Analysis of tumorigenesis in Drosophila reveals a tumor-suppressor role for Hedgehog signaling in the context of oncogenic Notch signaling.
Abstract
Fine-tuned Notch and Hedgehog signalling pathways via attenuators and dampers have long been recognized as important mechanisms to ensure the proper size and differentiation of many organs and tissues. This notion is further supported by identification of mutations in these pathways in human cancer cells. However, although it is common that the Notch and Hedgehog pathways influence growth and patterning within the same organ through the establishment of organizing regions, the cross-talk between these two pathways and how the distinct organizing activities are integrated during growth is poorly understood. Here, in an unbiased genetic screen in the Drosophila melanogaster eye, we found that tumour-like growth was provoked by cooperation between the microRNA miR-7 and the Notch pathway. Surprisingly, the molecular basis of this cooperation between miR-7 and Notch converged on the silencing of Hedgehog signalling. In mechanistic terms, miR-7 silenced the interference hedgehog (ihog) Hedgehog receptor, while Notch repressed expression of the brother of ihog (boi) Hedgehog receptor. Tumourigenesis was induced co-operatively following Notch activation and reduced Hedgehog signalling, either via overexpression of the microRNA or through specific down-regulation of ihog, hedgehog, smoothened, or cubitus interruptus or via overexpression of the cubitus interruptus repressor form. Conversely, increasing Hedgehog signalling prevented eye overgrowth induced by the microRNA and Notch pathway. Further, we show that blocking Hh signal transduction in clones of cells mutant for smoothened also enhance the organizing activity and growth by Delta-Notch signalling in the wing primordium. Together, these findings uncover a hitherto unsuspected tumour suppressor role for the Hedgehog signalling and reveal an unanticipated cooperative antagonism between two pathways extensively used in growth control and cancer.
Author Summary
Growth control mechanisms ensure that organs attain the correct final size, generally averting tumour growth. This control is often linked to spatially confined domains known as organizers (conserved signalling centres), established along the dorsal-ventral and anterior-posterior axes of the organ by the Notch and Hedgehog pathways, respectively. The organizers emit signals that dictate growth, cell fate specification, and differentiation. However, how the distinct organizing signals received are integrated by cells within a growing organ remains a mystery. By studying how Delta-Notch signalling drives tumorigenesis, we identified the conserved microRNA miR-7 as a co-operative element in tumorigenesis mediated by Delta. We found that the cooperation between the microRNA and Delta-Notch pathway converged on the silencing of two obligatory and functionally redundant Hedgehog receptors, interference hedgehog and brother of ihog. Downregulation of other hedgehog pathway genes via RNA interference or genetic mosaics revealed a tumour suppressor role for Hedgehog signalling in the context of the oncogenic Notch pathway. Given the conservation of miR-7, as well as of the Notch and Hedgehog pathways, the conclusions we have drawn from these studies on Drosophila may be applicable to some human cancers.
Introduction
A fundamental question in biology is what instructs cells to stop growing when the proper size is attained to commence terminal differentiation. Indeed, this issue is relevant not only to size regulation but also to cancer. One strategy that organisms use to promote the growth of organs involves the establishment of spatially confined domains called organizers, conserved signalling centres established along the dorsal-ventral (DV) and anterior-posterior (AP) axes of the organs, often involving members of the Notch (DV organizers) and Hedgehog (Hh) (AP organizers) families. Organizers act as a source of graded signals (e.g., Wingless/Wnts, and BMP/Dpp) that promote global organ growth and subsequently, or concurrently, cell fate specification along the DV or AP axes [1],[2]. Although how individual organizing pathways promote growth has been studied comprehensively (e.g., [3]–[5]), our understanding of how orthogonal organizers are integrated and of the cross-talk between them remains limited. Tumourigenesis may occur if the finely balanced growth-promotion and termination is disrupted. Yet little attention has been paid to the issue of how growth by organizers is terminated.
To discover mechanisms of Notch-induced tumourigenesis in an in vivo context, we used the fruit fly Drosophila melanogaster compound eye. This tissue provides a particularly powerful tool to define novel oncogenes and tumour suppressor networks via unbiased genome-wide screens. Particularly, the early stages of eye development seem to recapitulate molecular mechanisms in human NOTCH1-induced oncogenesis (e.g., [6]–[10]). Human NOTCH1 can function either as an oncogene or a tumour suppressor depending on the cellular context, which often reflects the physiological role of NOTCH1 in the particular stage or cell type. During early development of the fly eye, the pleiotropic Notch pathway plays a predominant role in growth promotion. Consequently, this tissue and stage is useful to identify contextual factors that may synergize with Notch to foster benign and/or invasive tumour growth in vivo.
The growth in the compound eye, which is derived from the centre of the eye imaginal disc, depends on a conserved DV Notch-mediated growth-promoting organizer, which is established early in the second larval instar by the asymmetric activation of the Notch receptor by its ligands Delta and Serrate (DLL1,2,4, and JAG1,2 in humans) along the DV boundary (reviewed in [11]). Downstream of the organizer, eyegone (eyg) gene is expressed specifically in the organizer cells and it controls global eye size [12],[13]. A similar DV organizer has been found in a variety of contexts, including the fly and vertebrate limbs, although the expression of eyg is restricted to the fly eye. Eyg is functionally related to the human PAX6(5a) oncogene [13] and acts as a transcriptional repressor [14],[15] though complementary patterns of expression of the organizer in developing eyes have never been reported.
Growth and retinal differentiation in the eye field is spatially and temporally coordinated. Retinal differentiation depends on a separate organizer, the AP organizer, which is associated with the morphogenetic furrow (MF). The MF begins to form at the posterior margin of the early third instar eye disc, and as it moves in an anterior direction, it leaves differentiated retinal cells in its wake. Just anterior to the MF, eye cells arrest in G1 of the cell cycle prior to the start of differentiation, and most cells then go through a synchronous round of cell division before they terminally exit the cell cycle [16]. The initiation and progression of the MF, and of G1 arrest, is positively regulated by Hh [17]–[24]. Though the initiation and progression of the MF in the developing eye disc follows that of the DV organizer [25], the expression of hh gene starts earlier in second instar [19] and hence overlaps in time with the DV growth-promoting organizer (Figure S1). Early studies of ectopic Hh signalling led to the idea that this signal ultimately contributes to retinal patterning and also directly regulate eye growth [18], although more recently it has been shown that when the Hh pathway is constitutively activated (via inactivation of downstream repressors) in cells confined to a clone, the surrounding wild-type cells overproliferate but the cells within the clone show growth disadvantage and eventually are eliminated by apoptosis [26]. The influence of Hh on growth in Notch-mediated growth regulation needs to be investigated by loss-of-function approaches in the appropriate context.
In both flies and humans, Hh signalling relieves the inhibition exerted by Patched (PTCH1 in humans) on the intermediate pathway component Smoothened (Smo/SMO), allowing Smo to stabilize full-length Cubitus interruptus (Ci), which acts as a transcriptional activator (Ci-155: Gli2,3 in mammals) and inhibiting the processing of Ci-155 to the truncated transcriptional repressor (Ci-75, in flies) [27]. In addition to these core components, two related members of the immunoglobulin/fibronectin type III–like superfamily have recently been identified as Hh co-receptors in Drosophila, with functionally overlapping roles: Interference hedgehog (Ihog) and Brother of Ihog (Boi) [28]–[32]. Indeed, the human counterparts of these proteins, CDO (named after CAM-related/down-regulated by oncogenes) and BOC (Brother of CDO), also act as obligatory co-receptors for Hh signalling [28],[32]–[41]. While overactive Hh signalling is unreservedly oncogenic, making Hh a prime target for therapeutic interventions, there is evidence that loss-of-function of some components of the Hh pathway may exert a tumour-suppressor role. A notable example is that of CDO and BOC, which were initially isolated on the basis of their downregulation by RAS oncogenes in transformed cells, and that were shown to act as tumour suppressors in vitro [42]. More recently, recurrent somatic mutations in the sonic Hh pathway were identified in human pancreatic cancers through global genomic studies, affecting GLI1, GLI3, and BOC [43]. However, the role of these mutations in cancer remains untested.
Here, we describe the identification of the conserved microRNA (miRNA) miR-7 as a gene that enhances Notch pathway-induced eye overgrowth in D. melanogaster. miRNAs are small noncoding RNAs that negatively regulate gene expression by binding to “seed” sequences in the untranslated regions (UTRs) and/or in the open reading frame of target messenger RNAs, thereby inhibiting translation and, at times, indirectly driving mRNA degradation. Although miRNAs are in the front line of cancer research, their role in cancer is often unconfirmed in vivo. We identified the ihog gene as a functionally relevant, direct target of miR-7 in Notch-mediated tumourigenesis in vivo. Further, we provide evidence that the microRNA mir-7 and Notch pathway cooperatively dampen Hh signal transduction via down-regulation of its receptors ihog and boi, respectively. As a consequence, we hypothesize that tumours form by the cooperation between the gain of Dl-Notch signalling and a deficiency to transduce Hh signal. We validated this hypothesis by showing that the inhibition of endogenous Hh core components similarly enhanced Dl-Notch-mediated organizing activity resulting in severe overgrowth both in the eye disc and the wing disc. Conversely, increasing Hh signal transduction pathway suppressed eye tumour-like growth by Dl and the microRNA. Given the conservation of these pathways, similar cooperative antagonistic interactions between oncogenic Notch and loss of Hh signalling might play a role in human cancers.
Results
MicroRNA miR-7 Cooperates with Delta to Trigger Severe Overgrowth in Drosophila Eye
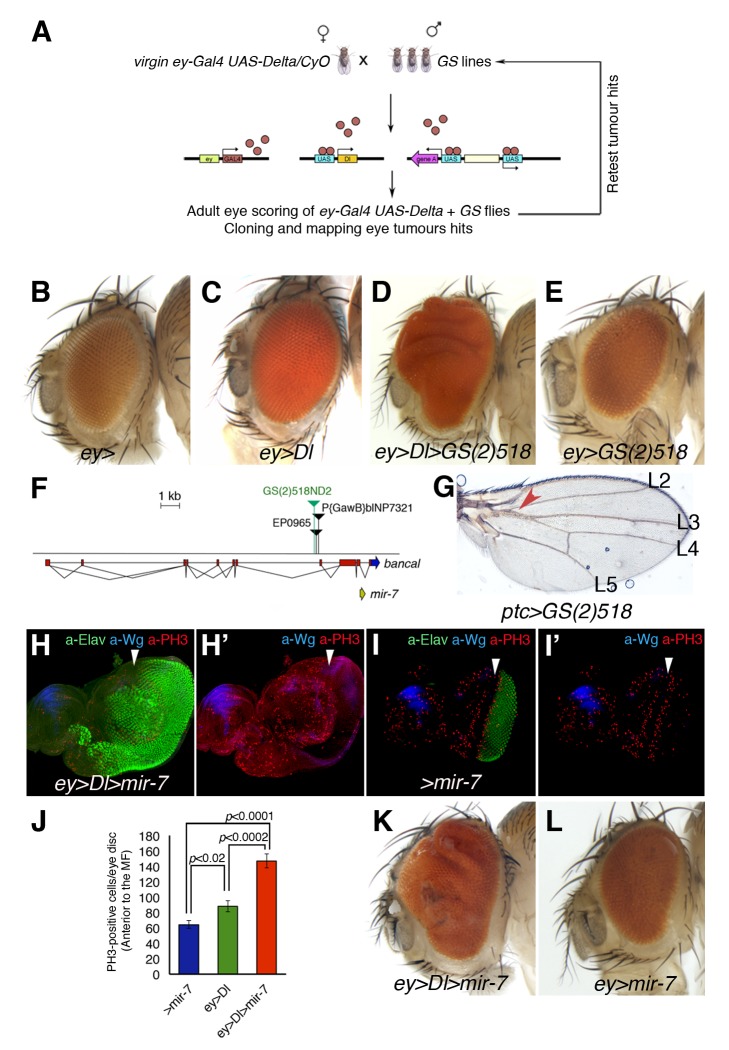
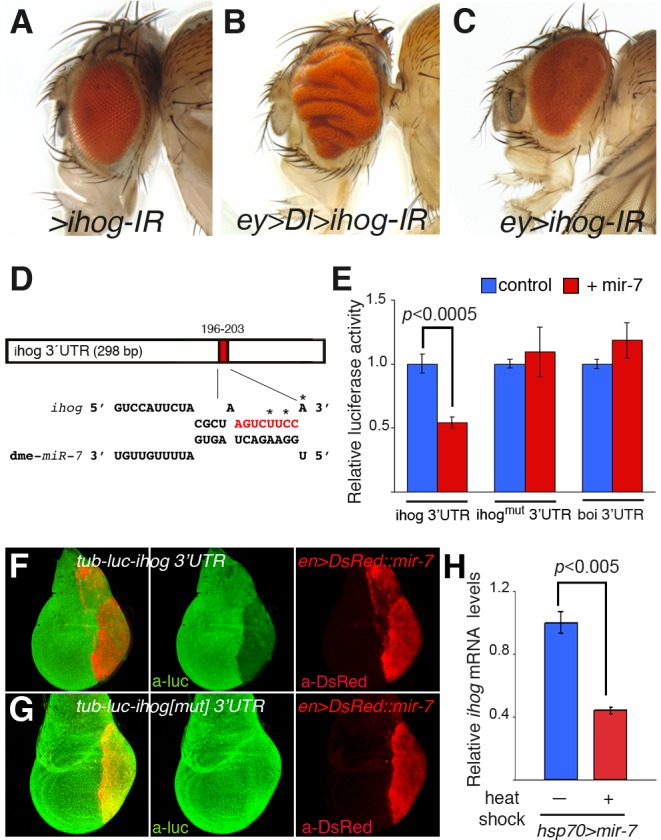
To identify endogenous genetic determinants that may limit Notch-driven tumourigenesis in vivo, we carried out an unbiased (genome-wide) gain-of-expression screen for loci that converted Dl-induced mild eye overgrowth into severe overgrowths (benign tumour-like growth: eye tissue is overgrown and folded) or metastatic tumours (provoke secondary eye growths throughout the body). A Gene Search (GS) transposon system was employed to systematically generate gain-of-expression mutations as in [44], using the eyeless (ey)-Gal4 to drive expression of UAS-containing transgenes and the GS lines in the imaginal disc cells of the growing eye (the precursors of the adult fly eye; Figure 1A–B). In this way, we identified a GS line (GS(2)518ND2) that converted Dl-induced modest eye overgrowth (Figure 1C; adult eyes are 130% bigger than control wild type eyes) into severely overgrown and folded eye tissue (ey-Gal4 UAS-Dl GS(2)518ND2, hereafter ey>Dl>GS(2)518) (250%–320% larger than wild-type eyes; 54% penetrance, n = 200 eyes; Figure 1D). Differentiation and growth defects of third instar eye discs of ey>Dl>GS(2)518 are shown in Figure S3. In the absence of Dl overexpression, the overexpression or misexpression of the gene(s) affected by GS(2)518ND2 did not increase eye size (ey>GS(2)518; Figure 1E).
Figure 1. The Conserved MicroRNA miR-7 co-operates with Notch in D. melanogaster oncogenesis.
(A) A schematic outline of the Gene Search (GS) gain-of-expression screen for Notch co-operating oncogenes in the developing Drosophila eye. (B–E and K–L) Adult heads of control female ey-Gal4 wild-type eye size (B) and combinations between GS line, UAS transgenes, and ey-Gal4 are shown. (C) Dl expression under the control of ey-Gal4 results in a mild overgrowth in the eye (130% larger than wild type size). (D) Introducing the GS(2)518ND2 line enhanced overgrowth by Dl (>320%, see also Figure S2). (E) The overexpression of gene(s) affected by the GS(2)518ND2 line alone causes no overt eye overgrowth. (F) Scheme of the GS(2)518ND2 insertion. (G) Overexpression of the GS(2)518ND2 line driven by ptc-Gal4 showed the typical wing vein L3–L4 fusion. (H–I′) Confocal images of third instar eye-antennal discs stained for the mitotic marker PH3 (red), Wg (blue) to define the DV axis, and the neuronal marker Elav (green) of the indicated genotypes. White arrowheads indicate the position of the MF. The co-expression of UAS-mir-7 with UAS-Dl causes eye disc overgrowth and a front of retinal differentiation highly disorganized (H, compare with control sibling eye disc in I). (J) Quantification of mitotic cells labelled by PH3 anterior to the MF of the genotypes: ey>Dl>mir-7 (red bar), ey>Dl (green bar), and wild-type sibling discs +/UAS-mir7 (>mir-7, blue bar). Data shown represent the mean ± s.e.m. of total PH3 measurement in 20 eye discs per genotype. P values were calculated by the unpaired Student's t test. (K–L) Adult heads overexpressing mir-7 driven by ey-Gal4 in the presence (K) or the absence (L) of the UAS-Dl transgene. See also Figures S2 to S4 for supplementary data.
The GS(2)518ND2 line carried an insertion 3.1 kb upstream of the mir-7 miRNA gene (Figure 1F), which is transcribed from an internal promoter within a 3′ intron of the bancal/heterogeneous nuclear ribonucleoprotein K (bl/hnRNP-K) gene [45]. A set of EP elements in the vicinity of GS(2)518ND2 has been previously described to cause mir-7 overexpression, and to induce proximal fusion of longitudinal (L) veins 3 and 4, as well as distal wing notching or bristle tufting [45]–[47]. Indeed, expressing GS(2)518ND2 along the AP compartment boundary in the wing imaginal disc using patched (ptc)-Gal4 caused similar L3-–L4 fusion as that reported following mir-7 overexpression in this domain (ptc>GS(2)518; Figure 1G). Conversely, the direct overexpression of mir-7 together with Dl (hereafter, ey>Dl>mir-7), using a mir-7 transgene that does not contain any bl sequences (UAS-mir-7), provoked overgrown larval eye discs ey>Dl>mir-7 (Figure 1H; compare with sibling wild type eye discs, Figure 1I) associated with significant increased cell proliferation (Figure 1J and Figure S4C–D,H), resulting in adult overgrown and folded eyes similar to that in the GS(2)518ND2 flies (70% of adult ey>Dl>mir-7 animals displayed eye benign tumour-like growth, n = 200; Figure 1K and Figure S2A–C). There was no increase in eye size when UAS-mir-7 alone was overexpressed by ey-Gal4 (ey>mir-7; Figure 1L).
Identification of Candidate Tumour Suppressor Targets of miR-7 by in Vivo RNAi Screening in the Delta Overexpression Model
In the wing disc, the miR-7 microRNA is thought to silence target genes of the Notch pathway [47],[48]. However, downregulation of Notch signalling alone might not explain the synergism between mir-7 and Dl overexpression in eye overgrowth as we did not detect reduction of the organizing signalling by Dl-Notch in these discs (Figure S3). Therefore, we sought to identify miR-7 target gene(s) that might be relevant to the cooperation with Dl-Notch signalling in eye overgrowth and tumourigenesis. As such, we systematically assayed a set of 39 D. melanogaster genes predicted to be miR-7 targets in silico (Table S1, [49]). We used RNA interference (RNAi) UAS-driven transgenes (UAS-IR) to downregulate candidate and previously validated miR-7 target genes in vivo. The UAS-IR transgenes silence specific mRNA transcripts by provoking their degradation, which is triggered by the generation of double-stranded RNA fragments complementary to the transcript driven by GAL4/UAS system [50],[51]. Here, we employed ey-Gal4 to drive simultaneously the overexpression of the UAS-IR and the UAS-Dl transgene (Table S1).
We hypothesized that mir-7 overexpression would be mimicked by endogenous downregulation of the functional relevant target gene(s) in the context of Dl overexpression. The assay would not, however, distinguish between a bona fide miR-7 target gene and those genes that are required normally for restricting tissue growth. To identify the former, we considered that a bona fide miR-7 target gene would not produce any effect when downregulated in the context of normal Notch signalling. Nevertheless, we took into consideration that RNAi silences mRNA more efficiently than microRNAs, and thus, we considered that UAS-IR lines of bona fide candidate genes would produce phenotypes similar to those of miR-7, or more severe. We tested candidate target genes predicted by several algorithms ([52]; see Materials and Methods) and that contain the conserved Drosophila miR-7 binding sites, which normally reduces the number of false positive target predictions.
Of the 39 candidate target genes assayed in conjunction with Dl overexpression, only reduction of two genes robustly cooperated with Dl-Notch signalling to provoke severely overgrown and folded eyes. A previously validated target of miR-7, hairy [48] was capable of converting Dl-induced mild overgrowth into tumour-like growth (Table S1). However, since miR-7 only very subtly reduces the expression of endogenous hairy and a GFP-3′UTR hairy sensor [48], we focused our interest on the gene, interference hedgehog (ihog), that when downregulated in Dl-overexpressing cells provoked robust overgrowth (Figure 2, Figure S4E–F,H, and Table S1).
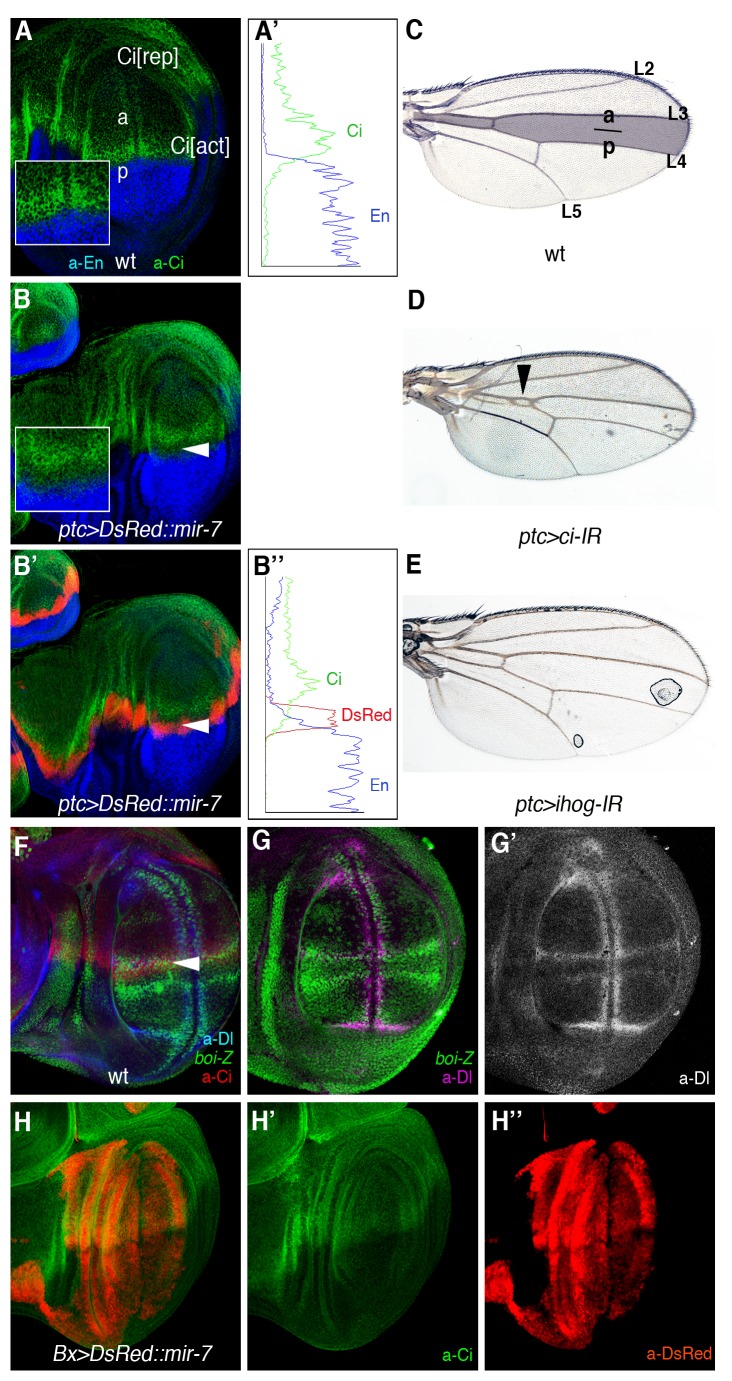
Figure 2. Tumourigenesis promoted by miR-7 via direct repression of interference hedgehog (ihog).
(A–C) Adult heads of female control UAS-ihog-IR (A) and combinations of UAS-ihog-IR and ey-Gal4 in the presence (B) or the absence (C) of the Dl transgene. (D) Computer predicted consequential pairing of ihog target region (top) and miRNA (bottom). The conserved seed match (8 mer) in the 3′UTR of ihog is in red. (E) Luciferase assay in Drosophila Schneider (S2) cells co-transfected with mir-7 (red bars) or the empty vector (blue bars), together with a firefly luciferase vector containing the ihog3′UTR (ihog3′UTR), or the luciferase vector with mutations in the seed sequence (asterisks in D, ihogmut3′UTR) or control boi3′UTR (boi3′UTR). Firefly luciferase activity was measured 48 h after transfection and normalized against Renilla luciferase. The values represent the mean ± s.e.m. of three or four independent experiments. Differences in ihog(mut) and boi luciferase levels were not statistically significant between treatments. (F–G) Confocal images of mid third instar wing discs carrying the tub-luc::ihog-3′UTR (F) or the tub-luc::ihogmut3′UTR sensor (G) and overexpression of mir-7 by en-Gal4 (en>DsRed::mir-7, red) and stained with anti-luciferase antibody (green). (H) Differences in ihog mRNA levels assessed by RT-qPCR between hsp70>mir-7 larvae subjected to heat shock treatment (red bar) or not (blue bar). Values represent the mean ± s.e.m. of three independent experiments. P values were calculated by the unpaired Student's t test.
Although not previously characterized as a target gene of miR-7, the downregulation of ihog by RNAi concomitant with the gain of Dl function consistently produced enlarged eye discs (Figure S4E–F) similar to that in eye discs co-expressing Dl and mir-7 (Figure S3I–J), resulting in adults with overgrown and folded eyes (ey>Dl>ihog-IR: 80% of severe overgrown eyes, n = 200; Figure 2B and Table S1). This phenotype was seen with the two independently generated ihog-IR transgenic lines available, both yielding identical results. Moreover, the expression of ihog RNAi alone during eye development did not alter the size or retinal patterning of this organ (ey>ihog-IR; Figure 2C). We confirmed that the ihog-IR transgenes inhibited ihog transcription 10-fold by quantitative reverse transcription-polymerase chain reaction (qRT-PCR; Figure S5A). Furthermore, the mRNA levels of brother of ihog (boi) were unaffected by these ihog-IR lines (Figure S5B). Thus, specific down-regulation of endogenous ihog, a predicted target of miR-7, facilitates overgrowth by Dl overexpression similar to those that develop when mir-7 is overexpressed in this context (Figure 1, Figure 2, and Figure S4H).
Validation of Interference Hedgehog as a Direct Target of miR-7 in Vitro and in Vivo
Since the ihog gene encodes a receptor of Hh in the embryo, including the imaginal eye disc [30], we assessed whether it is directly regulated by miR-7 in luciferase reporter-based cellular assays in vitro and in vivo (Figure 2). There is a single conserved miR-7 binding site in the 3′UTR of ihog (Figure 2D) and in Drosophila Schneider (S2) cells overexpressing mir-7, there was 45% less activity of a luciferase reporter containing the full-length ihog 3′ UTR downstream of the firefly luciferase coding region driven by the α-tubulin promoter (tub-luc::ihog-3′UTR Figure 2E and Figure S5C). By contrast, when the ihog 3′UTR construct carried point mutations in the miR-7 binding site (tub-luc::ihog(mut)-3′UTR), luciferase activity was the same as in control cells (Figure 2E). In addition, luciferase activity was unaffected by mir-7 overexpression in a control tub-luc::boi-3′UTR construct, indicative that the functional similar boi was not a target of miR-7 (Figure 2E).
In addition to the direct regulation of the ihog mRNA 3′UTR by miR-7 in vitro, there was specific in vivo repression of the tub-luc::ihog-3′UTR construct but not the ihog 3′UTR construct that carried the mutations in the seed sequence (Figure 2FG) and of an ihog 3′UTR eGFP sensor (tub-eGFP::ihog-3′UTR) but not a similar boi 3′UTR eGFP sensor (tub-eGFP::boi-3′UTR) (Figure S6AB) in the posterior compartment cells of third instar wing discs overexpressing mir-7 driven by engrailed (en)-Gal4. Finally, we demonstrated that endogenous ihog mRNA was inhibited by miR-7 in vivo as heat shock induction of mature mir-7 overexpression (hsp70-Gal4 UAS-mir-7) provoked a 55% reduction in ihog mRNA transcripts in larvae when assayed by qRT-PCR (Figure 2H and Figure S2D). Overall, these data provide convincing evidence that miR-7 is capable of directly repressing ihog, both in vitro and in vivo. Thus, the synergism between miR-7 and the Dl-Notch pathway activity in eye overgrowth would appear to be largely due to the silencing of ihog.
brother of ihog Is Negatively Regulated by Notch Signalling during Eye Growth
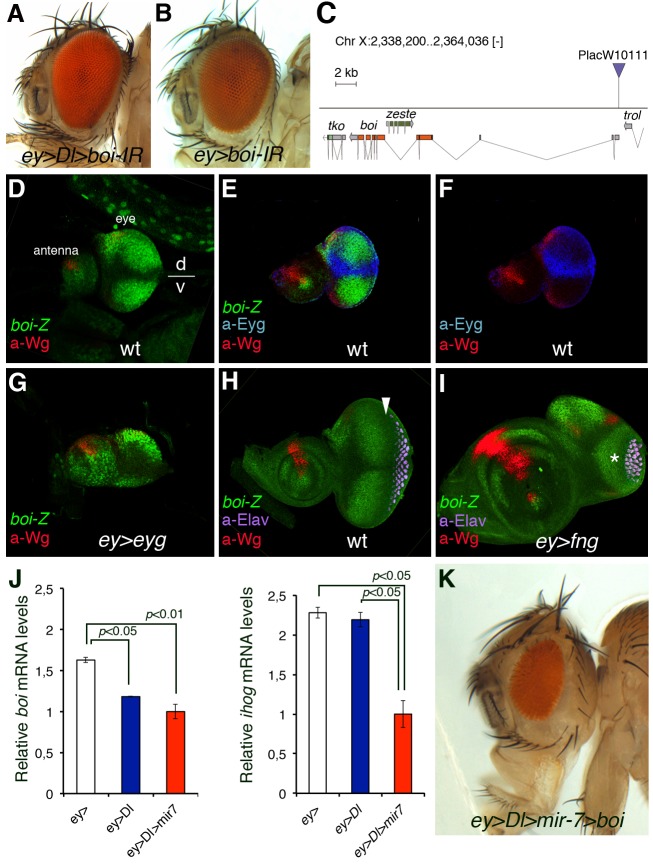
Although boi mRNA expression was not affected in the ihog-IR lines and Boi does not appear to be a target of miR-7, there is a well-documented functional overlap in the roles of Ihog and Boi. Moreover, genetic inactivation of both the boi and ihog genes is typically required to induce hh loss-of-function phenotypes [28],[30],[32]. However, unlike ihog-IR, we found that expressing an RNAi transgene against boi (boi-IR effectively reduces boi but not ihog mRNA levels by 65%; p = 0.0005; Figure S5A–B) did not enhance Dl-induced eye overgrowth (ey>Dl>boi-IR; Figure 3A–B and Table S2). Since only the concomitant loss of both ihog and boi leads to a loss of eye tissue [30], we reasoned that a similar situation might occur with respect to the ihog-IR-induced severe eye overgrowth (Figure 2B). Consequently, we verified the status of boi transcription in relation to eye disc growth. Interestingly, the spatial domain of boi in the developing eye disc in vivo using a ß-galactosidase enhancer trap inserted in boi (P-lacW stock 10111; Figure 3C) unveiled that boi is expressed nonuniformly in the region anterior to the MF with a weakest expression within the DV organizer (Figure 3D–E,H: the MF is denoted by an arrow in H). Indeed, in eye discs double labelled with anti-Eyg (a DV organizer-specific response gene and an obligatory Notch's effector in eye growth [13],[53]) and anti-ß-galactosidase (boi-lacZ in green), we found that the expression of Eyg precisely borders the “negative” domain of boi (Figure 3E–F). This led us to speculate that expression of boi is negatively regulated by Notch-Eyg at the growth-promoting organizer, which we investigated by monitoring the spatial domain of boi-lacZ in mutants of the DV organizer and by assessing boi mRNA levels by qRT-PCR analyses.
Figure 3. Notch signalling represses brother of ihog (boi) expression in the dorsal-ventral growth organizer in Drosophila eye.
(A–B) Adult heads of female flies overexpressing UAS-Dl and/or UAS-boi-IR and ey-Gal4. (C) Map of PlacW10111 P-element insertion (triangle) into the boi locus. (D–I) boi expression in wild-type (D, E, F, and H) and Notch pathway mutant (G and I) eye-antennal discs. The patterning gene wingless (a-Wg, in red) serves to orient the eye disc in the dorsal (D)/ventral (V) axis. Expression of Boi (green) Hh co-receptor at the early third larval stage is repressed along the DV organizer (D and E), as defined by the expression of the DV organizer gene eyg (blue, E and F). Retinal differentiation (neuronal marker a-Elav, magenta) is first detected at the posterior end of the eye disc (to the right) and progresses in an anterior direction (H). The arrow points to the MF. (G and I) Expression of boi-lacZ (boi-Z, green) and wingless (a-Wg, red) in ey>eyg (G) and ey>fng (I) eye discs. The discs in (H) and (I) are from the same stage and magnification. The enlarged antennal disc in (I) is an effect of the undergrowth of the eye disc, caused in part by defective Notch activation in the D/V organizer due to fng overexpression. (J) qRT-PCR analyses of boi (left) and ihog (right) in ey-Gal4 (white bar), ey>Dl (blue bar), and ey>Dl>mir-7 (red bar) late third instar eye discs. Two independent experiments of three replicates are shown in each case. Data were normalized to rp49. mRNA isolated from 50 pairs of eye-antennal discs per genotype. Data analysed by a two-tailed unpaired t test. Error bars represent s.e.m. of three replicates. (K) Adult fly head showing no eye overgrown induced by Dl and mir-7 when boi is overexpressed by a transgene (UAS-boi, 100% penetrance of rescue).
We assayed the ubiquitous expression of the Notch DV organizer transcriptional effector Eyg, which provokes a wider DV organizer domain [13],[53] and observed an extended domain lacking boi-lacZ expression under these conditions (Figure 3G). Conversely, the ubiquitous expression of the modulator fringe (fng) causes defective Notch receptor activation by its ligands and results in the thinning or loss of the DV organizer [53]–[55]. Under these conditions, the expression of boi was uniform throughout the eye disc due to the absence of the “central domain” that represses this gene in wild-type eye discs (Figure 3I). Thus, boi is negatively regulated by Notch's organizer activity or it at least reflects this activity negatively. Since Eyg encodes a transcriptional repressor [14],[15], it may directly repress boi transcription. This Hh co-receptor does contain a consensus Eyg-binding site for repression (TCACTGA [14]) at position chrX: 2.359.784, although we could not validate the direct binding of Eyg to the boi promoter region by chromatin immunoprecipitation (unpublished data). Nevertheless, it is possible that Eyg might bind through other nonconsensus sites. Furthermore, qRT-PCR analyses confirmed downregulated boi but not ihog transcripts in eye discs overexpressing Dl transgene alone by ey-Gal4 (ey>Dl; left in Figure 3J). Importantly, both boi and ihog mRNA levels were downregulated in eye discs that co-expressed Dl with the microRNA mir-7 (ey>Dl>mir-7; Figure 3J). boi and ihog RNA was isolated from whole eye-antennal disc complexes; thus, the mRNA levels are the sum of all regions of the discs, including the antenna, which is not affected by ey>Dl or ey>mir-7. Hence expression differences with control may be significant underestimations of the actual differences of each gene in the eye disc parts in the different genotypes. Nevertheless, the qRT-PCR comparisons between the different genotypes showed a trend in boi and ihog expression response to Dl overexpression that explains the cooperation between the miR-7 and Dl signalling, since there is the concomitant downregulation of the two functionally redundant Hh receptor genes, ihog and boi.
Animals homozygous for mutations in ihog and boi exhibit a phenotype typical of the loss of hh function (e.g., [30]). The defect in ihog− boi− animals can be rescued by expressing a UAS-ihog::myc transgene with weak constitutive expression in the absence of Gal4 activity [30]. Surprisingly, we could not overcome overgrowth by mir-7/Dl using this transgene (unpublished data). This may perhaps reflect that the elevated levels of Ihog expected by Gal4-induced expression of the transgene may exert a dominant negative effect on Hh signalling [31]. A boi transgene (UAS-boi) [56] fully suppressed the overgrowth induced by the combination of mir-7/Dl (Figure 3K, 100% penetrance, n = 100). The same result was obtained using the EP(X)1447(boi) that misexpresses endogenous boi gene (unpublished data).
Blocking Core Hedgehog Signalling Components or Expressing Ci/GLI Repressor Mimics the Effect of the MicroRNA in Delta-Induced Tumourigenesis
To confirm that silencing Hh signal transduction facilitates a tumorigenic response to Dl-Notch overactivation, we next assayed the effects of directly downregulating core Hh signalling elements with RNAi transgenes driven by ey-Gal4, including smo, ci, and hh itself. As noted above, Gal4 drives expression throughout early eye disc development anterior to the MF, a region of undifferentiated proliferating eye cells that act on signals from the Notch-mediated DV organizer, and Gal4 expression terminates before cells exit the cell cycle at the MF [54]. We down-regulated each of these Hh signalling components by RNAi, assaying several independent lines in which the use of ey-Gal4 avoided the possible effects of a loss of Hedgehog signal transduction on retinal differentiation that might confound the results (Table S2).
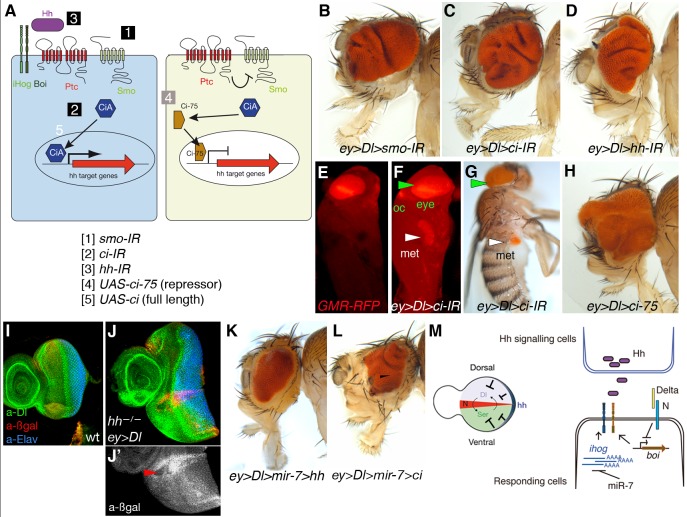
The downregulation of smo (80% flies exhibited eye tumour-like growth, n >200), ci (100%, n>200), or hh (30%–100%, n>200) in conjunction with Dl overexpression provoked a tumour phenotype similar to that of RNAi of ihog but stronger than the overexpression of mir-7 (compare Figure 1 and Figure 2 with Figure 4B–D; see also Table S2). Furthermore, downregulation of ci by RNAi (ci-IR) by ey-Gal4 stimulated a metastatic overproliferation of eye tissue in the context of the Dl gain of function, resulting in flies with secondary eye growths within the thorax and abdomen (Figure 4F–G and Table S2). This invasive overgrowth is also observed when Dl and the ci RNAi transgene are expressed in the wing imaginal discs by the dpp-Gal4 (Figure S7). Like the mir-7 and ihog-IR lines (Figure 1L and Figure 2C), none of the above RNAi lines were capable of inducing overgrowth by themselves.
Figure 4. Downregulation of elements in the Hh pathway or overexpression of the repressor form of ci co-operates with Dl overexpression to trigger tumour growth in the Drosophila eye.
(A) Schematic representation of Hh signalling and the UAS transgenes used to downregulate by RNAi (IR) or activate Hh pathway components. (B–D, H, and K–L) Representative adult heads of female flies of combinations of the indicated UAS transgenes and ey-Gal4 are shown. (E–F) Fluorescent images of Drosophila pupae of sibling control (ey>Dl, E) or ey>Dl>ci-IR (F). (G) Adult fly of ey>Dl>ci-IR with a metastatic (met) growth in the abdomen. Eye tissue in the endogenous site (green arrowheads) and distant site (white arrowheads) is labelled by the retinal-specific GMR-myrRFP marker (E, F) or the retinal-specific red pigments (G). (I–J′) Third instar wild type of sized eye disc (I) and ey>Dl eye disc carrying clones of hhAC labelled by the absence of arm-lacZ (ßgal, red in J and grey in J′). Arrowhead points to a clone and its associated twin spot (high red staining). (M) Model of antagonistic interaction between Hh and Notch signalling in normal eye imaginal disc (left) and model of regulatory interactions among the microRNA, Notch pathway, and the Hh receptors ihog and boi (right). Genotype in (J) is: yw ey-Flp; ey-Gal4 UAS-Dl/+; FRT82B hhAC/FRT82B arm-lacZ.
In all contexts, in the absence of Hh signal or its reception, the transcription factors of the Ci/Gli family (in Drosophila, full-length Ci-155) can be proteolytically processed into a truncated (N-terminal 75 kDa in Drosophila—Ci-75) transcriptional repressor of the Hh pathway (Ci, Gli3, and to a lesser extent Gli2) (Figure 4A). The bifunctional nature of Ci [57]–[59], and of the mammalian homologues Gli2 and Gli3, could fulfil oncogenic or tumour suppressor roles in function of the status of the Hh signalling. As ci-IR downregulates both activator and repressor forms, we next assessed the contribution of the truncated Ci repressor that forms in the absence of Hh signalling, testing the effect of overexpressing Dl with a transgene of the constitutive Ci repressor form (UAS-ci-75). Co-overexpression of Dl and ci-75 induced eye tumour-like growth in 75% of fly eyes (ey>Dl>ci75; n = 100; Figure 4H), in contrast to the overexpression of Ci full length (UAS-ci) that acts as an activator in Hh receiving cells and did not provoke eye tumour (unpublished data).
To further verify these findings with the RNAi transgenes, we generated marked clones of cells homozygous for hhAC (a null allele) in the ey>Dl background (hhAC/hhAC ey>Dl; Figure 4J). Eye discs carrying small patches of hhAC cells were 170% larger than control wild-type eye discs (Figure 4I) and 126% larger than ey>Dl without hhAC clones eye discs (see Figure S4B). Using the MARCM technique [60], we also examined GFP-labelled clones of cells overexpressing Dl and homozygous for smo3 (an amorphic allele) (smo3/smo3tub>Dl>GFP; Figure S8). Whereas clones of smo3 do not delay the MF [61] and clones of Dl-expressing cells normally cause autonomous advancement of the MF [62], we found that clones of smo3 Dl-expressing cells led to advancement of the MF also in surrounding wild-type cells (Figure S8B) and the disc was overall overgrown (unpublished data). The advanced MF is seen in ey>Dl eye discs with downregulation of Hh signalling via overexpression of mir-7 or direct downregulation via RNAi transgenes (Figures S3 and S4). Thus, interfering with Hh signalling exacerbates the organizing activity of Dl-Notch signalling in eye imaginal discs and can foster invasive tumour growth (Figure 4F–G, Figure S7C–D, and Table S2).
Increasing Hedgehog Signal Prevents Tumourigenesis by Delta and miR-7
In normal early eye development, when the Notch organizer induces a dramatic increase in cell proliferation in the disc, hh gene is expressed in a thin line of cells along the eye disc margin ([19],[20],[25]; see Figure S1). Previously, it has been shown that clones of eye disc cells lacking PKA, Ptc, or Cos2 proteins that normally prevent the inappropriate activation of Hh signal transduction exhibit within the clone a growth-disadvantage and are eliminated by apoptosis [26]. This negative influence of Hh signal was also hinted at by the small eye defect associated with overexpression of UAS-hh by ey-Gal4 [25] and is complementary to our findings.
The Ihog/Boc family proteins normally enhance Hh binding to Ptc, the 12-pass transmembrane protein involved in sensing extracellular Hh concentrations. Binding of Hh to Ptc relieves inhibition of Smo by Ptc and blocks the production of Ci repressor. Hence, the downregulation of ihog/boi levels by Dl/miR-7 (see Figure 3J) might reduce the interactions of Hh with Ptc. We therefore investigated whether increasing Hh signal via a UAS-hh transgene to counterbalance ihog/boi deficit could rescue the overgrowth by Dl/mir-7. Indeed, we detected significant reduction in eye size in flies ey>Dl>mir-7>hh (Figure 4K; 100% rescue, n>100; see Figure S9 for scheme of genetic test for rescuing experiment) and also in flies that expressed Ci full length (ey>Dl>mir-7>ci; Figure 4L). Note that when Ci full length is expressed in the context of Dl and mir-7 overexpression, although many eyes are substantially reduced in size they still exhibit abnormal patterned growth (see Figure 4L) and other exhibited enhanced tumorigenesis. We interpret these findings as Ci full length can be converted into the repressor form owing to the reduced Hh signalling caused by Dl and miR-7 depletion of ihog and boi.
Hh signal stimulates the maturation of Ci full length into a short-lived nuclear activator, while the PKA negative regulator opposes this event and when mutated results in constitutive Hh pathway activity. The undergrowth defect of knock-down of pka by RNAi expression in the Dl overexpressing eye discs (ey>Dl>pka-IR; Table S2) further support the tumour suppressor activity of Hh pathway in the context of gain of Dl-Notch signalling in the context of the eye primordium. We suggest here that in healthy flies the release of Hh by these eye disc marginal cells sets eye size in conjunction with the Dl-Notch organizer (Figure 4M, left scheme), and thereby dampening Hh signalling in the context of Dl overexpression (Figure 4M, right) fosters the developing eye tumours or overgrowth beyond the normal eye size.
Hedgehog Signal Transduction Also Attenuates Delta Signalling and Overgrowth in the Wing
Wing disc growth and patterning is also organized by Hh and Notch-mediated organizers [2], with Hh secreted by cells in the posterior (P) compartment inducing short-range targets in anterior (A) cells near the AP boundary (e.g., ptc, blue staining in Figure 5A) [63],[64]. Notch signalling is activated locally along the DV boundary by its ligands Dl and Serrate (Ser), and it induces symmetric expression of targets in boundary cells (e.g., wg, red staining in Figure 5A; reviewed in [2]). Hence, we investigated whether the antagonistic interaction between loss of Hh and gain of Notch apparent in the eye imaginal discs can also be applied to the wing discs.
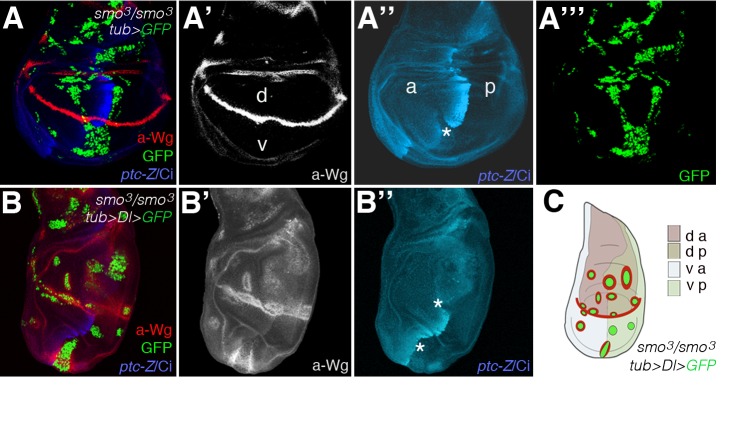
Figure 5. Failure to transduce the Hh signal due to mutations in smoothened enhances Dl-Notch signalling activity in the wing.
(A–B″) Confocal images of wing discs bearing MARCM GFP (green)-labelled clones homozygous for smo3 without (A) or with (B) Dl overexpression. Single channel images are also shown. Mosaic discs were stained for Wg (red in A and B, and grey in A′ and B′), and Ci (blue) and Ptc-lacZ (Ptc-Z, blue). (C) A schematic summary of clones in (B). Asterisks in (A″) and (B″) point to “posteriorly” situated clones that were of anterior origin as denoted by the failure to induce Ptc and the low levels of Ci protein (white line delineates the AP boundary in the discs in B). Clones were generated at 24–42 h after egg laying (AEL) by a 1 h heat shock at 37°C (n = 60 clones analysed). Genotypes: (A) yw hsp70-Flp tub-G4 UAS-GFP; tub-Gal80 FRT40A/smo3 FRT40A ptc-lacZ and (B) yw hsp70-Flp Tub-G4 UAS-GFP; Tub-Gal80 FRT40A/smo3 FRT40A ptc-lacZ; UAS-Dl/+.
Dl-expressing clones in the wing induce ectopic wg expression in D cells, where the fringe gene is expressed, whereas ventrally situated clones did not activate wg (e.g., [65]–[69]). Enhancing Dl activity by co-expressing Dl with the E3 ubiquitin ligase Neuralized, which promotes the endocytosis and signalling activity of Dl, can induce wg in ventrally situated clones [69]. Hence, we assayed ectopic induction of wg to examine Dl activity in smo3/smo3 clones. As shown in Figure 5, we found that ventrally situated A cells homozygous for smo3 and expressing Dl expressed high levels of Wg, similar to the levels of Wg induced by dorsally situated clones, in contrast with most smo3 Dl-expressing clones situated ventrally in P cells away from the boundary (Figure 5B–C) or clones of smo3 cells that do not overexpressed Dl (Figure 5A). Nonautonomous overgrowth is also evident in ventrally situated clones of smo3/smo3 Dl-expressing (Figure S8C). Clones of smo3 cells abutting the AP boundary often sort to the P compartment territory [70],[71]. MARCM clones do not label the twin spot (smo+/smo +); therefore, the inference that the clones at the AP boundary (asterisks in Figure 5A″–B′) are of anterior origin is supported by the finding that they retain anterior features (low levels of Ci protein). Loss of smo activity in A cells at the boundary fail to up-regulate Ci expression and do not induce ptc transcription. These clones cause an anterior shift in the distribution of ptc and up-regulated Ci non-cell-autonomously [64]. We occasionally found ambiguously positioned clones of smo3/smo3 tub>Dl cells in which the anterior part of the clone exhibited ectopic wg expression while the posterior of the clone did not (Figure S8D). Taken together, these findings show that Dl-expressing cells unable to transduce the Hh signal behave as they express hyperactivated Dl. Coupled with the analysis of RNAi transgenes, these results confirm that the loss of Hh signalling enhances Dl-Notch signalling activity.
Loss of Hedgehog Signalling in miR-7 Overexpression in the Wing
microRNAs are thought to regulate multiple target genes; however, often when tested in vivo, it is a subset or a given target that function as the major effector of the activity of the microRNAs in a given cellular context. We asked whether our identification of ihog as a key target of miR-7 during Dl-mediated tumorigenesis in the eye might reflect endogenous roles of the microRNA in other tissues. Previously, misexpression of mir-7 driven by ptc-Gal4 (ptc>mir-7) produces wing margin notches, and a reduction of the space between vein L3 and L4 ([48]; see [72]). Both of these phenotypes have been attributed to defects in Notch signalling [48],[73], although we noted that L3–L4 fusion is very reminiscent to the phenotype produced by hh loss-of-function mutations, including that associated with the ciCell mutation that produces a truncated form of Ci, which behaves as a constitutive repressor [59]. Indeed, we observed a clear downregulation of Ci protein levels in cells in ptc>mir-7 (Figure 6A–B″), which are precisely the cells receiving endogenous Hh signals and that upon normal Hh reception stabilize Ci protein levels and prevent the conversion of Ci-155 into truncated Ci repressor. Plots of fluorescence intensity profiles from the wild-type and ptc>mir-7 discs are shown in Figure 6A′ and B″. The weak downregulation of Ci by mild RNAi expression using ptc-Gal4 mimicked the L3–L4 fusion defect of ptc>mir7 (Figure 6C–D). Depleting ihog by RNAi driven by ptc-Gal4 did not produce a defect as mir-7 overexpression (Figure 6E). The lack of effect of ihog RNAi is almost certainly due to the activity of the other Hh co-receptor, boi, which is expressed at high levels in the wing margin and in the presumptive L3 vein territory (boi-lacZ in green; Figure 6F). These results raised the possibility that like ihog, ci is also a direct target of miR-7. Indeed, ci mRNA does contain a presumptive miR-7 binding site in the ci 3′UTR, although this site is not conserved across Drosophila species. Thus, the Ci low protein levels in ptc>mir-7 wing discs could reflect the direct repression of ci by the microRNA or the dampening of Hh signalling response by the miR-7-mediated downregulation of ihog or both. More consistently with indirect regulation of Ci by miR-7, we observed no change in Ci protein levels in wing discs ectopically expressing the mir-7 away from the normal Hh secreting cells (the P compartment cells marked by the absence of Ci (green) in Figure 6G). In this experiment, we used the Beadex (Bx)-Gal4 driver, with the Bx domain labelled by DsRed because of the UAS-DsRed::mir-7 transgene (Figure 6G). Therefore, either Ci is not a target of miR-7 or this regulation is context dependent. It is generally considered that when an individual miRNA affects the expression of various proteins in the same pathway, it does so in a rather mild manner [74]. Thus, the relevance of co-regulation of ihog and ci by miR-7 in Hh receiving cells deserves further analysis given that the human counterparts of these genes (CDO, BOC, and Gli3) also contain binding sites for human miR-7.
Figure 6. miR-7 silencing of Hh signalling explains the L3–L4 fusion defects in the wing.
(A) Ci protein (green) is distributed across the entire anterior (A) compartment of the discs. Hh signals from posterior (P) cells induce high levels of Ci in cells along the AP border, and they block Ci proteolysis into the repressor form (Ci[rep]), thereby allowing the Ci activator (C[act]) to accumulate. (B–B′) Overexpression of mir-7 denoted by red labelling (UAS-DsRed::mir-7) driven by patched (ptc)-Gal4 downregulates Hh signalling as visualized by low Ci levels (green; white arrowhead). Insets show magnifications. Engrailed (En) staining in blue serves to mark the P compartment in (A–B″). Plots of fluorescence intensity profiles of the anterior-posterior compartments from the WT (A) and ptc>DsRed::mir-7 (B′) discs are shown in (A′) and (B″), respectively. Green trace, Ci; blue trace, En; red trace, DsRed. (C) Adult wild-type wing. The shaded area denotes the domain of expression of the ptc-Gal4 reporter. (D) ci-IR expression by ptc-Gal4 mimicking the L3–L4 fusion defect seen in adult wings that is caused by mir-7 overexpression (compare with Figure 1G). (E) Adult wing expressing ihog-IR driven by ptc-Gal4. (F–F″) The expression of boi-lacZ (green) defines all longitudinal veins (L2–L5). Note the high boi-lacZ (green in F) expression along L3, marked by high Ci (red in F) and Dl (magenta in F″). (G–G″) Overexpression of mir-7 (in red) by Bx-Gal4 did not alter Ci protein levels (green, white arrowhead).
Discussion
A challenge to understand oncogenesis produced by pleiotropic signalling pathways, such as Notch, Hh, and Wnts, is to unveil the complex cross-talk, cooperation, and antagonism of these signalling pathways in the appropriate contexts. Studies in flies, mice, and in human cell cultures have provided critical insights into the contribution of Notch to tumourigenesis. These studies highlighted that Notch when acting as an oncogene needs additional mutations or genes to initiate tumourigenesis and for tumour progression, identifying several determinants for such co-operation (e.g., [7],[8],[10],[24],[44],[75]–[79]). The identification of these co-operative events has often been knowledge-driven, although unbiased genetic screens also identified known unanticipated tumour-suppressor functions. In this sense, we describe here a conserved microRNA that cooperates with Notch-induced overproliferation and tumour-like overgrowth in the D. melanogaster eye, miR-7. Alterations in microRNAs have been implicated in the initiation or progression of human cancers (e.g., [80]–[84]), although such roles of microRNAs have rarely been demonstrated in vivo (e.g., [85]–[88]). In addition, by identifying and validating functionally relevant targets of miR-7 in tumourigenesis, we also exposed a hitherto unsuspected tumour suppressor role for the Hh signalling pathway in the context of the oncogenic Notch pathway. Given the conservation of the Notch and Hh pathways, and the recurrent alteration of microRNAs in human cancers, we speculate that the genetic configuration of miR-7, Notch, and Hh is likely to participate in the development of certain human tumours.
In human cancer cells, miR-7 has been postulated to have an oncogene [89],[90] or a tumour suppressor functions [91]–[96] that may reflect the participation of the microRNA in distinct pathways, due to the regulation of discrete target genes in different cell types, such as Fos [97] in mouse, and Pak1 [91], IRS-2 [92], EGFR [92],[93], Raf-1 [93], α-synuclein [98], CD98 [99], IGFR1 [94], bcl-2 [100], PI3K/AKT [101],[102], and YY1 [103] in humans.
In Drosophila, multiple, cell-specific, targets for miR-7 have been previously validated via luciferase or in vivo eGFP-reporter sensors or less extensively via functional studies [47],[49],[73],[104]–[107]. Although microRNAs are thought to regulate multiple target genes, when tested in vivo it is a subset or a given target that predominates in a given cellular context. Indeed, of the 39 predicted miR-7 target genes tested by direct RNAi, only downregulating ihog with several RNAi transgenes (UAS-ihog-IR) fully mimicked the effect of miR-7 overexpression in the transformation of Dl-induced mild overgrowth into severe overgrowth and even tumour-like growth. Moreover, we confirmed that endogenous ihog is directly silenced by miR-7 and that this silencing involves direct binding of the microRNA to sequences in the 3′UTR of ihog both in vivo and in vitro.
Nevertheless, other miR-7 target genes may contribute to the cooperation with Dl-Notch pathway along with ihog, such as hairy and Tom. While miR-7 can directly silence hairy in the wing, this effect has been shown to be very modest [48], and thus, we consider that while hairy may contribute to such effects, it is unlikely to be instrumental in this tumour model. Indeed, the loss of hairy is inconsequential in eye development [108], although retinal differentiation is accelerated by genetic mosaicism of loss of hairy and extramacrochaetae [108]. hairy is a target of Hh [18],[21] that negatively sets the pace of MF progression. It is unclear how Hairy might contribute to Dl-induced tumourigenesis.
The RNAi against Tom produced overgrowth with the gain of Dl albeit inconsistently and with weak penetrance, where one RNAi line did not modify the Dl-induced overgrowth and the other RNAi line caused tumours in less than 40% of the progeny (Table S1). Tom is required to counteract the activity of the ubiquitin ligase Neuralized in regulating the Notch extracellular domain, and Dl in the signal emitting cells. These interactions are normally required to activate Notch signalling in the receiving cells through lateral inhibition and cell fate allocation [109]. However, although it remains to be shown whether similar interactions are active during cell proliferation and growth, the moderate enhancement of Dl that is induced when Tom is downregulated by RNAi suggests that miR-7-mediated repression of Tom may contribute to the oncogenic effects of miR-7 in the context of Dl gain of function, along with other targets such as ihog.
Conversely, while the target genes of the Notch pathway, E(spl)m3 and E(spl)m4 [48] as well as E(spl)mγ, Bob, E(spl)m5, and E(spl)mδ [60], have been identified as direct targets of miR-7 in the normal wing disc via analysis of 3′UTR sensors, there was no evidence that HLHm3, HLHm4, HLHm5, Bob, and HLHmγ are biological relevant targets of miR-7 in the Dl overexpression context. HLHmδ RNAi produced inconsistent phenotypes in the two RNAi transgenic lines available, causing tumour-like growth at very low frequency in only one of the lines (Table S1). We also did not obtain evidence that miR-7 provoked overgrowth by targeting the ETS transcription factor in the EGFR pathway AOP/Yan (Table S1), a functionally validated target of the microRNA miR-7 during retinal differentiation [47]. Neither had we obtained evidence that RNAi of atonal provoked eye tumours with Dl overexpression (Table S1), although a strong inhibition via expression of a fusion protein Atonal::EN that converts Atonal into a transcriptional repressor has been shown to be sufficient to trigger tumorigenesis together with Dl [24]. Thus, we reasoned that given that microRNA influenced target genes only subtly (even when using ectopic expression), it is possible that downregulation of atonal contributes to the phenotype along with the other targets.
In conclusion, we have identified cooperation between the microRNA miR-7 and Notch in the D. melanogaster eye and identified and validated ihog as a direct target of the miR-7 in this context and have identified boi as a target of Notch-mediated activity at the DV eye organizer, although it remains whether this regulation is direct or indirect. We also uncovered a hitherto unanticipated tumour suppressor activity of the endogenous Hh signalling pathway in the context of gain of Dl-Notch signalling (Figure 4) that is also apparent during wing development (Figure 5).
Hh tumour suppressor role is revealed when components of the Hh pathway were lost in conjunction with a gain of Dl expression in both the eye (Figure 4) and wing (Figure 5 and Figure S8) discs. Hh and Notch establish signalling centres along the AP and DV axes, respectively, of the disc to organize global growth and patterning. Where the organizer domains meet, the Hh and Notch conjoined activities specify the position of the MF in the eye disc and the proximodistal patterning in the wing disc [25],[47],[48]. We unveil here that in addition antagonistic interaction between the Hh and Notch signalling might help to ensure correct disc growth. Thus, we show that Hh signalling limits the organizing activity of Dl-Notch signalling (Figure 4, Figure 5, and Figure S8). Although it is often confounded whether Dl-Notch signalling instructs overgrowth by autonomous or nonautonomous (i.e., DV organizers) mechanisms, our findings uncover that loss of Hh signalling enhances a noncell autonomous oncogenic role of Dl-Notch pathway (Figure 4J and Figure S8D).
To date, Hh has not yet to be perceived as a tumour suppressor, although it is noteworthy that human homologs of ihog, CDO, and BOC were initially identified as tumour suppressors [42]. Importantly, both CDO and BOC are downregulated by RAS oncogenes in transformed cells [42] and their overexpression can inhibit tumour cell growth in vitro [42],[110],[111]. Since human RAS regulates tumourigenesis in the lung by overexpressing miR-7 in an ERK-dependent manner [90], it is possible that RAS represses CDO and BOC via this microRNA. Indeed, the 3′UTR of both CDO and BOC like Drosophila ihog contains predicted binding sites for miR-7 (www.targetscan.org). There is additional clinical and experimental evidence connecting elements of the Hedgehog pathway with tumour-suppression. The function of Growth arrest specific gene 1 (GAS1), a Hh ligand-binding factor, overlaps that of CDO and BOC [39],[41] and its downregulation is positively associated with cancer cells [94] and melanoma metastasis [112], while its overexpression inhibits tumour growth [113]. More speculative is the association of some cancer cells with the absence of cilium, a structure absolutely required for Hh signal transduction in vertebrate cells [27].
Given the pleiotropic nature of Notch, Wnts, BMP/TGFß, Ras, and Hh signalling pathways in normal development in vivo, we speculate that competitive interplay as that described here between Notch and Hh may not be uncommon among core growth control and cancer pathways that act within the same cells at the same or different time to exert multiple outputs (such as growth and cell differentiation). Moreover, context-dependent tumour suppressor roles could explain the recurrent, unexplained, identification of somatic mutations in Hh pathway in human cancer samples (e.g., [43]). Indeed, our findings stimulate a re-evaluation of the signalling pathways previously considered to be exclusively oncogenic, such as the Hh pathway.
Materials and Methods
Drosophila Husbandry
The GS(2)518ND2 line was isolated in a genetic screen for enhancers or suppressors of a mild overgrown eye phenotype induced by Dl overexpression when driven by the eye-specific ey-Gal4 driver (ey-Gal4 UAS-Dl). The PlacWP1O111 stock was a generous gift from Dr. C. Klambt (Munster University, Munster, Germany), and the other Drosophila stocks used here were: UAS-mir-7 and UAS-DsRed::mir-7 [47], UAS-boi [56], UAS-ci [57], and UAS-ci-75 [58],[59]. A detailed description of the stocks and transgenic flies used in this study can be found at http://flybase.org/ for ey-Gal4, ptc-Gal4, en-Gal4, hsp70-Gal4, Bx-Gal4, UAS-Dl, UAS-fng, UAS-hh, UAS-eyg, EP(X)1447 (boi), hhAC, and smo3or at http://flystocks.bio.indiana.edu/ and http://stockcenter.vdrc.at/control/main for the BDSC and VDRC RNAi stocks, respectively. Clones of hhAC surrounded by Dl-expressing tissue (Figure 4J) were generated by the ey-Flp in eye-antennal imaginal discs of the genotype: yw ey-Flp; ey-Gal4 UAS-Dl/+; FRT82B hhAC/FRT82B arm-lacZ. In Figure S8, the MARCM GFP-labelled clones of smo3/smo3 only or smo3/smo3tub-Gal4 UAS-Dl cells were induced by 1 h heat shock at 37°C at 48–72 h AEL in larvae: y w tub-Gal4 UAS-GFP hsp70-FLP122; smo3 FRT40A ptc-lacZ/tub-Gal80 FRT40A and y w tub-Gal4 UAS-GFP hsp70-FLP122; smo3 FRT40A ptc-lacZ/tub-Gal80 FRT40A; UAS-Dl/+, respectively.
All the combinations of Gal4, GS, and the different UAS transgenic lines and mutants were raised at 26.5°C.
GS-Element and PlacW Mapping
Genomic DNA flanking the P-element insertion in the GS(2)518ND2 and the PlacWP1O111stock were recovered by inverse PCR using the Pwht1/Plac1 and Plw3-1/Pry 4 primers, respectively (http://www.fruitfly.org/about/methods/inverse.pcr.html), and they were subsequently sequenced. A BLAST search with the sequence produced perfect matches to the genomic region on chr2R:16491078 for GS(2)518ND2 and on chrX: 2364036 for PlacWP1O111.
Quantitative Reverse Transcriptase PCR (qRT-PCR)
To assess the levels of ihog or boi mRNA when the mir-7 or RNAi lines were activated by Gal4, we performed qRT-PCR experiments using RNA isolated from wandering third instar larvae of the hsp70-Gal4 genotype crossed with transgenic lines (UAS-mir-7, UAS-ihog-IR, or UAS-boi-IR) directly or following heat shock (an hour at 37°C followed by 6 h at 25°C). Total RNA from 50 pairs of eye-antennal discs was extracted for experiments in Figure 3J. All tissue samples were stored in RNAlaterTissueProtect Tubes (Qiagen) until used and mature mir-7, ihog, or boi mRNA levels were assessed by qRT-PCR. Note that RNA was isolated from whole eye-antennal disc complexes; thus, the levels of boi and ihog mRNA expression are the sum of all regions of the discs, including the antennal disc part that might not be unaffected by the expression of ey-Gal4. Thus, expression differences between the control and Dl and/or mir-7 overexpressing eye-antennal disc complexes may be significant underestimations of the actual differences in the relevant eye disc part in each genotype. To analyse mature mir-7 expression, we used mir-7-specific primers from the TaqMan MicroRNA Assays (Applied Biosystems), together with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) and TaqMan Universal PCR Master Mix (Applied Biosystems). The mir-7 levels were normalized to U14 snRNA. To determine ihog and boi mRNA levels, we used SuperScript First-Strand Synthesis System for RT–PCR (Invitrogen) and SYBR Green PCR Master kit (Applied Biosystems), according to the manufacturer's instructions. The cDNAs were amplified using specific primers designed using the ProbeFinder software by Roche Applied Science, and rp49 was used as a house-keeping gene for normalization.
Primer sequences used in this study include the following: ihog, forward primer 5′-TCAGTCTAAAATCCCATAATAAGTGC-3, reverse primer 5′-AAACCGGAATTGCTTCGAG-3′; boi, forward primer 5′-TGCCTAAAGAGACGGGAAAA-3′, reverse primer 5′-ATGTGTTCCAATTGCGGTTT-3′; and rp49, forward primer 5′-TGTCCTTCCAGCTTCAAGATGACCATC-3′, reverse primer 5′-CTTGGGCTTGCGCCATTTGTG-3′.
In all cases, samples were tested in triplicate and qPCR reactions were run on a 7500 Real-Time PCR System (Applied Biosystems) following the manufacturer's protocol. The data shown are the mean ± s.e.m. of three experiments, and the relative expression was calculated using the comparative Ct method. The qPCR data were analysed by a two-tailed unpaired t test.
Immunofluorescence Staining
Third instar imaginal discs were fixed and stained by standard procedures using the following primary antibodies (dilutions, sources): anti-Eyg (1∶100, [98]), anti-Elav (1∶100, DSHB: Developmental Studies Hybridoma Bank), 4D4 (anti-Wg, 1∶100, DSHB), 4D9 (anti-En, 1∶100, DSHB), anti-phospho-H3 (anti-PH3; 1∶500, Sigma), anti-GFP (1∶1,000, Invitrogen), anti-β-galactosidase (1∶2,000, Cappel), anti-Cut (1∶5,000, DSHB), anti-DE-cad (1.50, DSHB), anti-Dac (1∶100, DSHB), anti-Ci (1∶5; a gift from Dr. Holgrem), anti-luciferase (luci27) (1∶200, Thermo Scientific), and anti-DsRed (1∶2,000, Clontech). The secondary antibodies used were conjugated to AlexaFluor-488, -555, -647 (Molecular Probes), and diluted at 1∶400. Discs were mounted in Fluoromount G (Southern Biotechnology), and the images were captured on a Leica TCS-NT Confocal microscope. The RGB Profile Plot function of ImageJ was employed for the intensity profile plots in Figure 6A′ and B″.
Construction of Sensor Transgenes
The tub-luc::ihog3′UTR or tub-luc::boi3′UTR constructs were generated by cloning the full-length 3′ UTR of the Drosophila ihog or boi genes into the 3′ end of the tub-firefly luciferase plasmid. To construct the tub-luc::ihogmut3′UTR reporter, three nucleotides of the predicted binding site for miR-7 in the ihog 3′UTR were mutated (AGTCTTCCA to AGTCATGCT) using the QuickChange Site-Directed Mutagenesis kit (Agilent Technologies Inc.). The tub-eGFP::ihog3′UTR or tub-eGFP::boi3′UTR constructs were generated by cloning the full-length 3′UTR of ihog or boi genes into the 3′ end of the tub-eGFP reporter vector (a gift from Dr. Cohen). The final constructs were verified by sequencing. Transgenic eGFP and luciferase sensor flies were generated on a w1118 background by standard transformation into Drosophila embryos (BestGene Inc.).
Luciferase Reporter Assays
For Drosophila S2 cell luciferase assays, cells were co-transfected in 24-well plates as described previously [7] with the Renilla luciferase plasmid (75 ng) for normalization and different combinations of the following plasmids: actin-Gal4 (400 ng), pUAS-mir-7 or empty pUAST (400 ng; [48]), tub-luc::ihog3′UTR, tub-luc::boi3′UTR, or tub-luc::ihogmut3′UTR (25 ng). The relative luciferase activity was measured 48 h after transfection using the Dual-Glo Luciferase Reporter Assay system (Promega) according to the manufacturer's instructions. The data shown are the mean ± s.e.m. of three independent experiments, which was analysed by a two-tailed unpaired t test.
Measurement of PH3 Positive Cells
Female virgin w; ey-Gal4 UAS-Dl/Cy0-GFP were crossed to males w; +/+; UAS-DsRed::mir-7 and their F1 progeny larvae (w; ey-Gal4, UAS-Dl/+; UAS-DsRed::mir-7/+) were selected by DsRed labelling in the pair of eye-antennal discs. The particle analysis function of ImageJ software was used to count PH3-positive nuclei of the confocal images of third instar imaginal discs to generate the data shown in Figure 1J. The analyses of the area of eye disc and antennal disc parts in Figure S4H was done using ImageJ, and data represent mean values of area of eye discs normalized against the antennal disc part in at least six discs per genotype.
Supporting Information
Hh signal along the disc AP axis and Notch-mediated DV growth promoting organizer starts long before the initiation of retinal differentiation. (A) Mid second larval instar (LII) eye disc carrying the enhancer trap line hhP30-lacZ and stained for ßgalactosidase (hh-Z, green), Wg (red), and Elav (blue). The absence of blue staining denotes that the MF has not yet initiated in this disc. (B) Mid-late LII eye disc carrying the eyg-lacZ enhancer trap line and stained for ßgal (blue) and Wg (red). Notch signalling target Eyg expression labels the growth organizer. Disc as in Figure 3F.
(TIF)
The conserved MicroRNA miR-7 and Dl-Notch pathway cooperatively induce eye overgrowth. (A–C) Illustrative images of adult eyes overexpressing Dl with the GS(2)518 line (A) or the UAS-mir-7 transgene (B–C) with ey-Gal4. (A) The overgrown, folded eye tissue often present areas of undifferentiated or poorly differentiated outgrowths (arrowhead) (10%, n = 200 in A). The undifferentiated outgrowths are seen also in flies co-expressing Dl with the UAS-mir-7 transgene (B and C). (D) Quantification of relative mature mir-7 RNA levels in larvae carrying hsp70>mir-7 after heat shock (red bar) or not (blue bar). P was calculated using the Student t test, and values represented the mean ± sem. of three independent experiments.
(TIF)
Overgrowth and abnormal neuronal differentiation progression in eye discs co-expressing Dl and the GS(2)518 line. Confocal images of eye discs of control wild type (ey>, A, C, E, and G) and eye discs overexpressing Dl and mir-7 by ey-Gal4 (ey>Dl>GS(2)518: B, D, F, H–J) and carrying the indicated enhancer trap lines to monitor DV patterning: expression of D marker mirror-lacZ (mirr-Z), ventral marker fringe-lacZ (fng-Z), DV organizer-specific marker Serrate-lacZ (Ser-Z), and eyegone-lacZ (Eq-Z). Eye discs are stained for ßgalactosidase (green), neuronal marker Elav (blue), or Wg (red). (I–J′) Eye discs are stained for Dac (pink) or DE-cadherin (DE-cad, green in I and J and grey in I′ and J′) to highlight the morphology of the front of retinal differentiation (MF) and cell shape changes the accompanied neuronal differentiation, respectively. Although it has been postulated that the microRNA mir-7 silences Notch signalling, the overexpression of mir-7 with Dl causes eye disc overgrowth associated with enhanced Dl-Notch signalling as detected by the misexpression of DV organizer-specific markers (F and H). Seldom the pattern of retinal differentiation is highly disrupted in the overgrown discs (F and H) and often the front of neuronal differentiation (arrowhead, I′) is highly irregular or advanced in discs co-expressing Dl and GS(2)518 line. Anterior is to the left. Scale bar, 2 mm.
(TIF)
Overgrowth and abnormal neuronal differentiation progression in eye discs co-expressing Dl and the microRNA mir-7 or the ihog-IR or ci-IR transgenes. Confocal images of mitotic marker PH3 (blue in A–E; pink in F and green in G), neuronal marker Elav (green, A–F and red in G), and Wg (red, A–D and pink in F) staining of third instar eye-antennal imaginal discs of wild-type ey-Gal4 (ey>, A–A′), ey-Gal4 UAS-Dl (ey>Dl, B–B′), ey-Gal4 UAS-Dl/+; UAS-mir-7/+ (ey>Dl>mir-7, C–D′), ey-Gal4UAS-Dl/+; UAS-ihog-IR/+ (ey>Dl>ihog-IR, E–F), and ey-Gal4 UAS-Dl/+; UAS-ci-IR/+ (ey>Dl>ci-IR, G). The asterisks point to undifferentiated outgrowth of the eye discs (C, F, and G). Disc in (C) is as in Figure 1H. Note that eye disc overgrowth is also accompanied by advanced or disorganized front of retinal differentiation. The ey-Gal4 transgene drives expression anterior to the MF (white arrowhead in A), where eye disc cells proliferate asynchronously. Posterior to the MF, subsets of cells start differentiating into photoreceptor neurons visualized by the neuronal marker Elav (green, A) and the remaining cells divide one last time synchronously (row of PH3 cells behind the MF). (H) Quantitation of the eye imaginal disc size of the indicated genotypes. The area for each disc was calculated in pixel using ImageJ and values were normalized with those of the corresponding antennal disc part. As expected, co-expressing Dl with the RNAi against ihog or ci with ey-Gal4 provoked overgrowth similar, but stronger than the misexpression of the mir-7. Anterior is to the left in all images, and dorsal is up.
(TIF)
Quantification of ihog and boi mRNAs and mature mir-7 levels. (A) Relative ihog mRNA levels in larvae. (B) Relative boi mRNA levels in larvae. (C) Relative miR-7 levels in S2 cells transfected with actGal5 plasmid and with (red bar) or without (blue bar) the UAS mir-7 plasmid. The values represented the mean ± s.e.m. of at least three independent experiments. Data analysed by a two-tailed unpaired t test.
(TIF)
Overexpression of DsRed::mir-7 by en-Gal4 in the wing disc also caused reproducible in vivo downregulation of eGFP in a tub-eGFP::ihog-3′UTR (A) but not in a tub-eGFP::boi-3′UTR sensor (B).
(TIF)
Invasive growth caused by co-expressing Dl and ci-IR in the wing primordium. (A) Wild-type third instar wing imaginal discs. Dpp-GAL4 (dpp>) drives expression of UAS-GFP (gree) in a narrow band of anterior cells along the AP compartment boundary. Expression of mitotic marker PH3 (blue) and En (red) are also shown. (B) Expression of the RNAi transgene against ci (dpp>ci-IR) led to anterior expansion of the dpp domain visualized by GFP (green) and ectopic P cells (grey in B′) in the A territory at the DV boundary, but the disc is not overgrown. (C, D) Co-expression of Dl along with ci-IR led to extensive overgrowths. Note that mutant A cells mix with wild-type P (En, positive) cells (arrowheads) in some parts, reminiscent of malignant growth. Expression of Ci (grey in the inset) is also shown in (D).
(TIF)
Blocking Hh signal transduction due to mutations in smoothened enhances organizing activity by Dl-Notch signalling in the mosaic eye and wing discs. (A) Control eye discs carrying MARCM GFP(green)-labelled smo3 clones and (B) GFP-labelled clones of smo3 that overexpress Dl and stained for ptc-lacZ (Ptc-Z, blue) and Ci (blue). Note that the smo3/smo3tub-Gal4 UAS-Dl clones cause nonautonomously advancement of the MF denoted by up-regulated Ci levels, similar to the effect seen in eye discs co-expressing Dl with the mir-7. (A′) and (B′) show single channel confocal images. (C) Wing discs carrying MARCM GFP-labelled clones of smo3 cells and staining for Wg (red, C and C″) and clones of smo3 that overexpress Dl (smo3/smo3 tub>Dl, D–D″). In (D–D″), arrowheads point to ventrally situated clones of anterior origin (visualized by ptc-lacZ, not shown). The asterisk points to a clone of ambiguous A origin with weak ectopic Wg only in the anterior portion of the clone. DAPI counterstaining (pink, C″′ and D″′) is shown to illustrate the stimulation of growth of the surrounding tissue by the smo3 tub-Dl clones. Genotype in (A and C) is yw tub-Gal4 UAS-GFP hsp70-Flp; smo3 FRT40A ptc-lacZ/tub-Gal80 FRT40A and in (B and D) is yw tub-Gal4 UAS-GFP hsp70-Flp; smo3 FRT40A ptc-lacZ/tub-Gal80 FRT40A; UAS-Dl/+.
(TIF)
General genetic scheme of crosses for rescuing experiments in Figure 4. Similar genetic schemes were following the rescue by the UAS-boi transgene in Figure 3J. Larvae carrying both the chromosomes with the transgenes ey-Gal4 UAS-Dl (2nd) and UAS-DsRed::mir-7 (3rd) were selected under a fluorescence binocular (MZFLIII, Leica) for expression of DsRed in the eye under the control of Gal4. The resulting adult males were crossed to female virgins of the genotype UAS-hh/CyO. Larvae resulting from the cross were again selected and the DsRed-positive were transferred to a new tube, and the eyes of the resulting non-CyO adults eyes (males and females) were analysed.
(DOCX)
Identification of candidate tumour-suppressor gene(s) of Drosophila in silico predicted miR-7 target genes in the gain of Dl context.
(DOCX)
Direct inhibition by RNAi expression of core Hedgehog pathway genes in the gain of Dl context.
(DOCX)
Acknowledgments
We thank Dr. R. Carthew, Dr. C. Klambt, Dr. A.M. O'Reilly, the Bloomington Stock Center, and the Vienna Drosophila Research Centre for fly stocks; Dr. N. Aspiazu for the anti-Eyg antibody; and Dr. Carthew and Dr. S. Cohen for the pUAS-mir-7 and the tub::eGFP plasmids, respectively. We also thank members of the lab for helpful comments, and I. Gutierrez-García and E. Ballesta-Illan for technical assistance.
Abbreviations
- anti-luc
antibody against Luciferase protein
- AP
anterior-posterior
- Bdx-Gal4
Beadex-Gal4
- BL
Bloomington Drosophila Stock Center
- boi
brother of ihog
- ci
cubitus interruptus
- Dac
Dacshund
- DE-cad
DE-cadherin
- Dl
Delta
- DV
dorsal-ventral
- eGFP
enhanced Green Fluorescent Protein
- Elav
Embryonic lethal abnormal visual system
- en-Gal4
engrailed-Gal4
- Eq-Z
Equatorial (eyegone)-lacZ
- ey-Flp
eyeless-Flippase
- eyeless-Gal4
ey-Gal4
- fng-Z
fringe-lacZ
- FRT
Flippase Recognition Target
- Hh
Hedgehog
- hsp70-Flp
heat shock promoter 70-Flippase
- ihog
interference hedgehog
- IR
interference RNA
- MF
morphogenetic furrow
- mirr-Z
mirror-lacZ
- PH3
phopho histone H3
- Ptc-Z
Patched-lacZ
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- RNAi
RNA interference
- s.e.m.
standard error of the mean
- Ser-Z
Serrate-lacZ
- smo
smoothened
- Tub-Gal4
αTubulin84B promoter -Gal4
- Tub-Gal80
αTubulin84B promoter -Gal80
- UAS
Upstream Activation Sequences
- UTR
untraslated region
- VDRC
Vienna Drosophila RNAi Center
- Wg
Wingless
- wt
wild type
Funding Statement
We acknowledge the funding received from the Ministerio de Ciencia e Innovación (BFU2009-09074 and MEC-CONSOLIDER CSD2007-00023), the Botin Foundation, the Asociación Española Contra el Cáncer (AECC), Generalitat Valenciana (PROMETEO-2008/134) and a European Union Research Grant UE-HEALH-F2-2008-201666. I.G.P. is a Spanish FPI fellow. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Day SJ, Lawrence PA (2000) Measuring dimensions: the regulation of size and shape. Development 127: 2977–2987. [DOI] [PubMed] [Google Scholar]
- 2. Irvine KD, Rauskolb C (2001) Boundaries in development: formation and function. Annu Rev Cell Dev Biol 17: 189–214 doi:10.1146/annurev.cellbio.17.1.189. [DOI] [PubMed] [Google Scholar]
- 3. Crickmore MA, Mann RS (2006) Hox control of organ size by regulation of morphogen production and mobility. Science 313: 63–68 doi:10.1126/science.1128650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Affolter M, Basler K (2007) The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. . Nat Rev Genet 8: 663–674 doi:10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 5. Wartlick O, Mumcu P, Kicheva A, Bittig T, Seum C, et al. (2011) Dynamics of Dpp signaling and proliferation control. Science 331: 1154–1159 doi:10.1126/science.1200037. [DOI] [PubMed] [Google Scholar]
- 6. Efstratiadis A, Szabolcs M, Klinakis A (2007) Notch, Myc and breast cancer. Cell Cycle 6: 418–429. [DOI] [PubMed] [Google Scholar]
- 7. Vallejo DM, Caparros E, Dominguez M (2011) Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO J 30: 756–769 doi:10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fre S, Pallavi SK, Huyghe M, Laé M, Janssen K-P, et al. (2009) Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci USA 106: 6309–6314 doi:10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, et al. (2007) Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 13: 1203–1210 doi:10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, et al. (2012) Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 18: 298–301 doi:10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dominguez M, Casares F (2005) Organ specification-growth control connection: new in-sights from the Drosophila eye-antennal disc. Dev Dyn 232: 673–684 doi:10.1002/dvdy.20311. [DOI] [PubMed] [Google Scholar]
- 12. Chao JL (2004) Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development 131: 3839–3847 doi:10.1242/dev.01258. [DOI] [PubMed] [Google Scholar]
- 13. Dominguez M, Ferres-Marco D, Gutierrez-Aviño FJ, Speicher SA, Beneyto M (2004) Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat Genet 36: 31–39 doi:10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- 14. Yao J-G, Sun YH (2005) Eyg and Ey Pax proteins act by distinct transcriptional mechanisms in Drosophila development. EMBO J 24: 2602–2612 doi:10.1038/sj.emboj.7600725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salvany L, Requena D, Azpiazu N (2012) Functional association between Eyegone and HP1a mediates wingless transcriptional repression during development. Mol Cell Biol 32: 2407–2415 doi:10.1128/MCB.06311-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baker NE (2007) Patterning signals and proliferation in Drosophila imaginal discs. Curr Opin Genet Dev 17: 287–293 doi:10.1016/j.gde.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 17. Ma C, Zhou Y, Beachy PA, Moses K (1993) The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 75: 927–938. [DOI] [PubMed] [Google Scholar]
- 18. Heberlein U, Singh CM, Luk AY, Donohoe TJ (1995) Growth and differentiation in the Drosophila eye coordinated by hedgehog. Nature 373: 709–711 doi:10.1038/373709a0. [DOI] [PubMed] [Google Scholar]
- 19. Dominguez M, Hafen E (1997) Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev 11: 3254–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Borod ER, Heberlein U (1998) Mutual regulation of decapentaplegic and hedgehog during the initiation of differentiation in the Drosophila retina. Dev Biol 197: 187–197 doi:10.1006/dbio.1998.8888. [DOI] [PubMed] [Google Scholar]
- 21. Pappu KS, Chen R, Middlebrooks BW, Woo C, Heberlein U, et al. (2003) Mechanism of hedgehog signaling during Drosophila eye development. Development 130: 3053–3062. [DOI] [PubMed] [Google Scholar]
- 22. Firth LC, Baker NE (2005) Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell 8: 541–551 doi:10.1016/j.devcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 23. Roignant J-Y, Treisman JE (2009) Pattern formation in the Drosophila eye disc. Int J Dev Biol 53: 795–804 doi:10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bossuyt W, De Geest N, Aerts S, Leenaerts I, Marynen P, et al. (2009) The atonal proneural transcription factor links differentiation and tumor formation in Drosophila. Plos Biol 7: e40 doi:10.1371/journal.pbio.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cavodeassi F, Diez Del Corral R, Campuzano S, Dominguez M (1999) Compartments and organising boundaries in the Drosophila eye: the role of the homeodomain Iroquois proteins. Development 126: 4933–4942. [DOI] [PubMed] [Google Scholar]
- 26. Christiansen AE, Ding T, Bergmann A (2012) Ligand-independent activation of the Hedgehog pathway displays non-cell autonomous proliferation during eye development in Drosophila. Mech Dev 129: 98–108 doi:10.1016/j.mod.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ingham PW (2012) Hedgehog signaling. Cold Spring Harbor Perspectives in Biology 4 doi:10.1101/cshperspect.a011221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yao S, Lum L, Beachy P (2006) The Ihog cell-surface proteins bind hedgehog and mediate pathway activation. Cell 125: 343–357 doi:10.1016/j.cell.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 29. McLellan JS, Yao S, Zheng X, Geisbrecht BV, Ghirlando R, et al. (2006) Structure of a heparin-dependent complex of Hedgehog and Ihog. Proc Natl Acad Sci USA 103: 17208–17213 doi:10.1073/pnas.0606738103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Camp D, Currie K, Labbé A, van Meyel DJ, Charron F (2010) Ihog and Boi are essential for Hedgehog signaling in Drosophila. Neural Dev 5: 28 doi:10.1186/1749-8104-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yan D, Wu Y, Yang Y, Belenkaya TY, Tang X, et al. (2010) The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development 137: 2033–2044 doi:10.1242/dev.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng X, Mann RK, Sever N, Beachy PA (2010) Genetic and biochemical definition of the Hedgehog receptor. Genes Dev 24: 57–71 doi:10.1101/gad.1870310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okada A, Charron F, Morin S, Shin DS, Wong K, et al. (2006) Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature 444: 369–373 doi:10.1038/nature05246. [DOI] [PubMed] [Google Scholar]
- 34. Tenzen T, Allen BL, Cole F, Kang J-S, Krauss RS, et al. (2006) The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell 10: 647–656 doi:10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 35. McLellan JS, Zheng X, Hauk G, Ghirlando R, Beachy PA, et al. (2008) The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature 455: 979–983 doi:10.1038/nature07358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cohen MM (2010) Hedgehog signaling update. Am J Med Genet A 152A: 1875–1914 doi:10.1002/ajmg.a.32909. [DOI] [PubMed] [Google Scholar]
- 37. Kavran JM, Ward MD, Oladosu OO, Mulepati S, Leahy DJ (2010) All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. J Biol Chem 285: 24584–24590 doi:10.1074/jbc.M110.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C (2010) Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev 24: 2001–2012 doi:10.1101/gad.1951710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Izzi L, Lévesque M, Morin S, Laniel D, Wilkes BC, et al. (2011) Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Dev Cell 20: 788–801 doi:10.1016/j.devcel.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang W, Hong M, Bae G-U, Kang J-S, Krauss RS (2011) Boc modifies the holoprosencephaly spectrum of Cdo mutant mice. Dis Model Mech 4: 368–380 doi:10.1242/dmm.005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Allen BL, Song JY, Izzi L, Althaus IW, Kang J-S, et al. (2011) Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev Cell 20: 775–787 doi:10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kang JS, Gao M, Feinleib JL, Cotter PD, Guadagno SN, et al. (1997) CDO: an oncogene-, serum-, and anchorage-regulated member of the Ig/fibronectin type III repeat family. J Cell Biol 138: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, et al. (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806 doi:10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ferres-Marco D, Gutierrez-Garcia I, Vallejo DM, Bolivar J, Gutierrez-Aviño FJ, et al. (2006) Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature 439: 430–436 doi:10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- 45. Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC (2005) Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA 102: 18017–18022 doi:10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Charroux B, Freeman M, Kerridge S, Baonza A (2006) Atrophin contributes to the negative regulation of epidermal growth factor receptor signaling in Drosophila. Dev Biol 291: 278–290 doi:10.1016/j.ydbio.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 47. Li X, Carthew RW (2005) A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123: 1267–1277 doi:10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 48. Stark A, Brennecke J, Russell RB, Cohen SM (2003) Identification of Drosophila MicroRNA targets. Plos Biol 1: E60 doi:10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW (2009) A MicroRNA imparts robustness against environmental fluctuation during development. Cell 137: 273–282 doi:10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dietzl G, Chen D, Schnorrer F, Su K-C, Barinova Y, et al. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 doi:10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 51. Ni J-Q, Zhou R, Czech B, Liu L-P, Holderbaum L, et al. (2011) A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Meth 8: 405–407 doi:10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mazière P, Enright AJ (2007) Prediction of microRNA targets. Drug Discov Today 12: 452–458 doi:10.1016/j.drudis.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 53. Tsai Y-C, Sun YH (2004) Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis 39: 141–153 doi:10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- 54. Dominguez M, de Celis JF (1998) A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature 396: 276–278 doi:10.1038/24402. [DOI] [PubMed] [Google Scholar]
- 55. Gutierrez-Aviñon FJ, Ferres-Marco D, Dominguez M (2009) The position and function of the Notch-mediated eye growth organizer: the roles of JAK/STAT and four-jointed. EMBO Rep 10: 1051–1058 doi:10.1038/embor.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hartman TR, Zinshteyn D, Schofield HK, Nicolas E, Okada A, et al. (2010) Drosophila Boi limits Hedgehog levels to suppress follicle stem cell proliferation. J Cell Biol 191: 943–952 doi:10.1083/jcb.201007142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dominguez M, Brunner M, Hafen E, Basler K (1996) Sending and receiving the hedgehog signal: control by the Drosophila Gli protein Cubitus interruptus. Science 272: 1621–1625. [DOI] [PubMed] [Google Scholar]
- 58. Aza-Blanc P, Ramírez-Weber FA, Laget MP, Schwartz C, Kornberg TB (1997) Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89: 1043–1053. [DOI] [PubMed] [Google Scholar]
- 59. Méthot N, Basler K (1999) Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96: 819–831. [DOI] [PubMed] [Google Scholar]
- 60. Lee T, Luo L (2001) Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24: 251–254. [DOI] [PubMed] [Google Scholar]
- 61. Dominguez M (1999) Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development 126: 2345–2353. [DOI] [PubMed] [Google Scholar]
- 62. Baonza AA, Freeman MM (2001) Notch signalling and the initiation of neural development in the Drosophila eye. Development 128: 3889–3898. [DOI] [PubMed] [Google Scholar]
- 63. Tabata T, Kornberg TB (1994) Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell 76: 89–102. [DOI] [PubMed] [Google Scholar]
- 64. Chen Y, Struhl G (1996) Dual roles for patched in sequestering and transducing Hedgehog. Cell 87: 553–563. [DOI] [PubMed] [Google Scholar]
- 65. Baonza A, Freeman M (2005) Control of cell proliferation in the Drosophila eye by notch signaling. Dev Cell 8: 529–539 doi:10.1016/j.devcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 66. Doherty D, Feger G, Younger-Shepherd S, Jan LY, Jan YN (1996) Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes Dev 10: 421–434 doi:10.1101/gad.10.4.421. [DOI] [PubMed] [Google Scholar]
- 67. de Celis JF, Garcia-Bellido A, Bray SJ (1996) Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development 122: 359–369. [DOI] [PubMed] [Google Scholar]
- 68. de Celis JFJ, Bray SS (1997) Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development 124: 3241–3251. [DOI] [PubMed] [Google Scholar]
- 69. Pitsouli C, Delidakis C (2005) The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development 132: 4041–4050 doi:10.1242/dev.01979. [DOI] [PubMed] [Google Scholar]
- 70. Rodriguez I, Basler K (1997) Control of compartmental affinity boundaries by hedgehog. Nature 389: 614–618 doi:10.1038/39343. [DOI] [PubMed] [Google Scholar]
- 71. Blair SS, Ralston A (1997) Smoothened-mediated Hedgehog signalling is required for the maintenance of the anterior-posterior lineage restriction in the developing wing of Drosophila. Development 124: 4053–4063. [DOI] [PubMed] [Google Scholar]
- 72. Bejarano F, Bortolamiol-Becet D, Dai Q, Sun K, Saj A, et al. (2012) A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development 139: 2821–2831 doi:10.1242/dev.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lai EC, Tam B, Rubin GM (2005) Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev 19: 1067–1080 doi:10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Uhlmann S, Mannsperger H, Zhang JD, Horvat E-Á, Schmidt C, et al. (2012) Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol 8: 570 doi:10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vidal M, Cagan RL (2006) Drosophila models for cancer research. Curr Opin Genet Dev 16: 10–16 doi:10.1016/j.gde.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 76. Liefke R, Oswald F, Alvarado C, Ferres-Marco D, Mittler G, et al. (2010) Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev 24: 590–601 doi:10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Herz H-M, Madden LD, Chen Z, Bolduc C, Buff E, et al. (2010) The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol 30: 2485–2497 doi:10.1128/MCB.01633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, et al. (2006) c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 20: 2096–2109 doi:10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pallavi SK, Ho DM, Hicks C, Miele L, Artavanis-Tsakonas S (2012) Notch and Mef2 synergize to promote proliferation and metastasis through JNK signal activation in Drosophila. EMBO J 31: 2895–2907 doi:10.1038/emboj.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866 doi:10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 81. Cho WC (2007) OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer 6: 60 doi:10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, et al. (2007) MicroRNA signatures in human ovarian cancer. Cancer Res 67: 8699–8707 doi:10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 83. Ma L, Weinberg RA (2008) Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet 24: 448–456 doi:10.1016/j.tig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 84. Lee YS, Dutta A (2009) MicroRNAs in cancer. Annu Rev Pathol Mech Dis 4: 199–227 doi:10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, et al. (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 doi:10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Voorhoeve PM, le Sage C, Schrier M, Gillis AJM, Stoop H, et al. (2006) A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124: 1169–1181 doi:10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 87. Nolo R, Morrison CM, Tao C, Zhang X, Halder G (2006) The bantam MicroRNA is a target of the hippo tumor-suppressor pathway. Curr Biol 16: 1895–1904 doi:10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 88. Thompson BJ, Cohen SM (2006) The hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126: 767–774 doi:10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 89. Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, et al. (2008) Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA 105: 13021–13026 doi:10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chou Y-T, Lin H-H, Lien Y-C, Wang Y-H, Hong C-F, et al. (2010) EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res 70: 8822–8831 doi:10.1158/0008-5472.CAN-10-0638. [DOI] [PubMed] [Google Scholar]
- 91. Reddy SDN, Ohshiro K, Rayala SK, Kumar R (2008) MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res 68: 8195–8200 doi:10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, et al. (2008) microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 68: 3566–3572 doi:10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 93. Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, et al. (2009) Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 284: 5731–5741 doi:10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 94. Jiang L, Liu X, Chen Z, Jin Y, Heidbreder CE, et al. (2010) MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J 432: 199–205 doi:10.1042/BJ20100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Erkan EP, Breakefield XO, Saydam O (2011) miRNA signature of schwannomas: possible role(s) of “tumor suppressor” miRNAs in benign tumors. Oncotarget 2: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Skalsky RL, Cullen BR (2011) Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS ONE 6: e24248 doi:10.1371/journal.pone.0024248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee H-J, Palkovits M, Young WS (2006) miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc Natl Acad Sci USA 103: 15669–15674 doi:10.1073/pnas.0605781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Junn E, Lee K-W, Jeong BS, Chan TW, Im J-Y, et al. (2009) Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci USA 106: 13052–13057 doi:10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nguyen HTT, Dalmasso G, Yan Y, Laroui H, Dahan S, et al. (2010) MicroRNA-7 modulates CD98 expression during intestinal epithelial cell differentiation. J Biol Chem 285: 1479–1489 doi:10.1074/jbc.M109.057141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Xiong S, Zheng Y, Jiang P, Liu R, Liu X, et al. (2011) MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int J Biol Sci 7: 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Fang YX, Xue J-L, Shen Q, Chen J, Tian L (2012) miR-7 inhibits tumor growth and metastasis by targeting the PI3K/AKT pathway in hepatocellular carcinoma. Hepatology 55: 1852–1862 doi:10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 102. Xu L, Wen Z, Zhou Y, Liu Z, Li Q, et al. (2013) MicroRNA-7 regulated TLR9 signaling enhanced growth and metastatic potential of human lung cancer cells by altering PIK3R3/Akt pathway. Mol Biol Cell 24: 42–55 doi:10.1091/mbc.E12-07-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang N, Li X, Wu CW, Dong Y, Cai M, et al. (2012) microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene doi:10.1038/onc.2012.526. [DOI] [PubMed] [Google Scholar]
- 104. Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM (2005) Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146 doi:10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 105. Yu JY, Reynolds SH, Hatfield SD, Shcherbata HR, Fischer KA, et al. (2009) Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells. Development 136: 1497–1507 doi:10.1242/dev.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tokusumi T, Tokusumi Y, Hopkins DW, Shoue DA, Corona L, et al. (2011) Germ line differentiation factor Bag of Marbles is a regulator of hematopoietic progenitor maintenance during Drosophila hematopoiesis. Development 138: 3879–3884 doi:10.1242/dev.069336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Pek JW, Lim AK, Kai T (2009) Drosophila maelstrom ensures proper germline stem cell lineage differentiation by repressing microRNA-7. Dev Cell 17: 417–424 doi:10.1016/j.devcel.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 108. Brown NL, Sattler CA, Paddock SW, Carroll SB (1995) Hairy and emc negatively regulate morphogenetic furrow progression in the Drosophila eye. Cell 80: 879–887. [DOI] [PubMed] [Google Scholar]
- 109. De Renzis S, Yu J, Zinzen R, Wieschaus E (2006) Dorsal-ventral pattern of Delta trafficking is established by a Snail-Tom-Neuralized pathway. Dev Cell 10: 257–264 doi:10.1016/j.devcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 110. Kang J-S, Mulieri PJ, Hu Y, Taliana L, Krauss RS (2002) BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J 21: 114–124 doi:10.1093/emboj/21.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kang J-S, Feinleib JL, Knox S, Ketteringham MA, Krauss RS (2003) Promyogenic members of the Ig and cadherin families associate to positively regulate differentiation. Proc Natl Acad Sci USA 100: 3989–3994 doi:10.1073/pnas.0736565100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gobeil S, Zhu X, Doillon CJ, Green MR (2008) A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev 22: 2932–2940 doi:10.1101/gad.1714608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. López-Ornelas A, Mejía-Castillo T, Vergara P, Segovia J (2011) Lentiviral transfer of an inducible transgene expressing a soluble form of Gas1 causes glioma cell arrest, apoptosis and inhibits tumor growth. Cancer Gene Ther 18: 87–99 doi:10.1038/cgt.2010.54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hh signal along the disc AP axis and Notch-mediated DV growth promoting organizer starts long before the initiation of retinal differentiation. (A) Mid second larval instar (LII) eye disc carrying the enhancer trap line hhP30-lacZ and stained for ßgalactosidase (hh-Z, green), Wg (red), and Elav (blue). The absence of blue staining denotes that the MF has not yet initiated in this disc. (B) Mid-late LII eye disc carrying the eyg-lacZ enhancer trap line and stained for ßgal (blue) and Wg (red). Notch signalling target Eyg expression labels the growth organizer. Disc as in Figure 3F.
(TIF)
The conserved MicroRNA miR-7 and Dl-Notch pathway cooperatively induce eye overgrowth. (A–C) Illustrative images of adult eyes overexpressing Dl with the GS(2)518 line (A) or the UAS-mir-7 transgene (B–C) with ey-Gal4. (A) The overgrown, folded eye tissue often present areas of undifferentiated or poorly differentiated outgrowths (arrowhead) (10%, n = 200 in A). The undifferentiated outgrowths are seen also in flies co-expressing Dl with the UAS-mir-7 transgene (B and C). (D) Quantification of relative mature mir-7 RNA levels in larvae carrying hsp70>mir-7 after heat shock (red bar) or not (blue bar). P was calculated using the Student t test, and values represented the mean ± sem. of three independent experiments.
(TIF)
Overgrowth and abnormal neuronal differentiation progression in eye discs co-expressing Dl and the GS(2)518 line. Confocal images of eye discs of control wild type (ey>, A, C, E, and G) and eye discs overexpressing Dl and mir-7 by ey-Gal4 (ey>Dl>GS(2)518: B, D, F, H–J) and carrying the indicated enhancer trap lines to monitor DV patterning: expression of D marker mirror-lacZ (mirr-Z), ventral marker fringe-lacZ (fng-Z), DV organizer-specific marker Serrate-lacZ (Ser-Z), and eyegone-lacZ (Eq-Z). Eye discs are stained for ßgalactosidase (green), neuronal marker Elav (blue), or Wg (red). (I–J′) Eye discs are stained for Dac (pink) or DE-cadherin (DE-cad, green in I and J and grey in I′ and J′) to highlight the morphology of the front of retinal differentiation (MF) and cell shape changes the accompanied neuronal differentiation, respectively. Although it has been postulated that the microRNA mir-7 silences Notch signalling, the overexpression of mir-7 with Dl causes eye disc overgrowth associated with enhanced Dl-Notch signalling as detected by the misexpression of DV organizer-specific markers (F and H). Seldom the pattern of retinal differentiation is highly disrupted in the overgrown discs (F and H) and often the front of neuronal differentiation (arrowhead, I′) is highly irregular or advanced in discs co-expressing Dl and GS(2)518 line. Anterior is to the left. Scale bar, 2 mm.
(TIF)
Overgrowth and abnormal neuronal differentiation progression in eye discs co-expressing Dl and the microRNA mir-7 or the ihog-IR or ci-IR transgenes. Confocal images of mitotic marker PH3 (blue in A–E; pink in F and green in G), neuronal marker Elav (green, A–F and red in G), and Wg (red, A–D and pink in F) staining of third instar eye-antennal imaginal discs of wild-type ey-Gal4 (ey>, A–A′), ey-Gal4 UAS-Dl (ey>Dl, B–B′), ey-Gal4 UAS-Dl/+; UAS-mir-7/+ (ey>Dl>mir-7, C–D′), ey-Gal4UAS-Dl/+; UAS-ihog-IR/+ (ey>Dl>ihog-IR, E–F), and ey-Gal4 UAS-Dl/+; UAS-ci-IR/+ (ey>Dl>ci-IR, G). The asterisks point to undifferentiated outgrowth of the eye discs (C, F, and G). Disc in (C) is as in Figure 1H. Note that eye disc overgrowth is also accompanied by advanced or disorganized front of retinal differentiation. The ey-Gal4 transgene drives expression anterior to the MF (white arrowhead in A), where eye disc cells proliferate asynchronously. Posterior to the MF, subsets of cells start differentiating into photoreceptor neurons visualized by the neuronal marker Elav (green, A) and the remaining cells divide one last time synchronously (row of PH3 cells behind the MF). (H) Quantitation of the eye imaginal disc size of the indicated genotypes. The area for each disc was calculated in pixel using ImageJ and values were normalized with those of the corresponding antennal disc part. As expected, co-expressing Dl with the RNAi against ihog or ci with ey-Gal4 provoked overgrowth similar, but stronger than the misexpression of the mir-7. Anterior is to the left in all images, and dorsal is up.
(TIF)
Quantification of ihog and boi mRNAs and mature mir-7 levels. (A) Relative ihog mRNA levels in larvae. (B) Relative boi mRNA levels in larvae. (C) Relative miR-7 levels in S2 cells transfected with actGal5 plasmid and with (red bar) or without (blue bar) the UAS mir-7 plasmid. The values represented the mean ± s.e.m. of at least three independent experiments. Data analysed by a two-tailed unpaired t test.
(TIF)
Overexpression of DsRed::mir-7 by en-Gal4 in the wing disc also caused reproducible in vivo downregulation of eGFP in a tub-eGFP::ihog-3′UTR (A) but not in a tub-eGFP::boi-3′UTR sensor (B).
(TIF)
Invasive growth caused by co-expressing Dl and ci-IR in the wing primordium. (A) Wild-type third instar wing imaginal discs. Dpp-GAL4 (dpp>) drives expression of UAS-GFP (gree) in a narrow band of anterior cells along the AP compartment boundary. Expression of mitotic marker PH3 (blue) and En (red) are also shown. (B) Expression of the RNAi transgene against ci (dpp>ci-IR) led to anterior expansion of the dpp domain visualized by GFP (green) and ectopic P cells (grey in B′) in the A territory at the DV boundary, but the disc is not overgrown. (C, D) Co-expression of Dl along with ci-IR led to extensive overgrowths. Note that mutant A cells mix with wild-type P (En, positive) cells (arrowheads) in some parts, reminiscent of malignant growth. Expression of Ci (grey in the inset) is also shown in (D).
(TIF)
Blocking Hh signal transduction due to mutations in smoothened enhances organizing activity by Dl-Notch signalling in the mosaic eye and wing discs. (A) Control eye discs carrying MARCM GFP(green)-labelled smo3 clones and (B) GFP-labelled clones of smo3 that overexpress Dl and stained for ptc-lacZ (Ptc-Z, blue) and Ci (blue). Note that the smo3/smo3tub-Gal4 UAS-Dl clones cause nonautonomously advancement of the MF denoted by up-regulated Ci levels, similar to the effect seen in eye discs co-expressing Dl with the mir-7. (A′) and (B′) show single channel confocal images. (C) Wing discs carrying MARCM GFP-labelled clones of smo3 cells and staining for Wg (red, C and C″) and clones of smo3 that overexpress Dl (smo3/smo3 tub>Dl, D–D″). In (D–D″), arrowheads point to ventrally situated clones of anterior origin (visualized by ptc-lacZ, not shown). The asterisk points to a clone of ambiguous A origin with weak ectopic Wg only in the anterior portion of the clone. DAPI counterstaining (pink, C″′ and D″′) is shown to illustrate the stimulation of growth of the surrounding tissue by the smo3 tub-Dl clones. Genotype in (A and C) is yw tub-Gal4 UAS-GFP hsp70-Flp; smo3 FRT40A ptc-lacZ/tub-Gal80 FRT40A and in (B and D) is yw tub-Gal4 UAS-GFP hsp70-Flp; smo3 FRT40A ptc-lacZ/tub-Gal80 FRT40A; UAS-Dl/+.
(TIF)
General genetic scheme of crosses for rescuing experiments in Figure 4. Similar genetic schemes were following the rescue by the UAS-boi transgene in Figure 3J. Larvae carrying both the chromosomes with the transgenes ey-Gal4 UAS-Dl (2nd) and UAS-DsRed::mir-7 (3rd) were selected under a fluorescence binocular (MZFLIII, Leica) for expression of DsRed in the eye under the control of Gal4. The resulting adult males were crossed to female virgins of the genotype UAS-hh/CyO. Larvae resulting from the cross were again selected and the DsRed-positive were transferred to a new tube, and the eyes of the resulting non-CyO adults eyes (males and females) were analysed.
(DOCX)
Identification of candidate tumour-suppressor gene(s) of Drosophila in silico predicted miR-7 target genes in the gain of Dl context.
(DOCX)
Direct inhibition by RNAi expression of core Hedgehog pathway genes in the gain of Dl context.
(DOCX)