Abstract
H4K20me1 is a critical histone lysine methyl modification in eukaryotes. It is recognized and “read” by various histone lysine methyl modification binding proteins. In this study, the function of MBTD1, a member of the Polycomb protein family containing four MBT domains, was comprehensively studied in mouse oocyte meiotic maturation. The results showed that depletion of MBTD1 caused reduced expression of histone lysine methyl transferase Pr-Set7 and H4K20me1 as well as increased oocyte arrest at the GV stage. Increased γH2AX foci were formed, and DNA damage repair checkpoint protein 53BP1 was downregulated. Furthermore, depletion of MBTD1 activated the cell cycle checkpoint protein Chk1 and downregulated the expression of cyclin B1 and cdc2. MBTD1 knockdown also affected chromosome configuration in GV stage oocytes and chromosome alignment at the MII stage. All these phenotypes were reproduced when the H4K20 methyl transferase Pr-Set7 was depleted. Co-IP demonstrated that MBTD1 was correlated with Pr-Set7 in mouse oocytes. Our results demonstrate that MBTD1 is associated with Pr-Set7 to stabilize H4K20me1 in mouse oocyte meiotic maturation.
Keywords: MBTD1, Pr-Set7, H4K20me1, oocyte, meiotic maturation
Introduction
Histone lysine methylation is a central epigenetic modification in eukaryotic chromosomes. There are five major positions for lysine methylation in the histone N termini, which are H3K4, H3K9, H3K27, H3K36 and H4K20. Each of these modifications has distinct regulatory functions to influence cellular behavior. The transcriptional repression mark H4K20 methylation is evolutionarily conserved from Schizosac charomyces pombe to man and is involved in constitutive heterochromosome formation, gene repression, X inactivation, and in DNA damage repair, mitotic chromosome condensation and gene regulation.1-3 The regulation of methylated H4K20 is complex due to the fact that lysine methylation can be present in three distinct states (mono, di or tri), which may have different biological readouts depending on the association with specific binding partners. H4K20me1 is exclusively induced by the Pr-Set7/KMT5A histone methyl transferase (HMTase), where it has been linked with transcriptional repression and X inactivation.4,5 More recently, genome-wide profiling of H4K20me1 also revealed enrichment of this mark across actively transcribed genes. H4K20me1 is highly dynamic throughout the cell cycle and becomes highly enriched during the S phase and peaks at the G2/M phase. Abrogation of H4K20me1 in mitosis results in severe chromosomal segregation defects, and H4K20me1 has been proposed to be an important mark for chromosomal memory.6 Studies in Drosophila melanogaster S2 cell lines have revealed that the depletion of Pr-Set7, which resulted in the decrease of H4K20me1 leads to chromosome condensation and DNA damage repair defects.7,8 Furthermore, the Pr-Set7 knockout could cause the failure to accumulate mitotic cyclin B by anaphase-promoting complex/cyclosome(APC/C)-dependent proteolysis and to activate Chk1. It has been well established that accumulation of mitotic cyclins and the activation of the Cdk-cyclin complexes are essential for entry into mitosis and formation of mitotic spindles, therefore aberrant expression of Pr-Set7 may cause abnormal mitosis.9,10 Taken together, H4K20me1 is essential for normal cellular progression.
Non-histone proteins which bind to the histone modification sites are also core factors that affect cellular progression. Malignant brain tumor (MBT) domain-containing protein, which is a member of methyl-lysine binding proteins in “Royal family” made up of Tudor, Agenet, chromo, PWWP, MBT, the WD40 repeat protein and the plant homeo domain (PHD), was first discovered in Drosophila melanogaster and has received increased attention in recent years.11-14 MBT domain constitutes a separate class of histone methyl-lysine reading modules that do not appear to have a high selectivity for their target sites. In vitro, diverse MBT domain regions from different proteins and organisms display specificity for mono- and di-methylated lysine residues over the unmodified and tri-methylated states. From a physiological perspective, MBT proteins are associated with chromosome condensation and act to repress the transcription of genes, ultimately affecting processes such as differentiation, mitotic progression and tumor suppression. For example, human L3mbtl is a negative regulator of E2F target genes, including c-myc and cyclin E1, suggesting that it plays a significant role in cell cycle and tumor repression.15,16 Malignant brain tumor domain-containing protein 1 (MBTD1), a newly identified member belonging to the MBT class, is a four MBT repeat protein comprising 628 amino acids and specifically binds to the mono- and di-methylation sites of H4K20 through one of its four MBT repeats utilizing a semi-aromatic cage.17 The crystal structure of MBTD1 has been thoroughly studied, however, what kind of function it exerts in cellular and chromosome progression is not yet known.
There has been evidence showing the vital function of H4K20me1 and MBT domain in mitosis,18-20 but roles in mammalian oocyte meiosis are not known. In this study, we started to clarify the role of MBTD1 in mouse oocyte meiotic maturation and the correlation and function of MBTD1, Pr-Set7 and H4K20me1 in chromosome condensation, DNA damage and meiosis progression.
Results
Expression and localization of MBTD1 and Pr-Set7 during mouse oocyte meiotic maturation
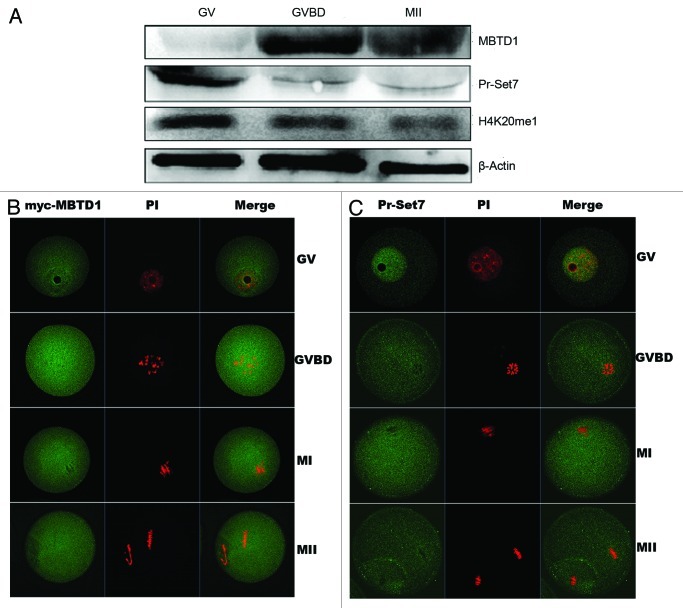
Oocytes were cultured in vitro for 0, 2 and 12 h, corresponding to GV, GVBD and MII stages, respectively. For protein extraction, 150 oocytes were collected for each stage. Using the extracted proteins, we performed western blotting and showed that MBTD1 was expressed in a low level at the GV stage, while it was significantly increased at the GVBD and MII stages, with a peak level at the GVBD stage. In contrast, Pr-Set7 was abundantly expressed in the GV stage, and its expression decreased after GVBD. The monomethylation level of H4K20 corresponded to the Pr-Set7 expression level (Fig. 1A).
Figure 1. Expression and localization of MBTD1 and Pr-Set7 in mouse oocyte in vitro meiotic maturation. (A) Expression of MBTD1 and Pr-Set7 during mouse oocyte meiotic maturation at 0, 2 and 12 h, corresponding to GV, GVBD, MII stage, respectively. (B) Confocal micrographs showing subcellular localization of immunostained myc-MBTD1 (green) and DNA (red) in mouse oocytes at GV, GVBD, MI and MII stages. Bar = 10 μm; (C) Confocal micrographs showing subcellular localization of immunostained Pr-Set7 (green) and DNA (red) in mouse oocytes at GV, GVBD, MI and MII stages. Bar = 10 μm.
Immunofluorescent confocal staining was employed to detect the subcellular distribution of MBTD1 and Pr-Set7 during mouse oocyte meiotic maturation. Because of the unavailability of MBTD1 antibody for immunofluorescence (IF), we microinjected myc-MBTD1 mRNA into GV stage oocytes. Anti-myc tagged domain examination verified the dispersed distribution of MBTD1 in mouse oocytes at various developmental stages (Fig. 1B). Pr-Set7 was concentrated in the nucleus at the GV stage, while no specific subcellular localization was observed after GVBD (Fig. 1C).
Depletion of MBTD1 and Pr-Set7 causes GV arrest and decreased monomethylation of H4K20 as well as reduced 53BP1 expression
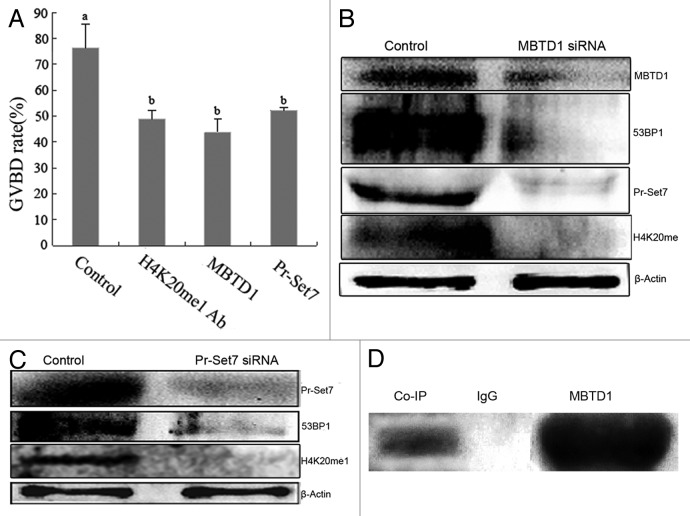
In order to investigate the role of MBTD1 in mouse oocyte meiotic maturation, we separately microinjected MBTD1 and Pr-Set7 siRNA into GV stage oocytes to deplete the expression of MBTD1 and Pr-Set7. When the protein was depleted, oocytes were cultured in M2 medium for 2 h, and the GVBD rate decreased significantly compared with the control group (scrambled siRNA injection group). The GVBD rate was 76.33% in the control group, while the GVBD rate decreased to 43.82% in the MBTD1-knockdown group, 53.25% in Pr-Set7-knockdown group and 48.74% in H4K20me1 antibody injection group, and the difference was significant (p < 0.05). (Fig. 2A). Western blotting was employed to detect the knockdown efficiency and the level of monomethylation of H4K20 in mouse oocytes. The results showed that when the MBTD1 was knocked down, the level of Pr-Set7 and monomethylation of H4K20 decreased significantly, and when Pr-Set7 was knocked down, the monomethylation of H4K20 decreased as well. Due to downregulation of H4K20me1 after MBTD1 and Pr-Set7 knockdown, it is assumed that 53BP1, a DNA damage repair checkpoint, would be downregulated, which influences DNA damage repair. We employed western blotting to determine the 53BP1 expression level, and the results showed that depletion of MBTD1 and Pr-Set7 could downregulate the expression level of 53BP1 (Fig. 2B and C).
Figure 2. Effects of MBTD1 or Pr-Set7 depletion in mouse oocyte meiotic maturation and correlation of MBTD1 and Pr-Set7. (A) Microinjection of MBTD1 siRNA, Pr-Set7 siRNA or H4K20me1 antibody caused significant decrease in the GVBD rate of mouse oocytes (p < 0.05). (B) Western blotting results demonstrated the decreased expression level of Pr-Set7, 53BP1 and H4K20me1 after MBTD1 depletion. (C) Western blotting results demonstrated the decreased expression of 53BP1 after Pr-Set7 depletion. (D) Co-IP result showing the MBTD1 correlating with Pr-Set7 to function in mouse oocyte.
MBTD1 is a histone methylation binding protein but not a histone methyl-transferase. We employed Co-IP and western blotting to detect the relationship between MBTD1 and Pr-Set7. The results showed that MBTD1 and Pr-Set7 formed a complex in mouse oocytes, indicating their direct interaction (Fig. 2D).
Depletion of MBTD1 and Pr-Set7 causes increased γH2AX foci
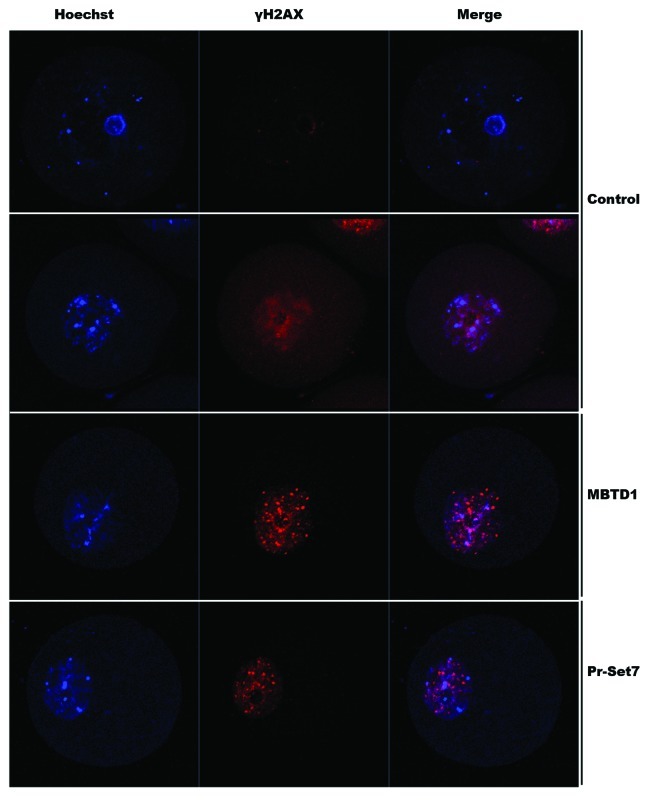
Like 53BP1, γH2AX is also a DNA damage repair checkpoint protein, and these two proteins form a complex at the damage site. 53BP1 is recruited under conditions of γH2AX foci formation and methylation of H4K20. 53BP1 has been downregulated due to the depletion of H4K20me1. In order to detect whether the depletion of MBTD1 and Pr-Set7, which caused H4K20mel decrease, could also cause γH2AX foci formation, we employed immunofluorescent confocal microscopy to detect γH2AX foci in MBTD1 or Pr-Set7 downregulated mouse oocytes at the GV stage. The results showed that γH2AX foci formation increased compared with the control group (Fig. 3).
Figure 3. Confocal micrographs showing increased γH2AX foci after MBTD1 or Pr-Ser7 depletion.
Depletion of MBTD1 and Pr-Set7 activated checkpoint protein Chk1 and downregulated cyclin B1 and cdc2
To explore why depletion of MBTD1 and Pr-Set7 affected GVBD in mouse oocytes, we used western blotting to detect the expression level of the cell cycle checkpoint proteins Chk1 and cyclin B1 in siRNA-microinjected mouse oocytes at the GV stage. The results showed that depletion of MBTD1 and Pr-Set7 could both activate Chk1 as indicated by its increased expression, while downregulating the expression of cyclin B1, resulting in the G2/M arrest of the meiotic cell cycle. We also detected the expression level of cdc2, the other subunit of the maturation-promoting factor (MPF). Our western blotting results showed that cdc2 expression was also decreased in MBTD1 and Pr-Set7 depleted oocytes (Fig. 4).
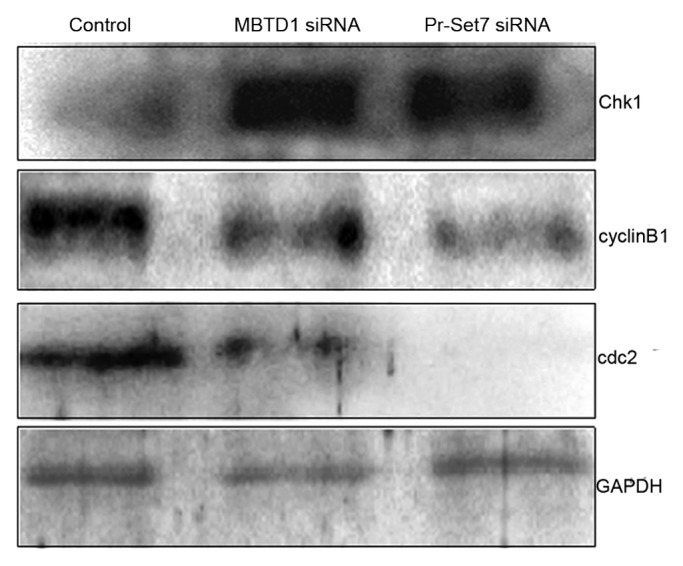
Figure 4. Effects of MBTD1 or Pr-Set7 depletion on cell cycle factors. Western blotting results showing that depletion of MBTD1 or Pr-Set7 activated Chk1 and ultimately decreased the expression level of MPF components: cdc2 and cyclin B1.
Depletion of MBTD1 and Pr-Set7 causes abnormal chromatin/chromosome configuration in mouse oocytes
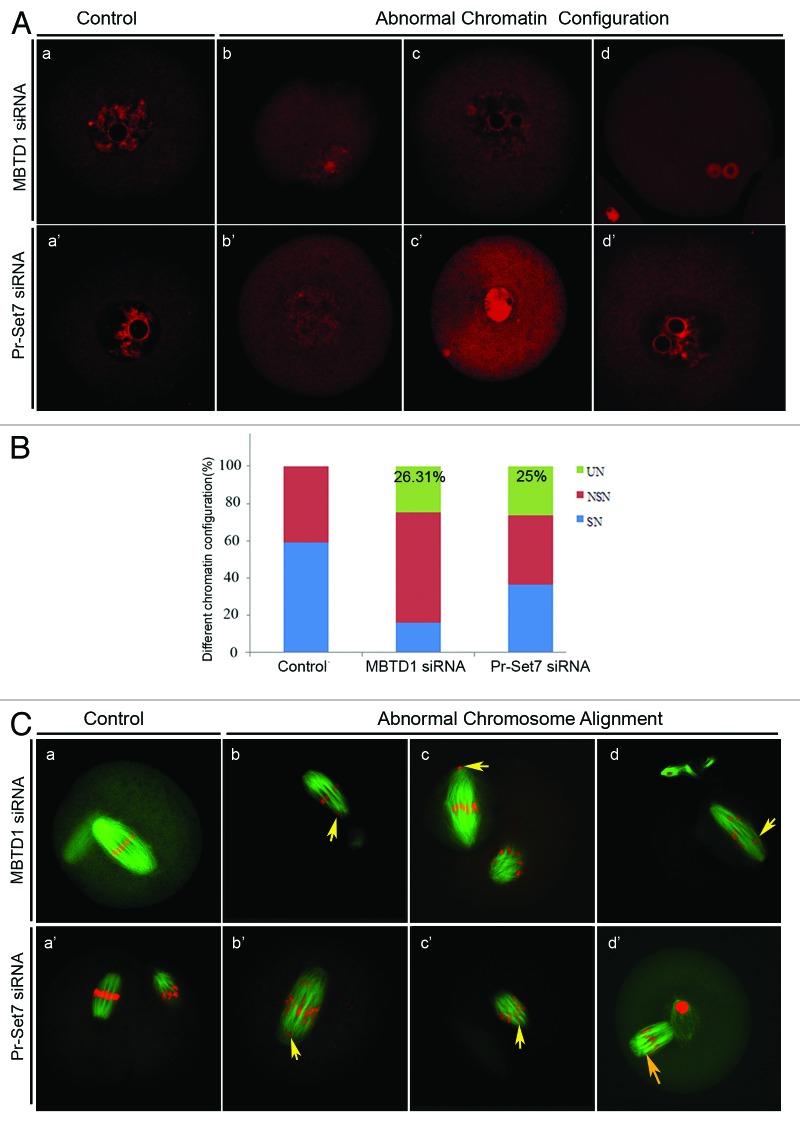
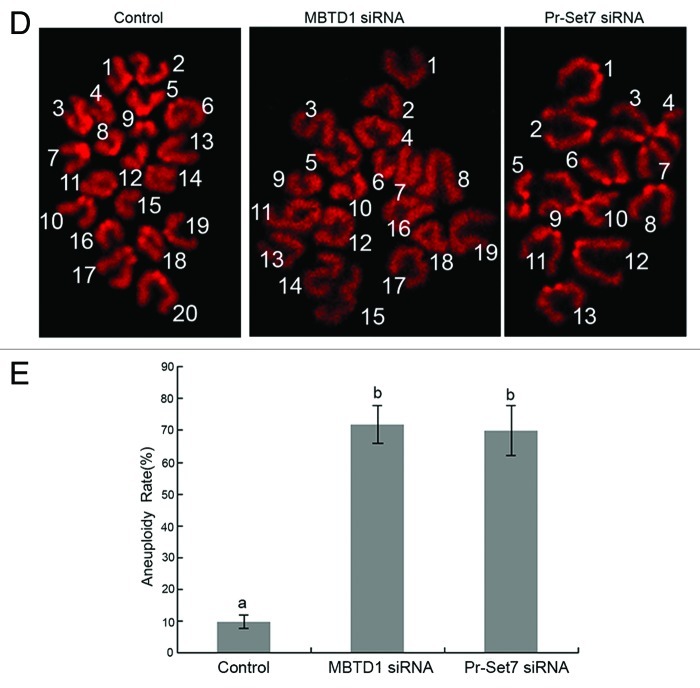
Due to the DNA damage and cdc2 downregulation in MBTD1 and Pr-Set7 depleted mouse oocytes, we assumed that the depletion of MBTD1 and Pr-Set7 could result in abnormal chromatin/chromosome configurations in mouse oocytes. Therefore, we compared the rate of SN (surrounding nucleolus) and NSN (non-surrounding nucleolus) oocytes at the GV stage in MBTD1 or Pr-Set7 depletion groups with controls. After 12 h of culture in M2 medium, chromosome spreading was employed to determine aneuploidy in MBTD1 and Pr-Set7 depletion oocytes. After PI staining, chromatin displayed abnormal configurations besides SN and NSN changes, which included a large nucleolus, condensed nucleus and chromatin de-condensation (Fig. 5A), which we termed UN (unidentified nucleolus). Oocytes with these abnormal chromatin configurations constituted about 26.31% (MBTD1 siRNA group) and 25% (Pr-Set7 siRNA group) of the tested oocytes (Fig. 5B). Most of the MBTD1 and Pr-Set7 knockdown oocytes showed disordered chromosome arrangements at MII stages, as detected by PI staining (Fig. 5C). However, spindle formation was not affected in these oocytes.
Figure 5A–C. Effects of MBTD1 or Pr-Set7 depletion on chromatin/chromosome configuration in mouse oocyte meiotic maturation. (A) PI staining micrographs showing abnormal chromatin after MBTD1 (b, c, d) or Pr-Set7 (b’, c’, d’) depletion; a and a’ are the controlled normal chromatin of MBTD1 and Pr-Set7, respectively.(B) Percent bars showing that 26.31% (MBTD1) and 25% (Pr-Set7) of the oocytes displayed abnormal chromosome configurations (UN, unidentified nucleolus) besides SN (surrounding nucleolus) and NSN (non-surrounding nucleolus) in mouse oocytes at the GV stage. (C) Confocal micrographs showing abnormal chromosome alignment in mouse oocytes at the MII stage after MBTD1 (b, c, d) or Pr-Set7 (b’, c’, d’) depletion; a and a’ are the controlled normal chromosomes alignment of MBTD1 and Pr-Set7, respectively. Spindle (green) and DNA (red). Arrows point out the abnormal aggregated chromosome. Bar = 10 μm.
Moreover, about 70% of the MBTD1 and Pr-Set7 depletion oocytes showed aneuploidy at the MII stage (Fig. 5D and E).
Figure 5D and E. (D) Chromosome spreading demonstrated aneuploidy at the MII stage in mouse oocytes. (E) Percent bars showing that there was 70% of aneuploidy in the MBTD1 knockdown group and 72% in Pr-Set7 knockdown group, while in the control group, the aneuploidy rate was only 12% (p < 0.05).
Discussion
In this study, we have revealed the role of a histone methylation binding protein, MBTD1, and its correlation with histone lysine methyl-transferase Pr-Set7 and H4K20me1 in mouse oocyte meiotic maturation. We demonstrated that MBTD1 associates with Pr-Set7 to stabilize H4K20me1 and, thus, regulates meiotic cell cycle progression, chromatin configuration and chromosome alignment/separation in mouse oocyte meiotic maturation.
MBTD1 does not specifically localize on chromosomes or in the nucleus; instead, it is distributed in all cellular components throughout the entire mouse oocyte meiotic maturation process, as shown by the antibody staining against myc-tagged MBTD1. However, the histone lysine methyl transferase Pr-Set7 solely localizes in the nucleus at the GV stage but is distributed in all cellular components after GVBD like MBTD1. It appears that along with germinal vesicle breakdown, all nuclear contents disperse into the oocyte. Up until now, there has been no evidence showing that proteins belonging to the “Royal family” can only localize in the nucleus, suggesting that they might have functions other than “reading” the histone modification.21 Regarding the expression of MBTD1 and Pr-Set7 in mouse oocyte meiotic maturation, these two proteins show opposite expression patterns. The MBTD1 expression level was at a minimum at the GV stage and at a maximum at the GVBD stage, while the Pr-Set7 expression level was high at the GV stage and decreased after GVBD. The H4K20me1 pattern was consistent with that of Pr-Set7. Because Pr-Set7 is the exclusive H4K20 mono-methyl transferase, and H4K20me1 has an essential function in chromosome repression and condensation,22 it is reasonable that Pr-Set7 is expressed at a higher level at the GV stage. Though at a minimum level at the GV stage, the sharp rise of MBTD1 along with GVBD implied that it might have an important role in the G2/M transition.
Previous studies of histone lysine methyl binding protein and Pr-Set7 in mitosis have revealed that these proteins are related to various cellular events, such as DNA damage, chromosome condensation, cell cycle and gene expression depression.23,24 We microinjected MBTD1 and Pr-Set7 siRNA into mouse oocytes at the GV stage to detect their roles in mouse oocyte meiotic progression. First, depletion of both MBTD1 and Pr-Set7 could cause an increase in GV stage arrest in mouse oocytes. The normal GVBD rate after 2 h of culture in M2 medium was about 78%, while the GVBD rate decreased significantly when MBTD1 or Pr-Set7 was depleted. Second, the H4K20me1 level decreased along with the knockdown of the MBTD1 or Pr-Set7. Western blotting results showed that knockdown of Pr-Set7 would not affect the expression of MBTD1 (data not shown). Although MBTD1 is only a histone methylation binding protein, it affects the H4K20me1 level similarly as H4K20me1 transferase Pr-Set7, suggesting that in mouse oocytes, MBTD1 may be correlated with Pr-Set7 to induce monomethylation at the H4K20 site. Therefore, Co-IP was employed to demonstrate the correlation of MBTD1 and Pr-Set7, which proved our assumption. Thus, in mouse oocytes, it can be concluded that MBTD1 is associated with Pr-Set7 to stabilize H4K20me1. Recently, a protein containing the PWWP domain was confirmed to regulate Set9-mediated methylation of H4K20 in Saccharomyces cerevisiae.25 Proteins containing the PWWP domain and MBT domain are both included in the “Royal family,” thus our results contribute new information on the functional understanding of the “Royal family.”
One of the most important consequences of H4K20me1 abrogation is to cause DNA damage, as indicated by γH2AX foci formation on chromosomes.26 Due to the unavailability of γH2AX antibody for western blotting, we used immunofluorescence confocal microscopy to detect γH2AX foci formation on mouse chromosomes at the GV stage. The results showed that both MBTD1 and Pr-Set7 knockdown caused increased γH2AX foci on chromosomes, indicating DNA damage on chromosomes. 53BP1, a TP53 binding protein, is a core DNA damage repair checkpoint in mammalian cells. It accumulates on the methylated H4K20 dock at the DNA damage site and co-localizes with γH2AX to form the DNA repair complex.27-29 However, in our study, when 53BP1 expression was decreased after MBTD1 or Pr-Set7 depletion, increased γH2AX foci were formed. We assumed that depletion of MBTD1 and Pr-Set7 would cause abrogation of H4K20me1, the substrate of H4K20me2 and H4K20me3 and binding sites of 53BP1,30,31 thus resulting in decreased expression of 53BP1. The DNA damage repair complex fails to form due to the lack of 53BP1; therefore, the mouse oocyte would be arrested at the GV stage.
Cell cycle events are regulated by sequential activation and deactivation of cyclin-dependent kinases (Cdks) and by proteolysis of cyclins. Chk1 is involved in these processes as regulators of Cdks. Chk1 functions as essential component in the G2 DNA damage checkpoint by phosphorylating Cdc25C in response to DNA damage. Phosphorylation inhibits Cdc25C activity, thereby blocking the G2/M transition.32-35 MBTD1 and Pr-Set7 can lead to DNA damage at the GV stage in mouse oocytes, thereby activating the checkpoint protein Chk1, which might influence GVBD through regulating the expression of CDK1. Western blotting results proved our hypothesis. The expression level of Chk1 was upregulated after depletion of MBTD1 or Pr-Set7, providing evidence that the mouse oocyte was arrested at the GV stage when MBTD1 or Pr-Set7 was abolished, and this might be achieved by regulating DNA damage-related Chk1 activation and MPF activity
In eukaryotic cells, the G2/M transition is initiated following activation of a protein kinase known as maturation-promoting factor (MPF),36 M-phase specific histone kinase or M-phase kinase. This protein kinase is composed of a catalytic subunit (cdc2), a regulatory subunit (cyclin B) and a low molecular weight subunit (p13-Suc 1).37,38 Cyclin B1 was downregulated due to the depletion of MBTD1 or Pr-Set7. Western blotting results also revealed the downregulation of cdc2, which is the catalytic subunit of MPF. MPF can first phosphorylate histone H1 and sequentially affect chromosome condensation. Second, MPF can catalyze nuclear lamina protein to break up the nuclear lamin layer structure resulting in GVBD. Lastly, MPF can also act on microtubules and affect the poleward movement of chromosomes.39-42 Therefore MPF has important functions in the meiotic progression. In our study, depletion of MBTD1 or Pr-Set7 both can downregulate expression of the MPF components cdc2 and cyclin B1, which arrest oocytes at the GV stage. Immunofluorescent staining showed that at the GV stage, the chromosomes displayed various configurations, such as de-condensation, enlargement and pyknosis of the nucleolus and muti-nucleoli. All of the phenotypes indicate cellular apoptosis.43,44 On the other hand, oocytes which can enter the MII stage after siRNA microinjection showed disordered chromosome arrangements, but the spindle assembled normally and was comparable with the control group, suggesting that the depletion of MBTD1 or Pr-Set7 did not influence spindle assembly. The chromosome-spreading assay revealed that most of the oocytes showed aneuploidy, providing further evidence for chromosome configuration defects.
In summary, our study for the first time demonstrates the role of histone lysine methyl binding protein MBTD1 in mouse oocyte meiotic maturation. MBTD1 was correlated with histone lysine methyl transferase Pr-Set7 to stabilize H4K20me1 in mouse oocytes, regulating meiotic cell cycle progression and chromosome configuration. In addition, our study provides new perspectives for the role of the “Royal family” and reinforces the importance of H4K20 methylation in mammalian oocyte meiotic maturation.
Materials and Methods
Mouse oocyte collection and in vitro maturation
The native breed mouse strain, ICR, was used in this study. Animal care and handling were conducted in accordance with policies regarding the care and use of animals issued by the ethical committee of the Institute of Zoology, Chinese Academy of Sciences. Immature oocytes at the germinal vesicle (GV) stage were collected from ovaries of 6–8-week-old female mice in M2 medium (Sigma). Oocytes used for microinjection were cultured in M2 medium supplemented with 2.5 μM milrinone to maintain the oocytes at the GV stage during microinjection. After microinjection, oocytes were washed thoroughly and further cultured in M2 medium covered with liquid paraffin oil at 37°C in an atmosphere of 5% CO2 in air until proceeding to the GV (0 h), GVBD (2 h), MI (8 h) and MII (12–14 h) stages.
Myc-MBTD1 plasmid construction and in vitro transcription
Total RNA was extracted from 150 GV oocyte with RNeasy micro purification kit (Qiagen). Then total cDNA was synthesized with cDNA synthesis kit (Invitrogen). The full length of MBTD1 cds was cloned by PCR with primers: MBTD1-F3, GTTGGCCGGCCGATGGACACTAGAAGTCACCCAAAGC; MBTD1-R3,GTTGGCGCGCCTCACCTTCAATCCTGCTACCTAAAC. After sequencing and blast, the full length of MBTD1 CDS was sub-cloned to the FseI/AscI restrict site of pSC2+ vector. The Myc-MBTD1-pCS2+ plasmid was linearized by SalI and purified by gel extraction kit (Promega). SP6 high-yield capped RNA tanscription kit (Ambion) was used for producing capped mRNA, and then the mRNA was purified with RNeasy cleanup kit (Qiagen). The concentration of myc-MBTD1 mRNA was detected with a Beckman DU 530 Analyzer and then diluted to a low concentration (0.4 mg/ml) for localization
Microinjection of MBTD1 and Pr-Set7 siRNA or mRNA
Microinjection was performed using an Eppendorf microinjector and completed within 1 h. Myc-MBTD1 mRNA solution was microinjected into cytoplasm of each GV stage oocyte for localization assays. The same amount of Myc mRNA without MBTD1 sequence was injected into control oocytes under the same conditions.
The MBTD1 and Pr-Set7 siRNA were synthesized by GenePharma Co., Ltd. The siRNA was diluted to 1 μM and microinjected into the GV oocyte. After microinjection, the GV stage oocytes were cultured for 24 h in M2 medium supplemented with 2.5 μM milrinone to maintain the oocytes at the GV stage and to ensure siRNA being able to deplete the target protein. Negative control siRNA was microinjected into the GV stage oocyte, and the oocyte was cultured under the same conditions.
Antibodies, immunofluorescence and western blotting
Antibody against MBTD1 was purchased from Abgent. Antibodies for detecting Pr-Set7, H4K20me1, cdc2, cyclin B1 were purchased from Santa Cruz Biotechnology. Antibodies against γH2AX and 53BP1 were purchased from Bioworld. Immunofluorescent staining and western blotting were performed using standard protocols. Briefly, for immunofluorescent staining, all oocytes were fixed in 4% paraformaldehyde for 30 min then treated with 0.5% Triton X-100 for 20 min. After blocking in 1% BSA for 1 h, oocytes were incubated in the first antibody (1:50) at 4°C overnight. After five washes in PBS with 0.05% Tween 20, the oocytes were incubated with the second antibody (1:200 in PBS with 0.05% Tween 20) for 1 h at room temperature. Then the oocytes were further washed four times in PBS with 0.05% Tween 20 and stained with anti-β-tubulin antibody (1:100 in PBS with 0.05% Tween 20, Sigma). After four washes in PBS with 0.05% Tween 20, the oocytes were incubated with PI/DAPI (propidium iodide/4, 6-diamino-2-phenyl indole) in PBS containing 0.05% Tween 20 for 10 min. Finally, the oocytes were mounted on glass slides and examined with a laser scanning confocal microscope (Zeiss LSM 510 and 710 META).
For western blotting, mouse oocytes were collected in SDS sample buffer and heated for 5 min at 100°C. The proteins were separated by SDS-PAGE and electrically transferred to polyvinylidene fluoride membrane, and then the membrane was blocked in TBST containing 5% BSA for 2 h, followed by incubation overnight at 4°C with the first antibody (1:300) and mouse monoclonal anti-β-actin antibody (1:1,000, Zhong Shan Jin Qiao Co.). After washing three times in TBST, each for 10 min, the membrane was incubated for 1 h at 37°C with peroxidase-conjugated second antibody (1:1,000, Zhong Shan Jin Qiao Co.). Finally, the membrane was processed using the SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific).
Co-immunoprecipitation
About 1,000 mouse oocytes were collected to perform Co-immunoprecipitation experiments. All procedures followed the instructions of the Pierce Co-immunoprecipitation Kit (Thermo Scientific).
Chromosome spreading and image analysis
Oocytes were left in hypotonic sodium citrate (1%, W/V) for 15 min at room temperature, and then each oocyte was placed on a glass slide. About 100 μl methanol:glacial acetic acid (3:1) was dropped onto the oocyte to break and fix the oocyte. Chromosomes were stained with PI (10 mg/ml). The specimen was examined with a Confocal Laser Scanning Microscope (Zeiss LSM 510 and 710 META).
Statistical analysis
All percentage data were subjected to arcsine transformation before statistical analysis. Data were analyzed by analysis of variance (ANOVA) with SPSS. Differences at p < 0.05 were considered significant.
Acknowledgments
This study was supported by Major Basic Research Program (2012CB944404, 2011CB944501) and National Natural Science Foundation of China (30930065) to Q.Y.S.
Glossary
Abbreviations:
- GV
germinal vesicle
- GVBD
germinal vesicle breakdown
- MI
first metaphase
- MII
second metaphase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24216
References
- 1.Schotta G, Lachner M, Peters AH, Jenuwein T. The indexing potential of histone lysine methylation. Novartis Found Symp. 2004;259(discussion 37-47, 163-9):22–37. doi: 10.1002/0470862637.ch3. [DOI] [PubMed] [Google Scholar]
- 2.Gu L, Wang Q, Sun QY. Histone modifications during mammalian oocyte maturation: dynamics, regulation and functions. Cell Cycle. 2010;9:1942–50. doi: 10.4161/cc.9.10.11599. [DOI] [PubMed] [Google Scholar]
- 3.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callén E, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–61. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carr SM, La Thangue NB. Cell cycle control by a methylation-phosphorylation switch. Cell Cycle. 2011;10:733–4. doi: 10.4161/cc.10.5.14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vielle A, Lang J, Dong Y, Ercan S, Kotwaliwale C, Rechtsteiner A, et al. H4K20me1 Contributes to Down regulation of X-Linked Genes for C. elegans Dosage Compensation. PLoS Genet. 2012;9:e1002933. doi: 10.1371/journal.pgen.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012;26:325–37. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu S, Rice JC. A new regulator of the cell cycle: the PR-Set7 histone methyltransferase. Cell Cycle. 2011;10:68–72. doi: 10.4161/cc.10.1.14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi A, Steward R. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J Cell Biol. 2007;176:155–62. doi: 10.1083/jcb.200607178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohouo PY, Smolka MB. The many roads to checkpoint activation. Cell Cycle. 2012;11:4495. doi: 10.4161/cc.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 11.Trojer P, Cao AR, Gao Z, Li Y, Zhang J, Xu X, et al. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol Cell. 2011;42:438–50. doi: 10.1016/j.molcel.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallette FA, Richard S. The fight of the Tudor domain “Royal family” for a broken DNA throne. Cell Cycle. 2012;11:1483–4. doi: 10.4161/cc.20124. [DOI] [PubMed] [Google Scholar]
- 13.Bantignies F, Cavalli G. Polycomb group proteins: repression in 3D. Trends Genet. 2011;27:454–64. doi: 10.1016/j.tig.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–28. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 16.Eryilmaz J, Pan P, Amaya MF, Allali-Hassani A, Dong A, Adams-Cioaba MA, et al. Structural studies of a four-MBT repeat protein MBTD1. PLoS ONE. 2009;4:e7274. doi: 10.1371/journal.pone.0007274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010;24:636–46. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koester-Eiserfunke N, Fischle W. H3K9me2/3 binding of the MBT domain protein LIN-61 is essential for Caenorhabditis elegans vulva development. PLoS Genet. 2011;7:e1002017. doi: 10.1371/journal.pgen.1002017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalo S, Blasco MA. Role of Rb family in the epigenetic definition of chromatin. Cell Cycle. 2005;4:752–5. doi: 10.4161/cc.4.6.1720. [DOI] [PubMed] [Google Scholar]
- 20.Trojer P, Reinberg D. Beyond histone methyl-lysine binding: how malignant brain tumor (MBT) protein L3MBTL1 impacts chromatin structure. Cell Cycle. 2008;7:578–85. doi: 10.4161/cc.7.5.5544. [DOI] [PubMed] [Google Scholar]
- 21.Schotta G. H4K20 monomethylation faces the WNT. Proc Natl Acad Sci USA. 2011;108:3097–8. doi: 10.1073/pnas.1100147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gadhia SR, Calabro AR, Barile FA. Trace metals alter DNA repair and histone modification pathways concurrently in mouse embryonic stem cells. Toxicol Lett. 2012;212:169–79. doi: 10.1016/j.toxlet.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Gargalionis AN, Piperi C, Adamopoulos C, Papavassiliou AG. Histone modifications as a pathogenic mechanism of colorectal tumorigenesis. Int J Biochem Cell Biol. 2012;44:1276–89. doi: 10.1016/j.biocel.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Lee TB, Moon YS, Choi CH. Histone H4 deacetylation down-regulates catalase gene expression in doxorubicin-resistant AML subline. Cell Biol Toxicol. 2012;28:11–8. doi: 10.1007/s10565-011-9201-y. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Reddy B, Thompson J, Wang H, Noma K, Yates JR, 3rd, et al. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell. 2009;33:428–37. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–33. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamelza P, Bhalla N. Histone methyltransferases MES-4 and MET-1 promote meiotic checkpoint activation in Caenorhabditis elegans. PLoS Genet. 2012;8:e1003089. doi: 10.1371/journal.pgen.1003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–63. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding SM, Bristow RG. Discordance between phosphorylation and recruitment of 53BP1 in response to DNA double-strand breaks. Cell Cycle. 2012;11:1432–44. doi: 10.4161/cc.19824. [DOI] [PubMed] [Google Scholar]
- 30.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–8. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–95. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canovas S, Cibelli JB, Ross PJ. Jumonji domain-containing protein 3 regulates histone 3 lysine 27 methylation during bovine preimplantation development. Proc Natl Acad Sci USA. 2012;109:2400–5. doi: 10.1073/pnas.1119112109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donzelli M, Draetta GF. Regulating mammalian checkpoints through Cdc25 inactivation. EMBO Rep. 2003;4:671–7. doi: 10.1038/sj.embor.embor887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhind N, Russell P. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J Cell Sci. 2000;113:3889–96. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–59. [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Chao SB, Wang ZB, Qi ST, Zhu XL, Yang SW, et al. Checkpoint kinase 1 is essential for meiotic cell cycle regulation in mouse oocytes. Cell Cycle. 2012;11:1948–55. doi: 10.4161/cc.20279. [DOI] [PubMed] [Google Scholar]
- 37.Le Gac G, Estève PO, Ferec C, Pradhan S. DNA damage-induced down-regulation of human Cdc25C and Cdc2 is mediated by cooperation between p53 and maintenance DNA (cytosine-5) methyltransferase 1. J Biol Chem. 2006;281:24161–70. doi: 10.1074/jbc.M603724200. [DOI] [PubMed] [Google Scholar]
- 38.Bogliolo L, Ledda S, Leoni G, Naitana S, Moor RM. Activity of maturation promoting factor (MPF) and mitogen-activated protein kinase (MAPK) after parthenogenetic activation of ovine oocytes. Cloning. 2000;2:185–96. doi: 10.1089/152045500454744. [DOI] [PubMed] [Google Scholar]
- 39.Thadani R, Uhlmann F, Heeger S. Condensin, chromatin crossbarring and chromosome condensation. Curr Biol. 2012;22:R1012–21. doi: 10.1016/j.cub.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 40.Osmani SA, Engle DB, Doonan JH, Morris NR. Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell. 1988;52:241–51. doi: 10.1016/0092-8674(88)90513-2. [DOI] [PubMed] [Google Scholar]
- 41.Fabian L, Troscianczuk J, Forer A. Calyculin A, an enhancer of myosin, speeds up anaphase chromosome movement. Cell Chromosome. 2007;6:1. doi: 10.1186/1475-9268-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakaguchi A, Joyce E, Aoki T, Schedl P, Steward R. The histone H4 lysine 20 monomethyl mark, set by PR-Set7 and stabilized by L(3)mbt, is necessary for proper interphase chromatin organization. PLoS ONE. 2012;7:e45321. doi: 10.1371/journal.pone.0045321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Ai JS, Idowu Ola S, Gu L, Zhang YZ, Chen DY, et al. The spatial relationship between heterochromatin protein 1 alpha and histone modifications during mouse oocyte meiosis. Cell Cycle. 2008;7:513–20. doi: 10.4161/cc.7.4.5356. [DOI] [PubMed] [Google Scholar]
- 44.Manosalva I, González A. Aging changes the chromatin configuration and histone methylation of mouse oocytes at germinal vesicle stage. Theriogenology. 2010;74:1539–47. doi: 10.1016/j.theriogenology.2010.06.024. [DOI] [PubMed] [Google Scholar]