Abstract
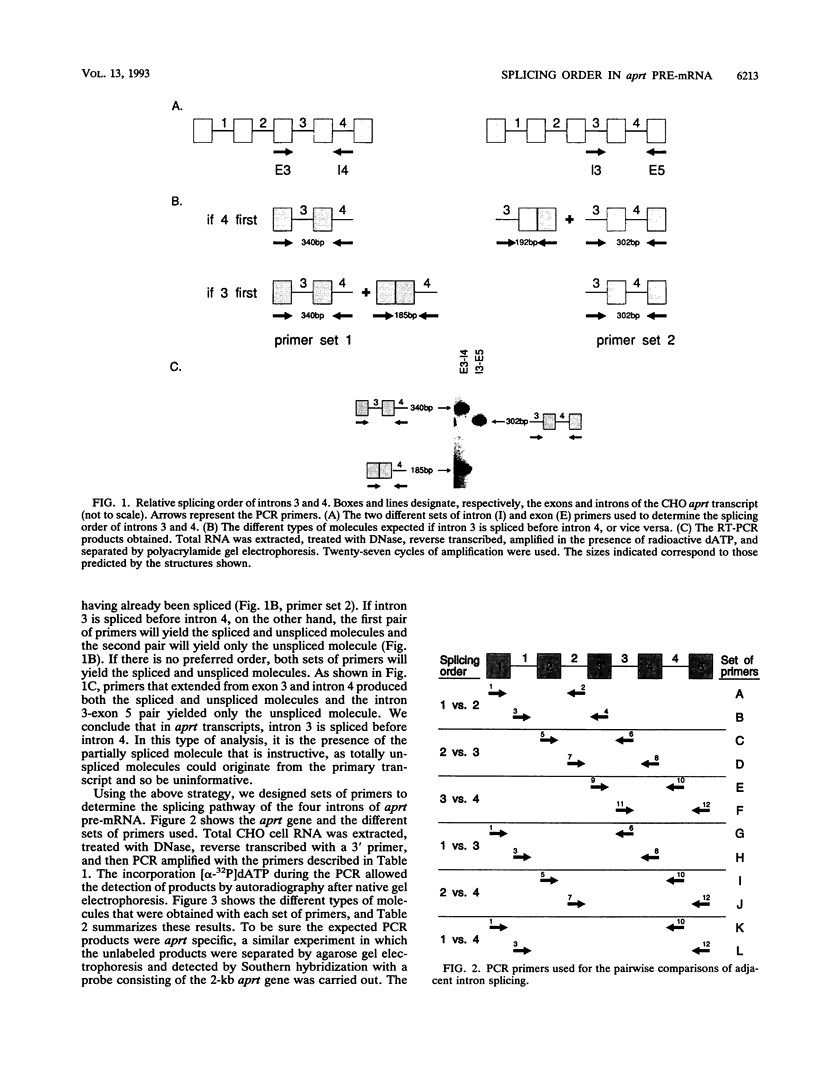
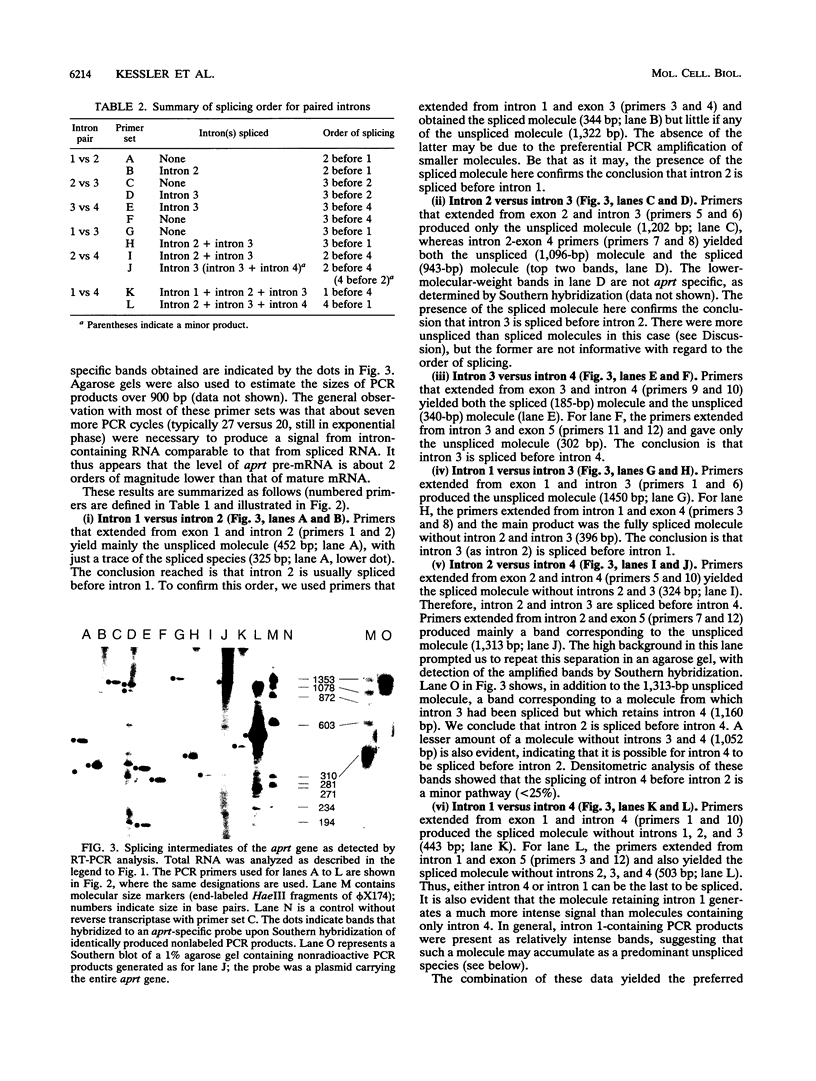
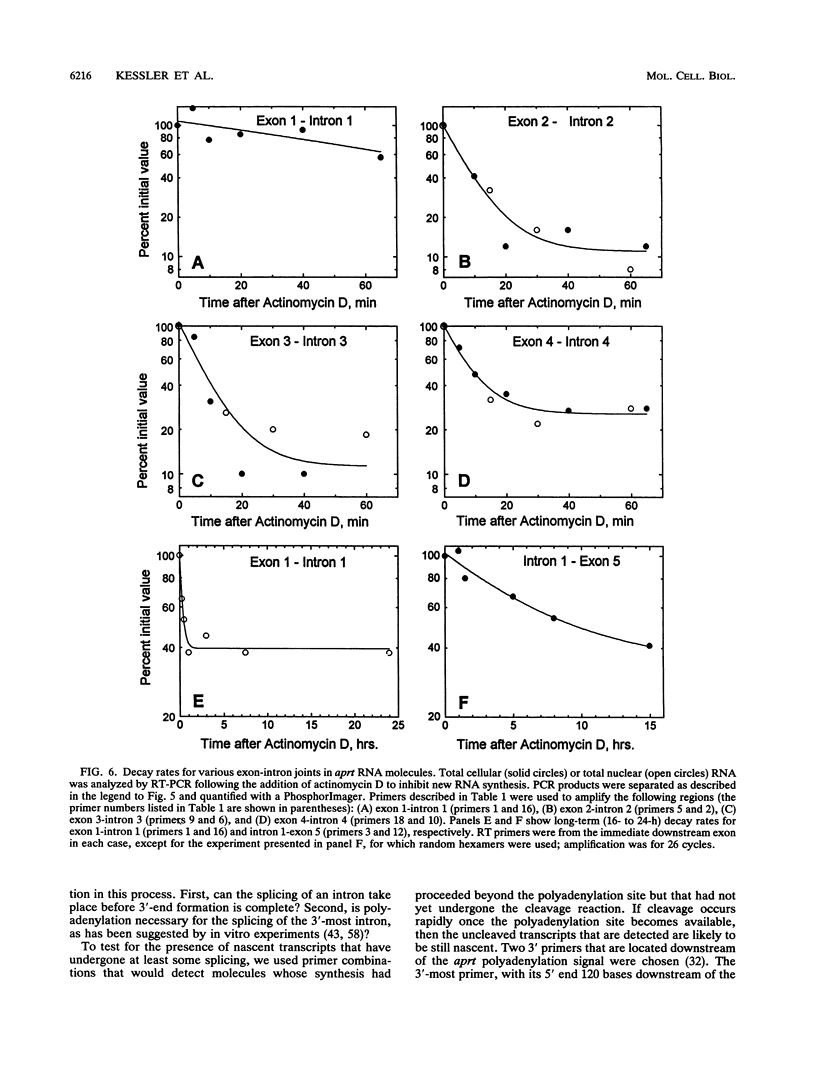
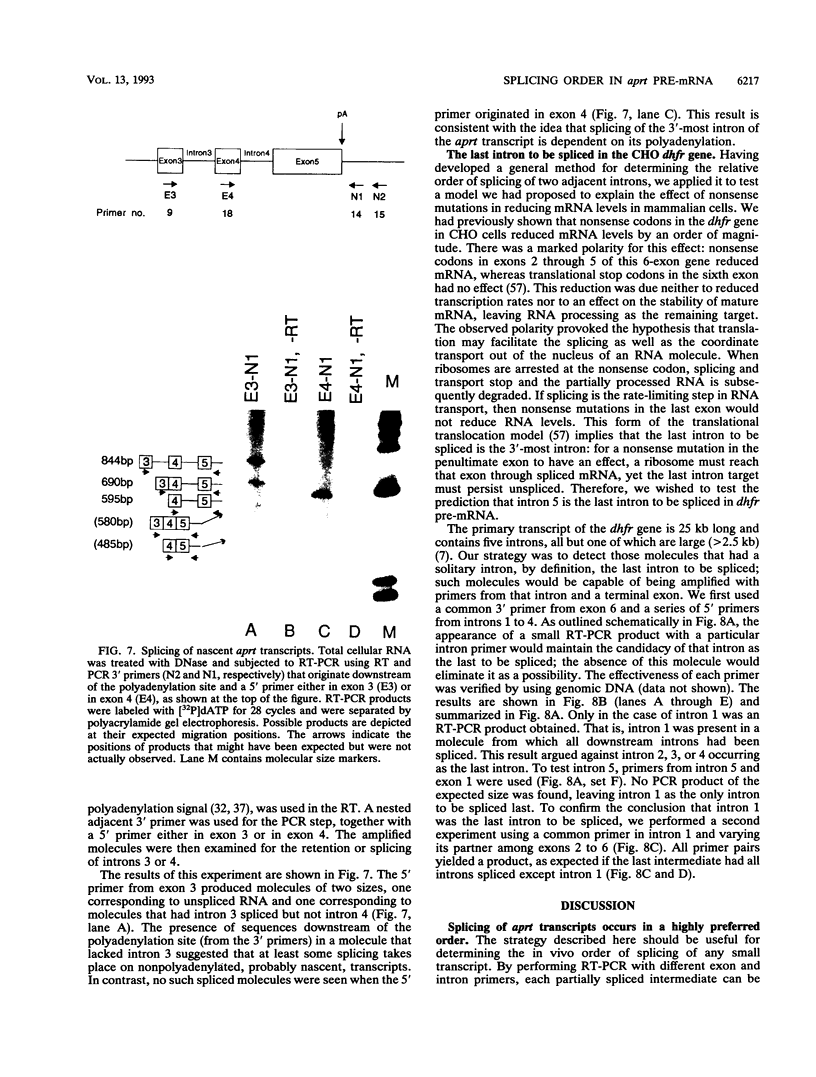
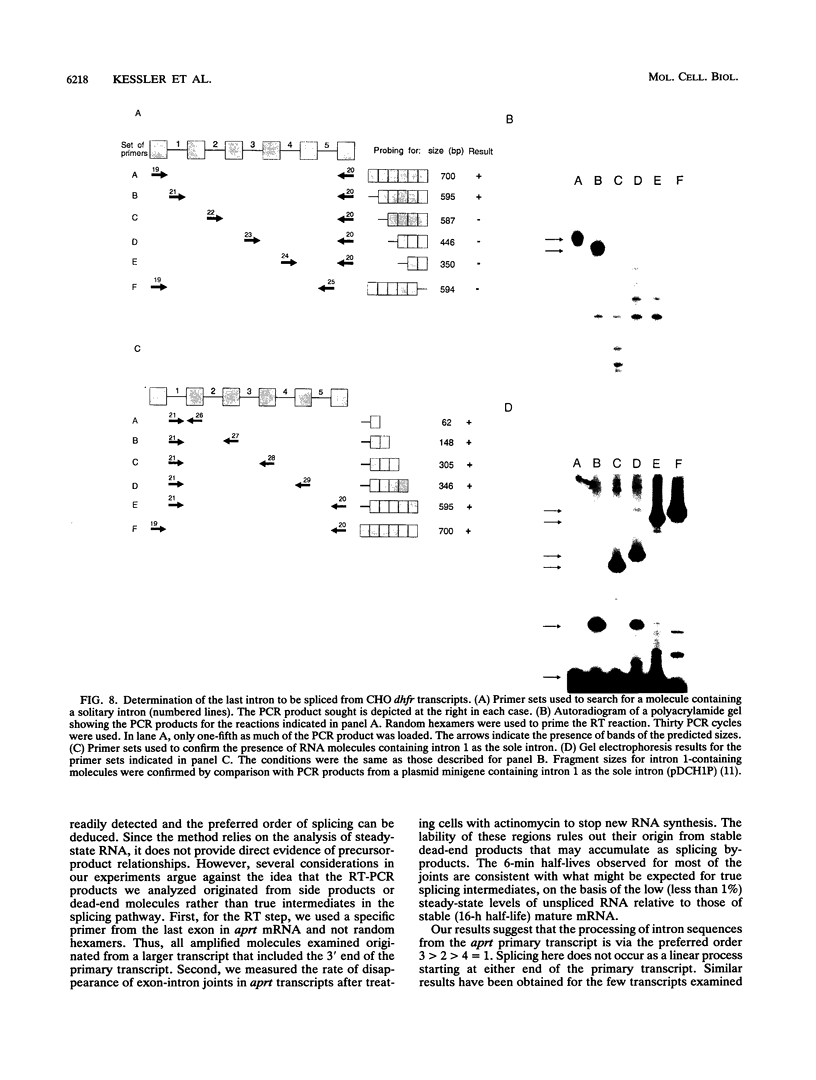
Using a strategy based on reverse transcription and the polymerase chain reaction, we have determined the order of splicing of the four introns of the endogenous adenine phosphoribosyltransferase (aprt) gene in Chinese hamster ovary cells. The method involves a pairwise comparison of molecules that retain one intron and have either retained or spliced another intron(s). A highly preferred order of removal was found: intron 3 > 2 > 4 = 1. This order did not represent a linear progression from one end of the transcript to the other, nor did it correlate with the conformity of the splice site sequences to the consensus sequences or to the calculated energy of duplex formation with U1 small nuclear RNA. By using actinomycin D to inhibit RNA synthesis, the in vivo rate of the first step in splicing was estimated for all four introns; a half-life of 6 min was found for introns 2, 3, and 4. Intron 1 was spliced more slowly, with a 12-min half-life. A substantial amount of RNA that retained intron 1 as the sole intron was exported to the cytoplasm. In the course of these experiments, we also determined that intron 3, but not intron 4, is spliced before 3'-end formation is complete, probably on nascent transcripts. This result is consistent with the idea that polyadenylation is required for splicing of the 3'-most intron. We applied a similar strategy to determine the last intron to be spliced in a very large transcript, that of the endogenous dihydrofolate reductase (dhfr) gene in Chinese hamster ovary cells (25 kb). Here again, intron 1 was the last intron to be spliced.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Baserga S. J., Benz E. J., Jr Nonsense mutations in the human beta-globin gene affect mRNA metabolism. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2056–2060. doi: 10.1073/pnas.85.7.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988 Jun;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Breimer L. H., Nalbantoglu J., Meuth M. Structure and sequence of mutations induced by ionizing radiation at selectable loci in Chinese hamster ovary cells. J Mol Biol. 1986 Dec 5;192(3):669–674. doi: 10.1016/0022-2836(86)90284-6. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Steigerwalt R. W., Urlaub G., Chasin L. A., Grunberger D. DNA base changes and RNA levels in N-acetoxy-2-acetylaminofluorene-induced dihydrofolate reductase mutants of Chinese hamster ovary cells. J Mol Biol. 1989 Aug 5;208(3):417–428. doi: 10.1016/0022-2836(89)90506-8. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Ellis N., Chasin L. A. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 1983 Apr 11;11(7):1997–2012. doi: 10.1093/nar/11.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin L. A., Urlaub G., Mitchell P., Ciudad C., Barth J., Carothers A. M., Steigerwalt R., Grunberger D. RNA processing mutants at the dihydrofolate reductase locus in Chinese hamster ovary cells. Prog Clin Biol Res. 1990;340A:295–304. [PubMed] [Google Scholar]
- Chen-Kiang S., Nevins J. R., Darnell J. E., Jr N-6-methyl-adenosine in adenovirus type 2 nuclear RNA is conserved in the formation of messenger RNA. J Mol Biol. 1979 Dec 15;135(3):733–752. doi: 10.1016/0022-2836(79)90174-8. [DOI] [PubMed] [Google Scholar]
- Cheng J., Maquat L. E. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol. 1993 Mar;13(3):1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciudad C. J., Urlaub G., Chasin L. A. Deletion analysis of the Chinese hamster dihydrofolate reductase gene promoter. J Biol Chem. 1988 Nov 5;263(31):16274–16282. [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Daar I. O., Maquat L. E. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol Cell Biol. 1988 Feb;8(2):802–813. doi: 10.1128/mcb.8.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattoni R., Keohavong P., Stévenin J. Splicing of the E2A premessenger RNA of adenovirus serotype 2. Multiple pathways in spite of excision of the entire large intron. J Mol Biol. 1986 Feb 5;187(3):379–397. doi: 10.1016/0022-2836(86)90440-7. [DOI] [PubMed] [Google Scholar]
- Goguel V., Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993 Mar 26;72(6):893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- Gudas J. M., Knight G. B., Pardee A. B. Ordered splicing of thymidine kinase pre-mRNA during the S phase of the cell cycle. Mol Cell Biol. 1990 Oct;10(10):5591–5595. doi: 10.1128/mcb.10.10.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzoglou M., Sekeris C. E., Hanson R. W. Processing of phosphoenolpyruvate carboxykinase (GTP) RNA in vivo. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4346–4350. doi: 10.1073/pnas.82.13.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B. E., Grabowski P. J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3' splice site by a network of interactions spanning the exon. Genes Dev. 1992 Dec;6(12B):2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- Jackson I. J. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 1991 Jul 25;19(14):3795–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Correct in vivo splicing of the mouse immunoglobulin kappa light-chain pre-mRNA is dependent on 5' splice-site position even in the absence of transcription. Genes Dev. 1988 Nov;2(11):1448–1459. doi: 10.1101/gad.2.11.1448. [DOI] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K. M., Spritz R. A. In vitro splicing pathways of pre-mRNAs containing multiple intervening sequences? Mol Cell Biol. 1987 Oct;7(10):3428–3437. doi: 10.1128/mcb.7.10.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K. M., Spritz R. A. RNA splice site selection: evidence for a 5' leads to 3' scanning model. Science. 1983 Jun 24;220(4604):1351–1355. doi: 10.1126/science.6304877. [DOI] [PubMed] [Google Scholar]
- LeMaire M. F., Thummel C. S. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol Cell Biol. 1990 Nov;10(11):6059–6063. doi: 10.1128/mcb.10.11.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear A. L., Eperon L. P., Wheatley I. M., Eperon I. C. Hierarchy for 5' splice site preference determined in vivo. J Mol Biol. 1990 Jan 5;211(1):103–115. doi: 10.1016/0022-2836(90)90014-D. [DOI] [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Lowy I., Pellicer A., Jackson J. F., Sim G. K., Silverstein S., Axel R. Isolation of transforming DNA: cloning the hamster aprt gene. Cell. 1980 Dec;22(3):817–823. doi: 10.1016/0092-8674(80)90558-9. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Carothers A. M., Han J. H., Harding J. D., Kas E., Venolia L., Chasin L. A. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5' region of the CHO dhfr gene. Mol Cell Biol. 1986 Feb;6(2):425–440. doi: 10.1128/mcb.6.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhar M. G., Johnson L. F. Delayed processing/export of messenger RNA under conditions of reduced protein synthesis. J Cell Physiol. 1988 Apr;135(1):115–121. doi: 10.1002/jcp.1041350116. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu J., Phear G. A., Meuth M. Nucleotide sequence of hamster adenine phosphoribosyl transferase (aprt) gene. Nucleic Acids Res. 1986 Feb 25;14(4):1914–1914. doi: 10.1093/nar/14.4.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim F. H., Spears P. A., Hoffmann H. M., Kuo H. C., Grabowski P. J. A Sequential splicing mechanism promotes selection of an optimal exon by repositioning a downstream 5' splice site in preprotachykinin pre-mRNA. Genes Dev. 1990 Jul;4(7):1172–1184. doi: 10.1101/gad.4.7.1172. [DOI] [PubMed] [Google Scholar]
- Nelson K. J., Haimovich J., Perry R. P. Characterization of productive and sterile transcripts from the immunoglobulin heavy-chain locus: processing of micron and muS mRNA. Mol Cell Biol. 1983 Jul;3(7):1317–1332. doi: 10.1128/mcb.3.7.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mechanism for cryptic splice site activation during pre-mRNA splicing. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6253–6257. doi: 10.1073/pnas.87.16.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988 Mar;2(3):319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Niwa M., Berget S. M. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 1991 Nov;5(11):2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- Noteborn M., Arnberg A., de Jonge M., Ab G., Gruber M. Splicing pathways of the chicken apo very low density lipoprotein II (pre)messenger RNA. FEBS Lett. 1986 Jan 1;194(1):151–156. doi: 10.1016/0014-5793(86)80067-9. [DOI] [PubMed] [Google Scholar]
- Nunberg J. H., Kaufman R. J., Schimke R. T., Urlaub G., Chasin L. A. Amplified dihydrofolate reductase genes are localized to a homogeneously staining region of a single chromosome in a methotrexate-resistant Chinese hamster ovary cell line. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5553–5556. doi: 10.1073/pnas.75.11.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Shiels B. R., Northemann W., Gehring M. R., Fey G. H. Modified nuclear processing of alpha 1-acid glycoprotein RNA during inflammation. J Biol Chem. 1987 Sep 15;262(26):12826–12831. [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. J., Ting A. C., Nordstrom J. L., Zimmer W., O'Malley B. W. Processing of high molecular weight ovalbumin and ovomucoid precursor RNAs to messenger RNA. Cell. 1980 Nov;22(1 Pt 1):219–230. doi: 10.1016/0092-8674(80)90170-1. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman K. M., Steitz J. A. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev. 1993 Apr;7(4):647–659. doi: 10.1101/gad.7.4.647. [DOI] [PubMed] [Google Scholar]
- Watakabe A., Tanaka K., Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993 Mar;7(3):407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Weil D., Brosset S., Dautry F. RNA processing is a limiting step for murine tumor necrosis factor beta expression in response to interleukin-2. Mol Cell Biol. 1990 Nov;10(11):5865–5875. doi: 10.1128/mcb.10.11.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]
- de Boer J. G., Drobetsky E. A., Grosovsky A. J., Mazur M., Glickman B. W. The Chinese hamster aprt gene as a mutational target. Its sequence and an analysis of direct and inverted repeats. Mutat Res. 1989 Aug;226(4):239–244. doi: 10.1016/0165-7992(89)90076-6. [DOI] [PubMed] [Google Scholar]