Abstract
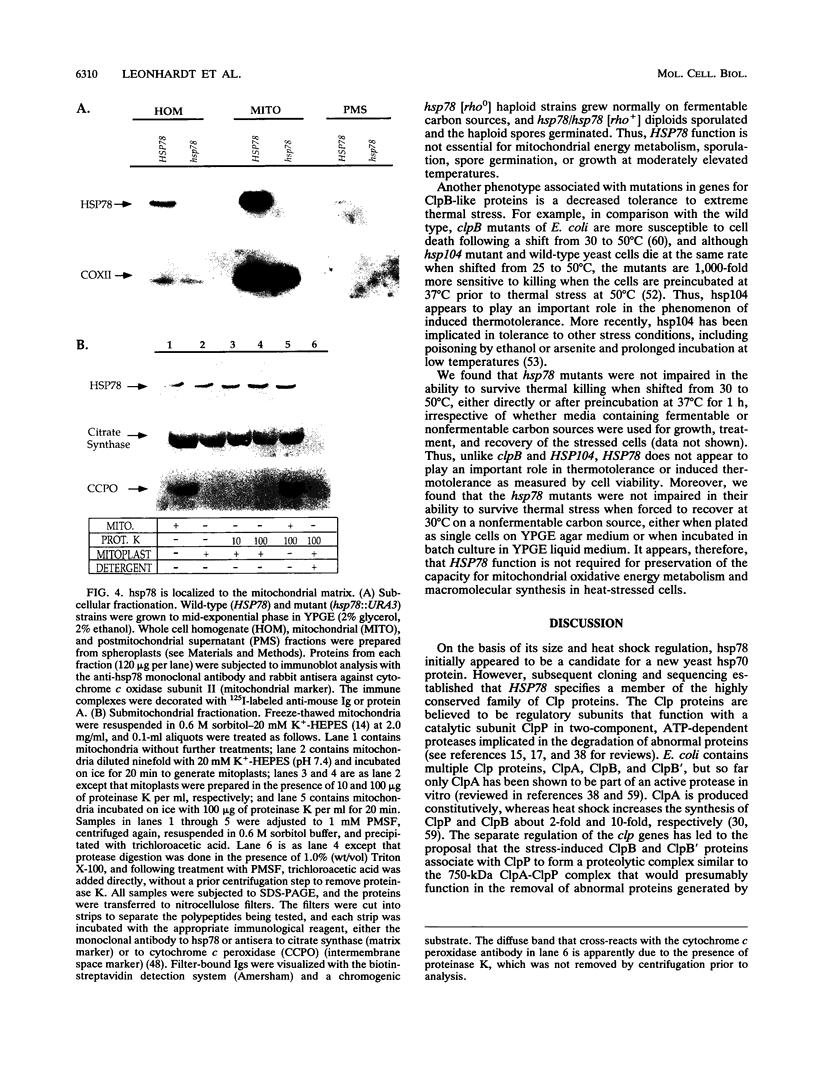
The Saccharomyces cerevisiae nuclear gene for a 78-kDa mitochondrial heat shock protein (hsp78) was identified in a lambda gt11 expression library through immunological screening with an hsp78-specific monoclonal antibody. Sequencing of HSP78 revealed a long open reading frame capable of encoding an 811-amino-acid, 91.3-kDa basic protein with a putative mitochondrial leader sequence and two potential nucleotide-binding sites. Sequence comparisons revealed that hsp78 is a member of the highly conserved family of Clp proteins and is most closely related to the Escherichia coli ClpB protein, which is thought to be an ATPase subunit of an intracellular ATP-dependent protease. The steady-state levels of HSP78 transcripts and protein varied in response to both thermal stress and carbon source with an approximately 30-fold difference between repressed levels in cells growing fermentatively on glucose at 30 degrees C and derepressed levels in heat-shocked cells growing on a nonfermentable carbon source. The response to heat shock is consistent with the presence of a characteristic heat shock regulatory element in the 5'-flanking DNA. Submitochondrial fractionation showed that hsp78 is a soluble protein located in the mitochondrial matrix. Cells carrying disrupted copies of HSP78 lacked hsp78 but were not impaired in respiratory growth at normal and elevated temperatures or in the ability to survive and retain mitochondrial function after thermal stress. The absence of a strong mitochondrial phenotype in hsp78 mutants is comparable to the nonlethal phenotypes of mutations in other Clp genes in bacteria and yeast. HSP78 is the third gene, with SSC1 and HSP60, known to encode a yeast mitochondrial heat shock protein and the second gene, with HSP104, for a yeast ClpB homolog.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amin J., Ananthan J., Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988 Sep;8(9):3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang D., Liberek K., Skowyra D., Zylicz M., Georgopoulos C. Biological role and regulation of the universally conserved heat shock proteins. J Biol Chem. 1991 Dec 25;266(36):24233–24236. [PubMed] [Google Scholar]
- Cheng M. Y., Hartl F. U., Martin J., Pollock R. A., Kalousek F., Neupert W., Hallberg E. M., Hallberg R. L., Horwich A. L. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989 Feb 16;337(6208):620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- Chung C. H., Goldberg A. L. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4931–4935. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E. A., Kramer J., Shilling J., Werner-Washburne M., Holmes S., Kosic-Smithers J., Nicolet C. M. SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol Cell Biol. 1989 Jul;9(7):3000–3008. doi: 10.1128/mcb.9.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G., Gasser S. M., Schatz G. Import of proteins into mitochondria. Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J Biol Chem. 1982 Nov 10;257(21):13075–13080. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon K., Mason T. L. Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP7, a protein of the large subunit of the mitochondrial ribosome. Mol Cell Biol. 1988 Sep;8(9):3636–3646. doi: 10.1128/mcb.8.9.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fry D. C., Kuby S. A., Mildvan A. S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci U S A. 1986 Feb;83(4):907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M. J., Sambrook J. Protein folding in the cell. Nature. 1992 Jan 2;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Glick B. S., Brandt A., Cunningham K., Müller S., Hallberg R. L., Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992 May 29;69(5):809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur J Biochem. 1992 Jan 15;203(1-2):9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Clark W. P., Maurizi M. R. The ATP-dependent Clp protease of Escherichia coli. Sequence of clpA and identification of a Clp-specific substrate. J Biol Chem. 1990 May 15;265(14):7886–7893. [PubMed] [Google Scholar]
- Gottesman S., Maurizi M. R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992 Dec;56(4):592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Squires C., Pichersky E., Carrington M., Hobbs M., Mattick J. S., Dalrymple B., Kuramitsu H., Shiroza T., Foster T. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc Natl Acad Sci U S A. 1990 May;87(9):3513–3517. doi: 10.1073/pnas.87.9.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs D. W., Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUNTER W. M., GREENWOOD F. C. Preparation of iodine-131 labelled human growth hormone of high specific activity. Nature. 1962 May 5;194:495–496. doi: 10.1038/194495a0. [DOI] [PubMed] [Google Scholar]
- Hwang B. J., Woo K. M., Goldberg A. L., Chung C. H. Protease Ti, a new ATP-dependent protease in Escherichia coli, contains protein-activated ATPase and proteolytic functions in distinct subunits. J Biol Chem. 1988 Jun 25;263(18):8727–8734. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. B., Fearon K., Mason T., Jindal S. Cloning and characterization of the yeast chaperonin HSP60 gene. Gene. 1989 Dec 14;84(2):295–302. doi: 10.1016/0378-1119(89)90503-9. [DOI] [PubMed] [Google Scholar]
- Jones E. W. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Juretić N., Theus M. Analysis of the polyadenylation consensus sequence context in the genes of nuclear encoded mitochondrial proteins. FEBS Lett. 1991 Sep 23;290(1-2):4–8. doi: 10.1016/0014-5793(91)81212-q. [DOI] [PubMed] [Google Scholar]
- Kang P. J., Ostermann J., Shilling J., Neupert W., Craig E. A., Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990 Nov 8;348(6297):137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- Katayama Y., Gottesman S., Pumphrey J., Rudikoff S., Clark W. P., Maurizi M. R. The two-component, ATP-dependent Clp protease of Escherichia coli. Purification, cloning, and mutational analysis of the ATP-binding component. J Biol Chem. 1988 Oct 15;263(29):15226–15236. [PubMed] [Google Scholar]
- Kitagawa M., Wada C., Yoshioka S., Yura T. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock sigma factor (sigma 32). J Bacteriol. 1991 Jul;173(14):4247–4253. doi: 10.1128/jb.173.14.4247-4253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozłowski M., Zagórski W. Stable preparation of yeast mitochondria and mitoplasts synthesizing specific polypeptides. Anal Biochem. 1988 Aug 1;172(2):382–391. doi: 10.1016/0003-2697(88)90459-9. [DOI] [PubMed] [Google Scholar]
- Kroh H. E., Simon L. D. The ClpP component of Clp protease is the sigma 32-dependent heat shock protein F21.5. J Bacteriol. 1990 Oct;172(10):6026–6034. doi: 10.1128/jb.172.10.6026-6034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler E., Steiner G., Barta A. Photoaffinity labeling of peptidyltransferase. Methods Enzymol. 1988;164:361–372. doi: 10.1016/s0076-6879(88)64055-9. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lindquist S., Craig E. A. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Maurizi M. R. ATP-promoted interaction between Clp A and Clp P in activation of Clp protease from Escherichia coli. Biochem Soc Trans. 1991 Aug;19(3):719–723. doi: 10.1042/bst0190719. [DOI] [PubMed] [Google Scholar]
- Maurizi M. R., Clark W. P., Katayama Y., Rudikoff S., Pumphrey J., Bowers B., Gottesman S. Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1990 Jul 25;265(21):12536–12545. [PubMed] [Google Scholar]
- Maurizi M. R. Proteases and protein degradation in Escherichia coli. Experientia. 1992 Feb 15;48(2):178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Moore T., Keegstra K. Characterization of a cDNA clone encoding a chloroplast-targeted Clp homologue. Plant Mol Biol. 1993 Feb;21(3):525–537. doi: 10.1007/BF00028809. [DOI] [PubMed] [Google Scholar]
- Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990 Sep;9(9):2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet C. M., Craig E. A. Inducing and assaying heat-shock response in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:710–717. doi: 10.1016/0076-6879(91)94052-e. [DOI] [PubMed] [Google Scholar]
- Parsell D. A., Sanchez Y., Stitzel J. D., Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991 Sep 19;353(6341):270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N., Neupert W. The mitochondrial protein import apparatus. Annu Rev Biochem. 1990;59:331–353. doi: 10.1146/annurev.bi.59.070190.001555. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Reid G. A., Yonetani T., Schatz G. Import of proteins into mitochondria. Import and maturation of the mitochondrial intermembrane space enzymes cytochrome b2 and cytochrome c peroxidase in intact yeast cells. J Biol Chem. 1982 Nov 10;257(21):13068–13074. [PubMed] [Google Scholar]
- Rendueles P. S., Wolf D. H. Proteinase function in yeast: biochemical and genetic approaches to a central mechanism of post-translational control in the eukaryote cell. FEMS Microbiol Rev. 1988 Feb;4(1):17–45. doi: 10.1111/j.1574-6968.1988.tb02706.x-i1. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Lindquist S. L. HSP104 required for induced thermotolerance. Science. 1990 Jun 1;248(4959):1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Taulien J., Borkovich K. A., Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992 Jun;11(6):2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger M. J. Heat shock proteins. J Biol Chem. 1990 Jul 25;265(21):12111–12114. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Squires C. L., Pedersen S., Ross B. M., Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991 Jul;173(14):4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C., Squires C. L. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol. 1992 Feb;174(4):1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Teem J. L., Rosbash M. Expression of a beta-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation of yeast. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4403–4407. doi: 10.1073/pnas.80.14.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias J. W., Shrader T. E., Rocap G., Varshavsky A. The N-end rule in bacteria. Science. 1991 Nov 29;254(5036):1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- Woo K. M., Kim K. I., Goldberg A. L., Ha D. B., Chung C. H. The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J Biol Chem. 1992 Oct 5;267(28):20429–20434. [PubMed] [Google Scholar]
- Xiao H., Lis J. T. Germline transformation used to define key features of heat-shock response elements. Science. 1988 Mar 4;239(4844):1139–1142. doi: 10.1126/science.3125608. [DOI] [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Conrad-Webb H., Liao X. S., Perlman P. S., Butow R. A. Functional expression of a yeast mitochondrial intron-encoded protein requires RNA processing at a conserved dodecamer sequence at the 3' end of the gene. Mol Cell Biol. 1989 Apr;9(4):1507–1512. doi: 10.1128/mcb.9.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]