Abstract
Age at menarche (AAM) is a complex trait involving both genetic and environmental factors. To identify the genetic factors associated with AAM, we conducted a large-scale meta-analysis of genome-wide association studies using more than 15,000 Japanese female samples. Here, we identified an association between SNP (single nucleotide polymorphism) rs364663 at the LIN28B locus and AAM, with a P-value of 5.49×10−7 and an effect size of 0.089 (year). We also evaluated 33 SNPs that were previously reported to be associated with AAM in women of European ancestry. Among them, two SNPs rs4452860 and rs7028916 in TMEM38B indicated significant association with AAM in the same directions as reported in previous studies (P = 0.0013 with an effect size of 0.051) even after Bonferroni correction for the 33 SNPs. In addition, six loci in or near CCDC85A, LOC100421670, CA10, ZNF483, ARNTL, and RXRG exhibited suggestive association with AAM (P<0.05). Our findings elucidated the impact of genetic variations on AAM in the Japanese population.
Introduction
Age at menarche (AAM), the onset of the first menstrual period in girls, is considered as a landmark of female pubertal development. Menarche generally occurs after a series of complex neuroendocrine events leading to full activation of the hypothalamic-pituitary-gonadal axis [1]. Menarche is associated with physical, emotional, and social development [2]. In addition, AAM was shown to be associated with the risk of various diseases. Early AAM is reported to be one of the significant risk factors for depression [3], eating disorders [4], obesity [5], diabetes [6], breast cancer [7], and coronary heart disease [8]. On the other hand, late AAM has been associated with osteoporosis [9] and taller adult stature [10]. Therefore, the identification of loci contributing to variation in AAM could lead to a better understanding of a wide range of phenotypes.
AAM is known to be a complex trait determined by an array of genetic and environmental variables [11]–[13]. Twin and family studies suggest a significant genetic contribution to AAM with a heritability of more than 50% [12], [14]. Several genetic variations within candidate genes such as the estrogen receptor genes (ESR1 and ESR2) [15], [16], CYP19A1 [17], and the SHBG gene [18] were shown to be associated with AAM. To date, a number of genome-wide linkage analyses [14], [19], [20] and genome-wide association studies (GWAS)[21]–[25] for genes underlying variation in AAM have been performed. In 2009, the association of genetic variations in LIN28B with AAM was identified by four independent groups. Currently, more than 30 loci have been shown to be significantly associated with AAM. However, most of these studies were conducted using women of European ancestry. Here we performed a large scale meta-analysis of GWAS using more than 15,000 Japanese female samples.
Results
A total of 15,495 Japanese female subjects from four GWAS using different SNP genotyping systems were enrolled in this analysis. Characteristics of samples and genotyping methods are summarized in Table 1 . All the subjects were of Japanese origin and obtained from the Biobank Japan Project [26]. Samples consist of patients that were classified into 33 disease groups. The average and S.D. of AAM in each disease cohort is shown in Table S1. Some diseases such as breast cancer and osteoporosis are likely to be associated with early or late AAM, as reported previously [7], [9]. We also found that AAM was negatively associated with birth year (p<0.0001). Thus we used disease status and birth year as covariates in this study. Genotyping was performed with over 500,000 SNP markers using Illumina HumanHap 550 Genotyping BeadChip, Illumina610-Quad Genotyping BeadChip, or Illumina Omni Express (Illumina, CA, USA). We applied stringent quality control criteria as mentioned in the methods section. We also conducted principal component analysis [27] to evaluate potential population stratification. To extend the genomic coverage and conduct meta-analysis, we subsequently performed a whole-genome imputation of the SNPs, using HapMap Phase II genotype data [28]. After the imputation, we performed SNP quality control (minor allele frequency≥0.01 and an imputation score (Rsq value by MACH software [29])≥0.7) and found that more than two million autosomal SNPs satisfied these criteria.
Table 1. Characteristics of study population.
| Cohorts | Number of Samples | Source | Platform | Inflation factor | SNP number | Age (S.D.) | Diseases |
| Cohort1 | 11,454 | BioBank Japa n | Illumina HumanHap 610 | 1.052 | 2,263,308 | 59.81+−13.25 | Cancer(Colorectal, breast, lung, gastric, pancreas, liver, cholangiocarcinoma), diabetes mellitus, myocardial infarction, brain infarction, arteriosclerosis obliterans, Arrythimia,drug eruption, liver cirrhosis, amyotrophic lateral sclerosis, osteoporosis, fibroid, Rheumatoid arthritis, and drug response |
| Cohort2 | 941 | BioBank Japan | Illumina HumanHap 550 | 1.001 | 2,220,799 | 47.01+−15.04 | Cancer (cervival, uterus, esophageal, hematopoietic, cholangiocarcinoma, ovarian, pancreas, liver), chronic hepatitis B, pulmonary tuberculosis, keloid, drug eruption, heat cramp |
| Cohort3 | 1957 | BioBank Japan | Illumina OmniExpress | 1.045 | 2,283,889 | 60.90+−9.47 | Cancer (esophageal, uterus) brain aneurysm, chronic obstractive lung disease, glaucoma |
| Cohort4 | 1,143 | BioBank Japan | Illumina HumanHap 550 | 1.008 | 2,220,799 | 37.86+−8.13 | Endometriosis |
| Metaanalysis | 15,495 | 1.039 | 2,310,762 | 57.56+−14.15 |
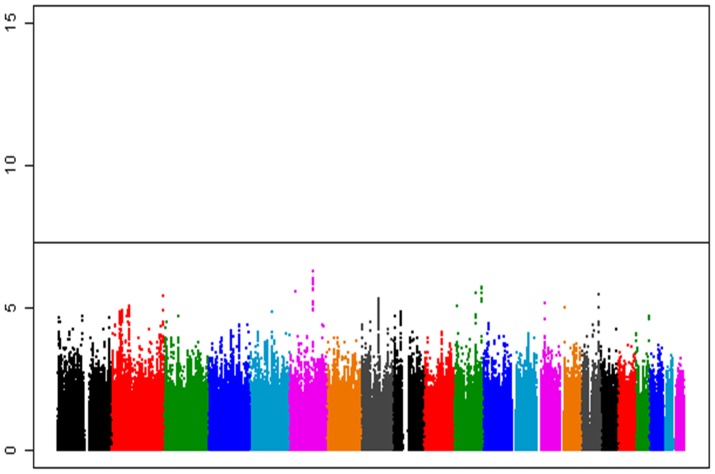
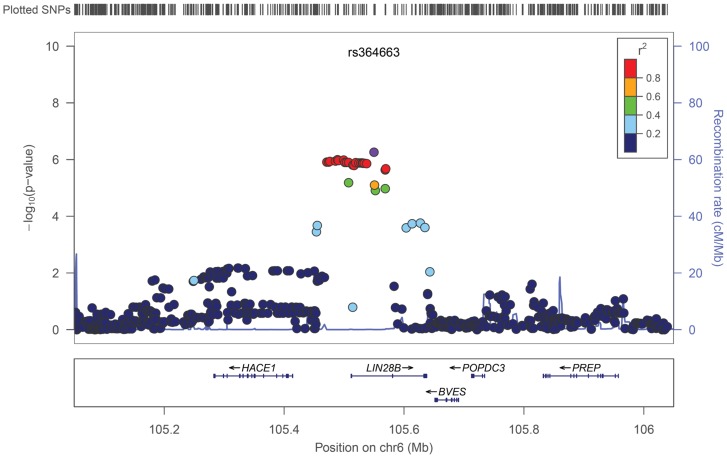
The associations of these imputed SNPs with AAM were evaluated using a linear regression model [30] and meta-analysis. Quantile-Quantile plots of P-values indicated the Inflation factors to be as low as 1.039 ( Figure 1 ), suggesting no substantial population stratification existed in our study population. Although we could not identify significant association that satisfied the genome-wide significant threshold (P<5×10−8), SNP rs364663 which is located within intron 2 of the LIN28B gene at 6q21 indicated the strongest association with a P-value of 5.49×10−7 ( Table 2 , Figure 2 ). SNP rs364663 exhibited the association with AAM in all four cohorts without significant heterogeneity in both effect sizes and directions (P-value for heterogeneity = 0.29; Table 3 ), suggesting that the observed associations at LIN28B are not the result of false-positives due to study-specific bias. Regional p-value plots indicated that all of the AMM-associated SNPs were clustered around the LIN28B locus ( Figure 2 ). Previously reported SNPs rs314263 and rs7759938 near the LIN28B locus 21,24 also associated with AAM (P = 1.03×10−6 and 1.12×10−6, respectively) in the same direction. In addition to the 6q21 locus, 17 SNPs in seven genomic regions exhibited suggestive association with a p-value of<1×10−5 ( Table 2 ). To further investigate the physiological role of these loci, the associations of these variations with body mass index (BMI) and height were evaluated using previously published results in 26,620 Japanese subjects [31]. SNP rs9404590 near the LIN28B locus associated with height with p-value of 0.0003 (Table S2), but we did not find significant association (P<0.01) between these loci and BMI (Table S2).
Figure 1. Results from meta-analysis of four genome-wide association studies.
A total of 15,495 female samples were analyzed in this study.
Table 2. The result of genome wide association analysis of age at menarche.
| SNP | Chr | Position | Gene | allele | Allele 1 freq. | Beta | SE | P | |
| 1 | 2 | ||||||||
| rs364663 | 6 | 105549882 | LIN28B | A | T | 0.71679 | −0.0893 | 0.01784 | 5.49×10−7 |
| rs12200251 | 6 | 105489108 | LIN28B | A | G | 0.71854 | −0.0868 | 0.01775 | 1.00×10−6 |
| rs314263 | 6 | 105499438 | LIN28B | T | C | 0.71852 | −0.0867 | 0.01775 | 1.03×10−6 |
| rs2095812 | 6 | 105490671 | LIN28B | G | C | 0.28544 | 0.08667 | 0.01777 | 1.08×10−6 |
| rs7759938 | 6 | 105485647 | LIN28B | T | C | 0.71771 | −0.0865 | 0.01777 | 1.12×10−6 |
| rs4946651 | 6 | 105476203 | LIN28B | A | G | 0.27974 | 0.08708 | 0.0179 | 1.15×10−6 |
| rs11156429 | 6 | 105471114 | LIN28B | T | G | 0.28116 | 0.08646 | 0.01782 | 1.22×10−6 |
| rs9391253 | 6 | 105474309 | LIN28B | A | T | 0.71867 | −0.0865 | 0.01782 | 1.22×10−6 |
| rs314262 | 6 | 105501314 | LIN28B | A | G | 0.71838 | −0.086 | 0.01775 | 1.26×10−6 |
| rs314280 | 6 | 105507530 | LIN28B | A | G | 0.28161 | 0.086 | 0.01775 | 1.26×10−6 |
| rs395962 | 6 | 105504111 | LIN28B | T | G | 0.28161 | 0.086 | 0.01775 | 1.26×10−6 |
| rs314274 | 6 | 105519625 | LIN28B | A | C | 0.28305 | 0.08279 | 0.01709 | 1.28×10−6 |
| rs1744206 | 6 | 105530624 | LIN28B | G | C | 0.28307 | 0.08271 | 0.01709 | 1.31×10−6 |
| rs314266 | 6 | 105528010 | LIN28B | T | C | 0.71684 | −0.0827 | 0.01709 | 1.31×10−6 |
| rs314268 | 6 | 105524671 | LIN28B | A | G | 0.71677 | −0.0827 | 0.01709 | 1.31×10−6 |
| rs314290 | 6 | 105533687 | LIN28B | A | G | 0.283 | 0.08263 | 0.01709 | 1.34×10−6 |
| rs314291 | 6 | 105531594 | LIN28B | T | C | 0.71699 | −0.0826 | 0.01709 | 1.34×10−6 |
| rs314289 | 6 | 105537627 | LIN28B | T | C | 0.71703 | −0.0825 | 0.01709 | 1.39×10−6 |
| rs314276 | 6 | 105514692 | LIN28B | A | C | 0.28276 | 0.08522 | 0.01775 | 1.57×10−6 |
| rs167539 | 6 | 105516741 | LIN28B | A | C | 0.71714 | −0.0851 | 0.01775 | 1.61×10−6 |
| rs7114000 | 11 | 126112790 | KIRREL3 | A | G | 0.06837 | 0.15025 | 0.03164 | 2.04×10−6 |
| rs3862645 | 11 | 126113656 | KIRREL3 | A | G | 0.06837 | 0.15019 | 0.03164 | 2.07×10−6 |
| rs314270 | 6 | 105569569 | LIN28B | T | C | 0.2829 | 0.0843 | 0.01777 | 2.10×10−6 |
| rs314272 | 6 | 105568697 | LIN28B | A | G | 0.71716 | −0.084 | 0.01777 | 2.26×10−6 |
| rs314273 | 6 | 105568575 | LIN28B | T | G | 0.28266 | 0.08396 | 0.01777 | 2.31×10−6 |
| rs1106419 | 11 | 126114297 | KIRREL3 | A | G | 0.06846 | 0.14829 | 0.0318 | 3.12×10−6 |
| rs12800752 | 11 | 99911850 | ARHGAP42 | T | C | 0.80147 | 0.09455 | 0.02028 | 3.12×10−6 |
| rs310008 | 16 | 80031150 | CMIP | G | C | 0.13392 | −0.1076 | 0.02322 | 3.57×10−6 |
| rs6431393 | 2 | 236204046 | AGAP1 | A | G | 0.4948 | 0.07757 | 0.01681 | 3.95×10−6 |
| rs4735738 | 8 | 77774180 | ZFHX4 | A | G | 0.49918 | 0.07283 | 0.01594 | 4.93×10−6 |
| rs3862642 | 11 | 126110244 | KIRREL3 | T | C | 0.06969 | 0.1438 | 0.03149 | 4.97×10−6 |
| rs6995390 | 8 | 77773567 | ZFHX4 | A | T | 0.49503 | 0.07271 | 0.01597 | 5.30×10−6 |
| rs3889461 | 11 | 126113915 | KIRREL3 | T | C | 0.93023 | −0.1426 | 0.03149 | 5.98×10−6 |
| rs9404590 | 6 | 105507706 | LIN28B | T | G | 0.77319 | −0.0847 | 0.01879 | 6.49×10−6 |
| rs7822501 | 8 | 77825351 | ZFHX4 | A | G | 0.51105 | 0.06865 | 0.01529 | 7.16×10−6 |
| rs7822914 | 8 | 77825593 | ZFHX4 | T | C | 0.48895 | −0.0687 | 0.01529 | 7.16×10−6 |
| rs6472982 | 8 | 77825957 | ZFHX4 | T | C | 0.48894 | −0.0685 | 0.01529 | 7.44×10−6 |
| rs2076751 | 14 | 36059170 | NKX2-1 | A | C | 0.23377 | −0.087 | 0.01941 | 7.45×10−6 |
| rs6472983 | 8 | 77834432 | ZFHX4 | T | G | 0.48902 | −0.0685 | 0.01529 | 7.56×10−6 |
| rs369065 | 6 | 105550751 | LIN28B | T | C | 0.65066 | −0.0721 | 0.01615 | 7.92×10−6 |
| rs1865294 | 8 | 77772597 | ZFHX4 | A | G | 0.50188 | 0.07121 | 0.01599 | 8.41×10−6 |
| rs7114467 | 11 | 15414043 | INSC | A | G | 0.45957 | −0.0713 | 0.01601 | 8.52×10−6 |
Genotyping result of 15,495 Japanese subjects were anlayzed in this study. Imputed SNPs with R2 of less than 0.7 were excluded from this analysis. A1 frequency of JPT was those from release 24 Hapmap JPT.
Effect size and SE of allele1 on age at menarche (year per allele) and P-values were obtained by inverse-variance method.
Figure 2. Regional association plot at rs364663.
Upper panel; P values of genotyped SNPs are plotted (as −log10 values) against their physical location on chromosome 6 (NCBI Build 36). Estimated recombination rates from HapMap JPT shows the local LD structure. Inset; Colors of other SNPs indicate LD with rs2596542 according to a scale from r 2 = 0 to r 2 = 1 based on pair-wise r 2 values from HapMap JPT. Lower panel; Gene annotations from the University of California Santa Cruz genome browser.
Table 3. Association of rs364663 with age at menarche.
| Groups | Chr | allele | Number of samples | Allele 1 Freq. | Beta* | SE* | P | |
| Position | 1 | 2 | ||||||
| Cohort1 | A | T | 11454 | 0.72 | −0.069 | 0.021 | 7.71×10−4 | |
| Cohort2 | 6 | 941 | 0.71 | −0.131 | 0.068 | 0.056 | ||
| Cohort3 | 1957 | 0.72 | −0.137 | 0.050 | 0.0064 | |||
| Cohort4 | 105549882 | 1143 | 0.71 | −0.163 | 0.060 | 0.0071 | ||
| Metaanalysis * | 15495 | 0.72 | −0.089 | 0.018 | 5.49×10−7 | |||
Effect size and S.E. of allele1 on age at menarche (year per allele) and P-values were obtained by inverse-variance method.
Since AAM is associated with various disease risks, we conducted separate analyses for each disease and then performed meta-analysis for the top 42 loci. As a result, some SNPs showed slightly stronger association, but none cleared the genome wide significant threshold (Table S3). Similar to the current study, our group had previously conducted QTL analyses using disease status as a covariate and successfully identified many QTL loci [31]–[35]. Therefore, different background due to disease status was unlikely to significantly affect the result of association analysis.
Additionally, we examined the loci already known to show significant association with AAM in women of European ancestry [22]–[25], [36]. We selected 37 SNPs for this candidate analysis and successfully obtained the genotyping results of 33 SNPs ( Table 4 ), and the risk allele was consistent with previous reports for 31 SNPs. In addition, eight SNPs in or near RXRG, CCDC85A, LOC100421670, TMEM38B, ZNF483, ARNTL, and CA10 indicated possible associations with AAM (P<0.05). Among them, rs4452860 and rs7028916 at the TMEM38B locus exhibited significant association even after Bonferroni's correction (P<0.0015 = 0.05/33). Taken together, these variations as well as LIN28B are likely to be common AAM loci, although their effect sizes are different between women of European ancestry and those of Japanese.
Table 4. Association results in Japanese woman of previously identified SNPs with age at menarche in Caucasian woman.
| SNP | Chr | Position | Gene | allele | Allele 1 freq. | Beta* | S.E.* | P | ref | concordance** | |
| 1 | 2 | ||||||||||
| rs466639 | 1 | 163,661,506 | RXRG | T | C | 0.204 | −0.0431 | 0.0196 | 0.028 | 1) | yes |
| rs633715 | 1 | 176,119,203 | SEC16B | T | C | 0.779 | 0.0312 | 0.0188 | 0.097 | 1) | yes |
| rs2947411 | 2 | 604,168 | TMEM18 | A | G | 0.096 | 0.0519 | 0.0268 | 0.053 | 1) | yes |
| rs17268785 | 2 | 56,445,587 | CCDC85A | A | G | 0.793 | −0.0561 | 0.0196 | 0.004 | 1) | yes |
| rs17188434 | 2 | 156,805,022 | NR4A2 | N.D. | 1) | N.D. | |||||
| rs12617311 | 2 | 199,340,810 | PLCL1 | A | G | 0.432 | −0.0026 | 0.0174 | 0.883 | 1) | yes |
| rs7617480 | 3 | 49,185,736 | KLHDC8B | N.D. | 1) | N.D. | |||||
| rs6762477 | 3 | 50,068,213 | RBM6 | A | G | 0.858 | 0.0219 | 0.0250 | 0.380 | 1) | yes |
| rs7642134 | 3 | 86,999,572 | VGLL3 | A | G | 0.463 | −0.0167 | 0.0173 | 0.334 | 1) | yes |
| rs6438424 | 3 | 119,057,512 | LOC100421670 | A | C | 0.370 | −0.0455 | 0.0162 | 0.005 | 1) | yes |
| rs6439371 | 3 | 134,093,442 | TMEM108, NPHP3 | A | G | 0.705 | −0.0080 | 0.0178 | 0.655 | 1) | yes |
| rs2002675 | 3 | 187,112,262 | TRA2B, ETV5 | A | G | 0.862 | −0.0128 | 0.0231 | 0.581 | 1) | yes |
| rs13187289 | 5 | 133,877,076 | PHF15 | G | C | 0.078 | 0.0440 | 0.0295 | 0.137 | 1) | yes |
| rs13357391 | 5 | 136,468,981 | SPOCK | T | C | 0.840 | −0.0248 | 0.0221 | 0.262 | 3) | yes |
| rs1859345 | 5 | 136,475,319 | SPOCK | T | C | 0.838 | −0.0213 | 0.0221 | 0.336 | 3) | yes |
| rs4840086 | 6 | 100,315,159 | PRDM13, MCHR2 | A | G | 0.619 | −0.0251 | 0.0161 | 0.118 | 1) | no |
| rs1361108 | 6 | 126,809,293 | C6orf173, TRMT11 | N.D. | 1) | N.D. | |||||
| rs1079866 | 7 | 41,436,618 | INHBA | G | C | 0.298 | 0.0150 | 0.0170 | 0.376 | 1) | yes |
| rs7821178 | 8 | 78,256,392 | PXMP3 | A | C | 0.436 | −0.0150 | 0.0170 | 0.380 | 1) | yes |
| rs4452860 | 9 | 107,965,210 | TMEM38B | A | G | 0.545 | 0.0513 | 0.0160 | 0.0013 | 2) | yes |
| rs7028916 | 9 | 107,966,889 | TMEM38B | A | C | 0.454 | −0.0512 | 0.0160 | 0.0013 | 2) | yes |
| rs7861820 | 9 | 107,976,495 | TMEM38B | T | C | 0.204 | 0.0363 | 0.0196 | 0.064 | 2) | yes |
| rs2090409 | 9 | 108,006,909 | TMEM38B | N.D. | 1) | N.D. | |||||
| rs12684013 | 9 | 108,037,935 | TMEM38B | T | C | 0.525 | −0.0265 | 0.0160 | 0.099 | 2) | yes |
| rs10980926 | 9 | 113,333,455 | ZNF483 | A | G | 0.633 | 0.0441 | 0.0161 | 0.006 | 1) | yes |
| rs4929923 | 11 | 8,595,776 | TRIM66 | T | C | 0.624 | 0.0068 | 0.0161 | 0.674 | 1) | yes |
| rs900145 | 11 | 13,250,481 | ARNTL | T | C | 0.489 | −0.0371 | 0.0160 | 0.020 | 1) | yes |
| rs10899489 | 11 | 77,773,021 | GAB2 | A | C | 0.439 | 0.0257 | 0.0160 | 0.109 | 1) | yes |
| rs6589964 | 11 | 122,375,893 | BSX | A | C | 0.549 | −0.0137 | 0.0183 | 0.453 | 1) | yes |
| rs6575793 | 14 | 100,101,970 | BEGAIN | T | C | 0.328 | −0.0279 | 0.0179 | 0.119 | 1) | yes |
| rs1659127 | 16 | 14,295,806 | MKL2 | A | G | 0.483 | 0.0085 | 0.0161 | 0.595 | 1) | yes |
| rs9939609 | 16 | 52,378,028 | FTO | A | T | 0.198 | −0.0191 | 0.0196 | 0.328 | 1) | yes |
| rs1364063 | 16 | 68,146,073 | NFAT5 | T | C | 0.864 | −0.0236 | 0.0231 | 0.307 | 1) | no |
| rs9635759 | 17 | 46,968,784 | CA10 | A | G | 0.435 | 0.0479 | 0.0172 | 0.005 | 1) | yes |
| rs1398217 | 18 | 43,006,236 | FUSSEL18 | G | C | 0.599 | −0.0225 | 0.0161 | 0.161 | 1) | yes |
| rs10423674 | 19 | 18,678,903 | CRTC1 | A | C | 0.705 | 0.0031 | 0.0171 | 0.856 | 1) | yes |
| rs852069 | 20 | 17,070,593 | PCSK2 | A | G | 0.776 | −0.0340 | 0.0187 | 0.069 | 1) | yes |
Genotyping result of 15,495 Japanese subjects were anlayzed in this study. Imputed SNPs with R2 of less than 0.7 were excluded from this analysis. A1 frequency of JPT were those from release 24 Hapmap JPT. N.D.; no data. References: 1 Elks et al Nat Genet 2010, 2 He et al Nature Genet 2009, 3 Liu et al Plos Genet 2009. P-value of 0.0015 (0.05/33) was set at the significant threshold for this candidate analysis.
:Effect size and S.E. of allele1 on age at menarche (year per allele) and P-values were obtained by inverse-variance method.
Concordance of association direction between this study and the previous report.
Discussion
AAM is a complex trait that is influenced by both genetic and environmental factors. In recent years, genome wide association analyses have become a standard method to identify genetic factors related with various diseases and phenotypes. In 2002 our group performed the first GWAS for myocardial infarction and successfully identified LTA as a disease susceptibility gene. Using this method, we have identified a number of loci associated with various phenotypes and common diseases [37]–[42].
Through the analysis of more than 15,000 Japanese female subjects from the Biobank Japan Project, we here report the association of LIN28B with AAM. In our analysis, SNP rs364663 within intron 2 of the LIN28B gene exhibited the strongest association. Previously reported SNPs rs314263, rs7759938, rs314280, and rs314276 near or in the LIN28B gene also showed equivalent association with p-values of 1.03−1.57×10−6. Although the directions of associations were consistent between women of European ancestry and those of Japanese, the effect sizes among Japanese (0.085–0.087) were less than those among European ancestry (0.09–0.14). Thus LIN28B is a common AAM loci, but its effects are relatively small among the Japanese population.
The lin-28 gene was originally identified through C. elegans mutants showing abnormality in developmental timing [43]. Deleterious mutations in lin-28 produce an abnormal rapid tempo of development through larval stages to adult cuticle formation [43]. Two mammalian homologs, Lin28a and Lin28b, possess similar biochemical activities [44], [45]. Transgenic mice expressing Lin28a exhibited increased body size, crown-rump length and delayed onset of puberty due to increased glucose metabolism and insulin sensitivity [46]. In humans, both LIN28B and LIN28A control preprocessing of the let7 microRNA family [45]. Lin28B is highly expressed in the majority of human hepatocellular carcinomas and embryonic stem cells [47], and regulate cell pluripotency [48] as well as cancer growth [47]. However the role of genetic variations in the LIN28B locus should be investigated in the future study.
Among the seven suggestive loci indentified in our GWAS, ZFHX4 and NKX2.1 loci were previously shown to be associated with height in the Caucasian population [49], [50]. These loci also exhibited association with height among the Japanese population (p = 0.053 and 0.023, respectively). The NKX2.1 gene encodes a thyroid-specific transcription factor that was shown to be associated with hypothyroidism [51]. Since delayed pubertal development is a common manifestation of hypothyroidism, variation at NKX2.1 locus might affect AAM through the regulation of serum thyroid hormone level. The ZFHX4 gene encodes a homeodomain-zinc finger protein. ZFHX4 mRNA is highly expressed in brain, liver, and muscle, however the molecular mechanism whereby variations within this genetic locus affect AAM is not clear. In addition, KIRREL3 locus was shown to be associated with breast size in women of European ancestry [52]. The KIRREL3 gene encode a member of the nephrin-like protein family. KIRREL3 was highly expressed in brain and kidney, and shown to be associated with mental retardation autosomal dominant type 4 [53] and neurocognitive delay associated with Jacobsen Syndrome [54]. Taken together, these three loci seem to be common development traits in women of both European ancestry and Japanese.
In our candidate analysis, the association of SNPs rs4452860 and rs7028916 on 9q31.2 with AAM was also validated. This locus was repeatedly shown to be associated with AAM [25], [55]as well as serum ALT (alanine transaminase) level[56]. SNPs rs4452860 and rs7028916 were located more than 300 kb away from the TMEM38B gene. In mice, TMEM38B is strongly expressed in brain, and TMEM38B deficient mice is neonatal lethal[57]. In humans, a homozygous mutation of TMEM38B was associated with autosomal recessive osteosgenesis imperfecta[58]. These findings also suggest an important role of TMEM38B in developmental processes.
In this study, we observed early AAM among breast cancer patients compared with overall samples (12.99 vs 13.44, Table S1). On the other hand, patients with osteoporosis exhibited late AAM (14.33 y.o.). These findings are concordant with previous reports. Since we do not have age-matched controls, the association of these diseases and AAM should be examined using more appropriate sample sets.
Although more than 15,000 samples were employed in this study, no SNP cleared the genome wide significance threshold (p<5×10−8). This could be explained by some limitations in our study. Firstly, the information about AAM was self-reported and might not be very accurate. Although this distinct event is often well recalled many years later [59], possible error due to age-associated memory impairment would increase the risk of false negative association. Secondly, we did not have a replication sample set, and the total sample size may be inadequate to detect variations with modest effects. At present, we unfortunately do not have sufficient samples for replication, since more than 2,900 additional samples are necessary to obtain a genome wide significant association for the top SNP rs364663. Previous studies comprising up to 17,510 women detected only one or two genome-wide significant signals [22]–[25]. However, 30 significant loci were identified by using 87,802 subjects in the screening stage [21]. Although no SNPs reached the genome wide significance in our study, analyzing an increased number of subjects in a relatively younger generation would improve statistical power as well as data accuracy, and subsequently enable us to identify other genetic factors with modest effects.
Methods
Samples
The subjects enrolled in the GWAS meta-analysis for age at menarche (AAM) (n = 15,495) consisted of female patients that were classified into 33 disease groups; cancer (lung, esophagus, stomach, colon, liver, cholangiocarcinoma, pancreas, breast, uterine cervix, uterine body, ovary, hematopoietic organ), diabetes, myocardial infarction, brain infarction, arteriosclerosis, arrhythmia, drug eruption, liver cirrhosis, amyotrophic lateral sclerosis, osteoporosis, fibroid, and drug response, rheumatoid arthritis. chronic hepatitis B, pulmonary tuberculosis, keloid, heat cramp, brain aneurysm, chronic obstractive lung disease, glaucoma, and endometriosis ( Tables 1 and 2 ). All subjects were collected under the support of the BioBank Japan Project [26], in which the individuals with any one of the 47 common diseases were enrolled between 2003 and 2008. Subjects under 17 years of age were not included in this study. Various life style information such as AAM was obtained through a face-to-face interview by trained medical coordinators using questionnaire sheets at each hospital. All participants provided written informed consent as approved by the ethical committees of each institute. For participants between the age of 17 and 20 years old, we obtained written informed consent from both the participant and her parents. AAM was collected by self-report on the questionnaire. The subjects with AAM between the ages of 10 and 17 were enrolled for the study. This project was approved by the ethical committees of the BioBank Japan Project [26] and the University of Tokyo.
Genotyping and quality control
In the four GWAS enrolled in the meta-analysis of AAM, all samples were genotyped at more than 500,000 loci using one of the following platforms: Illumina HumanHap550v3 Genotyping BeadChip, Illumina HumanHap610-Quad Genotyping BeadChip, or Illumina Omni express Genotyping BeadChip ( Table 1 ). Then we applied the following quality control for each GWAS separately: exclusion criteria; subjects with call rates<0.98, SNPs with call rates<0.99 or with ambiguous clustering of the intensity plots, or non-autosomal SNPs. We then excluded subjects whose ancestries were estimated to be distinct from East-Asian populations using principle component analysis performed by EIGENSTRAT version 2.0 [27]. Subsequently, the SNPs with MAF<0.01 or P-value of the Hardy-Weinberg equilibrium test<1.0×10−7 were excluded.
After the quality control criteria mentioned above were applied, genotype imputation was performed by MACH 1.0.16 [29] using the genotype data of Phase II HapMap JPT and CHB individuals (release 24)[28] as references, in a two-step procedure as described elsewhere [60]. In the first step of the imputation, recombination and error rate maps were estimated using 500 subjects randomly selected from the GWAS data. In the second step, imputation of the genotypes of all subjects was performed using the rate maps estimated in the first step. Quality control filters of MAF≥0.01 and Rsq values≥0.7 were applied for the imputed SNPs.
Statistical analysis
Associations of the SNPs with AAM were assessed by linear regression assuming the additive effects of the allele dosages on AAM, using mach2qtl software [29]. Years of birth and affection statuses of the diseases were used as covariates. Meta-analysis of all four GWAS was performed using an inverse-variance method from the summary statistics of beta and standard error (SE), using the Java source code implemented by the authors [61]. Genomic control correction was applied for each GWAS separately, and applied again for the results of GWAS meta-analysis. We set the P-values of 5.0×10−8 and 0.0015 ( = 0.05/33, Bonferroni's correction based on the numbers of the evaluated loci) as significance thresholds in GWAS and candidate gene analysis, respectively. Heterogeneity of the effect sizes among the studies was evaluated using Cochran's Q statistics.
Body mass index and Height QTL analysis
The associations of 42 SNPs with BMI and height were evaluated using previously published results, in which a total of 26,620 subjects with 32 diseases from Biobank Japan were enrolled [31]. Of the 26,620 subjects, 12,350 were also included in this AAM study. Genotyping was performed using the Illumina HumanHap610-Quad Genotyping BeadChip. Genotype imputation was performed using MACH 1.0. BMI was calculated based on self-reported body weight and height data. A rank-based inverse-normal transformation was applied to the BMI values of the subjects. Associations of the SNPs with transformed values of BMI were assessed by linear regression assuming additive effects of allele dosages (bound between 0.0 and 2.0) using mach2qtl software using gender, age, smoking history, the affection statuses of the diseases and the demographic classifications of the medical institutes in Japan where the subjects were enrolled were used as covariates.
Web resources
The URLs for data presented herein are as follows. BioBank Japan Project, http://biobankjp.org
MACH and mach2qtl software, http://www.sph.umich.edu/csg/abecasis/MACH/index.html International HapMap Project, http://www.hapmap.org PLINK software, http://pngu.mgh.harvard.edu/~purcell/plink/index.shtml EIGENSTRAT software, http://genepath.med.harvard.edu/~reich/Software.htm R statistical software, http://cran.r-project.org.
Supporting Information
Age at menarche in each disease cohort.
(DOCX)
The result of association analysis of TOP 42 SNPs from Japanese AAM study with body mass index and height from previous GWAS by Okada et al.
(DOCX)
The result of top 42 SNPs by separated analysis.
(DOCX)
Acknowledgments
We would like to thank all the subjects who donated their DNA for this work. We also thank the technical staff of the Laboratory for Genotyping Development, Center for Genomic Medicine, RIKEN for their technical support.
Funding Statement
This work was conducted as a part of the BioBank Japan Project that was supported by the Ministry of Education, Culture, Sports, Science and Technology of the Japanese government. This work was also supported by grant from the Ministry of Education, Culture, Sports, Science and Technology (C. Tanikawa and K. Matsuda) and Shiseido female researcher science grant (C. Tanikawa). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tena-Sempere M (2006) GPR54 and kisspeptin in reproduction. Hum Reprod Update 12: 631–639. [DOI] [PubMed] [Google Scholar]
- 2. Susman EJ, Nottelmann ED, Inoff-Germain GE, Dorn LD, Cutler GB Jr, et al. (1985) The relation of relative hormonal levels and physical development and social-emotional behavior in young adolescents. Journal of Youth and Adolescence 14: 245–264. [DOI] [PubMed] [Google Scholar]
- 3. Kaltiala-Heino R, Kosunen E, Rimpela M (2003) Pubertal timing, sexual behaviour and self-reported depression in middle adolescence. J Adolesc 26: 531–545. [DOI] [PubMed] [Google Scholar]
- 4. Kaltiala-Heino R, Rimpela M, Rissanen A, Rantanen P (2001) Early puberty and early sexual activity are associated with bulimic-type eating pathology in middle adolescence. J Adolesc Health 28: 346–352. [DOI] [PubMed] [Google Scholar]
- 5. Freedman DS, Khan LK, Serdula MK, Dietz WH, Srinivasan SR, et al. (2003) The relation of menarcheal age to obesity in childhood and adulthood: the Bogalusa heart study. BMC pediatrics 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kjaer K, Hagen C, Sandø S, Eshøj O (1992) Epidemiology of menarche and menstrual disturbances in an unselected group of women with insulin-dependent diabetes mellitus compared to controls. Journal of Clinical Endocrinology & Metabolism 75: 524–529. [DOI] [PubMed] [Google Scholar]
- 7. Velie EM, Nechuta S, Osuch JR (2005) Lifetime reproductive and anthropometric risk factors for breast cancer in postmenopausal women. Breast Dis 24: 17–35. [DOI] [PubMed] [Google Scholar]
- 8. Cooper GS, Ephross SA, Weinberg CR, Baird DD, Whelan EA, et al. (1999) Menstrual and reproductive risk factors for ischemic heart disease. Epidemiology 10: 255–259. [PubMed] [Google Scholar]
- 9. Fujiwara S, Kasagi F, Yamada M, Kodama K (1997) Risk factors for hip fracture in a Japanese cohort. Journal of Bone and Mineral Research 12: 998–1004. [DOI] [PubMed] [Google Scholar]
- 10. Onland-Moret N, Peeters P, Van Gils C, Clavel-Chapelon F, Key T, et al. (2005) Age at menarche in relation to adult height. Am J Epidemiol 162: 623–632. [DOI] [PubMed] [Google Scholar]
- 11. Anderson CA, Duffy DL, Martin NG, Visscher PM (2007) Estimation of variance components for age at menarche in twin families. Behav Genet 37: 668–677. [DOI] [PubMed] [Google Scholar]
- 12. Towne B, Czerwinski SA, Demerath EW, Blangero J, Roche AF, et al. (2005) Heritability of age at menarche in girls from the Fels Longitudinal Study. American journal of physical anthropology 128: 210–219. [DOI] [PubMed] [Google Scholar]
- 13. Treloar SA, Martin NG (1990) Age at menarche as a fitness trait: nonadditive genetic variance detected in a large twin sample. Am J Hum Genet 47: 137–148. [PMC free article] [PubMed] [Google Scholar]
- 14. Anderson CA, Zhu G, Falchi M, Van Den Berg SM, Treloar SA, et al. (2008) A genome-wide linkage scan for age at menarche in three populations of European descent. Journal of Clinical Endocrinology & Metabolism 93: 3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stavrou I, Zois C, Ioannidis JP, Tsatsoulis A (2002) Association of polymorphisms of the oestrogen receptor alpha gene with the age of menarche. Hum Reprod 17: 1101–1105. [DOI] [PubMed] [Google Scholar]
- 16. Stavrou I, Zois C, Chatzikyriakidou A, Georgiou I, Tsatsoulis A (2006) Combined estrogen receptor alpha and estrogen receptor beta genotypes influence the age of menarche. Hum Reprod 21: 554–557. [DOI] [PubMed] [Google Scholar]
- 17. Guo Y, Xiong DH, Yang TL, Guo YF, Recker RR, et al. (2006) Polymorphisms of estrogen-biosynthesis genes CYP17 and CYP19 may influence age at menarche: a genetic association study in Caucasian females. Hum Mol Genet 15: 2401–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xita N, Tsatsoulis A, Stavrou I, Georgiou I (2005) Association of SHBG gene polymorphism with menarche. Mol Hum Reprod 11: 459–462. [DOI] [PubMed] [Google Scholar]
- 19. Rothenbuhler A, Fradin D, Heath S, Lefevre H, Bouvattier C, et al. (2006) Weight-adjusted genome scan analysis for mapping quantitative trait Loci for menarchal age. The Journal of clinical endocrinology and metabolism 91: 3534–3537. [DOI] [PubMed] [Google Scholar]
- 20. Guo Y, Shen H, Xiao P, Xiong DH, Yang TL, et al. (2006) Genomewide linkage scan for quantitative trait loci underlying variation in age at menarche. The Journal of clinical endocrinology and metabolism 91: 1009–1014. [DOI] [PubMed] [Google Scholar]
- 21. Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, et al. (2010) Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet 42: 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, et al. (2009) Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet 41: 734–738. [DOI] [PubMed] [Google Scholar]
- 23. Ong KK, Elks CE, Li S, Zhao JH, Luan J, et al. (2009) Genetic variation in LIN28B is associated with the timing of puberty. Nature genetics 41: 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He C, Kraft P, Chen C, Buring JE, Pare G, et al. (2009) Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nature genetics 41: 724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, et al. (2009) Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet 41: 648–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakamura Y (2007) The BioBank Japan Project. Clin Adv Hematol Oncol 5: 696–697. [PubMed] [Google Scholar]
- 27. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 28. The International HapMap Consortium (2003) The International HapMap Project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- 29. Li Y, Willer C, Sanna S, Abecasis G (2009) Genotype imputation. Annu Rev Genomics Hum Genet 10: 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J, Guo YF, Pei Y, Deng HW (2012) The impact of imputation on meta-analysis of genome-wide association studies. PLoS One 7: e34486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Okada Y, Kubo M, Ohmiya H, Takahashi A, Kumasaka N, et al. (2012) Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat Genet 44: 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okada Y, Sim X, Go MJ, Wu JY, Gu D, et al. (2012) Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet 44: 904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okada Y, Hirota T, Kamatani Y, Takahashi A, Ohmiya H, et al. (2011) Identification of nine novel loci associated with white blood cell subtypes in a Japanese population. PLoS Genet 7: e1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okada Y, Kamatani Y, Takahashi A, Matsuda K, Hosono N, et al. (2010) A genome-wide association study in 19 633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci. Hum Mol Genet 19: 2303–2312. [DOI] [PubMed] [Google Scholar]
- 35. Okada Y, Kamatani Y, Takahashi A, Matsuda K, Hosono N, et al. (2010) Common variations in PSMD3-CSF3 and PLCB4 are associated with neutrophil count. Hum Mol Genet 19: 2079–2085. [DOI] [PubMed] [Google Scholar]
- 36. He C, Kraft P, Chasman DI, Buring JE, Chen C, et al. (2010) A large-scale candidate gene association study of age at menarche and age at natural menopause. Human genetics 128: 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kumar V, Kato N, Urabe Y, Takahashi A, Muroyama R, et al. (2011) Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet In press [DOI] [PubMed] [Google Scholar]
- 38. Kumar V, Kato N, Urabe Y, Takahashi A, Muroyama R, et al. (2011) Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet 43: 455–458. [DOI] [PubMed] [Google Scholar]
- 39. Tanikawa C, Urabe Y, Matsuo K, Kubo M, Takahashi A, et al. (2012) A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat Genet In press [DOI] [PubMed] [Google Scholar]
- 40. Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, et al. (2011) A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet 20: 3884–3892. [DOI] [PubMed] [Google Scholar]
- 41. Urabe Y, Tanikawa C, Takahashi A, Okada Y, Morizono T, et al. (2012) A Genome-Wide Association Study of Nephrolithiasis in the Japanese Population Identifies Novel Susceptible Loci at 5q35. 3, 7p14. 3, and 13q14. 1. PLoS Genet 8: e1002541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cui R, Okada Y, Jang SG, Ku JL, Park JG, et al. (2011) Common variant in 6q26–q27 is associated with distal colon cancer in an Asian population. Gut 60: 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ambros V, Horvitz HR (1984) Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226: 409–416. [DOI] [PubMed] [Google Scholar]
- 44. Heo I, Joo C, Cho J, Ha M, Han J, et al. (2008) Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Molecular cell 32: 276–284. [DOI] [PubMed] [Google Scholar]
- 45. Viswanathan SR, Daley GQ, Gregory RI (2008) Selective blockade of microRNA processing by Lin28. Science's STKE 320: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, et al. (2010) Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet 42: 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guo Y, Chen Y, Ito H, Watanabe A, Ge X, et al. (2006) Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 384: 51–61. [DOI] [PubMed] [Google Scholar]
- 48. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920. [DOI] [PubMed] [Google Scholar]
- 49. Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, et al. (2008) Many sequence variants affecting diversity of adult human height. Nat Genet 40: 609–615. [DOI] [PubMed] [Google Scholar]
- 50. Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, et al. (2008) Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet 40: 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krude H, Schutz B, Biebermann H, Von Moers A, Schnabel D, et al. (2002) Choreoathetosis, hypothyroidism, and pulmonary alterations due to human NKX2-1 haploinsufficiency. Journal of Clinical Investigation 109: 475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eriksson N, Benton GM, Do CB, Kiefer AK, Mountain JL, et al. (2012) Genetic variants associated with breast size also influence breast cancer risk. BMC Med Genet 13: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bhalla K, Luo Y, Buchan T, Beachem MA, Guzauskas GF, et al. (2008) Alterations in CDH15 and KIRREL3 in patients with mild to severe intellectual disability. Am J Hum Genet 83: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guerin A, Stavropoulos DJ, Diab Y, Chénier S, Christensen H, et al.. (2012) Interstitial deletion of 11q-implicating the KIRREL3 gene in the neurocognitive delay associated with Jacobsen syndrome. American Journal of Medical Genetics Part A. [DOI] [PubMed] [Google Scholar]
- 55. Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, et al. (2010) Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet 42: 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, et al. (2008) A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet 4: e1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yazawa M, Ferrante C, Feng J, Mio K, Ogura T, et al. (2007) TRIC channels are essential for Ca2+ handling in intracellular stores. Nature 448: 78–82. [DOI] [PubMed] [Google Scholar]
- 58. Volodarsky M, Markus B, Cohen I, Staretz-Chacham O, Flusser H, et al. (2013) A Deletion Mutation in TMEM38B Associated with Autosomal Recessive Osteogenesis Imperfecta. Hum Mutat [DOI] [PubMed] [Google Scholar]
- 59. Parent AS, Teilmann G, Juul A, Skakkebaek NE, Toppari J, et al. (2003) The timing of normal puberty and the age limits of sexual precocity: variations around the world, secular trends, and changes after migration. Endocr Rev 24: 668–693. [DOI] [PubMed] [Google Scholar]
- 60. Okada Y, Takahashi A, Ohmiya H, Kumasaka N, Kamatani Y, et al. (2011) Genome-wide association study for C-reactive protein levels identified pleiotropic associations in the IL6 locus. Hum Mol Genet 20: 1224–1231. [DOI] [PubMed] [Google Scholar]
- 61. Yamaguchi-Kabata Y, Nakazono K, Takahashi A, Saito S, Hosono N, et al. (2008) Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. The American Journal of Human Genetics 83: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Age at menarche in each disease cohort.
(DOCX)
The result of association analysis of TOP 42 SNPs from Japanese AAM study with body mass index and height from previous GWAS by Okada et al.
(DOCX)
The result of top 42 SNPs by separated analysis.
(DOCX)