Abstract
Protection provided by host bacterial microbiota against microbial pathogens is a well known but ill-understood property referred to as the barrier effect, or colonization resistance. Despite recent genome-wide analyses of host microbiota and increasing therapeutic interest, molecular analysis of colonization resistance is hampered by the complexity of direct in vivo experiments. Here we developed an in vitro-to-in vivo approach to identification of genes involved in resistance of commensal bacteria to exogenous pathogens. We analyzed genetic responses induced in commensal Escherichia coli upon entry of a diarrheagenic enteroaggregative E. coli or an unrelated Klebsiella pneumoniae pathogen into a biofilm community. We showed that pathogens trigger specific responses in commensal bacteria and we identified genes involved in limiting colonization of incoming pathogens within commensal biofilm. We tested the in vivo relevance of our findings by comparing the extent of intestinal colonization by enteroaggregative E. coli and K. pneumoniae pathogens in mice pre-colonized with E. coli wild type commensal strain, or mutants corresponding to identified colonization resistance genes. We demonstrated that the absence of yiaF and bssS (yceP) differentially alters pathogen colonization in the mouse gut. This study therefore identifies previously uncharacterized colonization resistance genes and provides new approaches to unravelling molecular aspects of commensal/pathogen competitive interactions.
Introduction
The mucosal surface of the intestinal tract is a complex ecosystem composed of gastrointestinal epithelium, immune cells and resident bacterial flora. In this environment, bacteria are either in contact with intestinal surfaces or embedded in host-produced mucus [1]–[3]. Genome-wide analyses performed on intestinal microbiota provided insights into beneficial metabolic activities following establishment of successful commensal or symbiotic relationships with the host [4]–[8]. These studies also showed that the absence of an intact microbiota drastically increases susceptibility to pathogens, underlining the fact that colonization of mucosa and competition with commensal bacterial flora is often the first step in most intestinal infections [8]–[10].
This long-known but ill-understood protection provided by commensals against pathogens is commonly described as being colonization resistance, the barrier effect, bacterial antagonism or bacterial interference [1], [10]–[13]. Several mechanisms have been proposed for explaining colonization resistance, including: direct competition for nutrients; prevention of access to adherence sites; limitation of pathogen proliferation through production of inhibitory substances or conditions; or stimulation of host natural immune defenses [10], [14], [15]. However, the complexity of bacterial interactions in the host and the absence of relevant models has severely hindered identification of molecular details on how commensal bacteria interfere with pathogens [13], [16]. Due to these shortcomings, analysis of competitive bacterial interactions that contribute to restricting pathogen establishment within the intestinal flora has almost exclusively focused on secreted inhibitory substances (colicins, microcins, toxins) produced in liquid or solid medium or brought to light in competition experiments performed many decades ago [13].
Recently, interest in bacterial group behavior drew attention to biofilms, swarms, aggregates and dense bacterial cultures as models for studying competitive and synergistic interactions [17]–[26]. Indeed, considering the biofilm-like structure of vertebrate bacterial flora, controlled biofilm communities could enable direct experimental investigations of some aspects of molecular events leading to pathogen establishment in a multispecies context [16], [27], [28].
Here we have developed an in vitro-to-in vivo approach to studying colonization resistance. We used dynamic and controlled mixed in vitro biofilm models to investigate how populations of commensal Escherichia coli, a predominant facultative anaerobe of the intestinal microbiota, are colonized by a pathogenic diarrheagenic enteroaggregative E. coli [9], [29], [30]. Gene expression profiling demonstrated that pathogen entry into commensal biofilm triggers specific genetic responses, some of them also induced upon colonization by an unrelated bacterial pathogen, Klebsiella pneumoniae. Systematic functional analysis led to identification of genes involved in preventing incoming pathogens from settling and growing within commensal biofilm. Finally, we explored the in vivo relevance of a subset of identified colonization resistance genes and demonstrated their implication in control of the commensal/pathogen ratio within the mouse gut environment. This study therefore provides new concepts and methods for investigating molecular responses that take place during colonization resistance and that may constitute an early signature in the infection process.
Materials and Methods
Bacterial strains and culture media
Bacterial strains are listed in Table 1. All experiments were performed in 0.4% glucose M63B1 minimal medium at 37°C. Antibiotics were added when required, at the following concentrations: ampicillin (100 µg ml−1), apramycin (30 µg ml−1), tetracycline (7.5 µg ml−1), kanamycin (50 µg ml−1) and streptomycin (100 µg ml−1).
Table 1. Strains used in this study.
| Strain | Relevant characteristics | References |
| MG1655 | λ-, rph-1 | Laboratory collection |
| MG1655 F′( = C in vitro) | MG1655 carrying the F′tet-ΔtraD::apra plasmid; ApraR, TetR | Laboratory collection |
| 55989a ′( = P in vitro) | E. coli 55989 λattampgfp: 55989 with ampgfp insertion at the λatt site. AmpR | (6) |
| 55989a-s( = P in vivo) | Spontaneous streptomycin-resistant mutant of E. coli 55989a; AmpR StrepR. | This study |
| KpLM21( = P in vitro) | K. pneumoniae clinical isolate; serogroup O25; AmpR | This study |
| KpLM21-s ( = P in vivo) | Spontaneous streptomycin-resistant mutant of KpLM21; AmpR, StrepR | This study |
| MG1655agaI F′ | ΔagaI::GB, KmR, ApraR, TetR | This study |
| MG1655cspF F′ | ΔcspF::GB, KmR, ApraR, TetR | This study |
| MG1655kduI F′ | ΔkduI::GB, KmR, ApraR, TetR | This study |
| MG1655rcsA F′ | ΔrcsA::GB, KmR, ApraR, TetR | This study |
| MG1655relF F′ | ΔrelF::GB, KmR, ApraR, TetR | This study |
| MG1655rzpD F′ | ΔrzpD::GB, KmR, ApraR, TetR | This study |
| MG1655sppA F′ | ΔsppA::GB, KmR, ApraR, TetR | This study |
| MG1655stfE F′ | ΔstfE::GB, KmR, ApraR, TetR | This study |
| MG1655yaeT F′ | ΔyaeT::GB, KmR, ApraR, TetR | This study |
| MG1655yafX F′ | ΔyafX::GB, KmR, ApraR,TetR | This study |
| MG1655ycbQ F′ | ΔycbQ::GB, KmR, ApraR, TetR | This study |
| MG1655yceP F′ | ΔyceP::GB, KmR, ApraR,TetR | This study |
| MG1655yciF F′ | ΔyciF::GB, KmR, ApraR, TetR | This study |
| MG1655ydfZ F′ | ΔydfZ::GB, KmR, ApraR, TetR | This study |
| MG1655yiaF F′ | ΔyiaF::GB, KmR, ApraR, TetR | This study |
| MG1655yiaV F′ | ΔyiaV::GB, KmR, ApraR, TetR | This study |
| MG1655yjcR F′ | ΔyjcR::GB, KmR, ApraR, TetR | This study |
| MG1655yjiY F′ | ΔyjiY::GB, KmR, ApraR,TetR | This study |
| MG1655ylcE F′ | ΔylcE::GB, KmR, ApraR, TetR | This study |
| MG1655yliE F′ | ΔyliE::GB, KmR, ApraR, TetR | This study |
| MG1655yliH F′ | ΔyliH::GB, KmR, ApraR, TetR | This study |
| MG1655ypjC F′ | ΔypjC::GB, KmR, ApraR, TetR | This study |
| MG1655PcLrbs-rcsA F′ | Constitutive expression of rcsA from the Km-PcLrbs cassette, KmR | This study |
| MG1655PcLrbs-stfE F′ | Constitutive expression of stfE from the Km-PcLrbs cassette, KmR | This study |
| MG1655PcLrbs-yiaF F′ | Constitutive expression of yiaF from the Km-PcLrbs cassette, KmR | This study |
| MG1655PcLrbs-yliE F′ | Constitutive expression of yliE from the Km-PcLrbs cassette, KmR | This study |
| MG1655PcLrbs-ypjC F′ | Constitutive expression of ypjC from the Km-PcLrbs cassette, KmR | This study |
| 55989a-yiaF | ΔyiaV::GB, AmpR, KmR | This study |
| 55989a -rcsA | ΔrcsA::GB, AmpR, KmR | This study |
| 55989a -yliE | ΔyliE::GB, AmpR, KmR | This study |
| MG1655-s F′( = C in vitro) | A streptomycin derivative of MG1655 F′, StrepR | This study |
| MG1655-s yceP F′ | ΔyceP::GB, KmR, ApraR, TetR, StrepR | This study |
| MG1655-s yliE F′ | ΔyliE::GB, KmR, ApraR, TetR, StrepR | This study |
| MG1655-s yiaF F′ | ΔyiaF::GB, KmR, ApraR, TetR, StrepR | This study |
Monospecific and mixed biofilm
Microfermentor experiments
Biofilms were produced in a continuous flow biofilm microfermentor at 37°C in minimal M63B1 medium supplemented with 0.4% glucose as in (www.pasteur.fr/recherche/unites/Ggb/biofilmfermenter.html) and [31]. Microfermentor inoculations were performed by placing the microfermentor internal spatula in a culture containing 2.108 bacteria/ml for 2 min. The glass slide was then briefly rinsed in minimal media and reintroduced into the microfermentor.
Biofilm colonization
After 6 h of continuous culture, biofilm formed on a microfermentor glass slide was re-inoculated by direct introduction of 109 bacteria of overnight cultures of E. coli MG1655 F′, E. coli 55989a or K. pneumoniae KpLM21 bacteria into the microfermentor. Mixed biofilm continuous flow culture was resumed for an additional 24 h (30 h total) with rapid dilution and evacuation of excess planktonic bacteria. For monospecies biofilms, no re-inoculation was performed. Mono- or mixed biofilms formed on the internal microfermentor glass slide were resuspended by vortexing and biofilm biomass was estimated by determining optical density at 600 nm (OD600 nm).
Colonization phenotype
To estimate the percentage of colonizing bacteria in mixed biofilms, serial dilutions of resuspended biofilm were plated onto LB (total count estimation) and LB with specific antibiotics, thus distinguishing commensal from colonizing exogenous bacteria. All experiments were repeated at least 6 times. Statistical significance of differences observed between colonization phenotypes was estimated by Student t-tests. Differences were considered statistically significant when p<0.05.
Macroarrays
Genomic expression profiles were performed on E. coli MG1655 F′ (C) and 55989a (P) grown as 24 h mono- or mixed biofilms. The equivalent of 15 OD600 nm of bacterial cells were collected, pelleted and rapidly frozen. Cells were then broken in a Fast Prep apparatus (Bio 101) and total RNA was extracted by Trizol (Gibco BRL) treatment. Genomic DNA was removed using RNase-free DNAse I (Roche Diagnostics). Radioactively labeled cDNAs, generated using E. coli K-12 CDS-specific primers (Sigma-GenoSys), were hybridized to E. coli K-12 panorama gene arrays containing duplicated spots for each of the 4,290 predicted E. coli K-12 open reading frames (ORFs; Sigma-GenoSys). The intensity of each dot was quantified with ArrayVision™ software (Imaging Research, Inc.). Experiments were carried out using three independent RNA preparations for each sample condition (C; C+C; C+P; P). Each hybridization with each independent sample was carried out with 1 µg and 10 µg of total RNA; 3 sets of arrays were used.
Statistical analysis of macroarray data
Genes that were statistically significantly over- or underexpressed were identified using T-test analysis followed by the non-parametric Wilcoxon rank sum test. For each gene, expression in monospecies MG1655 F′ or 55989a biofilm and in self-infected MG1655 F′ + MG1655 F′ or mixed MG1655 F′ + 55989a biofilms (n = 10 to 12 for each data set) were compared. Analyses were performed with one-tailed tests. Genes were considered statistically significantly over- or underexpressed when p<0.05. Low (less than 0.01) or negative levels of expression were removed from the analysis.
Molecular techniques and construction of deletion and expression mutants
The genome of E. coli 55989 was sequenced and annotated by the Coliscope Consortium at the end of the experimental work [32]. E. coli 55989 Sequence is deposited in GenBank (accession number NC_011748.1 and GI:218693476). Deletion mutants and introduction of constitutive promoter cassettes in front of described target genes were performed as described at (http://www.pasteur.fr/recherche/unites/Ggb/matmet.html) and in [33], [34] using primers presented in Table S6. DNA sequencing was performed using Eurofins MWG services.
RT-PCR in E. coli -K. pneumoniae mixed biofilms
Biofilm bacteria were directly resuspended in an equal volume of ice-cold RNAlater (Ambion). Total RNA was isolated and purified using an RNeasy mini-kit (Qiagen). After purification, RNA was treated with RNase-free DNase I to remove contaminating DNA and re-purified using Qiagen RNeasy columns. RNA samples were quantified spectrophotometrically at 260 nm and additionally checked by gel electrophoresis. Purified total RNA was precipitated with ethanol and stored at −80°C until further use. RNA was converted to cDNA using SuperScript II as described by the manufacturer (Invitrogen Life Technologies). cDNA was used directly as template for PCR using specific primers (Table S6). A negative control using the original RNA was consistently run in parallel to confirm the absence of contaminating DNA.
Mouse model of intestinal colonization
Female IOPS mice (Charles River Laboratories, OF1, 8 to 18 weeks old, 25 g) were used. They were given sterile water containing 5 g/L of streptomycin sulfate throughout the experiment and had ad libitum access to feed. After 24 h, 200 µl bacterial suspensions containing 106 cfu of either MG1655-s F′ or its mutants (MG1655-s yliE F′, MG1655-s yceP F′, MG1655-s yiaF F′) were given intragastrically. At least eight mice were infected with the wild-type strain and another eight mice with each mutant strain. At day 11, each animal was administrated intragastrically 102 cfu of the pathogenic strain, 55989a-s or 103 cfu KpLM21-s. These doses correspond to minimal inocula to detect pathogen colonization in feces. On day 3, and subsequently every other day after inoculation, feces were collected, homogenized in 0.9% saline, and serial dilutions were plated onto both tetracycline-containing media (detection of MG1655-s F′) and ampicillin-containing media (detection of the pathogen). The potential impact of initial colonization by wild-type MG1655-s F′ or its derivatives upon the capacity of the pathogens (EAEC 55989 or K. pneumoniae) to colonize was assessed by calculating the Pearson correlation coefficient using the number of cfus determined at days 10 and 12. A Mann-Whitney statistical test was then used to assess the colonization capacity of each pathogen (K. pneumoniae and EAEC) by comparing the number of cfus from D12 to D20 in feces of mice previously inoculated with the yliE, yceP or yiaF mutant to the number of pathogens observed in mice previously colonized with wild-type MG1655-s F′. A P value of <0.05 was considered statistically significant.
Ethics statement
Animal studies were performed in accordance with the European Community guiding in the care and use of animals (86/609/CEE). Furthermore, the models and protocols used in this study were all approved by the ethics committee of Auvergne (Comité Régional d'Ethique en Matière d'Expérimentation Animale Auvergne). Animals were housed under controlled environmental conditions and kept under a 12/12 h light/dark cycle, with food and water ad libitum.
Results
A new in vitro model of commensal biofilm colonization by exogenous pathogens
To identify the genetic responses triggered in a commensal biofilm upon entry of exogenous pathogens, we developed an in vitro model in which pathogenic bacteria were exogenously added to an already formed commensal biofilm. This procedure will be referred to as biofilm colonization throughout this study. As a biofilm-forming commensal bacterium (or C for commensal), we chose E. coli K12 MG1655 F′ carrying a conjugation-deficient derivative of the F conjugative plasmid (F′tetΔtraD) that rapidly forms biofilm under continuous flow microfermentor conditions [31]. The pathogenic strain (P) chosen to colonize MG1655 F′ commensal biofilm is an ampicillin-resistant derivative of E. coli 55989, a biofilm-forming enteroaggregative (EAEC) isolate originally isolated from diarrheagenic stools and causing acute and persistent diarrhea [9], [35], hereafter referred to as 55989a or P.
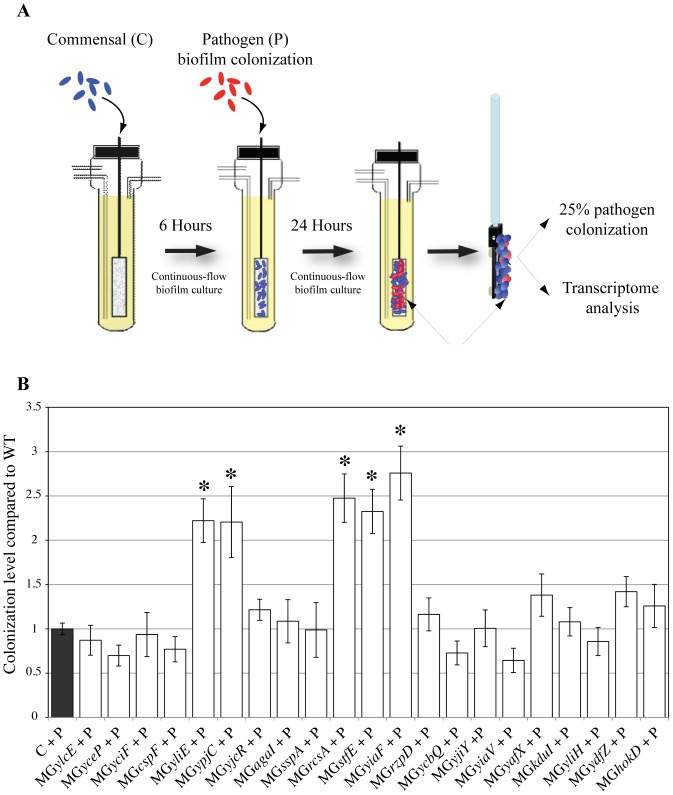
To establish conditions of MG1655 F′ colonization upon exogenous introduction of 55989a, we first produced MG1655 F′ biofilms formed for 6 to 24 h in continuous flow microfermentors. We then inoculated them with various titers of E. coli 55989a and allowed the resulting mixed biofilm to grow an additional 24 h. We defined E. coli 55989a colonization efficiency as the percentage of pathogens present in the resulting 24 h C+P mixed biofilm, as determined using the 55989a ampicillin antibiotic resistance marker (Table 1). At 24 h, a commensal colony-forming unit (cfu) had increased by a 2-log factor and the presence of the pathogen did not significantly alter development of the commensal biofilm, since C and C+P biofilm displayed similar biomass (data not shown). We found that the proportion of 55989a in C+P biofilm depended on both the 55989a initial inoculation titer and the age of MG1655 F′ biofilm. When MG1655 F′ 6 h biofilms were inoculated with a titer of 109 bacteria/ml of 55989a, we reproducibly obtained 25+/−5% of 55989a in 24 h C+P mixed biofilm; we used these experimental conditions throughout the rest of the study (Fig. 1A).
Figure 1. Identification of colonization resistance factors interfering with establishment of mixed pathogen/commensal biofilm.
A Experimental set-up: continuous flow biofilm growth in microfermentor. After initial inoculation of the microfermentor with E. coli MG1655 F′ commensal (C), biofilm develops for 6 h before re-inoculation (colonization) with exogenous pathogen E. coli 55989a (P). At 24 h post-colonization, mixed biofilm developing on the glass slide was resuspended and used for gene expression analysis and determination of colonization phenotype (% of pathogens in the mixed biofilm). B Microfermentors were inoculated with wild-type or mutant commensal (MG1655 F′ is abbreviated as MG) as indicated in the x-axis. After 6 h of growth, commensal biofilm was re-inoculated (colonized) with the 55989a (P). Colonization phenotype of each mixed biofilm was estimated and results are represented as ratio of colonization level in Cmutant+P mixed biofilms compared to wild-type C+P mixed biofilms. Black bar represents wild-type colonization level in C+P mixed biofilms arbitrarily set to one. White bars represent colonization level of Cmutant+P mixed biofilms. Results are averages of at least 6 replicates ± standard deviation of the mean. Stars indicate mutant mixed biofilm with a colonization level significantly different from that of wild-type C+P mixed biofilm, p<0.01.
Pathogen colonization of commensal biofilm triggers specific genetic responses
To investigate the genetic response of MG1655 F′ biofilm bacteria upon introduction of E. coli 55989a, we first compared the expression profile of monospecific MG1655 F′ biofilm (C) to that of both monospecific 55989a biofilm (P) and “self-mixed” biofilm, in which MG1655 F′ commensal biofilm was colonized alone (C+C). Comparison of monospecific biofilm (P/C), indicated that, among genes common to both strains, 545 exhibited differing expression profiles (Table 2). Analysis of “self-mixed” or self-colonization, abbreviated as (C+C/C), showed that 346 genes underwent significant transcription level changes between the two conditions, indicating that addition of an exogenous but identical commensal bacterium to commensal biofilm already induces changes in gene expression (see Tables 2, S2 and S3). We then compared bacterial gene expression in mixed MG1655 F′+55989a (C+P) biofilm with gene expression in monospecies commensal MG1655 F′ (C) biofilm. This analysis, abbreviated as (C+P/C), revealed the differential expression of 329 genes between the two biofilms (see Tables 2, S4 and S5).
Table 2. Summary of transcriptome analyses performed on biofilms colonized by different exogenous bacteria.
| Conditions | Number of genes repressed or overexpressede | Functional categories (COG) (% of total number) | |||||
| Total | ≥2 fold | Information storage and processing | Cellular processes | Metabolism | Unknownf | ||
| P/Ca | Repressed | 389 | 163 | 22.4 | 21.3 | 21.1 | 35.2 |
| Overexpressed | 156 | 18 | 16.7 | 19.9 | 28.2 | 35.2 | |
| All | 545 | ||||||
| C+C/Cb | Repressed | 185 | 60 | 14.1 | 12.4 | 19.5 | 54 |
| Overexpressed | 161 | 2 | 14.3 | 21.7 | 42.2 | 21.8 | |
| All | 346 | ||||||
| C+P/Cc | Repressed | 109 | 4 | 14.7 | 18.3 | 22.9 | 44 |
| Overexpressed | 220 | 4 | 12.7 | 23.2 | 38.2 | 25.9 | |
| All | 329 | ||||||
| C+P/C+Cd | Repressed | 61 | 1 | 21.3 | 9.8 | 26.2 | 42.6 |
| Overexpressed | 108 | 32 | 15.7 | 16.7 | 33.3 | 34.3 | |
| All | 169 | ||||||
: Monospecies pathogen E. coli 55989a biofilm (P) versus monospecies commensal E. coli K12 MG1655 F′ (C): comparison.
: Commensal biofilm infected by identical commensal bacteria (C+C) versus monospecies commensal biofilm (C): comparison.
: Commensal biofilm infected by E. coli 55989a (mixed biofilm, C+P) versus monospecies commensal biofilm (C): comparison.
: Commensal biofilm infected by E. coli 55989a (mixed biofilm, C+P) versus commensal biofilm infected by identical commensal bacteria (C+C): comparison.
: p<0.05.
: “Unknown” regroups poorly characterized and unknown function genes of the COG classification plus non-classified genes coding mainly for hypothetical proteins.
Comparison of gene expression upon self-colonization (C+C/C experiment) and upon pathogen colonization (C+P/C experiment) showed a common genetic response to entry of exogenous bacteria into commensal MG1655 F′ biofilm, with the same 89 overexpressed and 26 repressed genes in both situations (see Tables S4 and S5). Moreover, comparison of non-self versus self-colonized analyses (C+P/C+C comparison) indicated significant specific differential gene expression in response to 55989a colonization, with 61 repressed genes and 108 overexpressed genes. The distribution of these 169 genes in the different COG functional classes is comparable to that found in C+P/C transcription profile analysis, including 30 to 40% of poorly characterized genes (Tables 2, S4 and S5). Furthermore, several overexpressed and underexpressed genes overlapped C+P/C and C+P/C+C analysis, (Table S1), suggesting that these genes could be involved in specific responses to colonization of E. coli commensal biofilm by pathogenic bacteria.
Colonization responses of commensal biofilm bacteria reduce pathogen colonization
Although our analysis focused on E. coli MG1655 genes, most of these genes are also present on the E. coli 55989 genome and we could not rule out that the observed variation in transcription also reflected responses induced in the 55989a pathogen. We nevertheless hypothesized that the identified genetic responses could contribute to limiting pathogen colonization within MG1655 F′ commensal biofilm. To test this, we selected 22 genes differentially expressed by MG1655 F′ biofilm bacteria upon colonization by 55989a, by comparing the C+P/C and C+P/C+C versus C+C/C datasets. These 22 genes included genes coding for membrane and envelope proteins (36%), proteins of phage origin (27%), regulators implicated in biofilm-associated phenotype (13%) and genes of unknown function (14%), (Table 3 and Fig. S1). RT-PCR experiments on six of the most frequently expressed of these 22 genes (hokD, kduI, ylcE, yceP, yliH and yciF) indicated that, although ratios obtained with the two approaches differed, all tested genes displayed increased levels of transcription within mixed C+P biofilm compared to mixed C+C biofilm (data not shown). We then performed non-polar deletion of the 22 selected genes in commensal strain MG1655 F′. With the exception of yaeT (bamA), all mutants exhibited wild-type growth and biofilm formation ability (data not shown). Biofilms corresponding to the 21 remaining mutants were inoculated with wild-type 55989a using the procedure described in Fig. 1A. Determination of the percentage of pathogens in resulting mixed C+P biofilms showed that biofilms formed by MG1655 F′ mutants yliE, ypjC, rcsA, stfE and yiaF mutant were colonized by the pathogen 55989a at significantly higher levels than the wild-type strain (p<0.01) (Fig. 1B). yliE encodes a conserved inner membrane hypothetical protein of 90 kDa that contains an EAL domain characteristic of phosphodiesterase enzymes degrading c-di-GMP and often involved in transition between individual and community lifestyles [36]; ypjC encodes a hypothetical protein of 18 kDa; rcsA codes for a cofactor required for synthesis of colanic acid, capsular polysaccharide and curli synthesis in E. coli [37], [38]; stfE encodes a putative tail protein homolog from lambdoid prophage e14 and yiaF encodes a 25.6 kDa inner membrane protein exhibiting patchy distribution with polar and septal bias [39].
Table 3. Selection of genes differentially expressed in C+P/C and C+P/C+C, but not in C+C/C analysis.
| Gene | Gene function | Macroarray analysis (ratio)a | ||
| C+P/C | C+P/C+C | C+C/C | ||
| hokD * | (relF); cell killing | 4.85 | 7.52 | 0.4 |
| kduI | Homolog of pectin-degrading enzyme 5-keto 4-deoxyuronate isomerase | 2.88 | 2.56 | |
| ylcE * | (tfaX); tail fiber assembly predicted protein, DLP12 prophage | 2.55 | ||
| yceP | (bssS) biofilm regulator through signal secretion | 2.52 | 0.51 | |
| yliH | (bssR) putative receptor, biofilm regulator through signal secretion | 2.47 | 2.15 | |
| yciF | Putative structural protein | 2.01 | 0.61 | |
| cspF * | Cold shock protein homolog | 1.99 | ||
| ypjC | Hypothetical protein | 1.99 | ||
| yliE | Predicted cyclic-di-GMP phosphodiesterase, inner membrane protein | 1.99 | ||
| sppA | Protease IV, signal peptide peptidase | 1.90 | 1.92 | |
| agaI | 6-Phosphogluconolactonase/glucosamine-6-phosphate isomerase/deaminase | 1.89 | 2.37 | |
| rcsA | Positive regulatory gene for capsule (colanic acid) synthesis | 1.88 | ||
| ydfZ | Hypothetical protein, potential seleno-protein | 1.79 | 2.77 | 0.65 |
| yjcR | (sdsR); putative membrane protein of multidrug efflux pump | 1.74 | ||
| stfE * | Putative tail fiber protein, e14 prophage | 1.71 | ||
| yiaF | Inner membrane protein | 1.65 | ||
| rzpD * | Endopeptidase-like protein, DLP12 prpophage | 1.62 | ||
| yaeT | (bamA); integral β-barrel protein, outer membrane | 1.58 | ||
| ycbQ | Chaperone-usher fimbrial protein | 1.57 | ||
| yafX * | Hypothetical protein, CP4-6 prophage | 1.55 | ||
| yjiY | Inner membrane protein; putative carbon starvation protein | 1.53 | ||
| yiaV | Inner membrane protein, predicted component of efflux pump | 1.50 | ||
The increased pathogen colonization phenotype obtained for the 5 mutants correlated with a slight decrease in commensal bacteria cfu in mixed C+P biofilm (1.5-fold average for the rcsA, stfE, yliE and ypjC mutants and 2.3-fold for the yiaF mutant), associated with a 2-fold increase in pathogen cfu. However, estimation of bacterial cfu constituting the biofilm of MG1655 F′ yiaF, stfE and yliE mutants at the time of infection (6H) showed no significant difference from wild-type MG1655 F′, indicating that the increased 55989a pathogen colonization phenotype obtained with these mutants was not due to decreased ability to form the initial biofilm (Fig. S2). In contrast, MG1655rcsA F′ mutant biofilm biomass was reduced, while the number of bacteria recovered from MG1655ypjC F′ biofilms was 3.5-fold higher than in wild-type MG1655 F′ commensal biofilm (Fig. S2).
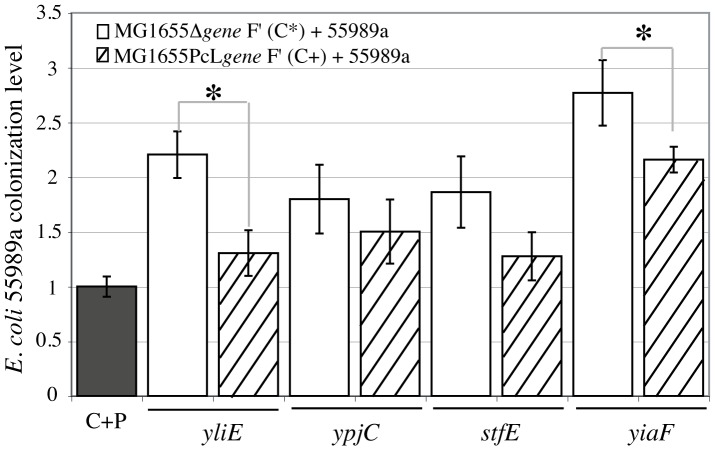
We then used a previously described plasmid-free expression strategy and placed a constitutive promoter in front of the yiaF, stfE, yliE and ypjC genes directly on the MG1655 F′ chromosome [34], [40]. This insertion had no effect on growth or biofilm formation for yiaF, stfE, yliE and ypjC mutant strains (data not shown). However, constitutive expression of rcsA (MG1655PcLrbs-rcsA F′) led to mucoidy and the inability to form biofilm (data not shown), and this mutant was not further analyzed. The colonization phenotype of the 4 remaining mutants was evaluated and compared to that of the corresponding deletion mutants. While constitutive expression of ypjC and stfE did not lead to significant 55989a colonization changes, yiaF and yliE constitutive expression in MG1655PcLrbs-yiaF F′ and MG1655PcLrbs-yliE F′, respectively, significantly reduced the ability of E. coli pathogen 55989a to colonize the resulting C+P mixed biofilm compared to colonization of the corresponding MG1655 F′ deletion mutants (Fig. 2).
Figure 2. Constitutive expression of potential colonization resistance genes.
Estimate of E. coli 55989a (P) colonization in mixed biofilms with wild-type E. coli MG1655 F′ (C), corresponding deletion mutants (MG1655Δgene: C*) or overexpressed (MG1655PcLgene: C+) derivative strains. Results are represented as ratio of colonization l of mutant mixed biofilms to wild-type mixed C+P biofilms. Black bar represents wild-type colonization in C+P mixed biofilms arbitrarily set to one. White bars represent colonization of pathogen in mixed CΔgene + P biofilms. Stripped bars show pathogen colonization in mixed CPcLgene + P biofilms with commensal overexpressing potential colonization resistance genes. Genes deleted or overexpressed are indicated under the bars. Results are averages of at least 12 replicates ± standard deviation of the mean. The extent of colonization in CΔgene + P mixed biofilm was significantly different from that of wild-type C+P biofilm p<0.05; asterisks indicate significant difference between extent of colonization in over-expressed and deletion mutants, p<0.05.
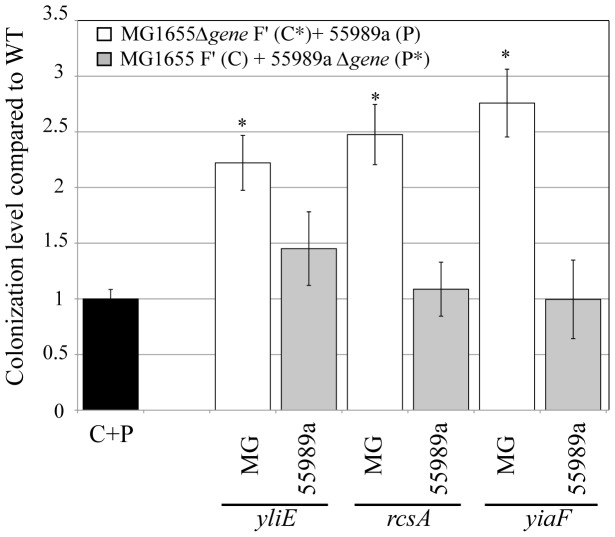
Finally, we tested whether yiaF, stfE, rcsA, yliE and ypjC could also play a reciprocal role in E. coli 55989a ability to colonize a commensal biofilm. While stfE was absent from the 55989 genome, a ypjC mutant could not be obtained despite repeated attempts. We therefore introduced only a yiaF, rcsA or yliE mutation in the 55989a strain, and we showed that none of these 3 mutations had a significant effect on colonization outcome, suggesting that the observed colonization phenotypes specifically affected pathogen colonization in commensal biofilm, but not the reverse (Fig. 3). Taken together, these results indicated that colonization of commensal MG1655 F′ biofilm by the diarrheagenic pathogenic strain 55989a triggers expression of commensal genes contributing to colonization resistance to the pathogen.
Figure 3. Colonization resistance genes are strain-specific.
Comparison of the effect on colonization of mutations introduced into commensal MG1655 F′ (C) or into pathogenic strain 55989a (P). Results are represented as ratio of colonization of mutant mixed MG1655 F′Δgene (C*) +P or C+55989aΔgene (P*) biofilms compared to wild-type mixed C+P biofilm. Black bar represents extent of wild-type colonization in C+P mixed biofilms arbitrarily set to one. White bars represent colonization levels of CΔgene +P mixed biofilm (mutation introduced into commensal and wild-type pathogens). Light gray bars represent colonization levels of C+PΔgene mixed biofilm formed by wild-type commensal and mutant pathogens. Names of deleted genes are indicated under the line. Results are averages of at least 6 replicates ± standard deviation of the mean. Asterisks indicate mutant mixed biofilm with a colonization level significantly different from that of wild-type MG1655 F′ + 55989a mixed C+P biofilm, P<0.05.
Common genetic responses of commensal biofilm upon colonization by distinct pathogens
We investigated whether in vitro commensal biofilm colonization by a bacterium other than E. coli 55989a could trigger similar genetic responses. We inoculated a pre-formed MG1655 F′ commensal biofilm with the biofilm-forming opportunistic enterobacterium Klebsiella pneumoniae strain LM 21 (hereafter referred to as KpLM21) responsible for a wide range of nosocomial infections, including pneumonia, bacteremia and urinary tract infections [41], [42]. K. pneumoniae can also be found in the intestine, where it can colonize the local microbiota. Conditions similar to those used with EAEC strain 55989a resulted in mixed biofilm composed of 75% MG1655 F′ and 25% KpLM21. We then monitored expression of 6 genes absent from the K. pneumoniae genome and expressed in MG1655 F′ in response to E. coli 55989a colonization. yiaF, yliE and stfE were chosen for their contribution to MG1655 F′ colonization resistance against 55989a, while yceP and yliH (bssR and bssS, respectively, for “regulator of biofilm through signal secretion”, see Discussion) were selected because they are overexpressed in mixed C+P biofilm compared to uninfected and/or self-infected biofilm (Table 3) [43], [44]. RT-PCR on RNA extracted from mixed 24 h C+P (E. coli + K. pneumoniae) biofilms showed that, although colonization of commensal MG1655 F′ biofilm by KpLM21 had no impact on stfE expression and reduced yliE expression, it led to induction of yiaF, yceP and yliH (Table 4 and Fig. S1)
Table 4. Gene expression level in mixed MG1655F′ + K. pneumoniae biofilms.
| Gene | Fold inductiona | T-test |
| yiaF | 1.62±0.13 | 0.034 |
| yliE | 0.72±0.04 | 0.043 |
| stfE | 0.83±0.09 | 0.902 |
| yliH | 1.41±0.11 | 0.047 |
| yceP | 1.69±0.20 | 0.037 |
Gene expression level was estimated by RT-PCR in single MG1655 F′ biofilm and mixed MG1655F′ + K. pneumoniae KpLM21 (MG+Kp). Gene expression level in MG+Kp biofilm was compared to gene expression in commensal biofilm set to 1. Results are averages of 3 replicates with triplicate measurements for each ± standard deviation of the mean.
These observations indicated the existence of common genetic responses upon colonization of E. coli commensal biofilm by two different exogenous bacterial pathogens.
Correlation between in vitro and in vivo reduction of pathogen colonization
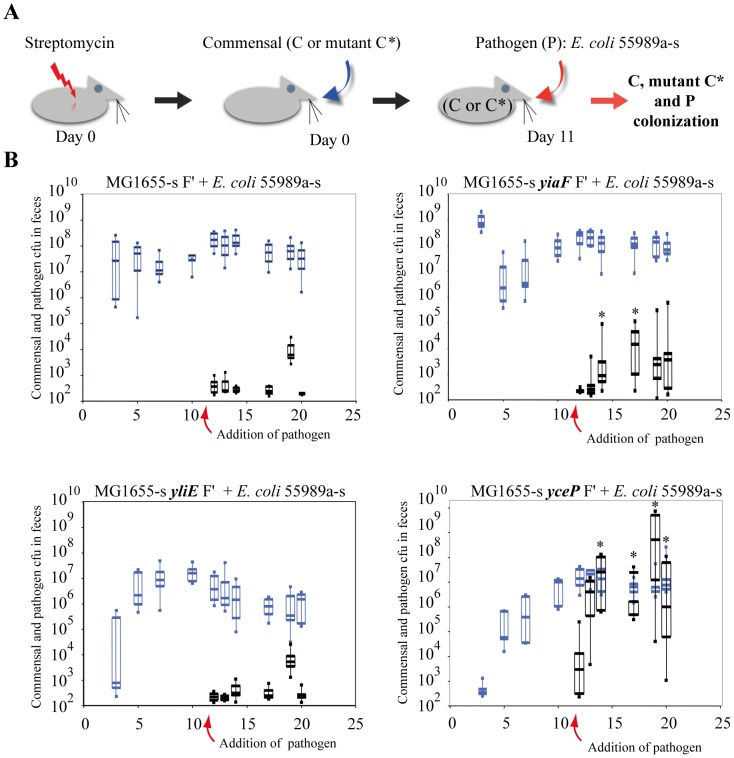
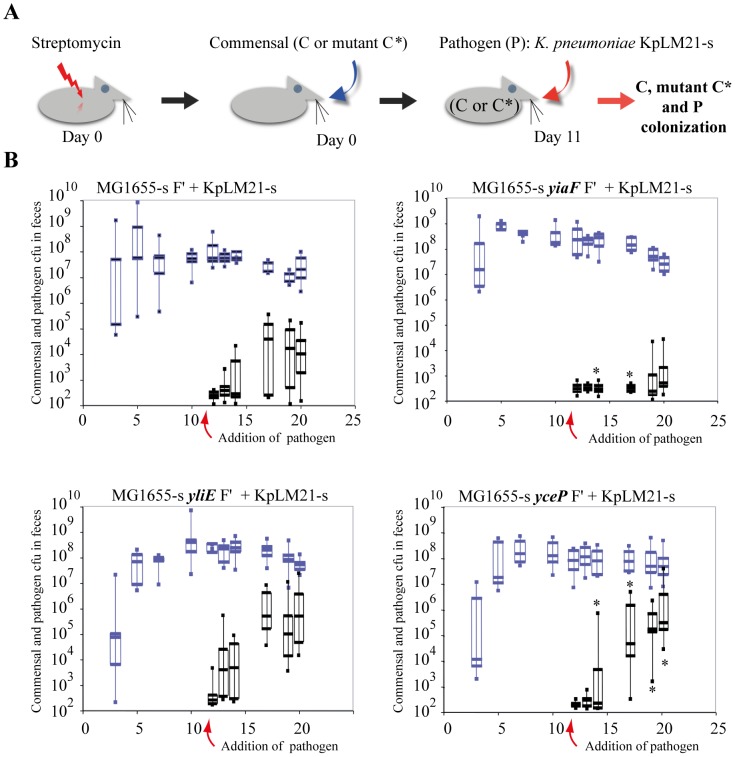
To test the in vivo role of colonization resistance genes identified via our approach, we used a mouse model of intragastric infection to compare the extent of intestinal colonization by 55989a and KpLM21 pathogens in streptomycin-treated mice, in which enterobacteria such as E. coli and Klebsiella were shown to colonize the same niches [45], [46]. Streptomycin-treated mice were pre-colonized with wild-type or mutant commensal E. coli MG1655 F′ and all three strains efficiently colonized the mouse intestine [47]–[49] (Fig. 4A). We chose to test the role of: i) yiaF, which affects in vitro colonization resistance to 55989a and was also induced upon KpLM21 colonization (Fig. 1B, and Table 4); ii) yliE, which was similarly involved in in vitro colonization resistance to 55989a, but was not induced upon KpLM21 colonization (Fig. 1B and Table 4 and Fig. S1); and iii) yceP, a gene induced in response to both pathogens, though without detectable in vitro effects on 55989a colonization (Fig. 1B and Table 4 and Fig. S1). To perform colonization experiments in streptomycin-treated mice, we first made E. coli 55989a and KpLM21 streptomycin-resistant derivatives (respectively, 55989a-s and KpLM21-s). We also introduced yiaF, yliE and yceP mutants into the same streptomycin-resistant derivative of MG1655 F′, (MG1655-s F′) and checked that these strains were not significantly affected in terms of growth and in vitro biofilm ability against this background (data not shown). We then determined bacterial counts in feces recovered from individually inoculated mice (n = at least 8 for each strain) and observed that wild-type and MG1655-s F′ yiaF, yliE and yceP mutants reached similar intestinal colonization capacity at day 10 (between 107 to 108 cfu/g of feces) prior to pathogen inoculation (Fig. 4B and 5B). At day 11 post-inoculation, streptomycin-treated mice colonized with wild-type MG1655-s F′ or corresponding yiaF, yliE and yceP mutants were inoculated intragastrically with either 55989a-s (Fig. 4) or KpLM21-s (Fig. 5). Determination of Pearson correlation coefficients indicated that there was no (−0.5<P<0.5) or only a moderate (0.5<P<0.8) correlation between colonization levels of wild-type MG1655-sF′ or its mutant derivatives and pathogens (KpLM21-s and 55989a-s) at days 10 and 12 post-inoculation by the commensal (wild-type or mutant) strains. Using the non-parametric Mann-Whitney test, comparison from day 12 to day 20 of the numbers of pathogen cfus in the feces of mice previously inoculated with wild-type MG1655-s or yliE, yceP or yiaF mutants indicated that pre-colonization of mice with MG1655-s F′ yceP, but not yliE, led to statistically significantly increased intestinal colonization by both pathogens (P = 2.3E10−7 and P = 0.19, respectively) (Fig. 4B and 5B). In addition, while mice pre-inoculated with MG1655-s F′ yiaF displayed lower level (P = 0.01) of KpLM21-s colonization, they showed higher levels (P = 0.01) of E. coli 55989a-s colonization compared to mice precolonized with wild-type MG1655-s control (Fig. 5B and 4B respectively)
Figure 4. In vivo colonization of E. coli commensal biofilm by enteroaggregative E. coli 55989 pathogen.
A Schematic representation of the experimental procedure. B Streptomycin-treated mice were first challenged intragastrically with commensal wild-type MG1655-s F′ (C) or its mutant ΔyceP, ΔyliE, and ΔyiaF derivatives (C*), followed on day 11 by administration of the E. coli 55989a-s pathogen. Numbers of commensal and pathogenic cfus recovered per gram of feces were determined every other day from day 3 to day 20. The lower limit of detection for bacteria was 102 cfu/g of feces. Box-and-whiskers plot indicates high and low values, median and interquartile ranges; each group contained between 8 and 12 mice. Pearson analysis of the bacterial count in faeces (impact of the initial colonization by the wild-type MG1655-s F′ or its derivatives on the capacity of the pathogen (Enteroaggregative E. coli 55989a-s) to colonize the mice intestine) and Mann-Whitney analysis of the number of the pathogen CFUs recovered (comparison of pathogen colonization level in mice precolonized with either MG1655-s F′ (control) or its derivatives (yliE, yceP or yiaF)) were performed. Statistically different results (P<0.05), are indicated by an asterisk.
Figure 5. In vivo colonization of E. coli commensal biofilm by K. pneumoniae KpLM21 pathogen.
A Schematic representation of the experimental procedure. B Streptomycin-treated mice were first challenged intragastrically with commensal wild-type MG1655-s F′ (C) or its mutant ΔyceP, ΔyliE, and ΔyiaF derivatives (C*), followed on day 11 by administration of the K. pneumoniae KpLM21-s pathogen. The numbers of commensal and pathogen cfus recovered per gram of feces were determined every other day from day 3 to day 20. The lower limit of detection for bacteria was 102 cfu/g of feces. Box-and-whiskers plots indicate high and low values, median and interquartile ranges; each group contained 8 to 12 mice. Pearson analysis of the bacterial count in faeces (impact of the initial colonization by the wild-type MG1655-s F′ or its derivatives on the capacity of the pathogen (K. pneumoniae KpLM21-s) to colonize the mice intestine) and Mann-Whitney analysis of the number of the pathogen CFUs recovered (comparison of pathogen colonization level in mice precolonized with either MG1655-s F′ (control) or its derivatives (yliE, yceP or yiaF)) were performed. Statistically different results (P<0.05), are indicated by an asterisk.
Discussion
Increased susceptibility to enteric infection after disruption of aerobic gastrointestinal microbiota in in vivo models led to the hypothesis that E. coli and other facultative aerobes contribute to colonization resistance [9], [10]. In streptomycin-treated mice, a protective effect associated with E. coli is partly attributed to production of antibacterial molecules such as colicins and microcins; however, non-producing strains still exhibit protection, suggesting involvement of other bacterial functions in colonization resistance [13]. Here we hypothesized that initial and virtually host-independent competitive interactions between commensal and pathogenic bacteria could be studied in simple in vitro experimental settings. We developed a model of commensal E. coli biofilms colonized by exogenous pathogens, a situation resembling the proximal intestine environment and the outcome of which partly determines the fate of many gastrointestinal infections.
Transcription profiling of commensal E. coli monospecies biofilm with commensal biofilm colonized alone or by the EAEC 55989a pathogen revealed variations in gene expression corresponding to a general response to colonization (self or non-self), potentially corresponding to growth disturbances and substrate competition occurring during introduction of exogenous bacteria into the bacterial community.
Our analysis also revealed variations in gene expression specifically triggered upon colonization by an enteroaggregative E. coli. Bacterial capacity to discriminate self from non-self in their environment was shown to result from secretion of quorum sensing molecules, expression of surface autotransporter adhesins enabling bacterial self-recognition and auto-aggregation, and other factors favoring clonality and enabling pathogens to maximize resource utilization and virulence potential [50]–[53]. Our data indicate that discrimination between self and non-self might also be at the origin of colonization resistance, potentially leading to induction of functions controlling bacterial intrusion or reinforcing commensal community cohesion.
We show that colonization of a commensal biofilm by an enteroaggregative E. coli induces expression of numerous genes coding for membrane and envelope proteins. One of them is YaeT, also known as BamA, a conserved member of the YaeT/Omp85 family of proteins required for biogenesis of β-barrel outer membrane proteins (OMPs) and involved in contact-dependent inhibition [54]. The strong growth defect displayed by a yaeT mutant did not enable us to meaningfully investigate the role of YaeT/BamA. However, yaeT expression in commensal bacteria upon pathogen colonization could be due to increased expression of membrane proteins in mixed biofilms. We also observed an intriguingly high proportion of genes located in regions corresponding to defective prophages, including e14 (stfE), DLP12 (ylcE, rzpD), Qin (hokD/relF, cspF) and CP4-6 (yafX). Defective prophages are usually considered to be in a state of mutational decay and have lost the ability to sustain a full phage replication cycle [55], [56]. Nevertheless, they often carry functional genes coding, for instance, for cell lysis functions or phage tail-like particles, a special group of bacteriocins composed of fragments of bacteriophages and produced by a number of Enterobacteriaceae and other Gram-negative bacteria [57], [58]. Expression of lytic genes carried by CP4-57- and DLP12-defective phages has recently been associated with biofilm development, suggesting that cell lysis may be an important aspect of E. coli biofilm physiology [59]. We show here that deletion of stfE, which encodes a putative tail-fiber protein, is involved in colonization resistance, as indicated by the increased colonization of E. coli 55989 in biofilm formed by stfE mutants. Given that other colonization-induced genes of phage origin are potentially associated with some cell lysis activity (hokD, cspD, ylcE, rzpD), this raises the possibility that StfE contributes to excluding incoming E. coli 55989a in commensal biofilm. Such a contribution to bacterial weaponry could represent a positive selective force for conservation of a defective prophage gene [60].
A general response to commensal biofilm colonization also involves YceP (BssS) and YliH (BssR), both previously associated with biofilm formation, regulation of indole production and uptake and export of AI-2 through a cAMP-dependent pathway [43], [44]. We observed that, while deletion of yceP did not lead to a significant reduction in EAEC pathogen commensal biofilm colonization in vitro, it significantly increased in vivo colonization of enteroaggregative E. coli 55989a-s and K. pneumoniae KpLM21-s in mice precolonized with E. coli MG1655ΔyceP F′. Since yceP is induced upon different stresses, including cold, heat shock and oxidative conditions, YceP could contribute to commensal protection in in vivo environments [61]–[64]. Finally, colonization of commensal biofilm by EAEC 59989a also leads to overexpression of yliE, which is involved in commensal capacity to prevent 55989a pathogen colonization in vitro. yliE codes for a conserved inner membrane hypothetical 90 kDa protein with an EAL domain associated with phosphodiesterase activity, involved in hydrolysis of the second messenger cyclic di-GMP (c-di-GMP), a key factor in the planktonic-to-biofilm lifestyle switch [36], [65]. Hence, expression of yliE in the commensal strain upon pathogen colonization could play a role in c-di-GMP-dependent cell-cell interactions resulting in reduced colonization by incoming pathogens.
Interestingly, while yliE and yiaF (encoding a conserved inner membrane protein of unknown function) specifically contributed to commensal colonization resistance to the EAEC pathogen in vitro, they were also differentially expressed in response to K. pneumoniae colonization, along with yliH and yceP. This suggests the existence of a common genetic response by MG1655 F′ commensal biofilm bacteria to colonization by non-self exogenous bacteria. However, analysis of the in vivo contribution of yliE and yiaF to commensal colonization resistance showed that, while a yliE mutation had no influence on EAEC 55989a-s colonization, mice precolonized with a yiaF commensal mutant showed increased EAEC 55989a-s colonization. In contrast, we observed decreased K. pneumoniae KpLM21-s capacity to be implanted in the intestine of a mouse precolonized with a ΔyiaF commensal strain. Hence, although yliE and yiaF are induced upon colonization of commensal biofilm by the two tested pathogens, they differentially contribute to the in vivo colonization phenotype depending on the pathogen. While the exact role of colonization resistance genes identified in vitro and in vivo currently remains under investigation, it should be noted that strains used in in vivo experiments are streptomycin-resistant derivatives of those used for in vitro biofilm experiments, thus potentially leading to differences in the colonization phenotype. In addition to genetic background differences, in vitro commensal biofilm colonization by pathogens could trigger responses that differ from those of in vivo gastrointestinal infection experiments, in which streptomycin-resistant flora might also contribute to regulating pathogen colonization.
In conclusion, the in vitro–to-in vivo approach described in this study provides a new strategy for studying colonization resistance and unravelling molecular aspects of commensal/pathogen interactions potentially leading to innovative prophylactic intervention in enteric infections.
Supporting Information
DNA-array data to in vivo test decision Flow-chart depicting the rational for selection of genes analyzed in the study.
(DOCX)
Estimate of biofilm biomass before inoculation with pathogen. Microfermentors were inoculated with commensal strain MG1655 F′ (C) or with indicated devivative mutants. After 6 h of growth, biofilm that developed on the glass slide was resuspended in 10 ml of minimal media and recovered bacterial count was estimated by serial dilution and cfu count. Results are average of at least 6 replicates ± standard deviation of the mean. Star indicates a mutant for which initial biofilm formation significantly differed from that of the wild type, P≤0.05.
(DOCX)
Genes over-expressed or repressed in response to colonization of MG1655 F′ biofilm by pathogenic 55989a.
(DOCX)
Genes induced upon self-colonization (C+C) of commensal biofilm.
(DOCX)
Genes repressed upon self-colonization (C+C) of commensal biofilm.
(DOCX)
Genes induced upon colonization by exogenous pathogen (C+P).
(DOCX)
Genes repressed upon colonization by exogenous pathogen (C+P).
(DOCX)
Primers used in this study.
(DOCX)
Acknowledgments
We thank Perrine Vasseur for preliminary animal experiments, Christophe De Champs for his help in statistical analyses, and P. Trieu-Cuot and O. Poupel respectively, for kindly providing laboratory facilities and technical assistance for some of the RT-PCR analyses presented herein. We are grateful to M. Mourez for critical reading of the manuscript.
Funding Statement
SDR was supported by Sanofi-Pasteur. JV was a Marie-Curie Fellow. This work was supported by grants from the Institut Pasteur, CNRS (Centre National de la Recherche Scientifique), URA (Unité de echerche Associée) 2172, the Network of Excellence EuroPathoGenomics; LSHB-CT-2005-512061, and the French Government's Investissement d'Avenir program, Laboratoire d'Excellence “Integrative Biology of Emerging Infectious Diseases” (grant n°ANR-10-LABX-62-IBEID). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rolfe RD (1984) Interactions among microorganisms of the indigenous intestinal flora and their influence on the host. Rev Infect Dis 6 (Suppl 1) S73–79. [DOI] [PubMed] [Google Scholar]
- 2. Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H (2005) Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol 43: 3380–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Hara AM, Shanahan F (2006) The gut flora as a forgotten organ. EMBO Rep 7: 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Savage DC (1977) Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31: 107–133. [DOI] [PubMed] [Google Scholar]
- 5. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI (2005) Host-bacterial mutualism in the human intestine. Science 307: 1915–1920. [DOI] [PubMed] [Google Scholar]
- 6. Sonnenburg JL, Chen CT, Gordon JI (2006) Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol 4: e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, et al. (2007) The human microbiome project. Nature 449: 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leser TD, Molbak L (2009) Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol 11: 2194–2206. [DOI] [PubMed] [Google Scholar]
- 9. Kaper JB, Nataro JP, Mobley HL (2004) Pathogenic Escherichia coli. Nat Rev Microbiol 2: 123–140. [DOI] [PubMed] [Google Scholar]
- 10. Stecher B, Hardt W-D (2008) The role of microbiota in infectious disease. Trends Microbiol 16: 107–114. [DOI] [PubMed] [Google Scholar]
- 11. Vollaard EJ, Clasener HA (1994) Colonization resistance. Antimicrob Agents Chemother 38: 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reid G, Howard J, Gan BS (2001) Can bacterial interference prevent infection? Trends Microbiol 9: 424–428. [DOI] [PubMed] [Google Scholar]
- 13. Lievin-Le Moal V, Servin AL (2006) The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev 19: 315–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, et al. (2003) Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol 21: 785–789. [DOI] [PubMed] [Google Scholar]
- 15. Marteau P, Seksik P, Lepage P, Dore J (2004) Cellular and physiological effects of probiotics and prebiotics. Mini Rev Med Chem 4: 889–896. [DOI] [PubMed] [Google Scholar]
- 16. Bourlioux P, Koletzko B, Guarner F, Braesco V (2003) The intestine and its microflora are partners for the protection of the host: report on the Danone Symposium “The Intelligent Intestine,” held in Paris, June 14, 2002. Am J Clin Nutr 78: 675–683. [DOI] [PubMed] [Google Scholar]
- 17. Tait K, Sutherland IW (2002) Antagonistic interactions amongst bacteriocin-producing enteric bacteria in dual species biofilms. J Appl Microbiol 93: 345–352. [DOI] [PubMed] [Google Scholar]
- 18. Boles BR, Thoendel M, Singh PK (2004) Self-generated diversity produces “insurance effects” in biofilm communities. Proc Natl Acad Sci U S A 101: 16630–16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. An D, Danhorn T, Fuqua C, Parsek MR (2006) Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc Natl Acad Sci U S A 103: 3828–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Valle J, Da Re S, Henry N, Fontaine T, Balestrino D, et al. (2006) Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc Natl Acad Sci U S A 103: 12558–12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansen SK, Rainey PB, Haagensen JA, Molin S (2007) Evolution of species interactions in a biofilm community. Nature 445: 533–536. [DOI] [PubMed] [Google Scholar]
- 22. Valle J, Da Re S, Schmid S, Skurnik D, D'Ari R, et al. (2008) The amino acid valine is secreted in continuous-flow bacterial biofilms. J Bacteriol 190: 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramsey MM, Whiteley M (2009) Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci U S A 106: 1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hibbing ME, Fuqua C, Parsek MR, Peterson SB (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rendueles O, Ghigo JM (2012) Multi-species biofilms: how to avoid unfriendly neighbors. FEMS microbiology reviews 36(5): 972–989. [DOI] [PubMed] [Google Scholar]
- 26. Rendueles O, Travier L, Latour-Lambert P, Fontaine T, Magnus J, et al. (2011) Screening of Escherichia coli species biodiversity reveals new biofilm-associated antiadhesion polysaccharides. mBio 2: e00043–00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hall-Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108. [DOI] [PubMed] [Google Scholar]
- 28. Macfarlane S, Dillon JF (2007) Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol 102: 1187–1196. [DOI] [PubMed] [Google Scholar]
- 29.Conway T, Krogfelt KA, Cohen PS (2004) The Life of Commensal Escherichia coli in the Mammalian Intestine. In: Curtiss R III, Böck A, Ingraham JL, Kaper JB, Neidhardt FC, et al., editors. Escherichia coli and Salmonella Cellular and Molecular Biology, online edition. Washington, DC: ASM Press. Chapter 8.3.1.2.
- 30. Beloin C, Roux A, Ghigo JM (2008) Escherichia coli biofilms. Curr Top Microbiol Immunol 322: 249–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghigo JM (2001) Natural conjugative plasmids induce bacterial biofilm development. Nature 412: 442–445. [DOI] [PubMed] [Google Scholar]
- 32. Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, et al. (2009) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5: e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chaveroche MK, Ghigo JM, d'Enfert C (2000) A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res 28: E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Da Re S, Le Quere B, Ghigo JM, Beloin C (2007) Tight modulation of Escherichia coli bacterial biofilm formation through controlled expression of adhesion factors. Appl Environ Microbiol 73: 3391–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bernier C, Gounon P, Le Bouguenec C (2002) Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect Immun 70: 4302–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jenal U, Malone J (2006) Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40: 385–407. [DOI] [PubMed] [Google Scholar]
- 37. Vianney A, Jubelin G, Renault S, Dorel C, Lejeune P, et al. (2005) Escherichia coli tol and rcs genes participate in the complex network affecting curli synthesis. Microbiology 151: 2487–2497. [DOI] [PubMed] [Google Scholar]
- 38. Stout V, Torres-Cabassa A, Maurizi MR, Gutnick D, Gottesman S (1991) RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol 173: 1738–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lai EM, Nair U, Phadke ND, Maddock JR (2004) Proteomic screening and identification of differentially distributed membrane proteins in Escherichia coli. Molecular microbiology 52: 1029–1044. [DOI] [PubMed] [Google Scholar]
- 40. Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, et al. (2007) Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun 75: 3233–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Podschun R, Ullmann U (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clinical microbiology reviews 11: 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Balestrino D, Ghigo JM, Charbonnel N, Haagensen JA, Forestier C (2008) The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ Microbiol 10: 685–701. [DOI] [PubMed] [Google Scholar]
- 43. Domka J, Lee J, Wood TK (2006) YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol 72: 2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jayaraman A, Wood TK (2008) Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu Rev Biomed Eng 10: 145–167. [DOI] [PubMed] [Google Scholar]
- 45. Poulsen LK, Lan F, Kristensen CS, Hobolth P, Molin S, et al. (1994) Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infection and immunity 62: 5191–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Favre-Bonte S, Joly B, Forestier C (1999) Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infection and immunity 67: 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, et al. (2004) Carbon nutrition of Escherichia coli in the mouse intestine. Proceedings of the National Academy of Sciences of the United States of America 101: 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coudeyras S, Nakusi L, Charbonnel N, Forestier C (2008) A tripartite efflux pump involved in gastrointestinal colonization by Klebsiella pneumoniae confers a tolerance response to inorganic acid. Infection and immunity 76: 4633–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martinez-Jehanne V, du Merle L, Bernier-Febreau C, Usein C, Gassama-Sow A, et al. (2009) Role of deoxyribose catabolism in colonization of the murine intestine by pathogenic Escherichia coli strains. Infection and immunity 77: 1442–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Walters M, Sperandio V (2006) Quorum sensing in Escherichia coli and Salmonella. International Journal of Medical Microbiology 296: 125–131. [DOI] [PubMed] [Google Scholar]
- 51. Gibbs KA, Urbanowski ML, Greenberg EP (2008) Genetic determinants of self identity and social recognition in bacteria. Science 321: 256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Blango M, Mulvey M (2009) Bacterial landlines: contact-dependent signaling in bacterial populations. Current Opinion in Microbiology 12 (2) 177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Girard V, Cote J-P, Charbonneau M-E, Campos M, Berthiaume F, et al. (2010) Conformation change in a self-recognizing autotransporter modulate bacterial cell-cell interaction. J Biol Chem 285 (14) 10616–10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, et al. (2008) Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Molecular microbiology 70: 323–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell A (1996) Cryptic prophages. In: Curtiss R III, Böck A, Ingraham JL, Kaper JB, Neidhardt FC, et al.., editors. Escherichia coli and Salmonella Cellular and Molecular Biology, online edition. Washington, DC: ASM Press. pp. 2041–2046.
- 56. Casjens S (2003) Prophages and bacterial genomics: what have we learned so far? Molecular microbiology 49: 277–300. [DOI] [PubMed] [Google Scholar]
- 57. Skurnik M, Strauch E (2006) Phage therapy: facts and fiction. International journal of medical microbiology : IJMM 296: 5–14. [DOI] [PubMed] [Google Scholar]
- 58. Daw MA, Falkiner FR (1996) Bacteriocins: nature, function and structure. Micron 27: 467–479. [DOI] [PubMed] [Google Scholar]
- 59. García-Contreras R, Zhang X-S, Kim Y, Wood TK (2008) Protein Translation and Cell Death: The Role of Rare tRNAs in Biofilm Formation and in Activating Dormant Phage Killer Genes. PLoS ONE 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Casjens S (2003) Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49: 277–300. [DOI] [PubMed] [Google Scholar]
- 61. Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, et al. (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 183: 4562–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Polissi A, De Laurentis W, Zangrossi S, Briani F, Longhi V, et al. (2003) Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res Microbiol 154: 573–580. [DOI] [PubMed] [Google Scholar]
- 63. Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA (2006) Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev 20: 1776–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. White-Ziegler CA, Um S, Perez NM, Berns AL, Malhowski AJ, et al. (2008) Low temperature (23 degrees C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154: 148–166. [DOI] [PubMed] [Google Scholar]
- 65. Hengge R (2009) Principles of c-di-GMP signalling in bacteria. Nature reviews Microbiology 7: 263–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA-array data to in vivo test decision Flow-chart depicting the rational for selection of genes analyzed in the study.
(DOCX)
Estimate of biofilm biomass before inoculation with pathogen. Microfermentors were inoculated with commensal strain MG1655 F′ (C) or with indicated devivative mutants. After 6 h of growth, biofilm that developed on the glass slide was resuspended in 10 ml of minimal media and recovered bacterial count was estimated by serial dilution and cfu count. Results are average of at least 6 replicates ± standard deviation of the mean. Star indicates a mutant for which initial biofilm formation significantly differed from that of the wild type, P≤0.05.
(DOCX)
Genes over-expressed or repressed in response to colonization of MG1655 F′ biofilm by pathogenic 55989a.
(DOCX)
Genes induced upon self-colonization (C+C) of commensal biofilm.
(DOCX)
Genes repressed upon self-colonization (C+C) of commensal biofilm.
(DOCX)
Genes induced upon colonization by exogenous pathogen (C+P).
(DOCX)
Genes repressed upon colonization by exogenous pathogen (C+P).
(DOCX)
Primers used in this study.
(DOCX)