Abstract
Two yeast manganese superoxide dismutases (MnSOD), one from Saccharomyces cerevisiae mitochondria (ScMnSOD) and the other from Candida albicans cytosol (CaMnSODc), have most biochemical and biophysical properties in common, yet ScMnSOD is a tetramer and CaMnSODc is a dimer or “loose tetramer” in solution. Although CaMnSODc was found to crystallize as a tetramer, there is no indication from the solution properties that the functionality of CaMnSODc in vivo depends upon the formation of the tetrameric structure. To elucidate further the functional significance of MnSOD quaternary structure, wild-type and mutant forms of ScMnSOD (K182R, A183P mutant) and CaMnSODc (K184R, L185P mutant) with the substitutions at dimer interfaces were analyzed with respect to their oligomeric states and resistance to pH, heat, and denaturant. Dimeric CaMnSODc was found to be significantly more subject to thermal or denaturant-induced unfolding than tetrameric ScMnSOD. The residue substitutions at dimer interfaces caused dimeric CaMnSODc but not tetrameric ScMnSOD to dissociate into monomers. We conclude that the tetrameric assembly strongly reinforces the dimer interface, which is critical for MnSOD activity.
Introduction
Manganese-containing superoxide dismutase (SOD) enzymes catalyze the disproportionation of superoxide (O2 −) into dioxygen and hydrogen peroxide (H2O2) and exist as either homodimers or homotetramers with a monomer size of ∼23,000 Da. Each MnSOD subunit is composed of two domains, a predominantly α-helical N-terminal domain and a mixed α/β C-terminal domain. The Mn ion is coordinated by four strictly conserved residues, two from the N-terminal domain and two from the C-terminal domain, as well as by one solvent molecule. The MnSOD functional unit is a dimer assembled in the fashion observed in prokaryotic MnSODs (Thermus thermophilus [1], Escherichia coli [2], [3], Deinococcus radiodurans [4]), while the MnSODs located in eukaryote mitochondria (human [5], Saccharomyces cerevisiae [6], Caenorhabditis elegans [7], Aspergillus fumigatus [8]) are homotetramers or dimers of dimers possessing dihedral symmetry (D 2). The structure of the dimers within the tetrameric MnSODs is similar to those of the dimeric bacterial enzymes.
In previous studies, we compared two yeast MnSODs that share 58.3% sequence identity, one from S. cerevisiae mitochondria (ScMnSOD), and an uncommon one from Candida albicans cytosol (CaMnSODc) [9], and we demonstrated that ScMnSOD and CaMnSODc both exhibit faster catalysis than MnSODs from human or bacteria [9], [10], likely through a novel mechanism involving six-coordinate Mn3+ species [11]. The two yeast enzymes are also similar in terms of spectroscopy and redox chemistry [9]. To our surprise, CaMnSODc was found to exist as a dimer in solution but as a tetramer when it was crystallized, whereas ScMnSOD was found to be a tetramer under all conditions [9]. Because ScMnSOD and CaMnSODc have most biochemical and biophysical properties in common and their dimer interfaces share >90% similarity, they together provide an opportunity to probe the significance of the differing quaternary structures of MnSODs.
The dimer interface plays a crucial role in maintaining MnSOD activity. Most MnSODs have a conserved arginine close to the strictly conserved DXWEHXXYL motif, while in yeast MnSODs it is a lysine (Figure 1, Lys182 in ScMnSOD and Lys184 in CaMnSODc). Replacement of this lysine by arginine was reported to cause loss of stability but not of catalytic activity in ScMnSOD [12]. This study, however, only measured the MnSOD activities at low levels of O2 −.
Figure 1. Alignment of MnSOD C-terminal Sequence.

Conserved residues and unconserved residues at dimer interface are highlighted in bold and shadowed in gray, respectively. The RP-mutations in ScMnSOD and CaMnSODc are highlighted in black.
Surprising findings that wild-type (WT) yeast MnSODs at high levels of O2 − are much faster enzymes than human and bacterial MnSODs have recently been reported [9], [10]. They gave impetus to our exploration of whether this discrepancy in catalytic behavior between yeast and human MnSOD is related to the different dimer interface structures. The side chain of the lysine in yeast MnSODs has a different conformation compared to that of the arginine in MnSODs from other organisms. This difference derives from the residue next to the lysine, which is an alanine or leucine in ScMnSOD and CaMnSODc, respectively, and a proline in all other MnSODs. To make yeast MnSODs resemble human MnSOD more closely and to investigate whether modifications of dimer interfaces has the same effects on tetrameric and dimeric MnSOD, we engineered the two yeast MnSODs by mutating the lysine to arginine and changing the residue next to the lysine to proline (K182R, A183P ScMnSOD and K184R, L185P CaMnSODc) (Figure 1). We call the mutant proteins RP-mutant MnSOD. We report here that, although the dimerization of the functional dimers to form a tetrameric assembly is not necessarily required for an eukaryotic MnSOD to function properly under physiological conditions, it preserves the dimeric functional unit and may protect MnSOD from deactivation and unfolding under harsh environments.
Results
The Mutations Create Holes at the Dimer Interface of RP-mutant ScMnSOD and RP-mutant CaMnSODc
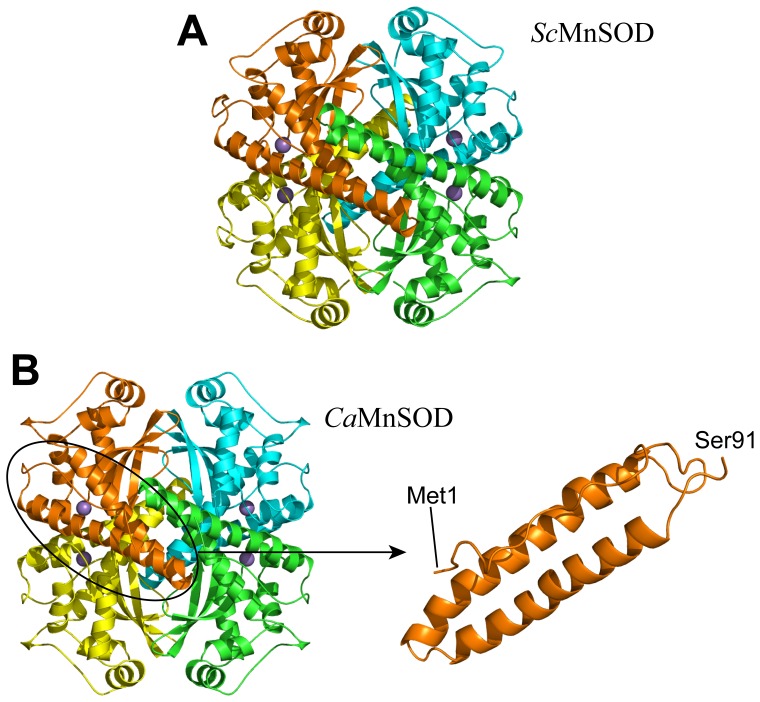
WT ScMnSOD and WT CaMnSODc and their RP-mutant proteins were overexpressed and purified from S. cerevisiae. Seeking clues as to why ScMnSOD was a tetramer and CaMnSODc a dimer or “loose tetramer” in solution, despite sharing a high sequence identity, we determined their crystal structures [9]. The crystal structure further confirmed that ScMnSOD was a homotetramer (Figure 2A). By contrast, CaMnSODc appeared as a homotetramer (Figure 2B) in crystal structures.
Figure 2. The tetramer interfaces are highly disordered, when CaMnSODc is in the tetramer form.
The ribbon diagram of ScMnSOD (PDB code: 3LSU) is shown in Panel A. The four subunits are colored in: A, yellow; B, orange; C, green; D, cyan. The ribbon diagram of tetrameric CaMnSODc (PDB code: 3QVN) and the N-terminal helical region (residues 1–91) of a CaMnSODc monomer are shown in Panel B. The four subunits are colored in: A, yellow; B, orange; C, green; D, cyan. Manganese ions are indicated as purple spheres.
As in other tetrameric MnSODs, the N-terminus of each subunit of both yeast MnSODs folds into a hairpin structure holding two long α-helices (Figure 2A, 2B). The N-terminal helical hairpin in each of the yeast MnSODs is much longer than those in dimeric MnSODs from bacteria (Figure S1-B), and, in ScMnSOD, it is longer than is found in any previously characterized tetrameric MnSOD (Figure S1-A). The buried surface areas at the tetramer interfaces are larger in ScMnSOD and tetrameric CaMnSODc (1417 and 1254 Å, respectively) than those (790–1000 Å2) in other MnSOD tetramers (human, A. fumigates, and C. elegans) (Table S2).
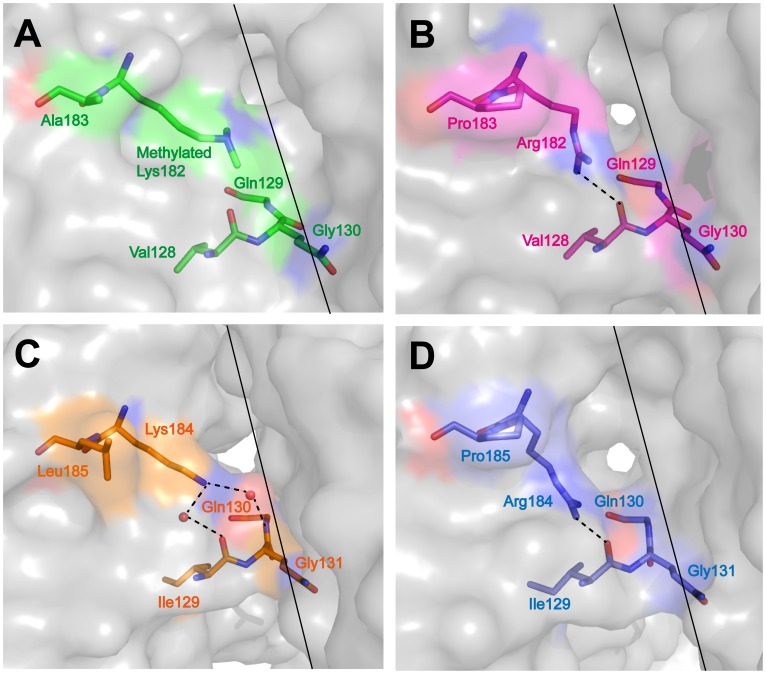
To investigate structural changes caused by the substitutions of Lys182 (Lys184) and Ala183 (Leu185) by arginine and proline, respectively, we solved the structures of the two RP-mutant proteins (Table 1). The tetrameric assemblies of WT and RP-mutant yeast MnSODs closely resemble each other (Figure S2). Superimpositions of all backbone atoms of the mutant subunit onto those of the WT subunit give root-mean-square deviations (RMSD) of 0.15 Å and 0.34 Å for ScMnSOD and CaMnSODc, respectively. The side chain of Arg182 (Arg184) of the mutants adopts a conformation different from that of Lys182 (Lys184) of the WT proteins (Figure 3). In two out of four chains in RP-mutant ScMnSOD, the chi2 and chi3 angles of the arginine shift by 5 and 127°, respectively (Figure 3A, 3B). In the other two chains, the chi1 angle of the arginine shifts by 132° (data not shown). These changes move Arg182 (Arg184) away from the dimer interface and thus open up a hole at the dimer interface of both RP-mutant ScMnSOD and RP-mutant CaMnSODc (Figure 3). The mutations also modify the hydrogen-bonding interactions surrounding residue 182 (184). Two hydrogen bonds, NZ(Lys184)···O(solv)···O(Ile129) and NZ(Lys184)···O(solv)···N(Gly131), are observed in WT CaMnSODc (Figure 3C), while in RP-mutant CaMnSODc the arginine is hydrogen bonded to Ile129 (Figure 3D).
Table 1. X-ray Data Collection and Refinement Statisticsa.
| K182R, A183P ScMnSOD | K184R, L185P CaMnSODc | |
| PDB code | 4F6E | 4GUN |
| X-Ray source | Rigaku FRE+ | Rigaku FRE+ |
| Detector | Rigaku HTC | Rigaku HTC |
| Wavelength (Å) | 1.5418 | 1.5418 |
| Resolution range (Å) | 37.65–1.60 | 53.02–1.94 |
| Rsym (%)b | 3.5 (17.0) | 9.1 (29.5) |
| Reflections observed | 400908 | 1206223 |
| Unique reflections | 105023 | 314901 |
| Redundancy | 3.8 (3.4) | 3.8 (2.8) |
| I/σ | 22.69 (6.79) | 9.94 (3.21) |
| Completeness (%) | 90.2 (70.8) | 91.3 (64.4) |
| Space group | P1 | P1 |
| Unit cell a, b, c (Å) | 65.54 66.25 66.62 | 129.33 73.80 134.29 |
| Unit cell α, β, γ (°) | 112.58 103.63 110.27 | 90.00 109.30 90.00 |
| Rwork (%)c | 16.4 (19.2) | 23.8 (23.5) |
| Rfree (%)c | 19.5 (23.3) | 26.6 (27.4) |
| Wilson B value (Å2) | 13.87 | 14.04 |
| Protein molecules in asymmetric unit | 4 | 16 |
| Number of protein atoms | 6665 | 25483 |
| Number of non-protein atoms | 573 | 1195 |
| RMSD bond Lengths (Å) | 0.006 | 0.007 |
| RMSD bond Angles (°) | 1.037 | 1.074 |
| Average B-factor for protein atoms (Å2) | 16.47 | 14.85 |
| Average B-factor for non-protein atoms (Å2) | 25.19 | 14.28 |
| Ramachandran angles | ||
| Most favored (%) | 91.8 | 92.2 |
| Additionally allowed (%) | 7.1 | 6.7 |
| Generously allowed (%) | 1.1 | 0.7 |
| Disallowed (%) | 0.0 | 0.4 |
Highest resolution shell shown in parenthesis.
Rsym = Σhkl |Ihkl−<Ihkl>|/ΣhklIhkl.
Rfactor = Σ||Fobs|−|Fcalc||/Σ|Fobs|. Rwork refers to the Rfactor for the data utilized in the refinement and Rfree refers to the Rfactor for 5% of the reflections randomly chosen that were excluded from the refinement.
Figure 3. Comparison of the dimer interface surface structure of K182R, A183P ScMnSOD and K184R, L185P CaMnSODc to the WT proteins.
The proteins are colored as: (A) WT ScMnSOD, green; (B) K182R, A183P ScMnSOD, red; (C) WT CaMnSODc, orange; (D) K184R, L185P CaMnSODc, blue. The dimer interfaces and hydrogen bonds are indicated as solid and dashed lines, respectively.
RP-Mutant Yeast MnSODs have WT Dismutase Activities
As-isolated proteins partially loaded with Mn (Table S1) were used in biophysical studies from this point, because the apoproteins of both WT and RP-mutant yeast MnSODs are unstable and titration of metal ions into as-isolated proteins has not been successful. As-isolated WT and RP-mutant ScMnSOD contain 0.70 and 0.71 Mn per monomer, respectively. As-isolated WT and RP-mutant CaMnSODc contain 0.59 and 0.43 Mn per monomer, respectively. SOD activities are reported on a per metal ion basis.
As noted earlier, the two yeast MnSODs were engineered through residue substitutions to imitate human MnSOD. To explore whether these residue substitutions affected the catalytic activity of yeast MnSODs, WT and RP-mutant yeast enzymes were pulse irradiated with various concentrations of O2 − and their dismutation efficiencies were compared. At neutral pH and room temperature, the disappearances of O2 − in the presence of WT and mutant proteins follow similar kinetics. Even when [O2 −] was high relative to enzyme concentration ([O2 −]:[MnSOD] = 41), the decay curves for O2 − disappearance catalyzed by WT or RP-mutant yeast MnSODs were superimposable (Figure S3), suggesting that the mutant proteins resemble the WT enzymes in displaying low degrees of product inhibition [9], [10]. Although our size exclusion chromatography experiments indicate that RP-mutant CaMnSODc could partially dissociate into monomers at 1.0 µM (see below), pulse radiolysis experiments suggest that it was more likely to stay predominantly as a dimer or “loose tetramer” at that concentration. Its rate constant determined at 1 µM (9.1×108 M−1s−1) was already near diffusion controlled and only slightly increased (9.9×108 M−1s−1) when the enzyme concentration was increased to 2.2 µM.
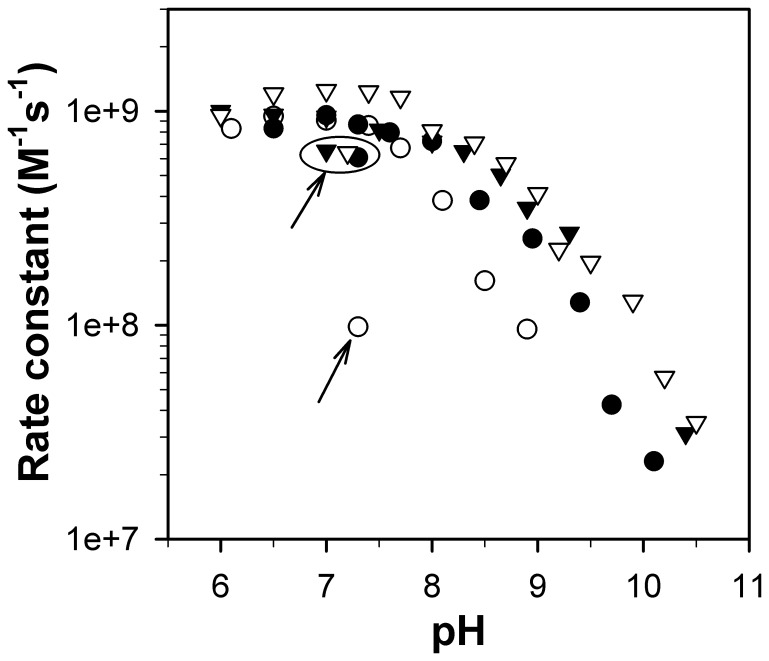
MnSOD catalytic activity (reactions 1 and 2) was measured when [O2 −]:MnSOD ratio ranged from 1–3 to exclude any effect of product inhibition. We previously showed that inactivation occurs at a significantly lower pH in yeast MnSODs than in human MnSOD, with pKs (the pH at which the SOD activity drops by 50%) of 8.5 and 10.5, respectively [9]. Here, although both yeast MnSODs were engineered to imitate human MnSOD, neither of the mutant proteins gained stability at higher pH compared to the WT enzymes (Figure 4). The profile of RP-mutant ScMnSOD activity as a function of pH closely resembles that of WT ScMnSOD (Figure 4). The same mutations on CaMnSODc, however, resulted in an enzyme even more sensitive to pH, with the pK decreasing from ∼8.5 in the wild type to ∼8 in the mutant protein (Figure 4). Loss of activity at high pH was found to be reversible in WT yeast enzymes as well as in RP-mutant ScMnSOD, with a restoration of ∼50% of their original activity (Figure 4). By contrast, when the pH of the sample solution was adjusted from basic (≥9) to neutral, no restoration of activity was observed for RP-mutant CaMnSODc (Figure 4).
Figure 4. RP-mutant CaMnSODc is more subject to inactivation by pH than the wild type.
Rate constants as a function of pH were determined by fitting the disappearances of low doses of O2 − ([O2 −]:[MnSOD] from 1–3) to first-order processes. The enzymes were WT ScMnSOD (solid triangle), K182R, A183P ScMnSOD (hollow triangle), WT CaMnSODc (solid circle) and K184R, L185P CaMnSODc (hollow circle). The data points circled and/or indicated with an arrow were measured after the pH was adjusted from 9–9.5 to neutral. The sample solutions contained 1 µM (in Mn) MnSOD in 10 mM potassium phosphate (pH 7), 10 mM sodium formate and 10 µM EDTA.
RP-mutant CaMnSODc is Inactivated by Heat, While RP-mutant ScMnSOD is not
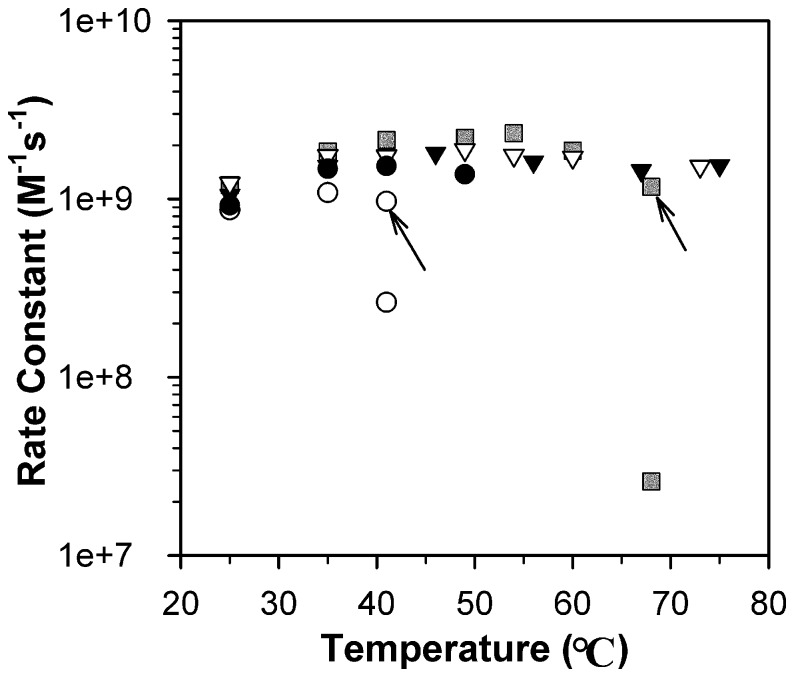
Yeast MnSODs showed full activity until the protein unfolding temperature. ScMnSOD was fully active up to 75°C, the highest temperature allowed in pulse radiolysis measurements. CaMnSODc, with a much lower thermostability than ScMnSOD (see below), was fully active as long as the enzyme stayed folded in solution. Protein aggregation was noticeable when the sample became more opaque to light at 260 nm. Aggregation of as-isolated CaMnSODc occurred at 50°C (see below), and the enzyme stayed fully active up to 49°C (Figure 5).
Figure 5. RP-mutant CaMnSODc is inactivated by heat like EcMnSOD.
Rate constants as a function of pH were determined by fitting the disappearances of low doses of O2 − ([O2 −]:[MnSOD] from 1–3) to first-order processes. The enzymes were EcMnSOD (grey rectangle), WT ScMnSOD (solid triangle), K182R, A183P ScMnSOD (hollow triangle), WT CaMnSODc (solid circle) and K184R, L185P CaMnSODc (hollow circle). The data points indicated with an arrow were obtained before the sample solution reached the desired temperature. All other data points were obtained after the sample solution was equilibrated to the desired temperature. The sample solutions contained 1 µM (in Mn) MnSOD in 10 mM potassium phosphate (pH 7), 10 mM sodium formate and 10 µM EDTA.
By contrast, MnSOD from E. coli (EcMnSOD) has been known for being inactivated by heat. Indeed, the reactivity of EcMnSOD decreased by ∼100 fold after the sample solution was allowed to equilibrate to 68°C (Figure 5, see method). Because there was no evidence of sample opacity at 260 nm, the dramatic inactivation by heat in EcMnSOD appears to result from some mechanism other than aggregation of the enzyme.
The substitutions of Lys182 (Lys184) and Ala 183 (Leu185) at dimer interfaces were found to affect the activities of ScMnSOD and CaMnSODc at elevated temperatures differently. Similar to WT ScMnSOD, RP-mutant ScMnSOD retained full activity up to 73°C (Figure 5). RP-mutant CaMnSODc, however, differed from the wild type but closely resembled the bacterial MnSODs with respect to its inactivation by heat. When RP-mutant CaMnSODc was heated at 41°C, a loss of ∼70% activity was observed (Figure 5). RP-mutant CaMnSODc started aggregating at ∼46°C (see below) but remained soluble at 41°C.
RP-mutant CaMnSODc is Susceptible to Dimer Dissociation
The oligomeric states of the as-isolated proteins were investigated by HPLC-SEC. When the protein concentration with respect to monomer was varied from 10 µM to 200 nM, WT and RP-mutant ScMnSOD both eluted solely as tetramers (Figure 6A), and WT CaMnSODc eluted solely as dimers (Figure 6B). The different oligomeric states of WT ScMnSOD and WT CaMnSODc are unlikely to result from differences in metallation states (Table S1), since WT ScMnSOD and WT CaMnSODc, when both metallated with ∼0.6 Mn per monomer, elute as tetramers and dimers, respectively [9].
Figure 6. RP-mutant CaMnSODc is susceptible to dimer dissociation.
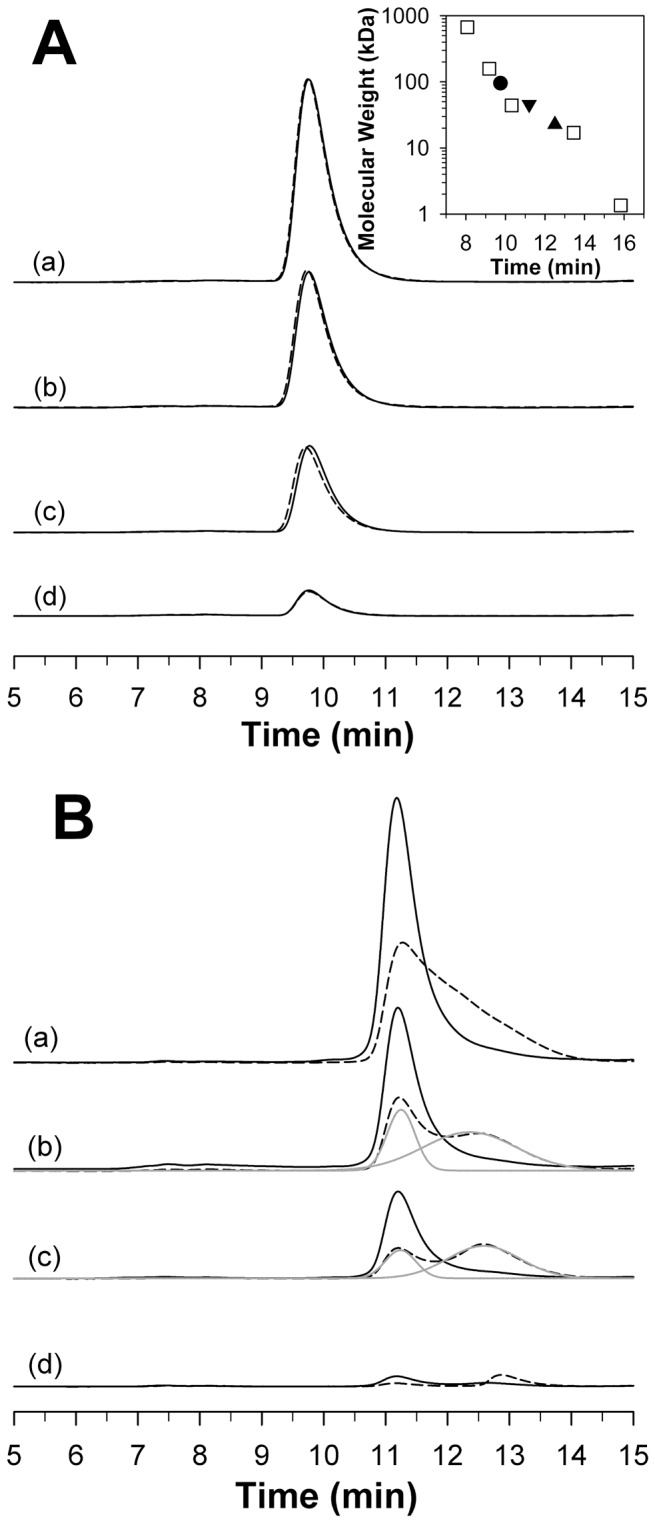
HPLC-SEC profiles of WT (solid line) and K182R, A183P (dashed line) ScMnSOD are shown in Panel A. Inset: The plot of the molecular weight of the five standards (square), ScMnSOD tetramer (circle) and CaMnSODc dimer (triangle down) and monomer (triangle up) versus their retention time. The column was calibrated using five standards: 1) bovine thyroglobulin (670 kDa), 2) bovine γ-globulin (158 kDa), 3) ovalbumin (44 kDa), 4) horse myoglobin (17 kDa), and 5) Vitamin B12 (1.35 kDa). HPLC-SEC profiles of WT (solid line) and K184R, L185P (dashed line) CaMnSODc are shown in Panel B. Deconvoluted peaks are shown in grey lines. The protein concentration relative to monomer was 1 µM (a), 750 nM (b), 500 nM (c) and 200 nM (d). The elution buffer contained 10 mM potassium phosphate (pH 6.7).
The elution profiles of RP-mutant CaMnSODc revealed two peaks corresponding to dimeric and monomeric forms when the protein concentration with respect to monomer was below 1 µM (Figure 6B). At 200 nM RP-mutant CaMnSODc, only the monomeric form was observed (Figure 6B). Based on the dimer-monomer equilibrium (Materials and Methods), K d of as-isolated RP-mutant CaMnSODc was determined to be 2.0±0.1 µM, with details of the calculation shown in Table S3. However, we cannot exclude the possibility that difference in metallation states (Table S1) might affect the dimer-monomer equilibrium in RP-mutant CaMnSODc.
WT and RP-mutant CaMnSODc are More Susceptible to Denaturant-Induced Protein Unfolding than WT and RP-mutant ScMnSOD
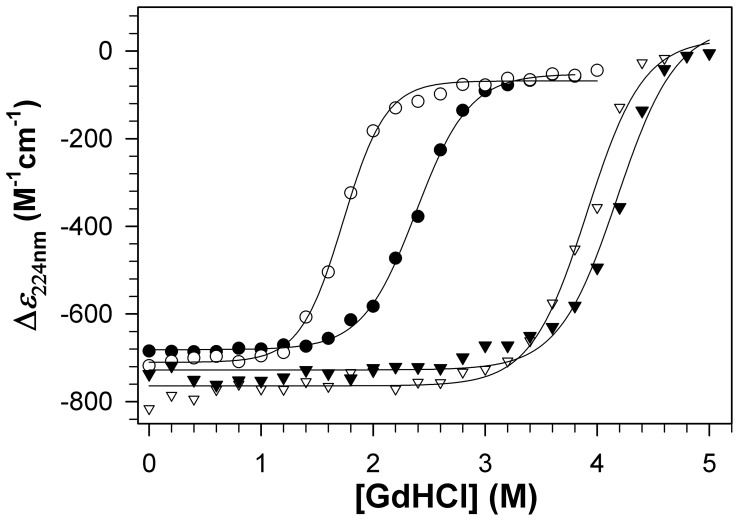
In order to understand the impact of the quaternary structure and the dimer interface on MnSOD stability, we used CD spectroscopy to monitor the denaturant-induced unfolding transitions of WT and RP-mutant yeast MnSODs (Figure S4). The molar CD at 224 nm was used to monitor changes in α-helical structure content as a function of [GdHCl] (Figure 7). Under these experimental conditions, ScMnSOD is a tetramer while CaMnSODc is a dimer or “loose tetramer”. The sharp decrease in helical structure content occurred at ∼3.4 M GdHCl in ScMnSOD and ∼1.6 M GdHCl in CaMnSODc (Figure 7). The unfolding of RP-mutant CaMnSODc occurred at a lower concentration (∼1.2 M) of GdHCl than that of WT CaMnSODc (Figure 7). By contrast, the unfolding profiles of WT and RP-mutant ScMnSOD are comparable to each other (Figure 7).
Figure 7. RP-mutant CaMnSODc is more subject to GdHCl-induced unfolding than the wild type.
The molar CD at 224 nm was used to monitor changes in α-helical structure content as a function of [GdHCl]. The enzymes were WT ScMnSOD (solid triangle), K182R, A183P ScMnSOD (hollow triangle), WT CaMnSODc (solid circle) and K184R, L185P CaMnSODc (hollow circle). The sample solutions contained 0.2 mg/mL (monomer concentration) MnSOD in 25 mM potassium phosphate (pH 7.4).
WT and RP-mutant CaMnSODc are Significantly Less Thermally Stable than WT and RP-mutant ScMnSOD
To investigate the impact of the quaternary structure and the residue substitutions at dimer interfaces on MnSOD thermostability, we monitored the unfolding transitions of WT and RP-mutant yeast MnSODs by DSC (Materials and Methods). Although the heat treatment of all WT and RP-mutant yeast MnSODs led to irreversible aggregation of the proteins, the DSC profiles were fitted using either a two-state irreversible model or a non-two-state reversible model, depending on which model yielded a better fitting. WT and mutant proteins are both partially loaded with Mn (Table S1). WT and RP-mutant ScMnSOD are loaded with 0.70 and 0.71 Mn per subunit, respectively. WT and RP-mutant CaMnSODc are loaded with 0.59 and 0.43 Mn per subunit, respectively. All four rest predominantly in the reduced (2+) state. The DSC profile of as-isolated ScMnSOD showed a single transition at 91°C, corresponding to one irreversible process (Figure 8A–a). This suggests cooperativity in the aggregation of apo and metallated subunits in ScMnSOD.
Figure 8. Thermostability of WT and RP-mutant ScMnSOD and CaMnSODc.
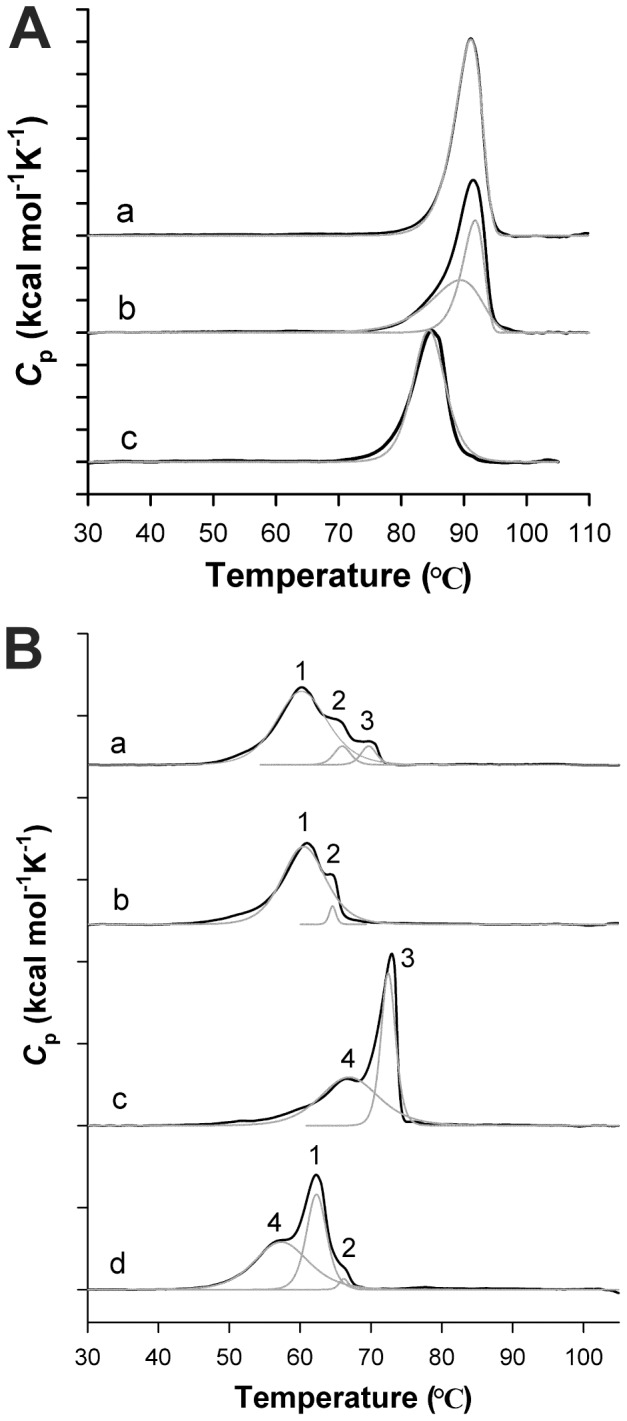
The S. cerevisiae enzymes in (A) are: (a) as-isolated ScMnSOD, (b) oxidized ScMnSOD and (c) as-isolated K182R, A183P ScMnSOD. The C. albicans enzymes in (B) are: (a) as-isolated CaMnSODc; (b) reduced CaMnSODc; (c) oxidized CaMnSODc and (d) as-isolated K184R, L185P CaMnSODc. Unfolding transitions are shown in black lines. The components (gray) were deconvoluted using a two-state irreversible model for WT ScMnSOD and a non-two-state reversible model for RP-mutant ScMnSOD, and WT and RP-mutant CaMnSODc. Reduced or oxidized enzymes were prepared by adding sodium hyposulfite or potassium permanganate to the sample solution prior to the DSC scan.
By contrast, three endotherms with much lower T ms were observed upon heat treatment of as-isolated CaMnSODc, assigned as Therm 1, 2, and 3 (Figure 8B–a, Table 2). In order to assign peaks with confidence, we measured the DSC profile for fully reduced CaMnSODc (Figures 8B–b). Therm 2 likely resulted from a small portion of Fe-containing SOD (Table S1) as it is retained in both fully reduced and as-isolated CaMnSODc. Because the activity of MnSOD is Mn-specific [13], [14], we assume that this small portion of Fe-substituted protein does not contribute to the activity of CaMnSODc. Since Therm 3 was missing in the thermal stability profile of reduced CaMnSODc (Figure 8B–b), it correlated with the aggregation of the oxidized Mn3+-CaMnSODc in the as-isolated enzyme. Therm 1 corresponded to the cooperative melting of apo- and Mn2+-containing CaMnSODc.
Table 2. Thermodynamic Parameters for Unfolding of Yeast MnSODs.
| Component Number | T m (°C) | ΔH a (kcal per mole of monomer) | |
| As-isolated ScMnSODb | 91.0 | 705 | |
| Oxidized ScMnSODb | 91.5 | 659 | |
| As-isolated CaMnSODcc | 1 | 60.3 | 431 |
| As-isolated CaMnSODcc | 2 | 66.0 | 36 |
| As-isolated CaMnSODcc | 3 | 69.7 | 33 |
| Reduced CaMnSODcc | 1 | 60.9 | 353 |
| Reduced CaMnSODcc | 2 | 64.5 | 13 |
| Oxidized CaMnSODcc | 4 | 66.5 | 325 |
| Oxidized CaMnSODcc | 3 | 72.9 | 254 |
| As-isolated K182R, A183P ScMnSODc | 84.4 | 563 | |
| As-isolated K184R, L185P CaMnSODcc | 1 | 57.4 | 288 |
| As-isolated K184R, L185P CaMnSODcc | 2 | 62.3 | 216 |
| As-isolated K184R, L185P CaMnSODcc | 3 | 66.2 | 12 |
ΔH are given per mole of tetramer or dimer.
The peaks of the DSC profile were deconvoluted using an irreversible two-state model.
The peaks of the DSC profile were deconvoluted using a reversible non-two-state model.
The stability profiles of yeast MnSODs are affected by the metal oxidation states. Chemically oxidized (3+) CaMnSODc gave a wide therm (4) prior to the emergence of Therm 3 (Figure 8B–c). To explore the origin of Therm 4, oxidized CaMnSODc was heated at 65°C for 20 min, and the metal content of the resulting supernatant was determined by ICP-MS. Because the proteins in the supernatant were nearly fully metallated with Mn, Therm 4 was likely associated with the aggregation of apo subunits in oxidized CaMnSODc. The separation of apoprotein endotherm from holoprotein endotherm was more subtle in oxidized ScMnSOD, although its main transition occurred at a lower temperature (∼75°C) than the as-isolated protein (∼80°C) and could well be deconvoluted to two irreversible processes (Figure 8A–b).
Like the WT enzyme, as-isolated RP-mutant ScMnSOD gave a single endotherm with a T m lowered to 84°C (Figure 8A–c). By contrast, the DSC profile of as-isolated RP-mutant CaMnSODc deviated more dramatically from that of WT CaMnSODc. Because nearly fully metallated protein remained in the supernatant upon heat treatment of RP-mutant CaMnSODc at 57°C, the endotherm at 57°C likely resulted from the aggregation of apo-RP-CaMnSODc, which, unlike in WT CaMnSODc, was separated from that of the aggregation of holo-RP-CaMnSODc (Figure 8B–d). The mutant protein maintained the two therms at 62 and 66°C, corresponding to the aggregation of Mn2+-containing and Fe-substituted CaMnSODc, respectively (Figure 8B–d, Table 2).
Discussion
Tetramerization is not Required for CaMnSODc to Function under Physiological Conditions
The two yeast MnSODs, one from S. cerevisiae mitochondria and the other from C. albicans cytosol, share 58.3% sequence identity. ScMnSOD is always a homotetramer like human MnSOD [5], [9]. To our surprise, CaMnSODc is a dimer or “loose tetramer” in solution (Figure 6B), and it is a tetramer when crystallized (Figure 2B). The biophysical characterization showing that CaMnSODc is significantly less stable than ScMnSOD (Figure 8) confirms that in solution ScMnSOD and CaMnSODc are tetrameric and dimeric, respectively. However, from their crystal structures (Figure 2) we cannot explain the discrepancy in their quaternary structures. Tetrameric MnSODs are in equilibrium between dimers and tetramers, and this kind of equilibrium is usually dependent on factors such as ionic strength, temperature, pH, and concentration of denaturant [15], [16]. Our results suggest that the tetramer-dimer equilibrium lies toward dimers in CaMnSODc, while it lies toward tetramers in other tetrameric MnSODs (MnSOD from S. cerevisiae, human, C elegans or A. fumigatus).
Because the helical hairpins contain two of the active site ligands, it was previously hypothesized that the tetramer interface played a role in stabilizing the helical hairpins and that only tetrameric enzymes would be active and stable [5]. Indeed, the I58T or L60F (tetramer interface residue) variants of human MnSOD have a much shorter half-life than the wild type at increased temperatures [17], [18]. Numerous other tetrameric proteins and enzymes with dihedral symmetry, such as malic enzyme, chaperone SecB, and RUNX1/ETO fusion protein, display impaired function when dissociating into dimers or monomers [19]–[21]. By contrast, CaMnSODc was in the dimeric form in our kinetics studies, yet its catalytic properties resemble those of tetrameric ScMnSOD [9]. Our studies suggest that the propensity for tetramerization found for eukaryotic but not for prokaryotic MnSODs is not related to optimization of SOD activity. A similar phenomenon has also been reported for aristolochene synthase from Aspergillus terreus, which functions as a dimer in solution, but is capable of tetramerization at high enzyme concentrations [22].
It is doubtful that tetrameric CaMnSODc significantly surpasses dimeric CaMnSODc in reactivity, because the latter is already near diffusion-controlled [9]. Dimeric CaMnSODc could be the active form that functions in vivo. However, since it is difficult to compare the composition of C. albicans cytosol to the in vitro conditions, we are unable to elucidate the in vivo quaternary structure of CaMnSODc.
The Tetramer Structure Reinforces the Dimer Interface
Although CaMnSODc is indistinguishable from ScMnSOD in terms of enzyme kinetics, spectroscopy, and redox properties [9], dimeric CaMnSODc is considerably less stable than tetrameric ScMnSOD. Compared to tetrameric ScMnSOD, the unfolding of dimeric CaMnSODc occurs at a much lower level of denaturant (GdHCl) (Figure 7), and the T m of the main components of dimeric CaMnSODc is lower by 30°C. These features suggest an important role of the tetramerization domain in enhancing the MnSOD enzyme’s resistance against unfolding in harsh environments.
Several studies suggest a significant role for the dimer interface in both catalysis and stability of MnSOD. Replacement of Glu170 in EcMnSOD, which spans the dimer interface and forms the double glutamate bridge, results in dissociation of the dimer, complete loss of catalytic activity, and a change in metal specificity [23]. Substitutions at Glu162 in human MnSOD, the counterpart of Glu170 in EcMnSOD, reduce the catalytic activity to 5–25% of that of the WT enzyme [24]. The Y166F mutant human MnSOD shows a significant decrease in catalytic activity and a major unfolding transition at a lower T m [25]. Replacement of Phe66 at the dimer interfaces of human MnSOD reduces the degree of product inhibition in the human enzyme and makes it resemble EcMnSOD [26], [27].
Here, two residues (Lys182 and Ala183 in ScMnSOD, Lys184 and Leu185 in CaMnSODc) were substituted at the dimer interface of the two yeast MnSODs. We show here that these two RP-mutant proteins resemble their WT enzymes in terms of SOD activity at room temperature and neutral pH (Figure S3) so that the roles of these two residues appear to be related to protein stability, not enzymatic function.
Even though the former is a tetramer and the latter is a dimer in solution, ScMnSOD and CaMnSODc, have >90% sequence similarity at the dimer interface and share most biochemical and biophysical characteristics. Based on our findings, we conclude that the dimer interface of RP-mutant CaMnSODc is considerably destabilized. The dimer dissociation constant, K d, which is too low to measure in WT CaMnSODc, significantly increases to 2.0±0.1 µM in the mutant protein. RP-mutant CaMnSODc also becomes more sensitive to high pH than the wild type, and this inactivation by increased pH becomes completely irreversible (Figure 4). In contrast to WT CaMnSODc, the RP-mutant protein exhibits loss of activity at high temperatures (Figure 5), and it is more subject to unfolding by GdHCl (Figure 7). These observations suggest that the mutations at the dimer interface cause CaMnSODc dimers to fall apart into monomers.
The destabilization of the dimer interface in RP-mutant CaMnSODc is also suggested by the DSC protein stability data. The oxidized form of CaMnSODc has a higher thermal stability than the reduced form (Figure 8B). The dependence of protein stability on the oxidation state was also reported in EcMnSOD, which has a greater affinity for Mn3+ [28]. Based on the DSC data in Figure 8B and Table 1, the molar energy required for aggregation of each enzyme species in WT CaMnSODc scales as Mn3+-containing CaMnSODc>Fe-substituted CaMnSODc>Mn2+-containing CaMnSODc ∼ apo subunits. Removal of the metal ions from WT or RP-mutant CaMnSODc causes precipitation of the proteins, suggesting that metal-free CaMnSODc is not stable. Therefore, since as-isolated WT and RP-mutant CaMnSODc contain ∼0.59 and ∼0.43 Mn per monomer (Table S1), respectively, each CaMnSODc dimer likely contains one metal ion. As the Mn,E-CaMnSODc dimer is heated, the apo subunits melt at 57–60°C (Table 2). The remaining manganese-bound subunits then self-associate to create the Mn,Mn-CaMnSODc dimer, the aggregation of which occurs at 65–73°C, depending on the oxidation state of the manganese (Table 2). This phenomenon has been reported for another type of SOD, copper-zinc SOD loaded with one or two zinc ions [29]. In RP-mutant CaMnSODc, the order of the molar energies changes to Fe-substituted CaMnSODc>Mn2+-containing CaMnSODc>apo subunits (Figure 8B–d, Table 2). The modifications of dimer interfaces lower the energy threshold for dimer dissociation and thus make aggregation of apo subunits occur more readily in RP-mutant CaMnSODc.
The mutations do not result in considerable perturbation in the subunit structure of the two yeast enzymes (Figure S2). Examination of the mutated residues, however, suggests that Arg182 (Arg184) in the mutants moves away from the dimer interface, which creates a hole at the dimer interface and would likely cause the destabilization of the dimer interface in both the mutants (Figure 3). Nevertheless, the same residue substitutions at dimer interfaces have much milder effects on tetrameric ScMnSOD, although WT ScMnSOD and WT CaMnSODc are similar in many ways [9]. Unlike RP-mutant CaMnSODc, which dissociates into monomers, RP-mutant ScMnSOD closely resembles the wild type in oligomeric state, and resistance to pH, heat, and denaturant (Figure 4–7, S3). The only sign of destabilization is the slightly lower thermal stability displayed by the mutant protein relative to the wild type (Figure 8A).
Assembly of proteins into oligomer structures has been proposed to confer several potential advantages, including greater folding efficiency, high stability, specific morphological functions, amenability to allosteric regulation, and greater error control in synthesis [15], [16], [30]. Tetramer interface has been reported to contribute to thermal and pH-dependent stability of numerous dihedral tetrameric enzymes, including malate dehydrogenase and serine hydroxymethyltransferase [31]–[33]. Specifically in tetrameric MnSODs, each side of the protein is encircled by one of the two 4-helix bundles at opposite ends of the dimer, which acts as a clamp, holding the dimers in place. In addition to the contribution to protein stability, our results here suggest that the dimer interface, which is critical for MnSOD activity, is strongly reinforced through tetramer formation.
In conclusion, the tetramer-dimer equilibrium of CaMnSODc lies toward dimers in solution, and its activity under physiological conditions does not rely upon tetramerization. Thus the significance of the tetramer structure may lie in the stabilization of protein assembly against harsh environments (heat and denaturant). More importantly, our results suggest that tetrameric assembly of the functional dimers strongly reinforces the functional dimer interface. As the functional dimer interface is critical for MnSOD activity, its reinforcement through tetramerization could be one of the reasons that MnSOD is tetrameric in higher organisms.
Materials and Methods
Samples
E. coli MnSOD (EcMnSOD) was purchased from a commercial source (Sigma-Aldrich). The protein was resuspended in 25 mM potassium phosphate (pH 7.4), washed with 1 mM EDTA in the same phosphate buffer for several times, and then purified through a G200 size exclusion column.
Construction of Plasmid for Expression of RP-mutant ScMnSOD and RP-mutant CaMnSODc
Site-directed mutagenesis [34] was carried out on an overexpression vector (YEp352-ScMnSOD) containing the URA3 selectable marker and a 2-kb genomic BamHI fragment containing the gene for ScMnSOD. The primers 5′-CAGTACCAAAACAAGAGACCCGACTACTTCAAAGC-3′ and 5′-GCTTTGAAGTAGTCGGGTCTCTTGTTTTGGTACTG-3′ were used to create the cDNA for K182R, A183P ScMnSOD.
The pVT102U-CaMnSODc (with URA3 and AMP marker) vector containing the complete coding sequence of CaMnSODc was generously given by Prof. Bourbonnais [35]. The primers 5′-CAAAATGTCAGGCCTGATTATTTCAAAGCAATTTGGAACGTG-3′ and 5′-GAAATAATCAGGCCTGACATTTTGATATTGCAAGTAGTACGC-3′ were used to create the cDNA for K184R, L185P CaMnSODc.
The PCR products were transformed into E. coli DH5α strain and screened by ampicillin selection. The purified vectors were transformed into S. cerevisiae sod2Δ strain (EG110).
Expression and Purification of WT and RP-mutant ScMnSOD and CaMnSODc
Yeast cells carrying YEp352-ScMnSOD (WT or mutant) were grown in YPEG media (1% yeast extract, 2% peptone, 3% glycerol, 2% ethanol, pH 4) supplemented with 0.5 mM Mn(II) sulfate at 30°C to O.D. >20. Yeast cells carrying pVT102U-CaMnSODc (WT or mutant) were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose, pH 4) media supplemented with 0.5 mM Mn(II) sulfate at 30°C to O.D. >10. Cells were harvested by centrifugation at 12,000×g for 10 min. Isolation of WT and RP-mutant ScMnSOD and CaMnSODc was performed as previously described [9].
Size Exclusion Chromatography
The mass weight of native proteins was determined by a HPLC (Agilent 1200 series) fitted with a size exclusion column (Tosoh Bioscience, TSK gel G2000SW) at a flow rate of 0.25–0.5 mL/min. The column was calibrated using five standards: bovine thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), ovalbumin (44 kDa), horse myoglobin (17 kDa), and vitamin B12 (1.35 kDa).
HPLC-SEC measurements were used to determine the oligomeric state of proteins. The equilibrium between dimers and monomers is shown as 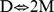 where D represents dimer and M represents monomer. The dimer dissociation constant (K
d) is calculated as
where D represents dimer and M represents monomer. The dimer dissociation constant (K
d) is calculated as
| (1) |
where [M] and [D] were calculated from area integrals of elution peaks (UV signal at 210 nm). The peak fitting was carried out in Origin 8.1 (OriginLab Corp.). The column buffer contained 10 mM potassium phosphate (pH 6.7). The protein concentration with respect to monomer was varied from 10 µM to 200 nM.
Crystallization of WT and RP-mutant ScMnSOD and CaMnSODc
The crystallization of both WT ScMnSOD and WT CaMnSODc, giving tetrameric structures, was described previously [9]. Reductive methylation of K182R, A183P ScMnSOD was carried out as described previously [36], in order to improve the diffraction of protein crystals. All free amino groups of the lysine residues and the N-terminus of each subunit of the two proteins were methylated as confirmed by mass spectrometry (Figure S5).
Methylated K182R, A183P ScMnSOD, containing 0.71 Mn per monomer (Table S1), was crystallized by hanging-drop vapor diffusion at 4°C against a well solution of 0.1 M tri-ammonium citrate (pH 7) in 20% (w/v) polyethylene glycol 3,350 with a protein concentration of 7 mg/mL. Unmodified K184R, L185P CaMnSODc, containing 0.43 Mn per monomer (Table S1), was crystallized by hanging-drop vapor diffusion at 4°C against a solution of 2 M ammonium sulfate with a protein concentration of 7 mg/mL. The protein crystals were cryo-protected in mother liquor solution containing 30% glycerol and flash frozen in liquid nitrogen prior to data collection. These two structures were deposited to PDB bank (see below).
Crystallography: Data Collection and Refinement
The crystallography information for WT ScMnSOD and WT CaMnSODc was reported previously [9]. The data of RP-mutant ScMnSOD and RP-mutant CaMnSODc were collected at 100 K at the UCLA X-ray diffraction facility, using a Rigaku FRE+ generator and a Rigaku HTC detector. The data was processed using XDS [37]. RP-mutant ScMnSOD and RP-mutant CaMnSODc were phased by molecular replacement using WT ScMnSOD (PDB code: 3LSU) and WT CaMnSODc (PDB code: 3QVN), respectively. All the molecular replacements were done using PHASER [38]. The models were built using COOT [39]. All model refinements were performed using REFMAC [40], PHENIX [41] and BUSTER [42].
Coordinates and structure factors have been deposited in the PDB database with accession numbers 4F6E for the K182R, A183P ScMnSOD structure and, 4GUN for the K184R, L185P CaMnSODc structure. Analysis of interface contacts was performed using the PISA server (PDBePISA Protein Interfaces, Surfaces and Assemblies [43].
Pulse Radiolysis
Pulse radiolysis experiments were carried out using the 2 MeV Van de Graaff accelerator at Brookhaven National Laboratory. Upon irradiation of water with a pulse of energetic electrons, hydrated electrons (eaq −), hydroxyl radicals (•OH) and, in lesser yield, hydrogen atoms (H•) are the primary radicals produced. Superoxide radical is then generated in air-saturated aqueous solution containing sodium formate through the following reactions: •OH+HCO2 − → H2O+CO2 •−, O2+ CO2 •− → O2 •−+CO2, eaq −+O2 → O2 •−, H+O2 → HO2 •.
The experiments to measure catalytic rates involved following the decay of various concentrations of O2 − at 260 nm using 1∶1 to 1∶50 ratios of [MnSOD]:[O2 −]. First-order rate constants were calculated by fitting the data using the kinetics program in PRWIN [44].
The thermal deactivation measurements were performed by heating the pulse radiolysis cell holder to the desired temperature. The sample cell containing a protein solution was then inserted into the holder and allowed to equilibrate for 4 minutes prior to irradiation of the sample solution.
All pulse radiolysis samples were prepared in 10 mM potassium phosphate, 10 mM sodium formate and 10 µM EDTA at 25°C. All MnSOD concentrations were taken as the ICP-measured concentration of manganese in the sample. The pH of the buffer was adjusted using ultrapure (Baker Ultrex) sodium hydroxide and sulfuric acid as needed.
Differential Scanning Calorimetry (DSC)
DSC scans were performed using a Nano II differential scanning calorimeter (Calorimetry Sciences Corp.). All buffers were degassed under vacuum and highly concentrated protein samples (∼24 mg/mL) were diluted with degassed buffers to a concentration of 2 mg/mL (600 µL) right before the DSC run. All samples were run at a rate of 1°C/min under 4 atm of pressure. A buffer (25 mM potassium phosphate, pH 7.4) base line was run prior to the analysis of protein samples with the same heating/cooling rates. Baseline subtraction was performed, and some of the peaks were fitted to non-two-state reversible transitions using Origin 8.0 (OriginLab Corp.).
The other peaks were fitted to two-state irreversible transitions, obeying pseudo-first-order kinetics [45]. The temperature dependence of the kinetic constant k obeys the Arrhenius equation,
| (2) |
where A is the preexponential factor and E is the activation energy of transition. The apparent excess heat capacity C p(T) at a temperature T is given by Sánchez-Ruiz’s methods as
| (3) |
where 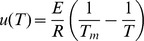 , Tm is the temperature at which Cp reaches its maximum value. DSC data were fitted to equation (3) using SigmaPlot 11.0 (Systat Software).
, Tm is the temperature at which Cp reaches its maximum value. DSC data were fitted to equation (3) using SigmaPlot 11.0 (Systat Software).
Miscellaneous Methods
Metal contents of the purified proteins were determined by inductive coupled plasma mass spectrometry (ICP-MS, Agilent 7500 series). UV-visible absorption spectra were collected on a Shimadzu UV-2501 PC spectrophotometer.
Circular dichroism (CD) measurements were performed on a JASCO J-715 spectropolarimeter at room temperature. The GdHCl concentration was varied from 0 to 5 M while the protein concentration was held constant at 0.2 mg/mL. Protein samples were mixed with GdHCl stock solutions and incubated at room temperature for 15 min prior to the scan.
Mass weight of the protein subunit was determined by electrospray ionization mass spectrometry (ESI-MS) either via a triple quadrupole instrument (API III, Applied Biosystems) or via a hybrid linear ion-trap mass spectrometer (LTQ, Thermo Electron). Mass spectra were processed and analyzed using MacSpec 3.3, Hypermass and BioMultiview 1.3.1 software for data obtained from API III, and ProMass for Xcalibur 2.8 for data obtained from LTQ.
Supporting Information
Superimposition of the subunit of ScMnSOD (green) over that of human (pink) and E. coli (orange) MnSOD. The two subunits are colored in: A, green; B, cyan.
(TIF)
Comparison of crystal structures between WT yeast MnSODs and their RP-mutant proteins. (A and B). Superimposition of the tetramer of K182R, A183P ScMnSOD (pink) and K184R, L185P CaMnSODc (blue) to that of WT ScMnSOD (green) and CaMnSODc (orange), respectively. (C and D). Superimposition of the monomer of K182R, A183P ScMnSOD (pink) and K184R, L185P CaMnSODc (blue) to that of WT ScMnSOD (green) and CaMnSODc (orange), respectively. Manganese atoms are shown in purple spheres.
(TIF)
Decay of 43 µM O2 − catalyzed by (A) WT ScMnSOD (solid, a), K182R, A183P ScMnSOD (dotted, b), (B) WT CaMnSODc (solid, a) and K184R, L185P CaMnSODc (dotted, b). The sample for pulse radiolysis contains 10 mM phosphate (pH 7), 10 mM sodium formate and 10 µM EDTA. The O2 − concentration in these figures was calculated from the absorbance at 260 nm and is slightly different from the O2 − dose given by the pulse radiolysis instrument.
(TIF)
CD spectra of WT CaMnSODc (A), K184R, L185P CaMnSODc (B), WT ScMnSOD (C) and K182R, A183P ScMnSOD (D) at increased concentrations of GdHCl. The solutions contained 25 mM potassium phosphate (pH 7.4). The measurements were carried out at room temperature.
(TIF)
Electrospray-ionization mass spectra of methylated RP-mutant ScMnSOD (left) and its reconstructed mass distribution profile (right). Ordinate units of intensity are arbitrary and the abscissa units of average molecular mass are in Daltons. The theoretical mass weight of methylated K182R, A183P ScMnSOD is 23,589 Da.
(EPS)
Metal contents of WT and RP-mutant ScMnSOD and CaMnSODc.
(DOC)
Interactions of Subunits at Dimer and Tetramer Interfaces in MnSODs from Different Organisms.
(DOC)
Calculation of K d for K184R, L185P CaMnSODc.
(DOC)
Acknowledgments
We thank Kevin Barnese for stimulating the ideas. We thank Jason Navarro for help with crystallization screens.
Funding Statement
This work was supported by grant DK46828 to J.S.V. Radiolysis studies were carried out at Center for Radiation Chemistry Research at BNL, which is funded under contract DE-AC02-98CH10886 with the United States Department of Energy and supported by its Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wagner UG, Pattridge KA, Ludwig ML, Stallings WC, Werber MM, et al. (1993) Comparison of the crystal structures of genetically engineered human manganese superoxide dismutase and manganese superoxide dismutase from Thermus thermophilus: differences in dimer-dimer interaction. Protein Sci 2: 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edwards RA, Baker HM, Whittaker MM, Whittaker JW, Jameson GB, et al. (1998) Crystal structure of Escherichia coli manganese superoxide dismutase at 2.1-angstrom resolution. Journal of Biological Inorganic Chemistry 3: 161–171. [Google Scholar]
- 3. Whittaker MM, Lerch TF, Kirillova O, Chapman MS, Whittaker JW (2011) Subunit dissociation and metal binding by Escherichia coli apo-manganese superoxide dismutase. Arch Biochem Biophys 505: 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abreu IA, Hearn A, An H, Nick HS, Silverman DN, et al. (2008) The kinetic mechanism of manganese-containing superoxide dismutase from Deinococcus radiodurans: a specialized enzyme for the elimination of high superoxide concentrations. Biochemistry 47: 2350–2356. [DOI] [PubMed] [Google Scholar]
- 5. Borgstahl GE, Parge HE, Hickey MJ, Beyer WF Jr, Hallewell RA, et al. (1992) The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell 71: 107–118. [DOI] [PubMed] [Google Scholar]
- 6. Ravindranath SD, Fridovich I (1975) Isolation and characterization of a manganese-containing superoxide dismutase from yeast. J Biol Chem 250: 6107–6112. [PubMed] [Google Scholar]
- 7. Trinh CH, Hunter T, Stewart EE, Phillips SE, Hunter GJ (2008) Purification, crystallization and X-ray structures of the two manganese superoxide dismutases from Caenorhabditis elegans. Acta Crystallogr Sect F Struct Biol Cryst Commun 64: 1110–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fluckiger S, Mittl PR, Scapozza L, Fijten H, Folkers G, et al. (2002) Comparison of the crystal structures of the human manganese superoxide dismutase and the homologous Aspergillus fumigatus allergen at 2-A resolution. J Immunol 168: 1267–1272. [DOI] [PubMed] [Google Scholar]
- 9. Sheng Y, Stich TA, Barnese K, Gralla EB, Cascio D, et al. (2011) Comparison of two yeast MnSODs: mitochondrial Saccharomyces cerevisiae versus cytosolic Candida albicans. Journal of the American Chemical Society 133: 20878–20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barnese K, Sheng Y, Stich TA, Gralla EB, Britt RD, et al. (2010) Investigation of the highly active manganese superoxide dismutase from Saccharomyces cerevisiae. Journal of the American Chemical Society 132: 12525–12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sheng Y, Butler Gralla E, Schumacher M, Cascio D, Cabelli DE, et al. (2012) Six-coordinate manganese(3+) in catalysis by yeast manganese superoxide dismutase. Proc Natl Acad Sci U S A 109: 14314–14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Borders CL Jr, Bjerrum MJ, Schirmer MA, Oliver SG (1998) Characterization of recombinant Saccharomyces cerevisiae manganese-containing superoxide dismutase and its H30A and K170R mutants expressed in Escherichia coli. Biochemistry 37: 11323–11331. [DOI] [PubMed] [Google Scholar]
- 13. Miller AF, Vance CK (2001) Novel insights into the basis for Escherichia coli superoxide dismutase’s metal ion specificity from Mn-substituted FeSOD and its very high E(m). Biochemistry 40: 13079–13087. [DOI] [PubMed] [Google Scholar]
- 14. Miller AF (2012) Superoxide dismutases: ancient enzymes and new insights. FEBS Lett 586: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goodsell DS, Olson AJ (2000) Structural symmetry and protein function. Annu Rev Biophys Biomol Struct 29: 105–153. [DOI] [PubMed] [Google Scholar]
- 16. Ali MH, Imperiali B (2005) Protein oligomerization: how and why. Bioorg Med Chem 13: 5013–5020. [DOI] [PubMed] [Google Scholar]
- 17. Borgstahl GE, Parge HE, Hickey MJ, Johnson MJ, Boissinot M, et al. (1996) Human mitochondrial manganese superoxide dismutase polymorphic variant Ile58Thr reduces activity by destabilizing the tetrameric interface. Biochemistry 35: 4287–4297. [DOI] [PubMed] [Google Scholar]
- 18. Hernandez-Saavedra D, Quijano C, Demicheli V, Souza JM, Radi R, et al. (2010) Thiol-sensitive mutant forms of human SOD2, L60F, and I58T: the role of Cys140. Free Radic Biol Med 48: 1202–1210. [DOI] [PubMed] [Google Scholar]
- 19. Hsieh JY, Chen SH, Hung HC (2009) Functional roles of the tetramer organization of malic enzyme. J Biol Chem 284: 18096–18105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muren EM, Suciu D, Topping TB, Kumamoto CA, Randall LL (1999) Mutational alterations in the homotetrameric chaperone SecB that implicate the structure as dimer of dimers. Journal of Biological Chemistry 274: 19397–19402. [DOI] [PubMed] [Google Scholar]
- 21. Wichmann C, Becker Y, Chen-Wichmann L, Vogel V, Vojtkova A, et al. (2010) Dimer-tetramer transition controls RUNX1/ETO leukemogenic activity. Blood 116: 603–613. [DOI] [PubMed] [Google Scholar]
- 22. Shishova EY, Di Costanzo L, Cane DE, Christianson DW (2007) X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate. Biochemistry 46: 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whittaker MM, Whittaker JW (1997) Mutagenesis of a proton linkage pathway in Escherichia coli manganese superoxide dismutase. Biochemistry 36: 8923–8931. [DOI] [PubMed] [Google Scholar]
- 24. Greenleaf WB, Perry JJ, Hearn AS, Cabelli DE, Lepock JR, et al. (2004) Role of hydrogen bonding in the active site of human manganese superoxide dismutase. Biochemistry 43: 7038–7045. [DOI] [PubMed] [Google Scholar]
- 25. Hearn AS, Fan L, Lepock JR, Luba JP, Greenleaf WB, et al. (2004) Amino acid substitution at the dimeric interface of human manganese superoxide dismutase. Journal of Biological Chemistry 279: 5861–5866. [DOI] [PubMed] [Google Scholar]
- 26. Abreu IA, Cabelli DE (2010) Superoxide dismutases-a review of the metal-associated mechanistic variations. Biochim Biophys Acta 1804: 263–274. [DOI] [PubMed] [Google Scholar]
- 27. Zheng J, Domsic JF, Cabelli D, McKenna R, Silverman DN (2007) Structural and kinetic study of differences between human and Escherichia coli manganese superoxide dismutases. Biochemistry 46: 14830–14837. [DOI] [PubMed] [Google Scholar]
- 28. Mizuno K, Whittaker MM, Bachinger HP, Whittaker JW (2004) Calorimetric studies on the tight binding metal interactions of Escherichia coli manganese superoxide dismutase. J Biol Chem 279: 27339–27344. [DOI] [PubMed] [Google Scholar]
- 29. Potter SZ, Zhu H, Shaw BF, Rodriguez JA, Doucette PA, et al. (2007) Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J Am Chem Soc 129: 4575–4583. [DOI] [PubMed] [Google Scholar]
- 30. Andre I, Strauss CE, Kaplan DB, Bradley P, Baker D (2008) Emergence of symmetry in homooligomeric biological assemblies. Proc Natl Acad Sci U S A 105: 16148–16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bjork A, Mantzilas D, Sirevag R, Eijsink VG (2003) Electrostatic interactions across the dimer-dimer interface contribute to the pH-dependent stability of a tetrameric malate dehydrogenase. FEBS Lett 553: 423–426. [DOI] [PubMed] [Google Scholar]
- 32. Bjork A, Dalhus B, Mantzilas D, Sirevag R, Eijsink VG (2004) Large improvement in the thermal stability of a tetrameric malate dehydrogenase by single point mutations at the dimer-dimer interface. J Mol Biol 341: 1215–1226. [DOI] [PubMed] [Google Scholar]
- 33. Appaji Rao N, Ambili M, Jala VR, Subramanya HS, Savithri HS (2003) Structure-function relationship in serine hydroxymethyltransferase. Biochim Biophys Acta 1647: 24–29. [DOI] [PubMed] [Google Scholar]
- 34. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-Directed Mutagenesis by Overlap Extension Using the Polymerase Chain-Reaction. Gene 77: 51–59. [DOI] [PubMed] [Google Scholar]
- 35. Lamarre C, LeMay JD, Deslauriers N, Bourbonnais Y (2001) Candida albicans expresses an unusual cytoplasmic manganese-containing superoxide dismutase (SOD3 gene product) upon the entry and during the stationary phase. J Biol Chem 276: 43784–43791. [DOI] [PubMed] [Google Scholar]
- 36. Walter TS, Meier C, Assenberg R, Au KF, Ren JS, et al. (2006) Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14: 1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kabsch W (2010) Xds. Acta Crystallogr D Biol Crystallogr 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. (2007) Phaser crystallographic software. Journal of Applied Crystallography 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- 40. Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography 53: 240–255. [DOI] [PubMed] [Google Scholar]
- 41. Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, et al. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954. [DOI] [PubMed] [Google Scholar]
- 42. Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, et al. (2012) Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr 68: 368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797. [DOI] [PubMed] [Google Scholar]
- 44.Schwarz H. BNL Pulse Radiolysis Program. Brookhaven National Laboratory.
- 45. Arroyo-Reyna A, Tello-Solis SR, Rojo-Dominguez A (2004) Stability parameters for one-step mechanism of irreversible protein denaturation: a method based on nonlinear regression of calorimetric peaks with nonzero deltaCp. Anal Biochem 328: 123–130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Superimposition of the subunit of ScMnSOD (green) over that of human (pink) and E. coli (orange) MnSOD. The two subunits are colored in: A, green; B, cyan.
(TIF)
Comparison of crystal structures between WT yeast MnSODs and their RP-mutant proteins. (A and B). Superimposition of the tetramer of K182R, A183P ScMnSOD (pink) and K184R, L185P CaMnSODc (blue) to that of WT ScMnSOD (green) and CaMnSODc (orange), respectively. (C and D). Superimposition of the monomer of K182R, A183P ScMnSOD (pink) and K184R, L185P CaMnSODc (blue) to that of WT ScMnSOD (green) and CaMnSODc (orange), respectively. Manganese atoms are shown in purple spheres.
(TIF)
Decay of 43 µM O2 − catalyzed by (A) WT ScMnSOD (solid, a), K182R, A183P ScMnSOD (dotted, b), (B) WT CaMnSODc (solid, a) and K184R, L185P CaMnSODc (dotted, b). The sample for pulse radiolysis contains 10 mM phosphate (pH 7), 10 mM sodium formate and 10 µM EDTA. The O2 − concentration in these figures was calculated from the absorbance at 260 nm and is slightly different from the O2 − dose given by the pulse radiolysis instrument.
(TIF)
CD spectra of WT CaMnSODc (A), K184R, L185P CaMnSODc (B), WT ScMnSOD (C) and K182R, A183P ScMnSOD (D) at increased concentrations of GdHCl. The solutions contained 25 mM potassium phosphate (pH 7.4). The measurements were carried out at room temperature.
(TIF)
Electrospray-ionization mass spectra of methylated RP-mutant ScMnSOD (left) and its reconstructed mass distribution profile (right). Ordinate units of intensity are arbitrary and the abscissa units of average molecular mass are in Daltons. The theoretical mass weight of methylated K182R, A183P ScMnSOD is 23,589 Da.
(EPS)
Metal contents of WT and RP-mutant ScMnSOD and CaMnSODc.
(DOC)
Interactions of Subunits at Dimer and Tetramer Interfaces in MnSODs from Different Organisms.
(DOC)
Calculation of K d for K184R, L185P CaMnSODc.
(DOC)