Abstract
CD4+ T helper (Th) cells play an instrumental role in orchestrating adaptive immune responses to invading pathogens through their ability to differentiate into specialized effector subsets. Part of this customized response requires the development of T follicular helper (Tfh) cells, which provide help to B cells for the generation of germinal centers and long-term protective humoral responses. Although initially viewed as terminally differentiated, we now recognize that Th cell subsets, including Tfh cells, display substantial flexibility and overlap in their characteristics. In this review, we highlight advances in our understanding of Tfh cell development, cytokine production and the potential plasticity that allows Tfh cells to possess characteristics of other effector Th cell populations.
Keywords: T follicular helper cells, Bcl-6, histone modification, plasticity
T helper cell subsets
Germinal centers (GCs) are specialized structures found within the B cell follicle of secondary lymphoid tissues where high affinity antibodies are generated and where B cells can differentiate into memory B cells and long-lived plasma cells. The development of GCs requires cognate interactions between antigen-specific B cells and a subpopulation of dendritic cell (DC)-primed CD4+ Th cells, known as T follicular helper (Tfh) cells [1]. This process is instrumental for host clearance of many invading pathogens and the development of subsequent protective humoral immunological memory.
Naïve CD4+ T cells receive instructions during their initial encounter with antigen-presenting cells that lead to the development of distinct effector subclasses. CD4+ Th cell subsets were initially described by Mosmann and Coffman, who designated Th1 and Th2 cells based on the mutually exclusive production of either IFN-γ or IL-4, respectively [2]. Cells that produce IL-17A were recently shown to represent yet another Th cell population; additional populations have also been described. Each specific subset has been associated with a ‘master’ transcriptional regulator: Th1 cells with T-bet, Th2 cells with Gata-3 and Th17 cells RORγt [3]. These ‘master’ transcription factors are required for the differentiation, cytokine production and acquisition of effector function of each distinct Th cell subset. Th1 cells, which produce IFN-γ, are important for protective response to intracellular viral and bacterial infections, Th2 cells which produce IL-4 and IL-13 are crucial for barrier protection and eliminating extracellular parasites including helminthes, whereas Th17 cells produce IL-17A and IL-17F and are essential for controlling fungal and bacterial infections [3].
Tfh cells were not initially described based on cytokine production and transcription factor expression patterns, but demarcated as a unique population of activated CD4+ T cells in the GCs of human tonsils that expressed the chemokine (C-X-C motif) receptor 5 (CXCR5) and that were proficient in providing B cell help [4, 5]. With subsequent gene expression and phenotypic analyses, Tfh cells were distinguished from other Th populations based on anatomical localization and elevated expression of cell-surface proteins including Programmed cell death-1 (PD-1), Inducible T-cell costimulator (ICOS), B- and T-lymphocyte attenuator (BTLA) and CD40L [1]. Tfh cells express the cytoplasmic adaptor protein Signal Lymphocyte Activation Molecule (SLAM)-associated protein (SAP), required for lymphocyte interactions, and the transcription factor B-cell lymphoma 6 (Bcl-6), required for Tfh cell differentiation, as well as secrete the cytokine IL-21 [1]. Through expression of these mediators, Tfh cells are essential for GC development; mutations that prevent Tfh cell differentiation or function prevent GC formation and the subsequent development of long-term humoral immunity [1]. However, recent data suggest that Tfh cells can express cytokines characteristic of other Th effector cells, including IL-4 and IFN-γ, consistent with their pivotal role in providing help for cytokine-driven patterns of immunoglobulin class switching [6]. Thus, Tfh cells exhibit overlapping characteristics with Th effector subsets, raising the possibility of flexibility between Tfh cells and other Th effector populations. In this review, we discuss Tfh cell development and the transcription factors that regulate this subset, with the perspective of understanding potential plasticity of this essential cell population.
Tfh cell development
Several lines of evidence suggest that Tfh cells require a multi-stage process for differentiation. DCs are crucial for CD4+ T cell priming and initial acquisition of Tfh cell characteristics, including the induction of Bcl-6 expression [7–9]. Antigen-primed CD4+ T cells in the T cell zone reduce CCR7 and upregulate CXCR5 expression, favoring re-positioning in close proximity to the B cell follicle where they are optimally located to support B cell expansion and differentiation [10–12]. Antigen-specific CD4+ T cells and cognate B cells can then stably interact, forming mobile conjugate pairs leading to T cell migration toward the follicle [13, 14]. Although Bcl-6 expression wanes in most dividing CD4+ T cells, a second wave of increased expression occurs in a subset coincident with interaction with antigen presenting B cells, which drives prolonged CXCR5 expression [7, 8]. These cognate T cell-B cell interactions are required for Tfh cell differentiation and in turn provide signals for B cells to enter into cell cycle and differentiate either extrafollicularly or along a follicular pathway. However, although B cell interactions are thought to be a distinct feature of Tfh cells, other cell populations such as Th2 cells also can require B cell interactions [15].
GC B cells die without T cell help, highlighting their requirement for extrinsic survival signals. In addition, there is clear consensus that B cells are instrumental for the maintenance and function of Tfh cells within the GC [7, 8, 16–19]. Tfh cell GC entry and retention is dependent on cognate interactions [14] such that GC B cells must bind, process and present antigen in an MHCII-dependent manner. Moreover, only pMHCII-binding T cell receptors (TCRs) with the highest affinity are found in Tfh cells within the B cell area, perhaps due to competition for binding to antigen-specific B cells [20]. Thus, GC B cells and Tfh cells are reciprocally dependent on each other for survival, proliferation and differentiation.
Depending on the infectious agent, cytokines produced by the innate immune system can influence Tfh cell development. Initial experiments provided evidence that IL-21 could promote Tfh cell development and IL-21 production [21–23]. However, in vivo analyses suggest that IL-21 acts principally as a paracrine factor for GC B cells, with a limited autocrine role in Tfh cells [24, 25]. Additionally, in vitro and in vivo analyses demonstrate that both IL-21 and IL-6 participate in promoting Tfh cell development and IL-21 production [1]. IL-27 has also been implicated in Tfh cell development, although this is likely via indirect effects on IL-21 production [26]. The redundant nature of these cytokines probably reflects signaling via the transcription factor, signal transducer and activator of transcription (STAT)3. Indeed, patients with STAT3 mutations have a reduction in Tfh cell frequency [27]. In contrast, IL-2 and the downstream activation of STAT5 can negatively regulate Tfh cell differentiation [28, 29]. Interestingly, IL-6 signal transduction in CD4+ T cells can also utilize STAT1 [30]. Notably, loss of the IFN-γR, which also activates STAT1, reduced Bcl-6 expression and improved the autoimmune manifestations observed in Roquinsan/san mice, which have increased Tfh cells and GCs [31]. In vitro analyses have also revealed a role for IL-12 (which activates STAT4) in driving human and murine CD4+ T cells to express Tfh cell markers and help B cells secrete immunoglobulins [32–34]. Together, these studies demonstrate that multiple cytokine pathways contribute to Tfh cell development and subsequent differentiation. Given the crucial role for Tfh cells in humoral immunity, this level of redundancy may have evolved to prevent the detrimental outcome of Tfh cell deficiency.
Cytokine production by Tfh cells
Although each specific Th cell subset has been associated with a particular cytokine profile, there is ample evidence in vivo that Th cells can produce additional cytokines in response to environmental cues. Th17 cells give rise to both IL-17A and IFN-γ producing cells under multiple conditions in vivo [35] and Th2 cells can express T-bet and IFN-γ during a viral infection [36]. It is now appreciated that conceptual Th cell designations oversimplifies the potential of these cell populations, thereby limiting our understanding of immune responses.
The cytokine most closely associated with Tfh cells is IL-21. IL-21 is required for the formation of extrafollicular antibody producing cells [37] and directs GC B cells to maintain proliferation [24, 25]. However, although Tfh cells produce abundant IL-21, Th1, Th2 and Th17 cells also produce IL-21 [1]. In addition, only 20–40% of cells expressing Tfh markers produce IL-21 [38] indicating heterogeneity within the population. Moreover, although the absence of IL-21 results in reduced early antibody production [37] and a reduction in GC B cell numbers [22, 24, 25, 39], the absence of Tfh cells leads to a profound defect in GC formation [40–42]. Thus, Tfh cells likely produce multiple factors that work in concert to induce GC formation and maintenance.
Although Tfh cells can be distinguished from effector Th cells based on cell-surface markers, a growing body of data demonstrates that Tfh cells can express cytokines (IFN-γIL-17, IL-4) characteristic of Th1, Th17 and Th2 effector populations, albeit often at lower levels. Indeed, Tfh cells isolated ex vivo can be induced to express these effector cytokines following secondary restimulation under polarizing conditions in vitro [21]. Moreover, although switch transcripts are expressed by B cells even prior to GC formation [43], directed cytokine secretion by Tfh cells in conjugates with GC B cells has been found to correlate with antibody isotype generation, suggesting a continued role for cytokine production by CD4+ T cells within the GC [6]. Following viral infection, cells with Tfh characteristics produced IFN-γ [38, 44]. IL-17 was identified as a central component of the GC response in the autoimmune prone BXD2 mouse strain [45], and cells displaying a Tfh phenotype produced IL-17 in an experimental autoimmune encephalomyelitis (EAE) model of multiple sclerosis (MS) [46] Whether cells with Tfh characteristics produce IL-17 under non-pathological conditions is not clear. However, both pathogenic and non-pathogenic Th17 cells exist in the repertoire [47] and IL-17 is critical for intestinal homeostasis and IgA secretion [48] as well as host responses to bacterial and fungal infections [3]. Notably, three studies using IL-4 reporter mice elegantly demonstrated that the majority of the IL-4 expressing CD4+ T cells in the lymph node expressed Tfh cell characteristics during a helminth infection [6, 49, 50]. Interestingly, these IL-4 producing Tfh cells localized to the B cell follicle and were functionally distinct from Th2 cells found in peripheral sites [6, 15]. Recently, Liang et al. utilized reporter mice to evaluate the spatial and temporal production of cytokines produced during a Th2-mediated helminth infection [51]. Th2 effector cells found in peripheral tissue produced abundant IL-4 and IL-13 and expressed high levels of Gata-3. In contrast, Tfh cells expressed lower levels of Gata-3, and although these cells produced IL-4, IL-13 was not detected [51]. The mechanism for the discordant activation of IL-4 and IL-13 in these Th cell subsets is not well defined, but suggests that cytokine regulation in Th2 and Tfh cells is divergent (see below). Interestingly, IL-4 production by GC Tfh cells is also seen during viral infection [52] and thus may be particularly important for Tfh cell function.
Tfh cells express multiple transcription factors
There is increasing evidence that Th effector cells not only secrete multiple cytokines but express many transcription factors that are integral for their development and function, beyond their ‘master’ regulators (Figure 1). Thus, modest changes in expression of transcription factors may influence availability to interact with binding partners and affect transcription. Several transcription factors impact Tfh cell differentiation including Bcl-6, basic leucine zipper transcription factor, ATF-like (BATF), c-musculoaponeurotic fibrosarcoma (maf), interferon regulatory factor (IRF)4 and STAT3. Additionally, Tfh cells can express transcription factors characteristic of effector Th cells that are likely to influence cytokine production in response to distinct pathogens.
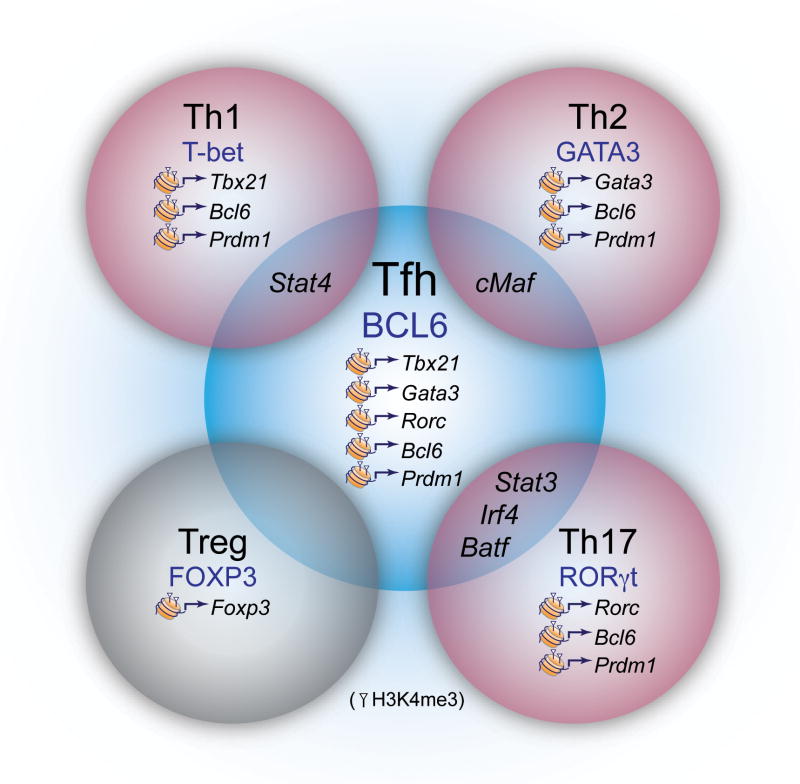
Figure 1.
Flexibility and plasticity of Tfh cells. Tfh cells express a constellation of transcription factors required for development including Bcl-6, BATF, STAT3, IRF4 and c-Maf, many of which are expressed by other Th effector subsets (indicated in the overlap of the cells). Tfh cells can also express transcription factors and Th effector cytokines (Gata-3 for IL-4 and T-bet for IFN-γ) depending on nature of stimulation. Although Th cell subsets express distinct transcription factors, epigenetic analysis has revealed that chromatin modifications may allow flexibility in the expression of ‘master regulator’ transcription factors (H3K4me3 modifications indicative of permissive active chromatin are indicated). Thus, rather than transcription factor expression viewed as ‘all or none’ it may be important to consider the balance of transcription factors expressed. This is likely advantageous for host defense, as both Th effectors and Tfh cell-driven B cell responses are often required for effective pathogen clearance.
Bcl-6 is a member of the BTB-POZ (bric-a-brac, tramtrack, broad complex-poxvirus zinc finger) family that directly binds specific DNA sequences to mediate transcriptional repression. Bcl-6 often functions by binding several co-repressors forming quaternary complexes [1]. Three independent loss-of-function studies demonstrated a T cell intrinsic Bcl-6 expression requirement for Tfh cell differentiation, whereas the development of other Th cell subsets was unaffected [40–42]. Bcl-6 was found to repress a microRNA gene cluster and thus, stabilize the expression of Tfh signature genes including Cxcr5 [42]. There are Bcl-6 binding sites within the Gata3, Tbx21 (encoding T-bet) and Rorc promoters [42, 53], and Bcl-6 overexpression repressed Th1, Th2 and Th17 cell differentiation [40–42]. Consistent with this idea, in Th1 cells, T-bet forms a complex with Bcl-6, masking the Bcl-6 DNA binding domain and preventing it from repressing target genes [54]. Interestingly, IL-12 induces an IL-21 and IFN-γ dual-producing population that expresses T-bet and Bcl-6. However, as T-bet and IFN-γ expression increased, Tfh characteristics declined [34], suggesting that IL-12 initially drives development of an early bipotential population. Indeed, following T. gondii and viral infections, Tbx21−/− mice demonstrate enhanced humoral responses due to a Tfh cell expansion [34, 44]. Thus, threshold levels of T-bet and Bcl-6 may dictate the balance of Th1 and Tfh cell development [34], supporting flexibility between or a succession of Tfh and Th1 populations whereby environmental cues alter Bcl-6 to T-bet ratios.
Bcl-6 induction and function within Tfh cells can also be regulated by ectopic expression of B lymphocyte-induced maturation protein-1 (Blimp-1, encoded by Prdm1), a reciprocal antagonist of Bcl-6. Initially the Bcl-6/Blimp-1 axis was recognized in B cells: GC B cells require Bcl-6, whereas antibody-secreting plasma cell development is contingent on Blimp-1 [1]. Although not absolute, this regulation is also apparent in T cells: ectopic Blimp-1 expression inhibited Tfh cell formation, whereas loss of Blimp-1 enhanced the number of Tfh cells generated during infection [40]. In Th1 cells, high concentrations of IL-2 reduced Bcl-6 expression and increased Blimp-1 expression [55, 56]. Blimp-1 expression was proposed to be responsible for the repression of Tfh signature genes in Th1 cells [56]. However, Blimp-1 also has a crucial role in maintaining immune homeostasis; it regulates Th cell differentiation by limiting IL-2 production (via a negative feedback loop), increasing IL-10 and promoting reactivation induced cell death [57]. Thus, Blimp-1 is multi-functional and is required for regulating many aspects of Th cell differentiation. Moreover, evidence suggests that the Blimp-1 locus is in an open chromatin conformation even in Tfh cells [21]. A simplistic ‘on-off’ expression pattern therefore is unlikely to reflect the complex relationship between these two antagonist proteins in Th cell differentiation. Recently the E3 ubiquitin ligase, Cullin3 was identified as an essential Bcl-6 binding factor. Strikingly, the loss of Cullin 3 had a differential effect on Tfh cell generation and GC formation providing [58] providing support for distinct Bcl-6 driven programs.
The Bcl-6 promoter has binding sites for STAT and Forkhead box O (Foxo) transcription factors. In Th1 polarized cells cultured in low IL-2 levels, STAT3 bound to the to the Bcl-6 promoter while with elevated IL-2, STAT5 bound to the to the Bcl-6 promoter [56]. Interestingly, STAT5 can displace STAT3 complexes and/or recruit repressive complexes [59, 60], providing insight into how IL-2 signaling via STAT5 could restrain Tfh cell differentiation [28, 29]. In contrast, when IL-2 is limiting, Foxo1 and Foxo3 bind to the Bcl-6 promoter and activate transcription [56]. These observations are consistent with data showing a bifurcation between cells expressing elevated levels of the high affinity IL-2 receptor, CD25, and those expressing Bcl-6 [16]. Moreover, during Th1 in vivo responses, CD25 expression inversely correlated with Bcl-6, creating a continuum of Th1 central memory, Tfh and Th1 effector cells [61].
In addition to Bcl-6, several other transcription factors have been implicated in Tfh cell development, including those important for the development of other Th effector cell populations (Figure 1). BATF is basic leucine zipper protein in the AP-1 family that was initially implicated Th17 cell differentiation [62]. Although Batf is only moderately increased in Tfh cells compared to other Th effector populations [63], Batf−/− mice fail to generate Tfh cells [64, 65]. BATF functions in a cell intrinsic manner to directly regulate the expression of Bcl-6 and c-Maf, by binding promoter regions encoding these genes. Overexpression of both Bcl-6 and c-Maf in Batf−/− T cells improved Tfh cell development and GC responses, albeit not to the level of BATF reconstitution [65].
c-Maf is a bZIP transcription factor that binds to the Il4 promoter and plays an instrumental role in Th2 cell development [66]. c-Maf deficiency also results in reduced IL-17 production [46]. Strikingly, ICOS-mediated c-Maf expression is required for IL-21 production and the expansion of Tfh cells [46, 67]. Similarly, STAT3 and IRF4 also bind to the Il21 promoter and are important for the generation of Tfh cells [27, 68]. Similar to BATF, both STAT3 and IRF4 play fundamental roles in Th17 cell development [69–71]. Although IRF4 binds with low affinity to IFN response elements, IRF4 and BATF bind cooperatively to structurally divergent AP-1/IRF composites sites [72, 73]. Whether IRF4/BATF complexes synergize with Bcl-6 and c-Maf to induce a Tfh gene expression signature is not known.
Plasticity of Tfh cells
Although traditionally Th cell subsets were defined as terminally differentiated lineages, there is increasing evidence of flexibility between given Th cell populations. Strikingly, transcription factors thought to be ‘master’ regulators of Th cell lineages, including Gata-3 and T-bet, can also be expressed Tfh cells [38, 44, 50, 51]. These coinciding patterns of cytokine production and transcription factor expression strengthen the concept of flexibility between Tfh cells and other Th cell subsets.
Epigenetic analyses have also provided support for flexibility between Th cell populations (Box 1). Analyses of lysine 4 trimethylation of histone H3 (H3K4me3), which is associated with permissive active chromatin conformations and H3K27 trimethylation (H2K27me3) which is associated with repressive chromatin modifications, have been used to evaluate the chromatin state in a variety of Th cell populations. Genes encoding cytokines typically show permissive marks in the Th subset where they are expressed (ie Ifng in Th1 cells), with repressive marks found in alternate subsets (Th2, Th17) [74]. These modifications may reflect the activation of transcription factors: T-bet not only directly trans-activates Ifng, it recruits jumonji-domain containing histone demethylase3 to remove repressive H3K27me3 modifications as well as recruits the methyltransferase SET7 to induce H3K4me2, creating permissive markings on the Ifng locus [75].
Box 1. Epigenetic Analyses.
The chromatin structure of gene loci can impact gene expression via regulating the accessibility of transcriptional machinery to binding sites in proximal gene promoters and cis-regulatory DNA elements [92, 93]. The N-terminal tails of histone proteins can be covalently marked by the addition or removal of acetyl, methyl, phosphate, ubiquitin, sumoyl or ADP-ribose groups. Such post-translational modifications relax or condense the chromatin structure to activate or repress gene transcription [94, 95] and these epigenetic changes can reflect both the transcriptional patterns as well as the transcriptional potential of DNA. Global analyses of histone modifications, in particular, lysine 4 trimethylation of histone H3 (H3K4me3), which is associated with permissive active chromatin conformations and H3K27 trimethylation (H2K27me3) which is associated with repressive chromatin modifications, have provided support for plasticity between Th cell populations [94, 95]. These studies demonstrated that the genes encoding the master transcription factors for specific subsets showed H3K4me3 markings in multiple Th cell populations, suggesting that they were “poised” for transcription.
DNA can also be methylated on cytosines in CpG dinucleotides and affect gene transcription by influencing binding of transcription factors to promoters and regulatory elements. DNaseI hypersensitivity sites (HS) are defined as regions of chromatin less than a few hundred base pairs that are more sensitive to digestion by DNaseI than bulk chromatin. Many cis-regulatory elements have been identified by the detection of evolutionary conserved sequences and HS sites [78, 96]. Again, the study of these elements has provided insight into the regulation of cytokine transcription in Th cell populations.
Interestingly, ‘lineage specific’ transcription factors do not exhibit such modes of regulation. The Tbx21 gene (encoding T-bet) is associated with accessible marks in Th1 cells. However, in Th2 and Th17 cells, both repressive and, surprisingly, activating marks are found on Tbx21 [21, 74]. Although such data needs to be evaluated carefully given potential heterogeneity within in vitro generated Th cell populations, these observations provide evidence that different Th cell transcription factors may be in a poised state for expression and readily induced in populations that appear committed to a particular subset. Such patterns are also seen in stem cell populations with potential for activating multiple transcription factors [76] and are in accordance with observations of plasticity in Th cell populations.
Consistent with low expression of Th effector cytokines, both in vitro and ex vivo Tfh cells displayed repressive histone markings with low to no detectable positive marks on the genes encoding Il4, Ifng and Il17a compared to conventional Th2, Th1 and Th17 cells, respectively, and positive markings on the Il21 locus [21, 74]. However, surprisingly, Tfh cells maintained accessible marks on Tbx21, Gata3 and Rorc genes (Figure 1). Importantly, both in vitro generated as well as isolated ex vivo derived Tfh cells could produce Th1, 2 and 17 effector cytokines when cultured in polarizing cytokines, while retaining the ability to produce IL-21 [21]. These findings are consistent with the ability of Tfh cells to produce effector cytokines contingent on the infectious agent [6, 15, 38, 49, 50]. Utilizing an IL-21-GFP reporter mouse to track Tfh cells, Luthje et al. demonstrated that Tfh cells gave rise to memory T cells capable of differentiating into both Th effector and Tfh cells in a recall response [38]. Thus, both in vitro and in vivo data establish that Tfh cells maintain the flexibility to differentiate into other Th effector cells upon subsequent stimulation [9, 21, 38]. This feature may allow Tfh cells to preserve facets of Th effector cells depending on the nature of the antigen encountered and contribute to protective immunity.
As anticipated, positive histone modifications were detected at the Bcl6 locus in vitro generated cells with Tfh characteristics as well as ex vivo Tfh cells. However, the Bcl6 locus also displayed permissive marks from in vitro differentiated Th1, Th2 and Th17 populations and these subsets could acquire Tfh characteristics, including increased IL-21 production under the appropriate conditions [21]. Furthermore, although Prdm1 was the most downregulated gene in Tfh cells [40], both positive and repressive modifications of Prdm1 were detected in all Th cell subsets including Tfh cells [21] (Figure 1). The detection of positive histone alterations on both Bcl6 and Prdm1 in diverse Th cell populations reinforces the potential for reprogramming between Tfh and Th effector cell subsets and suggests that a complex regulation of Blimp-1 and Bcl-6 expression occurs during Th cell differentiation.
Regulation of expression of effector cytokines
The extended murine Th2 cytokine locus spanning the Il4, Il5, Il13 and Rad50 genes contain multiple cis-regulatory elements [77, 78] (Box 1). The DNaseI hypersensitivity site (HS)V overlaps with a highly conserved noncoding sequence (CNS2) and has recently been identified as a putative distal enhancer located 3′ of the Il4 gene [79]. Mice deficient in either HSV or CNS2 show a profound reduction in IgG1 and IgE post-immunization, consistent with predicted effects of decreased IL-4 expression. Further examination revealed that although Tfh cells developed, this region was crucial for Tfh cell Il4 expression. In striking contrast, Th2 cells, basophils and eosinophils were less dependent on HSV or CNS2 for IL-4 production [79, 80]. Interestingly, mice deficient in SLAM, which is important for IL-4 production by GC-localized Tfh cells during a viral response [52], displayed reduced permissive modifications of the CNS2 enhancer [80]. These experiments indicate that Th2 and Tfh cells utilize unique yet overlapping mechanisms to regulate Il4 transcription, suggesting that although both populations express IL-4, they still retain distinct characteristics.
Tfh characteristics exploited by other CD4+ T cells
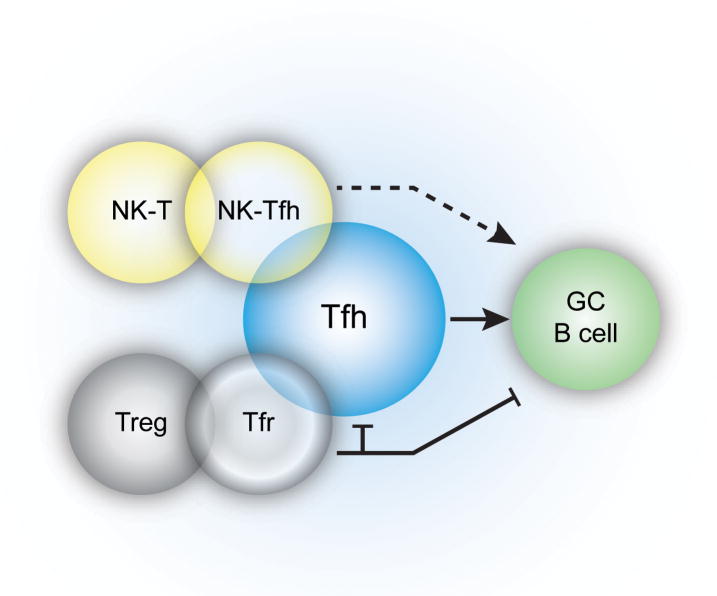
Regulatory T (Treg) cells can express transcription factors and markers associated with effector Th cell populations, which confer upon the Treg cells the ability to suppress specific subsets [81]. Tregs express miR-10a, which functions to limit their plasticity by targeting Bcl-6 and the corepressor, Ncor2, thereby preventing Treg-Tfh cell conversion [82]. However, a subset of Treg cells can localize to the B cell follicle [83, 84], upregulate Bcl-6 and express surface markers CXCR5, PD-1 and ICOS, consistent with a Tfh cell phenotype. These T follicular regulatory (Tfr) cells retain expression of the Treg ‘master’ transcription factor, Foxp3, and suppressive function [85–87]. Tfr cells specifically repress Tfh cells and GC numbers, and consequently prevent the outgrowth of non-antigen specific B cells, perhaps averting the selection of autoreactive B cells [85–88]. Although Tfr cells appear specialized for limiting GC reactions, previous data demonstrated that Foxp3+ cells could convert into functional Foxp3− Tfh cells in Peyer’s patches [84] (Figure 2). However, it is not clear whether these Foxp3+ cells were Treg cells or effectors cells transiently expressing Foxp3 as an activation marker.
Figure 2.
Multiple CD4+ T cell populations can adopt Tfh characteristics. Foxp3+ T regulatory cells and NKT cells can express markers consistent with Tfh cell populations. During immunization with lipid antigen, a subset of NKT cells can express Tfh cell markers and provide B cell help. However, NKTfh cells are not able to induce memory B cell formation and long-lived plasma cells (indicated by the dotted line). Following protein immunization, Foxp3+ Tregs can increase expression of the transcription factor Bcl-6 and express CXCR5, and migrate into the GC where these cells can regulate the GC response. Additionally, Foxp3+ cells found within the intestine, can also upregulate Tfh markers and reduce Foxp3 expression, migrate into the B cell follicle of Peyer’s Patches and function as bone fide Tfh cells.
The majority of splenic natural killer T (NKT) cells patrol the marginal zone where they can be activated by antigen-presenting subcapsulary sinus macrophages, in close proximity to the B cell follicle [89]. Interestingly, a subset of NKT cells has recently been identified that display Tfh cell characteristics (NKTfh cells) and are capable of prolonged contacts with B cells [90, 91] (Figure 2). These NKTfh cells produced IL-21 and provided help to B cells for both extrafollicular and GC responses [90, 91]. However, despite GC formation, neither memory B cells nor long-lived plasma cells were generated [90, 91]. Therefore, NKT cells can access the follicles and adopt a Tfh phenotype, yet remain distinct from conventional CD4+ Tfh cells.
Concluding remarks
As we learn more about the development and characteristics of Tfh cells, it is evident that these cells can acquire characteristics overlapping with other Th effector cells. Such considerations may be particularly important for the field of T cell therapeutics, as we learn that polarized T cells may not maintain specific characteristics when transferred to new hosts.
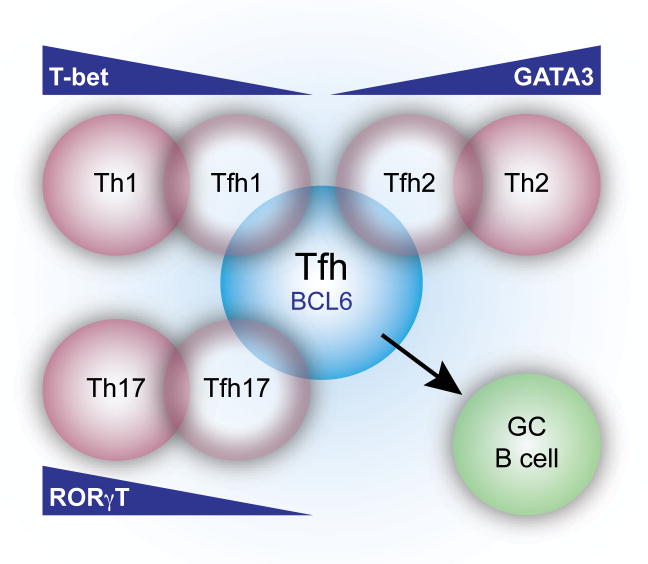
However, many questions remain regarding Tfh cell plasticity. Although Tfh cells can produce defining cytokines of effector Th cell populations, whether Tfh cells acquire other characteristics of these subsets is less clear. Furthermore, the identification of Treg cells and NKT cells that localize to B cell follicles and influence GC formation raises the question of whether plasticity is a feature of multiple CD4+ T cell subsets. Alternatively, another interpretation of these data is that there may be numerous subtle differences in the flavors of T cell populations, so as regulatory T cells adopt Treg1, Treg2 or Tfr cell phenotypes, Tfh cells may assume Tfh1, Tfh2 or Tfh17 cell characteristics depending on the context of activation and type of immune response required (Figure 3). In either model, our definitions of effector CD4+ Th cell subsets need to be viewed dynamically, with the ability to express characteristics of other effector Th cell populations depending on the ratio and timing of transcription factors expressed. This ability to maintain subset fluidity may be crucial for responding to specific types of infection and the acquisition of distinct features required for effective immune responses.
Figure 3.
Diversity of Tfh cells. Tfh cells can further specialize through upregulation or activation of transcription factors in response to different environmental stimuli. For example, during a helminth infection, Tfh cells can express the transcription factor Gata-3 and express IL-4. However, Th2 effector cells preferentially found in peripheral sites express higher levels of Gata-3 (Gata-3 gradient: Th2>Tfh2>Tfh) and produce IL-4 as well as other effector cytokines including IL-13. Thus, we propose a model whereby CD4+ T cells may receive pathogen and milieu specific signals to induce different levels of expression of Th cell subset specific transcription factors, enabling a customized response critical for the development of protective humoral immunity. This model would suggest that Tfh cells are not terminally differentiated, but retain the ability to acquire characteristics of other Th cell subsets upon subsequent challenge.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 2.Mosmann TR, et al. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 3.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol Rev. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitfeld D, et al. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaerli P, et al. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhardt RL, et al. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumjohann D, et al. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J Immunol. 2011;187:2089–2092. doi: 10.4049/jimmunol.1101393. [DOI] [PubMed] [Google Scholar]
- 8.Kitano M, et al. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–972. doi: 10.1016/j.immuni.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. J Exp Med. 2012 doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ansel KM, et al. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J Exp Med. 1999;190:1123–1134. doi: 10.1084/jem.190.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardtke S, et al. Balanced expression of CXCR5 and CCR7 on follicular T helper cells determines their transient positioning to lymph node follicles and is essential for efficient B-cell help. Blood. 2005;106:1924–1931. doi: 10.1182/blood-2004-11-4494. [DOI] [PubMed] [Google Scholar]
- 12.Haynes NM, et al. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 13.Okada T, et al. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3:e150. doi: 10.1371/journal.pbio.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi H, et al. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leon B, et al. Regulation of TH2 development by CXCR5+ dendritic cells and lymphotoxin-expressing B cells. Nat Immunol. 2012;13:681–690. doi: 10.1038/ni.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi YS, et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goenka R, et al. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J Immunol. 2011;187:1091–1095. doi: 10.4049/jimmunol.1100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerfoot SM, et al. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 2011;34:947–960. doi: 10.1016/j.immuni.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poholek AC, et al. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol. 2010;185:313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fazilleau N, et al. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat Immunol. 2009;10:375–384. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu KT, et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 2011;35:622–632. doi: 10.1016/j.immuni.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suto A, et al. Development and characterization of IL-21-producing CD4+ T cells. J Exp Med. 2008;205:1369–1379. doi: 10.1084/jem.20072057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linterman MA, et al. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J Exp Med. 2010;207:353–363. doi: 10.1084/jem.20091738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zotos D, et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J Exp Med. 2010;207:365–378. doi: 10.1084/jem.20091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batten M, et al. IL-27 supports germinal center function by enhancing IL-21 production and the function of T follicular helper cells. J Exp Med. 2010;207:2895–2906. doi: 10.1084/jem.20100064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma CS, et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood. 2012;119:3997–4008. doi: 10.1182/blood-2011-11-392985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballesteros-Tato A, et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnston RJ, et al. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotson AN, et al. The T cell STAT signaling network is reprogrammed within hours of bacteremia via secondary signals. J Immunol. 2009;182:7558–7568. doi: 10.4049/jimmunol.0803666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SK, et al. Interferon-gamma Excess Leads to Pathogenic Accumulation of Follicular Helper T Cells and Germinal Centers. Immunity. 2012;37:880–892. doi: 10.1016/j.immuni.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Ma CS, et al. Early commitment of naive human CD4+ T cells to the T follicular helper (TFH) cell lineage is induced by IL-12. Immunol Cell Biol. 2009;87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt N, et al. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 2009;31:158–169. doi: 10.1016/j.immuni.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayamada S, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirota K, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegazy AN, et al. Interferons direct Th2 cell reprogramming to generate a stable GATA-3+T-bet+ cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Lee SK, et al. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J Exp Med. 2011;208:1377–1388. doi: 10.1084/jem.20102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luthje K, et al. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat Immunol. 2012;13:491–498. doi: 10.1038/ni.2261. [DOI] [PubMed] [Google Scholar]
- 39.Vogelzang A, et al. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 2008;29:127–137. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Toellner KM, et al. T helper 1 (Th1) and Th2 characteristics start to develop during T cell priming and are associated with an immediate ability to induce immunoglobulin class switching. J Exp Med. 1998;187:1193–1204. doi: 10.1084/jem.187.8.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marshall HD, et al. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4+ cell properties during viral infection. Immunity. 2011;35:633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu HC, et al. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat Immunol. 2008;9:166–175. doi: 10.1038/ni1552. [DOI] [PubMed] [Google Scholar]
- 46.Bauquet AT, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao AT, et al. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol. 2012;189:4666–4673. doi: 10.4049/jimmunol.1200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.King IL, Mohrs M. IL-4-producing CD4+ T cells in reactive lymph nodes during helminth infection are T follicular helper cells. J Exp Med. 2009;206:1001–1007. doi: 10.1084/jem.20090313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaretsky AG, et al. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J Exp Med. 2009;206:991–999. doi: 10.1084/jem.20090303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang HE, et al. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat Immunol. 2012;13:58–66. doi: 10.1038/ni.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yusuf I, et al. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kusam S, et al. Inhibition of Th2 differentiation and GATA-3 expression by BCL-6. J Immunol. 2003;170:2435–2441. doi: 10.4049/jimmunol.170.5.2435. [DOI] [PubMed] [Google Scholar]
- 54.Oestreich KJ, et al. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med. 2011;208:1001–1013. doi: 10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gong D, Malek TR. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J Immunol. 2007;178:242–252. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 56.Oestreich KJ, et al. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 58.Mathew R, et al. BTB-ZF factors recruit the E3 ligase cullin 3 to regulate lymphoid effector programs. Nature. 2012;491:618–621. doi: 10.1038/nature11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mandal M, et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat Immunol. 2011;12:1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang XP, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pepper M, et al. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schraml BU, et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rasheed AU, et al. Follicular B helper T cell activity is confined to CXCR5hiICOShi CD4 T cells and is independent of CD57 expression. Eur J Immunol. 2006;36:1892–1903. doi: 10.1002/eji.200636136. [DOI] [PubMed] [Google Scholar]
- 64.Betz BC, et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J Exp Med. 2010;207:933–942. doi: 10.1084/jem.20091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ise W, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho IC, et al. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 67.Kroenke MA, et al. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwon H, et al. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brustle A, et al. The development of inflammatory TH-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 70.Ma CS, et al. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205:1551–1557. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Milner JD, et al. Impaired TH17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glasmacher E, et al. A Genomic Regulatory Element That Directs Assembly and Function of Immune-Specific AP-1-IRF Complexes. Science. 2012;338:975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li P, et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490:543–547. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller SA, et al. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stemberger C, et al. Stem cell-like plasticity of naive and distinct memory CD8+ T cell subsets. Semin Immunol. 2009;21:62–68. doi: 10.1016/j.smim.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 77.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 78.Ansel KM, et al. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 79.Vijayanand P, et al. Interleukin-4 production by follicular helper T cells requires the conserved Il4 enhancer hypersensitivity site V. Immunity. 2012;36:175–187. doi: 10.1016/j.immuni.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harada Y, et al. The 3′ enhancer CNS2 is a critical regulator of interleukin-4-mediated humoral immunity in follicular helper T cells. Immunity. 2012;36:188–200. doi: 10.1016/j.immuni.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 81.Josefowicz SZ, et al. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takahashi H, et al. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol. 2012;13:587–595. doi: 10.1038/ni.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fields ML, et al. CD4+ CD25+ regulatory T cells inhibit the maturation but not the initiation of an autoantibody response. J Immunol. 2005;175:4255–4264. doi: 10.4049/jimmunol.175.7.4255. [DOI] [PubMed] [Google Scholar]
- 84.Tsuji M, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 85.Chung Y, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Linterman MA, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wollenberg I, et al. Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. J Immunol. 2011;187:4553–4560. doi: 10.4049/jimmunol.1101328. [DOI] [PubMed] [Google Scholar]
- 88.Sage PT, et al. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol. 2012 doi: 10.1038/ni.2496. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barral P, et al. The location of splenic NKT cells favours their rapid activation by blood-borne antigen. Embo J. 2012;31:2378–2390. doi: 10.1038/emboj.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chang PP, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol. 2012;13:35–43. doi: 10.1038/ni.2166. [DOI] [PubMed] [Google Scholar]
- 91.King IL, et al. Invariant natural killer T cells direct B cell responses to cognate lipid antigen in an IL-21-dependent manner. Nat Immunol. 2012;13:44–50. doi: 10.1038/ni.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kanno Y, et al. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilson CB, et al. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 96.Wilson CB, et al. DNA methylation and the expanding epigenetics of T cell lineage commitment. Semin Immunol. 2005;17:105–119. doi: 10.1016/j.smim.2005.01.005. [DOI] [PubMed] [Google Scholar]