4.1% silt, and 6.9% clay, with a pH level of 8.3; for a more detailed description of soil properties see [24]) in a 35 ml Pyrex test tube. Prior to inoculation Nevada soil was sifted with 1 mm2 screen. Inoculation resulted in a wetting event. Soil water content throughout the experiment varied from fully saturated conditions (0 kPa) to permanent wilting point (-1500 kPa). Tubes were capped. Growth and persistence in soil depends on functional DapB (Figure 1). Strains that grow in soil carry promoters in the genomic fragment which activate dapB transcription, thus rescuing the no-growth phenotype. To carry out two rounds of seven- day soil exposure, a soil sample of 1 g from inoculated soil was recovered, suspended in 9 mL dH2O, and 1mL of suspension was used to inoculate a further 5 g of soil. Bacteria were allowed to grow in this soil for an additional 7 days.
Figure 1.
Growth and persistence in Nevada arid soil of P. fluorescens Pf0-1 carrying mutations in arid soil-induced genes relative to wild-type Pf0-1 and Pf0-1ΔdapB. A. When inoculated at relatively high density, the sif2 (Pfl01_2143) mutant fails to maintain the population density reached by wild-type Pf0-1 while the sif10 (Pfl01_5595) mutant shows no aberrant phenotype. B. When inoculated at relatively lower density, the sif10 (Pfl01_5595) mutant fails to establish the same population level as wild-type Pf0-1, whereas the sif2 (Pfl01_2143) mutant is indistinguishable from wild-type. In both panels, error bars represent 4 replications. Error bars represent standard errors. Anova for these experiments indicates significant values at P ≤0.01. For the experiments in 1A, difference values between any two means that were greater than 0.11 (day1), 0.05 (day3) and 0.08 (day7) denoted statistical significance. For the experiments in 1B, difference values between any two means that were greater than 0.07 (day1), 0.07 (day3) and 0.11 (day7) denoted statistical significance.
After the second 7-day period, a suspension was made from 1 g of soil (as described above), diluted, and plated onto Pseudomonas minimal medium supplemented with diaminopimelic acid (DAP) and X-gal, and ampicillin and tetracycline to select IVET strains. Control plates indicated that these conditions were effective at inhibiting growth of indigenous bacteria. White colonies presumed to contain soil-activated promoters fused to dapB were chosen for further study. We surmised that blue colonies carry fusions active in both soil and laboratory; these were not studied further.
Sequence and promoter analysis
DNA sequences from the 30 soil induced fragments (sif) were blasted against the Pf0-1 annotated genome. Based on their match to the annotated genome, sifs were grouped into metabolism, transport, regulation and poorly characterized genes categories (Table 3). In addition to BLAST analysis, promoter scans of the regions upstream of sifs were conducted using PromScan (http://molbiol-tools.ca/promscan/), which searches for σ54 (σN) consensus sequences [25].
Table 3.
Arid soil-induced coding sequences
| Soil-induced fragment | Locus tag | Annotated product | COG ID | Grouping |
|---|---|---|---|---|
| Nutrition and transport | ||||
| 28ab |
Pfl01_2547 |
Putative 4-alpha-glucanotransferase |
COG1640 |
Carbohydrate transport and metabolism |
| 29 |
Pfl01_0225 |
Amino acid ABC transporter, permease protein |
COG0765 |
Amino acid transport and metabolism |
| 2b |
Pfl01_2143 |
Putative glutamine synthetase |
COG1629 |
Amino acid transport and metabolism |
| Secretion | ||||
| 10 |
Pfl01_5595 |
type VI secretion protein TssB2 |
COG3516 |
T6SS |
| Regulation | ||||
| 11a |
Pfl01_5642 |
Transcriptional Regulator, RpiR family |
COG1737 |
Regulation of phosphosugarmetabolism |
| 9a |
Pfl01_3972 |
Putative diguanylate phosphodiesterase (EAL domain-containing protein) |
COG2200 |
Signal transduction mechanisms |
| 18 |
Pfl01_0719 |
Transcriptional Regulator, LysR family |
COG0583 |
Transcriptional regulation |
| 24 |
Pfl01_2366 |
Transcriptional Regulator, XRE family |
COG1709 |
Translation, ribosomal structure and biogenesis |
| Defense | ||||
| 4 |
Pfl01_2660 |
Putative 5-Methylcytosine-specific restriction enzyme |
COG1401 |
Defense Mechanism |
| Poorly Characterized and uncharacterized | ||||
| 16 |
Pfl01_1075 |
Conserved hypothetical with extensin-like domain |
COG3921 |
Function unknown |
| 23 |
Pfl01_3777 |
Hypothetical protein |
COG0596 |
General function prediction only |
| 19 |
Pfl01_0609 |
Hypothetical protein |
|
|
| 27a |
Pfl01_2750 |
Hypothetical protein |
|
|
| 20 |
Pfl01_2901 |
Xylose isomerase-like TIM barrel |
|
|
| Antisensec | ||||
| 13a |
Pfl01_3287 |
Putative Rho-binding antiterminator |
COG4568 |
Transcription |
| 8a |
Pfl01_5547 |
Ribonuclease PH |
COG0689 |
Transcription |
| 7 |
Pfl01_4448 |
Pyruvate Kinase |
COG0469 |
Carbohydrate transport and metabolism |
| 12a |
Pfl01_4455 |
Putative insecticidal Toxin Protein (TccC) |
|
|
| 25 |
Pfl01_4265 |
Cytochrome C family protein |
|
|
| 30a |
Pfl01_3916 |
alkanesulfonate monooxygenase |
|
|
| 1 |
Pfl01_0250 |
TonB-dependent receptor |
|
|
| 21 |
Pfl01_2744 |
Putative Thiolase |
|
|
| 26 |
Pfl01_0911 |
Putative Fumarylacetoacetase |
|
|
| 3 |
Pfl01_5256 |
Putative alginate lyase |
|
|
| 14 | Pfl01_5509 | Hypothetical protein | ||
(a) indicates the absence of a sigma 70 promoter; (b) indicates that the region was recovered twice in independent assays; (c) for antisense loci, the annotated product refers to the coding sequence found opposite the IVET-recovered antisense sequence. Locus tag is NCBI identification number for the P. fluorescens coding sequences.
Construction of mutant strains
To construct genetic variants defective in the genes expressed in arid soil conditions, internal sequences (varying from 300 to 700 bp) of sif2, sif4, sif9 and sif10 were amplified using Pf0-1 genomic DNA template and primers shown in Table 2, and cloned in pGEM®-T Easy (Promega, WI). The internal fragments of sif2, sif4, sif9 and sif10 were released from pGEM®-T Easy with EcoRI, and cloned into the EcoRI site of pKNOCK [22]. The resulting clones (pKNOCK/EcoRI: sif2, pKNOCK/EcoRI: sif4, pKNOCK/EcoRI:: sif9 and pKNOCK/EcoRI: sif10) were used to transform E. coli DH5αλpir, and subsequently transferred to Pf0-1 by conjugation in the presence E. coli carrying the helper plasmid pRK2013. Transconjugants from each mating were selected for ampicillin and kanamycin resistance, which gave rise to Pf0-1: pKNOCK sif2, Pf0-1: pKNOCK sif4, Pf0-1: pKNOCK sif9 and Pf0-1: pKNOCK sif10 respectively. These four strains were subject to the arid soil assay (described below).
Complementation
The primer pairs fFr2com/rFr2com and fFr10com/rFr10com (Table 2) were used to amplify Pfl01_2143 (sif2) and Pfl01_5593 (sif10) from the Pf0-1 genome, respectively. Purified PCR products were digested with either AflIII and NotI (sif2), or EcoRI and NotI (sif10) and cloned into the AflIII/NotI or EcoRI/NotI sites of pJB866 respectively, yielding the complementation plasmids pJB866:: sif2 and pJB866:: sif10. The complementation plasmids were transferred by conjugation into Pf0-1::pKNOCK sif2 and Pf0-1::pKNOCK sif10 (triparental matings with pRK2013 helper), generating Pf0-1::pKNOCK sif2+ sif2 and Pf0-1::pKNOCK sif10+ sif10. The two complemented strains were subject to colonization of arid soil.
Nevada soil growth and survival assays
Growth and survival of mutant strains in arid Nevada desert soil was carried out essentially as described in the section detailing the screening of the IVET library, with some modifications. Individual strains were grown for 20 h in PMM prior to dilution to an OD550 value of 0.01 or 0.001, and used to inoculate 5 g soil. Populations were monitored by periodic sampling and plating of dilutions as outlined above. The different inoculation densities were used to more fully explore colonization and persistence traits in the face of competition from indigenous microbes.
Massachusetts soil growth and competition assays
The soil used in these experiments was a gamma irradiated fine loam from Sherborn, Massachusetts, as described [26]. Bacterial strains were grown for 16 h in PMM with appropriate antibiotics, after which cells were diluted to approximately 1×105 cfu/mL in sterile distilled H2O (sdH2O). Soil growth and competition assays were carried out as described previously [14], but with the addition of 0.5% (w/w) CaCO3 to increase the pH to approximately 7. For soil growth experiments, 1mL of diluted cell suspension was mixed with 5 g of soil, achieving a water holding capacity of approximately 50%. For competition experiments, cultures were adjusted to equal OD600 values prior to dilution, and then 500 μL of each diluted competing strain were combined, and mixed with soil as for the survival experiments. Note that the OD600 here does not differ significantly from the OD550 used in the arid soil experiments. Inoculated soil samples were transferred to 15 mL polypropylene conical tubes. After 30 minutes, the initial recoverable population was established by removal of 0.5 g of soil, and recovery of and enumeration of bacteria from each sample, as we have described previously [11]. The initial populations of wild-type and mutant strains were approximately equal. The population of each member in a competing pair was monitored over time by extraction of bacteria from the soil, and determination of numbers by counting colonies on appropriate selective media [14]. The wild-type strain in competition experiments was Pf0-1Smr. In wild-type vs wild-type controls, Pf0-1Smr was competed with Pf0-1Kmr. Previous work has shown that these selective markers do not influence fitness [13,14]. The competitive index is the ratio of mutant: wild-type at a given time point divided by the initial mutant: wild-type ratio.
Statistical tests
Statistical analyses were carried out using Microsoft Excel and GraphPad Prism v5 (GraphPad Software Inc). Specific tests are indicated in the figures in which data are presented. For the arid soil experiments, the statistical tests performed were based on ANOVAs between the strain treatments and total variance. A student′s t test with an alpha value of 0.05 was used to calculate the least significant difference between means. For competition experiments, an unpaired T-test was used, with p<0.05 used to define statistically significant differences.
Results and discussion
IVET selection of Pf0-1 promoters induced in arid Nevada desert soil
A library of DNA fragments, covering 94% of the P. fluorescens genome, was used to trap promoters induced during growth in arid Nevada desert soil, a non-native soil for Pf0-1, essentially as described previously in IVET studies of agricultural soil [11]. After two rounds of growth and enrichment in soil, bacteria which survived the soil environment were examined for expression of the fusions in vitro by plating onto medium containing X-gal. Thirty white colonies of the 3000 that were recovered (about 1%) contained dapB-lacZ fusions transcriptionally activated in soil conditions but repressed in laboratory media were chosen for further study.
The pIVETdap-based plasmids excise from the Pf0-1 genome at a low frequency, allowing recovery from the 30 strains of interest by plasmid isolation and subsequent transformation of E. coli. The Pf0-1 sequence fused to dapB in each recovered IVET plasmid was identified by DNA sequencing using the pdap primer, followed by comparison to the Pf0-1 genome sequence [27]. Sequences obtained matched predicted genes or expressed sequences antisense to predicted genes, as has been reported in previous IVET studies [for examples see [12,27-29]. Three genes, including one ‘antisense’ sequence, were recovered twice in independent selection experiments, which validated the use of IVET.
Analysis of arid soil-activated genes
Among the 30 IVET-identified sequences isolated were representatives of several major functional groups (Table 3). Although the IVET-identified genes fell into similar broad functional categories, none of the sequences recovered here matched those results from a previous study of loam soil [11]. The lack of overlap suggests that either one or both screens provided incomplete coverage of the Pf0-1 genome, or that the transcriptional response of Pf0-1 upon introduction to arid sandy soil is distinct from that seen in loam soil. Genes we have identified as up-regulated in arid soil are predicted to have roles in metabolism, transport, and regulation. Eleven expressed sequences were reverse complements to annotated genes in the Pf0-1 genome, further supporting the suggestion that antisense regulation is widespread and important in bacteria [30-33]. Five poorly characterized/hypothetical genes were identified. The identification of novel genes induced in soil suggests that these novel functions may need to be investigated in the context of complex non-laboratory environments where their expression is induced.
We have not experimentally determined the factors in soil which induce expression of the sif genes, but some insight is possible from analysis of putative promoters. The antisense sequences sif12 and sif30 are both predicted to be preceded by sigma54-dependent promoters. In other organisms, the σ54-mediated response is at least in part because of nitrogen limitation, suggesting the possibility that low nitrogen levels in soil trigger expression of these antisense genes as repressors. This suggestion fits with that for sif2 (discussed below).
Nutrient use and transport
Two of the soil-induced fragments (fragments 2 and 29; Table 3) are predicted to be related to amino acid production or transport. The sif2 locus is predicted to encode a glutamine synthetase. Mutation of sif2 does not result in glutamine auxotrophy under laboratory conditions, possibly because of the presence of the multiple genes for glutamine synthetases predicted in the Pf0-1 genome, and as has been noted previously in Rhizobium meliloti[34]. Amino acid transporters and a glutamine synthetase ortholog were identified in an IVET study of Burkholderia multivorans[8], supporting the general importance of such systems in soil, and possibly implicating nitrogen homeostasis as a critical factor for optimal growth and persistence in soil (see discussion below). Soil-induced fragment 28 is predicted to encode a 4-alpha-glucanotransferase, similar to MalQ of E. coli. MalQ is important in the metabolism of maltose and maltodextrins [35], possibly suggesting that maltose or maltodextrins, derived from partial hydrolysis of plant starch, are used as carbon or energy sources in arid soil.
Regulation
Four sequences showing similarity to members of different regulatory families were identified in our IVET screen in arid soil (Table 3). As the bacteria passage from the laboratory to the soil environment, numerous environmental parameters are altered. Many of these changes will necessitate an adaptive response by the bacterium to enable competitive survival. Up-regulation of a range of regulatory proteins has been observed previously in studies of P. fluorescens strains in soil and on plant surfaces [11,27] and in a study of soil-induced genes in B. multivorans[8]. While the sequence similarities of the sifs are indicative of how DNA transcription is induced, our ability to predict the nature of the environmental regulatory responses is limited by the fact that the up-regulated regulator genes do not appear to encode orthologs of functionally characterized proteins.
Antisense
Several IVET screens have yielded fusions to the reporter in which the annotated gene in the fusion appears to be transcribed away from the reporter [for example [8,11,29,36-38]. In the present study, 11 of 25 unique fusions were in the reverse fusion ‘antisense’ category. It has been suggested that these reverse fusions identify transcribed sequences which function as cis-acting antisense regulators of the annotated genes [28,29,39]. There are at least two cases showing biological relevance for cis-acting antisense elements in soil environments [13,40]. The reverse fusions found in this study may indicate antisense transcripts involved in controlling a range of processes: insecticidal toxin production (sif12); antitermination of transcription (sif13); pyruvate kinase (sif7); sulfur scavenging (sif30); tRNA maturation/processing (sif8); transport of iron or perhaps other substrates (sif1) [41]; degradation of alginate (sif3), beta oxidation of fatty acids (sif21), and phenylalanine or tyrosine (sif26). The relevance of these for colonization of soil and long term persistence remains to be explored, but it is possible to suggest a role for controlling these processes in soil. For example, it seems reasonable to speculate that cells benefit from controlling degradation of large molecules such as alginate which may have been costly to produce and could be necessary or important for survival.
Evidence for transcription of regions that produce RNA antisense to predicted genes has accumulated from genetic studies similar to this [for example [11,28,38,42], and more recently from strand-specific transcriptome sequencing [for example [43-46]. Most of these antisense RNA (asRNA) molecules are of unknown function, and are thought-provoking because they support the concept that bacterial genomes have ‘dark matter’, functional regions not easily detectable with standard gene-finding algorithms [47]. Recent functional studies have begun to assign roles to asRNA molecules [for example [13,40,44,48], and those uncovered in this study provide a rich resource for future experiments which will further expand our understanding of the genetics of soil survival and persistence.
Soil-induced genes influence survival in arid soil
Four IVET-identified genes representing different functional classes were chosen for mutational studies. Using pKNOCK-km [22] we generated mutants of sif2, 4, 9, and 10, and tested these for colonization of and persistence in arid soil. The mutations in sif4 and sif9 did not alter colonization or survival of Pf0-1 in arid soil (data not shown). In contrast, disruption of both sif2 and sif10 resulted in small but significant changes in the performance of Pf0-1 in arid soil.
When soil was inoculated with the sif2 mutant diluted to above 105 cfu/mL, the mutant was slightly impaired in its ability to persist over a seven day period relative to Pf0-1 (Figure 1A). The predicted role for sif2 in nitrogen metabolism suggests that maintenance of a high population depends on the ability to assimilate sufficient nitrogen, and the sif2 mutant is reduced in this function in soil. Under the same conditions, the sif10 mutant showed no such defect. In contrast, when soil was inoculated with 10-fold fewer cells, the sif10 mutant was depressed in soil colonization while the sif2 mutant reached a similar population to the wild-type (Figure 1B). We suggest that sif2 is important in the maintenance of high population density in soil, while the role of sif10 is in the establishment of high density. Thus, sif2 appears to have no effect when the inoculation is low (Figure 1B), because under these conditions Pf0-1 does not reach the density at which sif2 is required (>6 log cfu/g of soil). Conversely, sif10 is not necessary at higher inoculation levels (Figure 1A) because the population threshold below which sif10 is important (<5 log cfu/g of soil) has already been surpassed. The effects of the sif2 and sif10 mutations were reversed by complementation (not shown).
It is important to note that the effects of sif2 and sif10 inactivation on soil colonization/persistence are small but significant. This was observed in independent replicate experiments that included the complemented strains (P≤0.01). The sif2 and sif10 regions were identified based on induction of expression and may contribute additively to arid soil colonization/persistence. The fact that one sif-defective strain fails to compete against the parental strain in a different environment (see section on agricultural soil) supports the notion that effects observed in arid soil were not experimental artifacts.
These two genes which were upregulated during growth in arid soil are important for optimal performance of Pf0-1 in that environment and represent attractive targets to improve persistence in bacteria applied to natural environments as biocontrol or bioremediation agents. Alternatively, identification of these sequences which contribute to fitness could add to a catalog of desirable traits which can be sought when prospecting for new biocontrol/bioremediation strains.
The sif10 sequence identifies Pfl01_5595 as being induced in arid soil, and important for colonization of arid soil. Pfl01_5595 is predicted to be part of an HSI-II type six secretion system (T6SS) gene cluster encoded by Pfl01_5577-Pfl01_5596 [49]. T6SSs translocate effectors from the secreting cell into both eukaryote and prokaryote targets (depending on the T6SS system in question) in a contact-dependent manner reviewed in [50]. For example, P. aeruginosa has three T6SS gene clusters, at least two of which have distinct functions [51]. The gene Pfl01_5595 is a predicted ortholog of the P. aeruginosa gene PA1657 which specifies the T6SS component termed TssB, which along with TssC is thought to assemble into structures similar to bacteriophage sheath which may be contractile. This ‘sheath’ is found around a phage tail filament-like structure, and mediates the secretion of effectors into target cells [50]. T6S has been implicated in virulence toward eukaryotic hosts [for example [51-53].
Although sif10 has not yet been experimentally confirmed to participate in T6S, we suggest that in soil sif10 could participate in effector translocation, negatively impacting the recipient cell. In the live arid soil used here it is possible that sif10 helps to reduce the fitness of competing bacteria by actively suppressing their growth. Many bacteria secrete antibacterial compounds into the milieu, which may inhibit competitors from a distance. However, the potential implication of T6S in fitness points toward an additional more intimate way by which bacteria may interact with and inhibit their neighbors in natural environments such as soil.
Previous studies of genes specifically induced within a given environment have yielded similar data in terms of the importance of those genes for survival or fitness. Selected environmentally induced genes from P. fluorescens isolates have been shown to be important in soil colonization [11] phyllosphere colonization [12], and a subset of V. cholerae genes induced in an infant mouse model of cholera were important for colonization [38]. The cholera study and our own unpublished data for P. fluorescens in agricultural soil indicate that only a subset of environmentally induced genes are necessary for full fitness in those environments, as has also been shown in the present study. It seems likely that the majority of important environmental functions have some level of functional redundancy.
Arid soil survival genes have varied importance in agricultural soil
We noted the absence of overlap between the Pf0-1 genes found to be upregulated in arid soil and those identified as upregulated in agricultural loam soil [11]. This difference could be because of limited sampling, or because of specific requirements for colonization of, and persistence in, different soil types. The soils used in these experiments differ considerably in content [24,26], and thus it might not be unexpected for different traits to be required by Pf0-1. To examine these possibilities, we tested the sif2 and sif10 mutants for colonization and competitive fitness in sterile agricultural loam soil as we have done in previous studies [11,14]. Neither mutant showed a colonization or persistence defect relative to Pf0-1 when inoculated alone into the sterile loam soil (not shown). However, when in competition with Pf0-1 the sif2 mutant showed a significant competitive defect (Figure 2) while the sif10 continued to show no discernible phenotypic difference from Pf0-1 in the agricultural soil (not shown).
Figure 2.
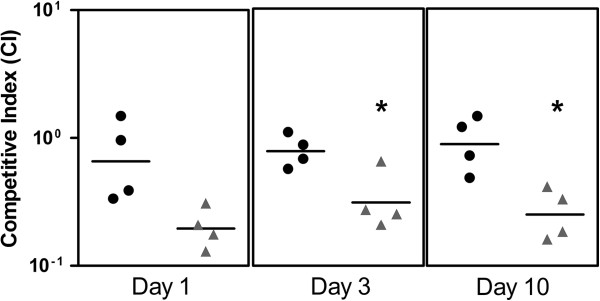
Competitive fitness of the sif2 (Pfl01_2143) mutant relative to wild-type Pf0-1 in Massachusetts agricultural loam soil. Black circle symbols represent the competitive index of the control experiment where differently marked wild-type Pf0-1 strains are competed against each other. Each data point represents the result from an independent experiment (four trials total). Neither strain has a competitive advantage. Grey triangle symbols represent the competitive index of the sif2 mutant relative to Pf0-1. When differently marked mutant and wild-type strains are used to co-inoculate soil, the mutant is outcompeted by the wild-type. The competitive index was calculated by dividing the ratio of mutant:wildtype on a particular day by the initial ratio at the beginning of the experiment. An asterisk indicates the differences at day 3 and day 10 are significant (p<0.05; unpaired T-test).
The importance of sif2 in both soil types suggests that its function in soil relates to a characteristic common to the arid and agricultural loam soils. In terms of composition, these soils are not generally similar. Physical parameters differ greatly between them, as does mineral content [24,26]. However, low inorganic nitrogen content is common between these, and probably many other soil types. The arid desert soil has a nitrate content of 15 ppm, and the agricultural loam soil used contains 69 ppm nitrate. These levels are far below those added to defined growth media used in laboratory culture such as M9 medium [17] or PMM [18]. The sif2 sequence is predicted to specify one of several glutamine synthetases in Pf0-1. Glutamine is central to nitrogen flow in cellular metabolism, making nitrogen available for many biosynthetic reactions reviewed in [54]. Glutamine synthetases are critical players in the assimilation of nitrogen.
In E. coli glutamine synthetase, encoded by glnA, is intricately involved in nitrogen assimilation. In nitrogen-limiting conditions, expression of glnA is increased, thereby increasing glutamine synthetase-mediated assimilation of ammonia. Glutamine is then transformed by glutamate synthase into glutamate, which makes glnA the first step in ammonia assimilation. Inactivation of glnA renders E. coli auxotrophic for glutamine in conditions in which ammonia, the preferred source of inorganic nitrogen in E. coli, is the sole N source. Further, in N-limiting conditions the glutamine synthetase-dependent ammonia-assimilation pathway provides close to 100% of the N required in the cell. Expression of glutamine synthetase is controlled by NtrB, NtrC and GlnK, which sense glutamine levels in the cell [55].
In Synechocystis PCC6804, two glutamine synthetases are responsive to nitrogen availability, but differently so. The glnN gene is up-regulated greatly during nitrogen starvation compared to the expression level during growth in the presence of nitrate or ammonium [56].
Conclusions
Pseudomonas fluorescens Pf0-1 upregulates many genes upon encountering natural environments such as soil. It is thus essential to characterize this and other bacteria in the context of their natural environments in order to gain a full appreciation of the mechanisms they use to survive and thrive. P. fluorescens Pf0-1 has specific genetic responses to different soil types, but also general mechanisms required for persistence. Our observation that sif2 is important in two distinct soil types points to a general phenomenon in which bacterial responsiveness to nitrogen and its shunting into central metabolism via glutamine in situ is critical for fitness. This concept is further supported by the observation that several of soil-activated sequences are associated with putative σ54 promoters. Thus, a general key element in bacterial adaptation to soils is to maintain nitrogen homeostasis.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
KV carried out the IVET screen and subsequent experiments in arid soil, and contributed to the writing of the manuscript; LC carried out experiments in agricultural soil, performed statistical tests, and contributed to manuscript writing. MS and ER designed and oversaw the study and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Katila Varivarn, Email: katila2000@hotmail.com.
Lindsey A Champa, Email: lchampa@umassd.edu.
Mark W Silby, Email: mark.silby@umassd.edu.
Eduardo A Robleto, Email: eduardo.robleto@unlv.edu.
Acknowledgements
This work was supported in part by the Agriculture and Food Research Initiative Competitive Grant 2010-65110-20392 from the USDA’s National Institute of Food and Agriculture, Microbial Functional Genomics Program.
References
- Chin-A-Woeng TFC, Bloemberg GV, van der Bij AJ, van der Drift KMGM, Schripsema J, Kroon B, Scheffer RJ, Keel C, Bakker PAHM, Tichy HV. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant Microbe Interact. 1998;11:1069–1077. doi: 10.1094/MPMI.1998.11.11.1069. [DOI] [Google Scholar]
- Thomashow LS, Weller DM. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DS, Stein JI, Torkiewitz NR, Morse AM, Howell CR, Pachlatko JP, Becker JO, Ligon JM. Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomonas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl Environ Microbiol. 1994;60:78–85. doi: 10.1128/aem.60.1.78-85.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laville J, Blumer C, Von Schroetter C, Gaia V, Defago G, Keel C, Haas D. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J Bacteriol. 1998;180:3187–3196. doi: 10.1128/jb.180.12.3187-3196.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza JT, Weller DM, Raaijmakers JM. Frequency, Diversity, and Activity of 2,4-Diacetylphloroglucinol-Producing Fluorescent Pseudomonas spp. in Dutch Take-all Decline Soils. Phytopathology. 2003;93:54–63. doi: 10.1094/PHYTO.2003.93.1.54. [DOI] [PubMed] [Google Scholar]
- Fenton AM, Stephens PM, Crowley J, O’Callaghan M, O’Gara F. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl Environ Microbiol. 1992;58:3873–3878. doi: 10.1128/aem.58.12.3873-3878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell CR, Stipanovic RD. Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology. 1980;70:712–715. doi: 10.1094/Phyto-70-712. [DOI] [Google Scholar]
- Nishiyama E, Ohtsubo Y, Nagata Y, Tsuda M. Identification of Burkholderia multivorans ATCC 17616 genes induced in soil environment by in vivo expression technology. Environ Microbiol. 2010;12:2539–2558. doi: 10.1111/j.1462-2920.2010.02227.x. [DOI] [PubMed] [Google Scholar]
- Winstanley C, Langille MGI, Fothergill JL, Kukavica-Ibrulj I, Paradis-Bleau C, Sanschagrin Fo, Thomson NR, Winsor GL, Quail MA, Lennard N. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 2009;19:12–23. doi: 10.1101/gr.086082.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio CG, Crawford JA, Michalski J, Martinez-Wilson H, Kaper JB, Camilli A. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect Immun. 2005;73:972–980. doi: 10.1128/IAI.73.2.972-980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby MW, Levy SB. Use of IVET to identify genes important in growth and survival of Pseudomonas fluorescens Pf0-1 in soil: discovery of expressed sequences with novel genetic organization. J Bacteriol. 2004;186:7411–7419. doi: 10.1128/JB.186.21.7411-7419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal M, Preston GM, Massey RC, Spiers AJ, Rainey PB. Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol Ecol. 2003;12:3109–3121. doi: 10.1046/j.1365-294X.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- Silby MW, Levy SB. Overlapping Protein-Encoding Genes in Pseudomonas fluorescens Pf0-1. PLoS Genet. 2008;4:e1000094. doi: 10.1371/journal.pgen.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby MW, Nicoll JS, Levy SB. Requirement of Polyphosphate by Pseudomonas fluorescens Pf0-1 for Competitive Fitness and Heat Tolerance in Laboratory Media and Sterile Soil. Appl Environ Microbiol. 2009;75:3872–3881. doi: 10.1128/AEM.00017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, Rigden DJ. Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol. 2012;14:2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Kirner S, Krauss S, Sury G, Lam ST, Ligon JM, Van Pee KH. The non-haem chloroperoxidase from Pseudomonas fluorescens and its relationship to pyrrolnitrin biosynthesis. Microbiology. 1996;142:2129–2135. doi: 10.1099/13500872-142-8-2129. [DOI] [PubMed] [Google Scholar]
- Kolter R, Inuzuka M, Helinski DR. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Compeau G, Al-Achi BJ, Platsouka E, Levy SB. Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl Environ Microbiol. 1988;54:2432–2438. doi: 10.1128/aem.54.10.2432-2438.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatny JM, Brautaset T, Winther-Larsen HC, Karunakaran P, Valla S. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid. 1997;38:35–51. doi: 10.1006/plas.1997.1294. [DOI] [PubMed] [Google Scholar]
- Alexeyev MF. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques. 1999;26:824–826. doi: 10.2144/99265bm05. [DOI] [PubMed] [Google Scholar]
- Matthews M, Roy CR. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect Immun. 2000;68:3971–3982. doi: 10.1128/IAI.68.7.3971-3982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus JH, Nowak RS, Smith SD. Soil resource heterogeneity in the Mojave Desert. J Arid Environ. 2002;52:269–292. doi: 10.1006/jare.2002.1010. [DOI] [Google Scholar]
- Studholme DJ, Dixon R. Domain Architectures of σ54-Dependent Transcriptional Activators. J Bacteriol. 2003;185:1757–1767. doi: 10.1128/JB.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastropaolo MD, Silby MW, Nicoll JS, Levy SB. Novel Genes Involved in Motility and Biofilm Formation in Pseudomonas fluorescens Pf0-1. Appl Environ Microbiol. 2012;78:4318–4329. doi: 10.1128/AEM.07201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby MW, Cerdeno-Tarraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, Zhang X-X, Moon CD, Gehrig SM, Godfrey SAC. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 2009;10:R51. doi: 10.1186/gb-2009-10-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby MW, Rainey PB, Levy SB. IVET experiments in Pseudomonas fluorescens reveal cryptic promoters at loci associated with recognizable overlapping genes. Microbiology. 2004;150:518–520. doi: 10.1099/mic.0.26871-0. [DOI] [PubMed] [Google Scholar]
- Mahan MJ, Slauch JM, Mekalanos JJ. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- Lasa I, Toledo-Arana A, Dobin A, Villanueva M, Mozos IR Dl, Vergara-Irigaray M, Segura V, Fagegaltier D, Penadés JR, Valle J. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci USA. 2011;108:20172–20177. doi: 10.1073/pnas.1113521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornenburg JE, DeVita AM, Palumbo MJ, Wade JT. Widespread Antisense Transcription in Escherichia coli. mBio. 2010;1:e00024–10. doi: 10.1128/mBio.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Hess WR. cis-Antisense RNA, Another Level of Gene Regulation in Bacteria. Microbiol Mol Biol Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Vosz B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn FJ, Rossbach S, Schneider M, Ratet P, Messmer S, Szeto WW, Ausubel FM, Schell J. Rhizobium meliloti 1021 has three differentially regulated loci involved in glutamine biosynthesis, none of which is essential for symbiotic nitrogen fixation. J Bacteriol. 1989;171:1673–1682. doi: 10.1128/jb.171.3.1673-1682.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos W, Shuman H. Maltose/Maltodextrin System of Escherichia coli: Transport, Metabolism, and Regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamir-Ariel D, Navon N, Burdman S. Identification of Genes in Xanthomonas campestris pv. vesicatoria Induced during Its Interaction with Tomato. J Bacteriol. 2007;189:6359–6371. doi: 10.1128/JB.00320-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PB. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ Microbiol. 1999;1:243–257. doi: 10.1046/j.1462-2920.1999.00040.x. [DOI] [PubMed] [Google Scholar]
- Camilli A, Mekalanos JJ. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio CG, Camilli A. Hidden Dimensions of Vibrio cholerae Pathogenesis. ASM News. 2003;69:396–401. [Google Scholar]
- Silby MW, Nicoll JS, Levy SB. Regulation of Polyphosphate Kinase Production by Antisense RNA in Pseudomonas fluorescens Pf0-1. Appl Environ Microbiol. 2012;78:4533–4537. doi: 10.1128/AEM.07836-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: do we only see the tip of the iceberg? Trends Biochem Sci. 2008;33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Marco ML, Legac J, Lindow SE. Pseudomonas syringae genes induced during colonization of leaf surfaces. Environ Microbiol. 2005;7:1379–1391. doi: 10.1111/j.1462-2920.2005.00825.x. [DOI] [PubMed] [Google Scholar]
- Flaherty B, Van Nieuwerburgh F, Head S, Golden J. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics. 2011;12:332. doi: 10.1186/1471-2164-12-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H, Harwood CS, Pechter KB, Schaefer AL, Greenberg EP. Antisense RNA that affects Rhodopseudomonas palustris quorum-sensing signal receptor expression. Proc Natl Acad Sci USA. 2012;109:12141–12146. doi: 10.1073/pnas.1200243109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JM, Livny J, Lawrence MS, Kimball MD, Waldor MK, Camilli A. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucl Acids Res. 2009;37:e46. doi: 10.1093/nar/gkp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, Schweitzer P, Wang W, Schroth GP, Luo S. Transcriptome analysis of Pseudomonas syringae identifies new genes, ncRNAs, and antisense activity. J Bacteriol. 2010;192:2359–2372. doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Duhring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci USA. 2006;103:7054–7058. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret M, Egan F, Fargier E, Morrissey JP, O’Gara F. Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology. 2011;157:1726–1739. doi: 10.1099/mic.0.048645-0. [DOI] [PubMed] [Google Scholar]
- Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and Regulation of the Type VI Secretion System. Annu Rev Microbiol. 2012;66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, Engel J, Filloux A, Bleves S. The Second Type VI Secretion System of Pseudomonas aeruginosa Strain PAO1 Is Regulated by Quorum Sensing and Fur and Modulates Internalization in Epithelial Cells. J Biol Chem. 2012;287:27095–27105. doi: 10.1074/jbc.M112.376368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, Lipscomb L, Kim HS, Mrázek J, Nierman WC. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick MJ, Edwards RA. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Muro-Pastor MI, Florencio FJ. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J Bacteriol. 1997;179:2678–2689. doi: 10.1128/jb.179.8.2678-2689.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]