Abstract
Parkin, an E3 ubiquitin ligase well known for its role in the pathogenesis of juvenile Parkinson disease, has been considered as a candidate tumor suppressor in certain types of cancer. It remains unknown whether parkin is involved in the development of pancreatic cancer, the fourth leading cause of cancer-related deaths worldwide. Herein, we demonstrate the downregulation and copy number loss of the parkin gene in human pancreatic cancer specimens. The expression of parkin negatively correlates with clinicopathological parameters indicating the malignancy of pancreatic cancer. In addition, knockdown of parkin expression promotes the proliferation and tumorigenic properties of pancreatic cancer cells both in vitro and in mice. We further find that parkin deficiency increases the proportion of cells with spindle multipolarity and multinucleation. Parkin-depleted cells also show a significant increase in spindle misorientation. These findings indicate crucial involvement of parkin deficiency in the pathogenesis of pancreatic cancer.
Keywords: parkin, pancreatic cancer, cell proliferation, spindle multipolarity, spindle misorientation
Introduction
Parkin is a multifaceted E3 ubiquitin ligase linked to the development of early-onset Parkinson disease.1,2 The parkin gene is located at chromosome 6q25.2-27 within FRA6E, one of the most common fragile sites in the human population.3 At the cellular level, parkin participates in a variety of cellular activities, primarily through its ubiquitination of target proteins, resulting in proteasome-mediated degradation.4 Parkin also plays an important role in the biogenesis of mitochondria and is required for the repair of mitochondrial DNA and the autophagy of damaged mitochondria.5-7 In addition, parkin has been implicated in the binding and stabilization of the microtubule cytoskeleton.8 In addition to the involvement in Parkinson disease, the loss of parkin function contributes to the development of a wide spectrum of common cancers, such as ovarian, breast, lung, liver and colorectal cancers, glioblastoma and leukemia.9-17
Pancreatic cancer is the fourth most common cause of cancer-related mortality in the world.18 Over the last two decades, our understanding of the molecular mechanisms of pancreatic cancer has been improved, and it is now well recognized that pancreatic cancer is fundamentally a genetic disease caused by the alteration of genes, especially oncogenes and tumor suppressor genes.19-21 In addition, it has been revealed that genes such as k-ras, p16ink4a/cdkn2a, tp53 and smad4/dpc4 are altered in pancreatic cancer, accompanied by a substantial compendium of genomic and transcriptomic alterations that facilitate cell cycle deregulation, cell survival, invasion and metastasis.19-21 Despite the above progress, advances in the diagnosis, therapeutic intervention and survival benefit of pancreatic cancer are still poor. Therefore, there is an urgent demand to have a better understanding of the molecular mechanisms that underlie the pathogenesis of this aggressive malignancy. Recently, two overlapping out-of-frame deletions of exon 6 of the parkin gene have been identified in two patients with metastatic pancreatic cancer,20 prompting us to investigate its potential involvement in the development of pancreatic cancer.
Results
Parkin expression is downregulated in human pancreatic cancer specimens
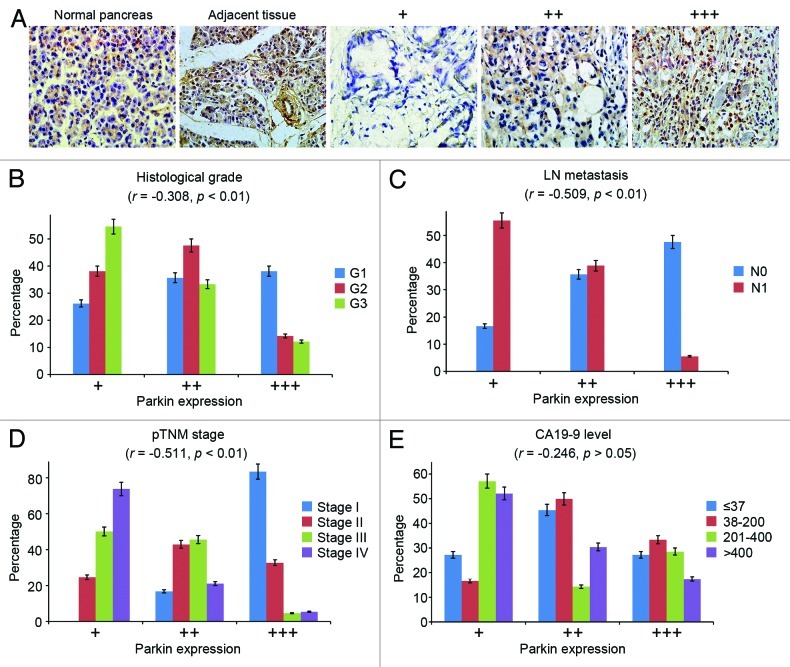
To study the potential role of parkin in pancreatic tumorigenesis, we first examined by immunohistochemistry its expression in human pancreatic adenocarcinomas, tissues adjacent to adenocarcinomas and normal pancreas tissues obtained from patients who underwent distal pancreatectomy for diseases other than pancreatic cancer. We observed high expression of parkin in normal pancreas and tissues adjacent to adenocarcinomas. In contrast, the majority of tumor samples exhibited low or medium levels of parkin expression; out of the 96 samples examined, only 23 samples showed high parkin expression (Fig. 1A).
Figure 1. Parkin expression in human pancreatic adenocarcinoma. (A) Representative images of parkin expression in normal pancreas, pancreatic adenocarcinomas and tissue adjacent to pancreatic adenocarcinoma. For pancreatic adenocarcinoma samples, parkin expression was scored as low (+), medium (++) and high (+++) levels based on immunohistochemical staining intensity. Correlations between parkin expression and clinicopathological parameters indicating the malignancy of pancreatic cancer were analyzed, including the histological grade (B), lymph node (LN) metastasis (C), pathological tumor node metastasis (pTNM) stage (D) and CA19-9 level (E). Correlation coefficient (r) and p values were calculated by Spearman’s rank correlation test. N0 and N1 refer to the absence or presence of lymph node metastasis.
To further investigate the involvement of parkin in pancreatic cancer, we analyzed the correlation between parkin expression and several clinicopathological parameters, indicating the malignancy of pancreatic cancer. We observed a significant negative correlation between parkin expression and the histological grade of pancreatic cancer, with a correlation coefficient (r) of −0.308 (p < 0.01) (Fig. 1B). In addition, parkin expression negatively correlated with the incidence of lymph node metastasis (r = −0.509, p < 0.01) and the pathological tumor node metastasis stage (r = −0.511, p < 0.01) (Fig. 1C and D). There was no significant correlation between parkin expression and the level of CA19-9 (r = −0.246, p > 0.05) (Fig. 1E), the standard serum marker of pancreatic cancer.22 Collectively, these results demonstrate the downregulation of parkin expression in pancreatic cancer specimens.
Gene copy number loss contributes to parkin deficiency in pancreatic cancer
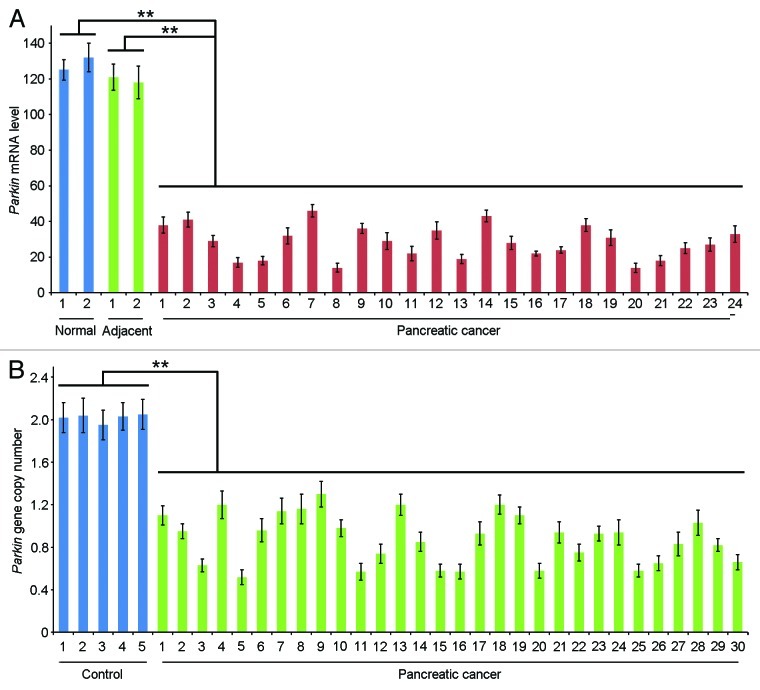
To gain mechanistic insight into the downregulation of parkin expression in pancreatic cancer, we measured the level of parkin mRNA by quantitative real-time RT-PCR. We found a reduction of parkin mRNA expression in all the 24 pancreatic adenocarcinoma samples examined, relative to the normal pancreas or tissues adjacent to pancreatic adenocarcinomas (Fig. 2A). The level of parkin mRNA in pancreatic adenocarcinomas was 4.4-fold lower on average than in normal pancreas or tissues adjacent to pancreatic adenocarcinomas (Fig. 2A).
Figure 2. Examination of parkin mRNA level and gene copy number. (A) Quantitative real-time RT-PCR detection of parkin mRNA expression in normal pancreas, pancreatic adenocarcinomas and tissues adjacent to pancreatic adenocarcinomas. (B) Analysis of parkin gene copy number with quantitative real-time PCR. DNA isolated from blood leukocytes of healthy individuals was utilized as a control. ** p < 0.01.
Deletions of the parkin gene have been revealed in several types of cancer9-14 and have recently been reported in metastatic pancreatic cancer.20 Given the frequent gene copy number loss in pancreatic cancer,23,24 we sought to analyze whether pancreatic cancer has alterations in parkin gene copy number. Quantitative real-time PCR was performed using DNA isolated from pancreatic adenocarcinomas, with DNA from blood leukocytes of healthy individuals as a control. In the control group, parkin gene copy number showed the standard diploid type (Fig. 2B). In contrast, all the 30 adenocarcinoma samples examined exhibited alterations of parkin gene copy number, and both heterozygous and homozygous loss of parkin was found in the tumor samples (Fig. 2B). These results indicate that gene copy number loss contributes significantly to parkin deficiency in pancreatic cancer.
Depletion of parkin stimulates pancreatic cancer cell proliferation and tumorigenic properties
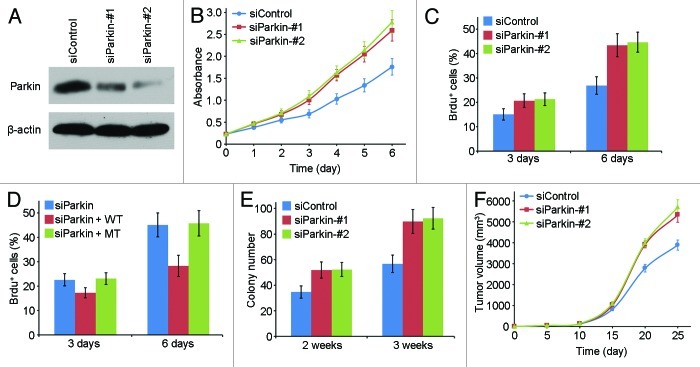
We then examined the effect of parkin deficiency on the proliferation of pancreatic cancer cells. We used two specific siRNAs targeting parkin, and both of them could efficiently reduce parkin expression in EPP85 cells (Fig. 3A). By sulforhodamine B staining assay, we found that transfection of parkin siRNAs resulted in a substantial increase in the rate of cell proliferation (Fig. 3B). BrdU incorporation assay, which reflects the index of DNA synthesis, further confirmed that transfection of parkin siRNAs could promote pancreatic cancer cell proliferation (Fig. 3C). To exclude the off-target effects of the parkin siRNAs, we checked whether ectopic expression of parkin could rescue the effect of the parkin siRNAs on cell proliferation. As shown in Figure 3D, wild type parkin, but not the ubiquitin ligase-dead mutant, rescued the stimulating effect of the parkin siRNA on cell proliferation, indicating that the function of parkin in the regulation of pancreatic cancer cell proliferation is tightly associated with its ubiquitin ligase activity.
Figure 3. Depletion of parkin stimulates pancreatic cancer cell proliferation and tumorigenic properties. (A) Western blot analysis of parkin and actin expression in cells transfected with control and parkin siRNAs. (B) Cells were transfected with control and parkin siRNAs, and cell proliferation was examined by sulphorhodamine B staining. (C) Cells were transfected with control and parkin siRNAs, and the percentage of BrdU-positive cells was examined. (D) Cells were transfected with parkin siRNA and wild type (WT) or ubiquitin ligase-dead mutant (MT) parkin, and the percentage of BrdU-positive cells was then examined. (E) Cells transfected with control or parkin siRNAs were cultured in agarose for 2 or 3 wk, and the number of colonies formed was counted. (F) Cells transfected with control or parkin siRNAs were injected subcutaneously into nude mice, and the tumor volume was measured at the indicated time points.
Subsequently, we investigated whether parkin is involved in the tumorigenic properties of pancreatic cancer cells. We found that depletion of parkin significantly increased the ability of EPP85 cells to form colonies in soft agar (Fig. 3E), suggesting that parkin could suppress anchorage-independent cell growth. We next investigated the role of parkin in pancreatic tumor growth in vivo. EPP85 cells transfected with control or parkin siRNAs were injected subcutaneously into athymic nude mice, and the tumor volume was then measured. There was no significant difference in the tumor volume between the parkin siRNA groups and the control siRNA group by 15 d post-injection (Fig. 3F). However, the tumor volume in the parkin siRNA groups was significantly larger than that in the control siRNA group after this time point (Fig. 3F). Taken together, these findings reveal that depletion of parkin could stimulate the proliferation and tumorigenic properties of pancreatic cancer cells.
Knockdown of parkin expression increases the frequency of spindle multipolarity and multinucleation
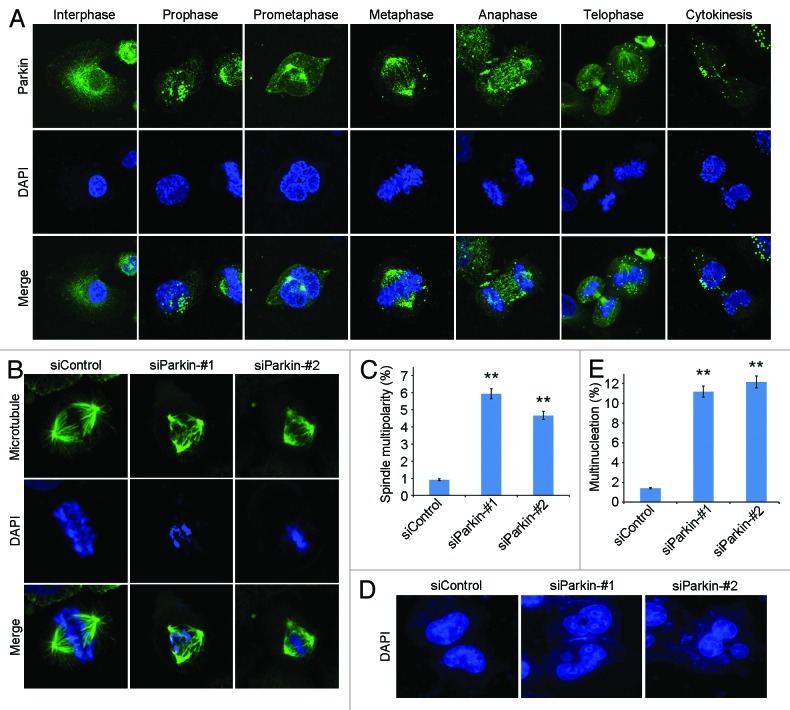
To understand the molecular mechanisms underlying the role of parkin in pancreatic cancer, we analyzed its subcellular localization during the cell cycle. Consistent with previous findings,25,26 we found that in interephase cells parkin had punctate distribution in the cytosol and prominent accumulation in the perinuclear region (Fig. 4A). In mitotic cells, the majority of parkin was localized to the spindle poles and was also observed at the central spindle during anaphase and at the midbody during telophase and cytokinesis (Fig. 4A).
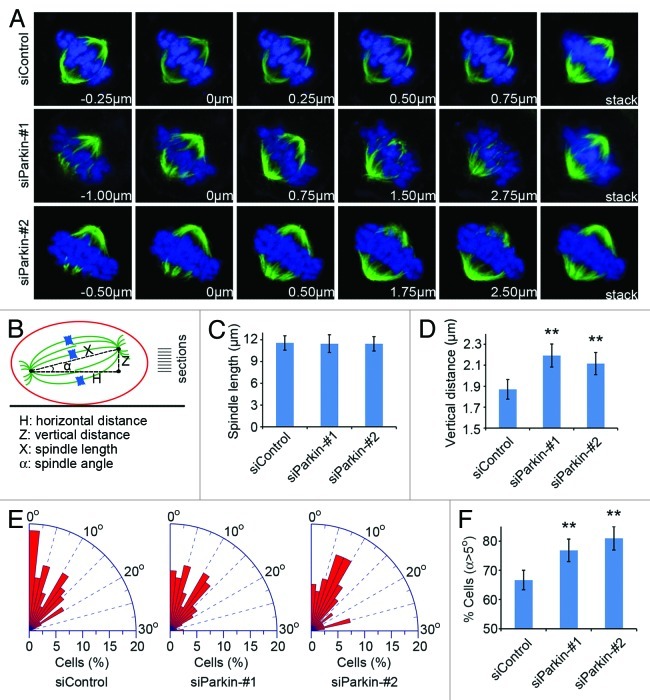
Figure 4. Knockdown of parkin expression increases the frequency of spindle multipolarity and multinucleation. (A) Immunofluorescence microscopic analysis of the subcellular localization of parkin during the cell cycle. Cells were stained with anti-parkin antibody and the DNA dye DAPI. (B) Transfection of parkin siRNAs leads to the formation of multipolar spindles. Cells transfected with control or parkin siRNAs were stained with anti-α-tubulin antibody and DAPI. (C) Experiments were performed as in (B), and the percentage of mitotic cells with multipolar spindles was calculated. (D) Parkin siRNAs result in multinucleation. Cells transfected with control or parkin siRNAs were stained with DAPI. (E) Experiments were performed as in (D), and the percentage of multinucleated cells was determined. ** p < 0.01 vs. control.
We then examined spindle morphology in parkin siRNA-transfected cells to study its potential role in mitosis. By immunofluorescence microscopy, we found that transfection of parkin siRNAs resulted in a significant increase in the percentage of cells with multipolar spindles, whereas control mitotic cells showed normal bipolar spindles (Fig. 4B and C). In addition, knockdown of parkin expression dramatically increased the proportion of multinucleated cells (Fig. 4D and E), indicating cytokinesis defects.
Parkin deficiency leads to spindle misorientation
To further study the effect of parkin on mitosis, we analyzed the Z-stage stacks of the mitotic spindle by confocal microscopy and measured various parameters of the mitotic spindle (Fig. 5A and B). We found that knockdown of parkin expression did not obviously affect the spindle length (Fig. 5C). However, the vertical distance between the two spindle poles was significantly increased in cells transfected with parkin siRNAs; the vertical distance was 1.87, 2.18 and 2.09 µm, respectively in cells transfected with control and the two parkin siRNAs (Fig. 5A and D). We then calculated the angle between the spindle axis and the substratum plane with the inverse trigonometric function. As shown in Figure 5E and F, depletion of parkin led to a remarkable increase in the percentage of mitotic cells with spindle angles larger than 5°, indicating spindle misorientation.
Figure 5. Parkin deficiency leads to spindle misorientation. (A) Cells were transfected with control or parkin siRNAs and stained with anti-α-tubulin antibody (green) and DAPI (blue) and the image series of mitotic cells were shown. The position of the Z stage of the mitotic spindle is indicated in µm, and stack refers to the projected image. (B) Scheme describing the method used for analysis of various parameters of the mitotic spindle. The vertical distance (Z) and the horizontal distance (H) between the two spindle poles were measured directly with the LASAF software. The corrected spindle length (X) was calculated based on the pythagorean theorem. The angle (α) between the spindle axis and the substratum plane was determined with the inverse trigonometric function. (C–E) Experiments were performed as in (A), and the spindle length, the vertical distance between the two spindle poles and the spindle angle were measured as described in (B). (F) Quantification of the percentage of mitotic cells with spindle angles over than 5°. ** p < 0.01 vs. control.
Eg5 is involved in the role of parkin in pancreatic cancer development
The defects in spindle polarity and orientation in parkin siRNA-transfected cells suggested that parkin might function in pancreatic tumorigenesis through its regulation of protein(s) critical for spindle properties. A potential target is the microtubule-dependent mitotic kinesin Eg5, a key regulator of spindle assembly;27 moreover, parkin has been shown to negatively regulate Eg5 expression, and ectopic expression of Eg5 causes spindle multipolarity and is associated with pancreatic tumorigenesis.28-30
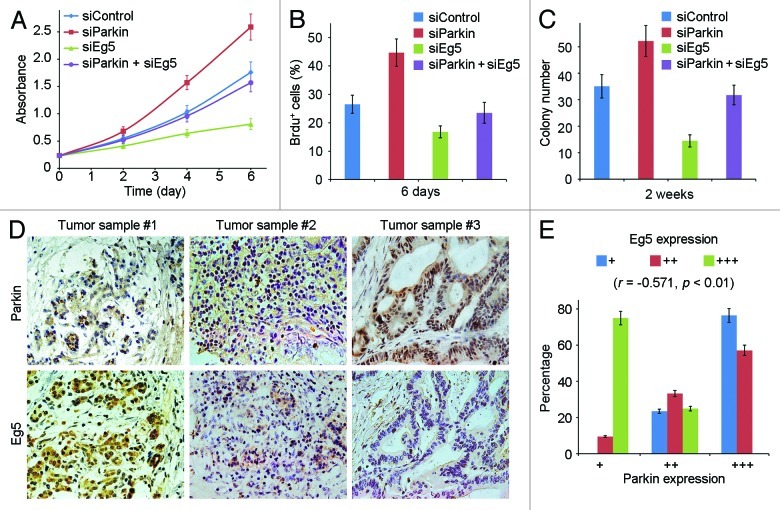
We thus investigated whether Eg5 contributes to the function of parkin in pancreatic cancer development. As shown in Figure 6A–C, transfection of Eg5 siRNA significantly compromised the stimulating effect of the parkin siRNA on the proliferation and tumorigenic properties of pancreatic cancer cells. To evaluate the clinical relevance of the parkin-Eg5 axis in pancreatic cancer, we examined by immunohistochemistry the expression of parkin and Eg5 in serial sections of 48 pancreatic adenocarcinoma samples (Fig. 6D). We found a significant negative correlation between the levels of parkin and Eg5 in the tumor samples (r = −0.571, p < 0.01). These findings suggest that parkin may function in pancreatic tumorigenesis through negative regulation of Eg5.
Figure 6. Eg5 is involved in the role of parkin in pancreatic cancer development. (A) Cells were transfected with the indicated siRNAs, and cell proliferation was examined by sulphorhodamine B staining. (B) Cells were transfected with the indicated siRNAs, and the percentage of BrdU-positive cells was examined. (C) Cells transfected with the indicated siRNAs were cultured in agarose for 2 wk, and the number of colonies formed was counted. (D) Immunohistochemical staining of parkin and Eg5 on serial sections of pancreatic adenocarcinomas. (E) Experiments were performed as in (D), and the expression of parkin and Eg5 was scored as low (+), medium (++) and high (+++) levels. Correlation coefficient (r) and p values were calculated by Spearman’s rank correlation test.
Discussion
Pancreatic cancer is a notoriously aggressive disease with a grim prognosis, due to its late diagnosis, propensity to rapidly metastasize and therapeutic resistance.23,24 The underlying molecular events remain poorly defined. In this study, we provide several lines of evidence indicating a critical involvement of parkin deficiency in the pathogenesis of pancreatic cancer: (1) Human pancreatic cancer specimens exhibit parkin gene downregulation and copy number loss; (2) Parkin expression level negatively correlates with clinicopathological parameters, indicating the malignancy of pancreatic cancer; (3) Silencing of parkin expression promotes pancreatic cancer cell proliferation and tumorigenic properties. These findings, in line with previous reports showing parkin gene mutations or deletions in other cancer types,9-17 substantiate the role of parkin as a tumor suppressor in a variety of common cancers, in addition to its role in the pathogenesis of Parkinson disease.
How does parkin deficiency lead to the development of pancreatic cancer? It has previously been shown that parkin could accumulate in the centrosome in response to misfolded proteins.26 In addition, parkin has been implicated in the spindle assembly checkpoint, although the molecular details remain to be uncovered.25 In the present study, our data reveal that in mitosis, parkin is mainly localized at the spindle poles, with modest distribution at the central spindle and the midbody. Consistent with the localization pattern of parkin, silencing of its expression results in a remarkable increase in the percentage of cells with multipolar spindles and also increases the frequency of multinucleation. Spindle multipolarity is known to contribute to chromosomal instability, allowing cells to overcome critical barriers to deregulated growth and thereby leading to malignant transformation.31-33 It is, therefore, tempting to hypothesize that parkin deficiency may promote pancreatic cancer development by inducing spindle multipolarity and subsequent chromosomal instability.
Another intriguing phenotype observed in parkin-knockdown cells is the increased occurrence of spindle misorientation. Spindle orientation is known to be tightly associated with epithelial (stem) cell fate determination and tissue architecture and morphogenesis.34,35 Several important tumor suppressor proteins, including adenomatous polyposis coli, von Hippel-Lindau and E-cadherin, have recently been shown to participate in the regulation of spindle orientation, establishing a connection between spindle misorientation and cancer.36-38 Deficiency in parkin expression may contribute to pancreatic cancer development by the induction of spindle misorientation, simulating pancreatic tissue disorganization and hypertrophy and/or expanding the pancreatic cancer stem cell pool. Additional studies are warranted, however, to demonstrate whether spindle misorientation induced by the deficiency of parkin or other proteins is a causal event in tumorigenesis.
The functions of parkin in cells are performed mainly through its E3 ubiquitin ligase activity. In addition to the polyubiquitination of proteins leading to their degradation through the proteasome pathway,4 parkin can catalyze the monoubiquitination of certain proteins and thereby regulate protein-protein interactions and signal transduction events.39-41 It has been shown previously that through multiple monoubiquitination of Hsp70, parkin inactivates c-Jun NH2-terminal kinase, resulting in decreased phosphorylation of c-Jun; as a result, Eg5 gene transcription is repressed due to decreased c-Jun binding to the activator protein 1 site present in the Eg5 promoter.28 In agreement with this finding, the data presented in our study show that the stimulating effect of parkin deficiency on pancreatic cancer cell proliferation and tumorigenic properties is compromised by the suppression of Eg5. Moreover, the levels of parkin and Eg5 negatively correlate in pancreatic cancer specimens. Our findings are also consistent with the observation that overexpression of Eg5 causes spindle multipolarity.29,3029,30 Plus that elevated activity or expression of Eg5 has been implicated in several types of cancer, including pancreatic cancer;29,30,42 our data suggest that parkin deficiency may contribute to pancreatic tumorigenesis through the deregulation of Eg5 expression and subsequent defects in spindle behavior. However, given that parkin has numerous substrates and interacting partners, it would not be surprising if an Eg5-independent mechanism were identified in the future.
Materials and Methods
Materials
Sulforhodamine B, 4’-6-diamidino-2-phenylindole (DAPI), bromodeoxyuridine (BrdU) and anti-β-actin antibody were purchased from Sigma-Aldrich. Antibodies against parkin, α-tubulin, BrdU and Eg5 were from Abcam. Horseradish peroxidase-conjugated secondary antibodies were from Amersham Biosciences, and fluorescein-conjugated secondary antibody was from Jackson ImmunoResearch Laboratories. Small-interfering RNAs (siRNAs) against parkin, Eg5 and luciferase control were synthesized by Dharmacon and transfected into cells with the lipofectamine 2000 reagent (Invitrogen), and plasmids were transfected with the polyethyleneimine reagent (Sigma-Aldrich).
Cells and tumor samples
EPP85 human pancreatic cancer cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum at 37°C in a humidified atmosphere with 5% CO2. Pancreatic tumor samples were obtained from patients undergoing surgical resection at Tianjin Cancer Hospital. Normal pancreatic tissues were obtained from patients undergoing distal pancreatectomy for diseases other than pancreatic cancer. Use of human samples in this study was approved by the Ethics Committee of Tianjin Cancer Hospital.
Western blot analysis
Proteins were resolved by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were blocked in Tris-buffered saline containing 0.1% Tween 20 and 5% fat-free dry milk and incubated first with primary antibodies and then with horseradish peroxidase-conjugated secondary antibodies as described previously.43 Specific proteins were visualized with the enhanced chemiluminescence detection reagent (Pierce Biotechnology).
Sulforhodamine B staining
Cells were seeded at 5 × 104 cells per well in 24-well tissue culture plates. After several days, cells were fixed with 50% trichloroacetic acid and stained with 0.4% sulforhodamine B dissolved in 1% acetic acid. Cells were then washed with 1% acetic acid to remove unbound dye. The protein-bound dye was extracted with 10 mM Tris base to determine the optical density at 490-nm wavelength.
BrdU incorporation assay
Cells grown on glass coverslips were incubated with 10 μM BrdU for 45 min and fixed with 70% ethanol. Cellular DNA was denatured with HCl. Cells were then stained with anti-BrdU antibody and rhodamine-conjugated secondary antibody. The percentage of BrdU-positive cells was examined by fluorescence microscopy.
Immunohistochemistry
Paraffin-embedded tissue sections were cut, deparaffinized and rehydrated with xylene and graded alcohols. Antigen retrieval was performed in 5 mM citrate buffer. After inactivation of endogenous peroxidase with 3% H2O2, the sections were blocked with goat serum and incubated with primary antibody. The sections were then incubated with biotinylated secondary antibody and streptavidin-biotin-peroxidase, and diaminobenzidine was used as a chromogen substrate. The sections were counterstained with hematoxylin. Protein expression was graded based on the intensity of staining and the percentage of stained cells as described previously.44
Immunofluorescence microscopy
Cells grown on glass coverslips were fixed with cold methanol for 5 min. Cells were blocked with bovine serum albumin and incubated in succession with primary and secondary antibodies, followed by staining with DAPI as described.45 Coverslips were mounted with 90% glycerol and examined with a Zeiss fluorescence microscope or Leica TCS SP5 confocal microscope.
Quantitative real-time RT-PCR and PCR
Total RNA was isolated using the TRIzol reagent (Invitrogen), and DNA was isolated using the Extract-N-Amp Tissue PCR kit (Sigma-Aldrich). Quantitative real-time PCR was performed using the SYBR Premix Ex Taq reagent (Takara) according to the manufacturer’s instruction.
Colony formation assays
Cells were cultured in media containing 0.3% low melting point agarose (Invitrogen) for 2 or 3 wk. Visible colonies with greater than 50 cells were then counted.
Mouse experiments
Cells (5 × 105) were injected subcutaneously into the right flanks of female athymic nude mice (eight mice per group). Tumor volume was measured every 5 d with a vernier caliper and calculated with the following formula: V = π/6 × length × width.2 The mice were sacrificed 25 d post injection. Tumors were then isolated from mice, photographed and weighed.
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (2012CB945002 and 2010CB912204), the National Natural Science Foundation of China (31130015 and 31170820) and the 111 project of the Ministry of Education of China (B08011).
Glossary
Abbreviations:
- DAPI
4’,6-diamidino-2-phenylindole
- BrdU
bromodeoxyuridine
- siRNA
small interfering RNA
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24215
References
- 1.Shimura H, Hattori N, Kubo Si, Mizuno Y, Asakawa S, Minoshima S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–5. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 2.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 3.Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012;28:22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka K, Suzuki T, Hattori N, Mizuno Y. Ubiquitin, proteasome and parkin. Biochim Biophys Acta. 2004;1695:235–47. doi: 10.1016/j.bbamcr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 5.Kuroda Y, Mitsui T, Kunishige M, Shono M, Akaike M, Azuma H, et al. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum Mol Genet. 2006;15:883–95. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- 6.Rothfuss O, Fischer H, Hasegawa T, Maisel M, Leitner P, Miesel F, et al. Parkin protects mitochondrial genome integrity and supports mitochondrial DNA repair. Hum Mol Genet. 2009;18:3832–50. doi: 10.1093/hmg/ddp327. [DOI] [PubMed] [Google Scholar]
- 7.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 8.Yang F, Jiang Q, Zhao J, Ren Y, Sutton MD, Feng J. Parkin stabilizes microtubules through strong binding mediated by three independent domains. J Biol Chem. 2005;280:17154–62. doi: 10.1074/jbc.M500843200. [DOI] [PubMed] [Google Scholar]
- 9.Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc Natl Acad Sci USA. 2003;100:5956–61. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denison SR, Wang F, Becker NA, Schüle B, Kock N, Phillips LA, et al. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 2003;22:8370–8. doi: 10.1038/sj.onc.1207072. [DOI] [PubMed] [Google Scholar]
- 11.Wang F, Denison S, Lai JP, Philips LA, Montoya D, Kock N, et al. Parkin gene alterations in hepatocellular carcinoma. Genes Chromosomes Cancer. 2004;40:85–96. doi: 10.1002/gcc.20020. [DOI] [PubMed] [Google Scholar]
- 12.Picchio MC, Martin ES, Cesari R, Calin GA, Yendamuri S, Kuroki T, et al. Alterations of the tumor suppressor gene Parkin in non-small cell lung cancer. Clin Cancer Res. 2004;10:2720–4. doi: 10.1158/1078-0432.CCR-03-0086. [DOI] [PubMed] [Google Scholar]
- 13.Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci USA. 2010;107:15145–50. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, et al. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Letessier A, Garrido-Urbani S, Ginestier C, Fournier G, Esterni B, Monville F, et al. Correlated break at PARK2/FRA6E and loss of AF-6/Afadin protein expression are associated with poor outcome in breast cancer. Oncogene. 2007;26:298–307. doi: 10.1038/sj.onc.1209772. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara M, Marusawa H, Wang HQ, Iwai A, Ikeuchi K, Imai Y, et al. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27:6002–11. doi: 10.1038/onc.2008.199. [DOI] [PubMed] [Google Scholar]
- 17.Agirre X, Román-Gómez J, Vázquez I, Jiménez-Velasco A, Garate L, Montiel-Duarte C, et al. Abnormal methylation of the common PARK2 and PACRG promoter is associated with downregulation of gene expression in acute lymphoblastic leukemia and chronic myeloid leukemia. Int J Cancer. 2006;118:1945–53. doi: 10.1002/ijc.21584. [DOI] [PubMed] [Google Scholar]
- 18.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 19.Koorstra JB, Hustinx SR, Offerhaus GJ, Maitra A. Pancreatic carcinogenesis. Pancreatology. 2008;8:110–25. doi: 10.1159/000123838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iacobuzio-Donahue CA. Genetic evolution of pancreatic cancer: lessons learnt from the pancreatic cancer genome sequencing project. Gut. 2012;61:1085–94. doi: 10.1136/gut.2010.236026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. 2007;33:266–70. doi: 10.1016/j.ejso.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Ghaneh P, Costello E, Neoptolemos JP. Biology and management of pancreatic cancer. Gut. 2007;56:1134–52. doi: 10.1136/gut.2006.103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–49. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Fang ST, Yeh PC, Yang HH, Chen SY, Chang CJ, et al. The C-terminus of PARK2 is required for its self-interaction, solubility and role in the spindle assembly checkpoint. Biochim Biophys Acta. 2012;1822:573–80. doi: 10.1016/j.bbadis.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, Ren Y, Jiang Q, Feng J. Parkin is recruited to the centrosome in response to inhibition of proteasomes. J Cell Sci. 2003;116:4011–9. doi: 10.1242/jcs.00700. [DOI] [PubMed] [Google Scholar]
- 27.Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–8. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 28.Liu M, Aneja R, Sun X, Xie S, Wang H, Wu X, et al. Parkin regulates Eg5 expression by Hsp70 ubiquitination-dependent inactivation of c-Jun NH2-terminal kinase. J Biol Chem. 2008;283:35783–8. doi: 10.1074/jbc.M806860200. [DOI] [PubMed] [Google Scholar]
- 29.Liu M, Wang X, Yang Y, Li D, Ren H, Zhu Q, et al. Ectopic expression of the microtubule-dependent motor protein Eg5 promotes pancreatic tumourigenesis. J Pathol. 2010;221:221–8. doi: 10.1002/path.2706. [DOI] [PubMed] [Google Scholar]
- 30.Castillo A, Morse HC, 3rd, Godfrey VL, Naeem R, Justice MJ. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res. 2007;67:10138–47. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- 31.Saunders W. Centrosomal amplification and spindle multipolarity in cancer cells. Semin Cancer Biol. 2005;15:25–32. doi: 10.1016/j.semcancer.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–87. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morin X, Bellaïche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21:102–19. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Pease JC, Tirnauer JS. Mitotic spindle misorientation in cancer--out of alignment and into the fire. J Cell Sci. 2011;124:1007–16. doi: 10.1242/jcs.081406. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–50. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- 37.den Elzen N, Buttery CV, Maddugoda MP, Ren G, Yap AS. Cadherin adhesion receptors orient the mitotic spindle during symmetric cell division in mammalian epithelia. Mol Biol Cell. 2009;20:3740–50. doi: 10.1091/mbc.E09-01-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thoma CR, Toso A, Gutbrodt KL, Reggi SP, Frew IJ, Schraml P, et al. VHL loss causes spindle misorientation and chromosome instability. Nat Cell Biol. 2009;11:994–1001. doi: 10.1038/ncb1912. [DOI] [PubMed] [Google Scholar]
- 39.Joch M, Ase AR, Chen CX, MacDonald PA, Kontogiannea M, Corera AT, et al. Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol Biol Cell. 2007;18:3105–18. doi: 10.1091/mbc.E05-11-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore DJ, West AB, Dikeman DA, Dawson VL, Dawson TM. Parkin mediates the degradation-independent ubiquitination of Hsp70. J Neurochem. 2008;105:1806–19. doi: 10.1111/j.1471-4159.2008.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chew KC, Matsuda N, Saisho K, Lim GG, Chai C, Tan HM, et al. Parkin mediates apparent E2-independent monoubiquitination in vitro and contains an intrinsic activity that catalyzes polyubiquitination. PLoS ONE. 2011;6:e19720. doi: 10.1371/journal.pone.0019720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen GM, Justice MJ. Activation of Hex and mEg5 by retroviral insertion may contribute to mouse B-cell leukemia. Oncogene. 1999;18:6531–9. doi: 10.1038/sj.onc.1203023. [DOI] [PubMed] [Google Scholar]
- 43.Sun X, Shi X, Liu M, Li D, Zhang L, Liu X, et al. Mdp3 is a novel microtubule-binding protein that regulates microtubule assembly and stability. Cell Cycle. 2011;10:3929–37. doi: 10.4161/cc.10.22.18106. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Liu B, Zhang C, Peng G, Liu M, Li D, et al. Parkin regulates paclitaxel sensitivity in breast cancer via a microtubule-dependent mechanism. J Pathol. 2009;218:76–85. doi: 10.1002/path.2512. [DOI] [PubMed] [Google Scholar]
- 45.Shi X, Liu M, Li D, Wang J, Aneja R, Zhou J. Cep70 contributes to angiogenesis by modulating microtubule rearrangement and stimulating cell polarization and migration. Cell Cycle. 2012;11:1554–63. doi: 10.4161/cc.19954. [DOI] [PubMed] [Google Scholar]