Abstract
TCR-mediated activation of the Ras signaling pathway is critical for T cell development in the thymus and function in the periphery. However, which members of a family of Ras GTPase-activating proteins (RasGAPs) negatively-regulate Ras activation in T cells is unknown. In this study we examined a potential function for the neurofibromin 1 (NF1) RasGAP in the T cell lineage with the use of T cell-specific NF1-deficient mice. Surprisingly, on an MHC class I-restricted TCR transgenic background, NF1 was found to promote thymocyte positive selection. By contrast, NF1 neither promoted nor inhibited the negative selection of thymocytes. In the periphery, NF1 was found to be necessary for the maintenance of normal numbers of naïve CD4+ and CD8+ T cells but was dispensable as a regulator of TCR-induced Ras activation, cytokine synthesis, proliferation and differentiation and death. These findings point to a novel unexpected role for NF1 in T cell development as well as a regulator of T cell homeostasis.
Keywords: T cells, Signal transduction, Ras GTPase-activating proteins, Neurofibromin
1. Introduction
TCR-mediated activation of the Ras signaling pathway is essential for most T cell responses. In the thymus, Ras signaling is required for pre-TCR-induced maturation of thymocytes from the CD4−, CD8− double-negative (DN)1 to the CD4+, CD8+ double-positive (DP) stage of development (Crompton et al., 1996; Fischer et al., 2005). Furthermore, Ras signaling is necessary for thymocyte positive selection during which DP cells receive survival signals and differentiate into single positive (SP) CD4+ or CD8+ T cells depending upon the specificity of their TCR for MHC class II or I respectively (Alberola-Ila et al., 1995; Alberola-Ila and Hernandez-Hoyos, 2003; Fischer et al., 2005; Swan et al., 1995). In the peripheral immune system, Ras is required for T cell cytokine secretion, proliferation and differentiation into effector cells (Dong et al., 2002; Rincon et al., 2001).
Ras is an inner membrane-tethered small GTP-binding protein that in quiescent cells exists predominantly in an inactive GDP-bound state (Wennerberg et al., 2005). The TCR promotes activation of Ras through its ability to recruit to membranes and activate mammalian son of sevenless (mSOS) and Ras guanine nucleotide releasing protein 1 (RasGRP1) molecules (Das et al., 2009; Ebinu et al., 2000; Kortum et al., 2012; Lapinski and King, 2012; Priatel et al., 2002; Roose et al., 2005; Roose et al., 2007). Both proteins function as Ras guanine nucleotide exchange factors that eject GDP from the Ras guanine nucleotide-binding pocket thereby allowing to Ras to bind to GTP resulting in its activation. In its GTP-bound state Ras triggers different downstream signaling events that include the activation of MAPK that drive T cell responses (Chang and Karin, 2001; Genot and Cantrell, 2000).
While much is understood of the mechanisms involved in the activation of Ras in T cells, less is known of how Ras is inactivated in this cell type. Efficient Ras inactivation requires physical interaction with Ras GTPase activating proteins (RasGAPs) that increase the ability of Ras to hydrolyse GTP by several orders of magnitude (Donovan et al., 2002; King et al., 2013). RasGAPs contain a catalytically active GAP domain together with one or more modular binding domains that allow RasGAP targeting to membranes and juxtaposition to Ras (Bernards, 2003; Donovan et al., 2002; King et al., 2013). Ten different RasGAPs have now been reported and most are expressed in T cells. Therefore, which RasGAPs are required for inactivation of Ras in T cells is currently uncertain.
Two prototypical members of the RasGAP family are p120 RasGAP (RASA1) and neurofibromin 1 (NF1). To address the importance of RASA1 in T cells we recently generated T cell-specific RASA1-deficient mice (Lapinski et al., 2011). On a non-TCR transgenic (Tg) background, pre-TCR selection and positive selection were not affected by the loss of RASA1. However, on an MHC class II-restricted TCR (Tg) background, enhanced positive selection was observed that was associated with increased activation of the Ras-MAPK pathway in DP thymocytes. In addition, in the periphery of non-TCR Tg and TCR Tg mice, reduced numbers of naïve CD4+ and CD8+ T cells were observed in RASA1-deficient mice that could be explained by impaired naïve T cell survival. However, RASA1 was found to be dispensable for peripheral T cell activation and function induced by full agonist MHC-peptide complexes. Thus, while RASA1 appears to be important for T cell development and homeostasis, other RasGAPs are likely to participate in these processes and to control T cell activation in the periphery.
In humans, germline mutations in NF1 cause the autosomal dominant disorder neurofibromatosis 1 that is characterized by the development of benign dermal neurofibromas, skin pigmentation abnormalities, skeletal defects and learning disabilities (Cawthon et al., 1990; Viskochil et al., 1990; Wallace et al., 1990). In addition, neurofibromatosis 1 patients show increased susceptibility to a variety of other benign and malignant tumors including myeloid leukemia (Hope and Mulvihill, 1981). Mice that are homozygous for an Nf1 null mutation show impaired cardiac development and die in utero at E14 whereas heterozygote NF1-deficient mice show age-related susceptibility to a variety of tumors (Brannan et al., 1994; Jacks et al., 1994). To examine a potential role for NF1 in the development and function of T cells, Ingram et al, transferred bone marrow (BM) from NF1-deficient mice into immunocompromised RAG2-deficient mice (Ingram et al., 2002). Recipients exhibited thymic and splenic hyperplasia as a result of an increase in the number of all thymic and splenic T cell subsets. Thymocytes showed elevated levels of Ras-GTP and proliferated spontaneously in vitro. However, both thymocytes and splenic T cells proliferated less in response to TCR stimulation or to the combination of TCR plus IL-2 stimulation compared to wild type T cells. These findings are consistent with a negative regulatory role for NF1 in T cell development and a potential positive regulatory role for this RasGAP in peripheral T cells. However, it is unclear if the T cell developmental and functional defects observed in these experiments result from loss of NF1 in T cells or are secondary to loss of NF1 in a non-T cell hematopoietic compartment. This is particularly important in the context that mice that receive NF1-deficient hematopoietic precursors also develop myeloproliferative disorders (Bollag et al., 1996; Largaespada et al., 1996).
To address the question of a T cell intrinsic role for NF1 we have used a conditional NF1 allele to generate T cell-specific NF1-deficient mice (Zhu et al., 2002; Zhu et al., 2001). Contrary to previous findings, expansion of the T cell compartment was not observed in these mice. Instead, similar to T cell-specific RASA1-deficient mice, diminished numbers of peripheral naïve T cells were found in T cell-specific NF1-deficient mice. Furthermore, T cell activation and differentiation proceeded normally in these mice. However, on an MHC class I-restricted TCR Tg background, an important function for NF1 in thymocyte positive selection was revealed. These studies clarify and provide further insight into the role of NF1 within the T cell compartment.
2. Materials and Methods
2.1 Mice
Nf1fl/fl mice have been described (Zhu et al., 2001). Mice were crossed with plck-cre Tg mice (Taconic) to generate Nf1fl/fl plck-cre mice and littermate NF1fl/fl controls. Nf1fl/fl plck-cre mice were crossed with AND and HY TCR Tg mice (JAX and Taconic respectively) to generate AND and HY TCR Tg Nf1fl/fl plck-cre mice and littermate AND and HY TCR Tg Nf1fl/fl controls. Genotype of mice was determined by PCR of tail genomic DNA using PCR primers described previously (Zhu et al., 2001). All NF1 mutant mice used in this study are on a mixed 129S6/Sv × C57BL/6 (H-2b) background and were 2–3 mo of age at the time of experiments. C57BL/6 (H-2b) and B10.BR (H-2k) mice were purchased from JAX. All experiments were performed in compliance with University of Michigan guidelines and were approved by the University Committee on the Use and Care of Animals.
2.2 Flow cytometry
Subpopulations of thymocytes, splenocytes and lymph node (LN) cells were enumerated by cell counting and flow cytometry using fluorochrome-conjugated HY TCR, TCR Vα 11 and Vα 3, CD4, CD8, CD44, CD24, CD25, CD90.2 and CD69 mAb (Becton Dickinson). Cell viability was determined by staining with fluorochrome-coupled annexin V and 7-amino-actinomycin D (7AAD) (Becton Dickinson). To examine MAPK activation in female HY TCR Tg DP thymocytes, 1.5 × 106 total thymocytes were mixed with an equal number of splenic adherent cells (APC) from female C57BL/6 mice that had been pre-pulsed with HY peptide (10 µM) for 1 h. Cells were co-pelleted and incubated at 37°C for different times before fixation and permeabilization and analysis of MAPK activation by flow cytometry using a fluorochrome-coupled phospho-ERK MAPK mAb (Cell Signaling) as described (Lapinski et al., 2011). All cell staining was analyzed on a FACSCanto (Becton Dickinson).
For flow cytometric sorting of thymocytes for quantitative PCR analysis, cells were stained with CD4, CD8, CD44, CD24, CD25 and CD90.2 mAb. DN1 (CD90.2+, CD24hi, CD4−, CD8−, CD44+, CD25−), DN2 (CD90.2+, CD24hi, CD4−, CD8−, CD44+, CD25+), DN3 (CD90.2+, CD24hi, CD4−, CD8−, CD44lo, CD25+), DN4 (CD90.2+, CD24hi, CD4−, CD8−, CD44−, CD25−), DP (CD90.2+, CD4+, CD8+), CD4 SP (CD90.2+, CD4+, CD8−) and CD8 SP (CD90.2+, CD4−, CD8−) were sorted on an iCyt Synergy FACS machine (Sony Biotechnology).
2.3 Cell isolation
Thymocytes from female HY TCR Tg mice were depleted of CD8 SP cells by positive selection using CD4 mAb-coated immunobeads (Miltenyi). Peripheral pan-T cells, CD4+ and CD8+ T cells from non-TCR Tg mice, CD8+ T cells from female HY TCR Tg mice and naïve CD4+CD44− T cells from AND TCR Tg mice were isolated from spleen and LN by negative selection using immunobeads (Miltenyi or StemCell Technologies). To generate CD4+ T cell blasts, CD4+ T cells from non-TCR Tg mice were stimulated in complete medium (RPMI 1640 containing FCS and antibiotics) in 24 well plates that had been pre-coated with CD3 mAb (1 µg/ml; eBioScience). Soluble CD28 mAb (1 µg/ml; eBioScience) plus IL-2 (100 ng/ml; R&D Systems) were included in the culture medium. After 6 d, cells were washed and recultured in IL-2 (100 ng/ml) in complete medium for a further 2 d. Cells were washed and cultured overnight in complete medium before use in assays.
2.4 Quantitative PCR
Genomic DNA was prepared from sorted thymocyte populations using a Qiagen Blood and Tissue DNA kit. Efficiency of Nf1 gene deletion was determined by qPCR using a primer/probe set located at the Nf1 intron 31-exon 32 border (Applied Biosystems). A transferrin receptor primer/probe set was used as an internal control for all samples (Applied Biosystems). For each thymocyte population, the relative amount of Nf1 target in Nf1fl/fl plck-cre mice compared to Nf1fl/fl mice was calculated, as described (Oliver et al., 2012; Schmittgen and Livak, 2008).
2.5 Western blotting
CD8 SP-depleted female HY TCR Tg thymocytes were stimulated with HY peptide-pulsed APC as indicated in flow cytometry. CD4+ and CD8+ T cells and CD4+ T cell blasts from non-TCR Tg mice were stimulated with CD3 and CD28 mAb in U-bottomed 96 well plates without IL-2 (1 × 106/well) as above. After different times of stimulation cells were lysed and activation of the Ras-MAPK pathway was determined by Western blotting using a phospho-ERK antibody (Cell Signaling).
2.6 Apoptosis assays
Thymocytes were cultured in 96 well flat-bottomed plates at 2 × 105 cells/well in complete medium in the presence of the indicated concentrations of CD3 and CD28 mAb, dexamethasone or staurosporine (Sigma). After 20 h culture, cells were harvested and viability was assessed by flow cytometry.
Activation induced death (AICD) of peripheral T cells was assessed as before (Lapinski et al., 2008). Briefly, purified splenic CD4+ T cells were stimulated with CD3/CD28 mAb plus IL-2 for 72 h, washed and recultured in IL-2 in complete medium for 48 h as above. After a further wash, cells were cultured in IL-2 in complete medium or in complete medium alone for an additional 48 h at which point cell viability was assessed by flow cytometry.
2.7 T cell proliferation
Pan T cells were labeled with CFSE (10 µg/ml) and stimulated in 96 well U-bottomed plates with CD3 and CD28 mAb in complete medium as indicated in Western blotting. Dilution of CFSE fluorescence was determined by flow cytometry after 3 d of culture.
2.8 T cell cytokine synthesis
Peripheral CD8+ T cells from female HY TCR Tg mice and CD4+, CD44− T cells from AND TCR Tg mice were incubated with HY peptide-pulsed APC from female C57BL/6 mice or with PCC peptide-pulsed APC from B10.BR mice respectively in complete medium in 96 well U-bottomed plates (2 × 105 responders and 1 × 106 APC/well). Concentrations of IL-2 and IFN-γ in culture supernatants were determined by ELISA after 48 h of culture (R&D Systems).
2.9 T cell polarization
T cell polarization was performed as described (Bauler et al., 2008). Briefly, CD44−, CD4+ T cells from non-TCR Tg mice were stimulated with CD3 and CD28 mAb in complete medium with IL-2 (100 ng/ml) as indicated in Western blotting. For Th1 skewing, IL-12 (10 ng/ml; R&D Systems) and IL-4 neutralizing mAb (10 µg/ml; BD Pharmingen) were added to the culture medium. For Th2 skewing, IL-4 (10 ng/ml; R&D Systems) and IFN-γ neutralizing mAb (10 µg/ml; BD Biosciences) were added to the culture medium. After 3 d, cells were transferred to 48 well plates and cultures were supplemented with IL-2 (100 ng/ml). After a further 2 d, cells were washed and 1 × 106 cells were restimulated in complete medium in 96 well U-bottomed plates that had been pre-coated with CD3 mAb (1 µg/ml). Concentrations of IFN-γ and IL-4 in supernatants were determined by ELISA.
3. Results
3.1 T cell development in T cell-specific NF1-deficient mice
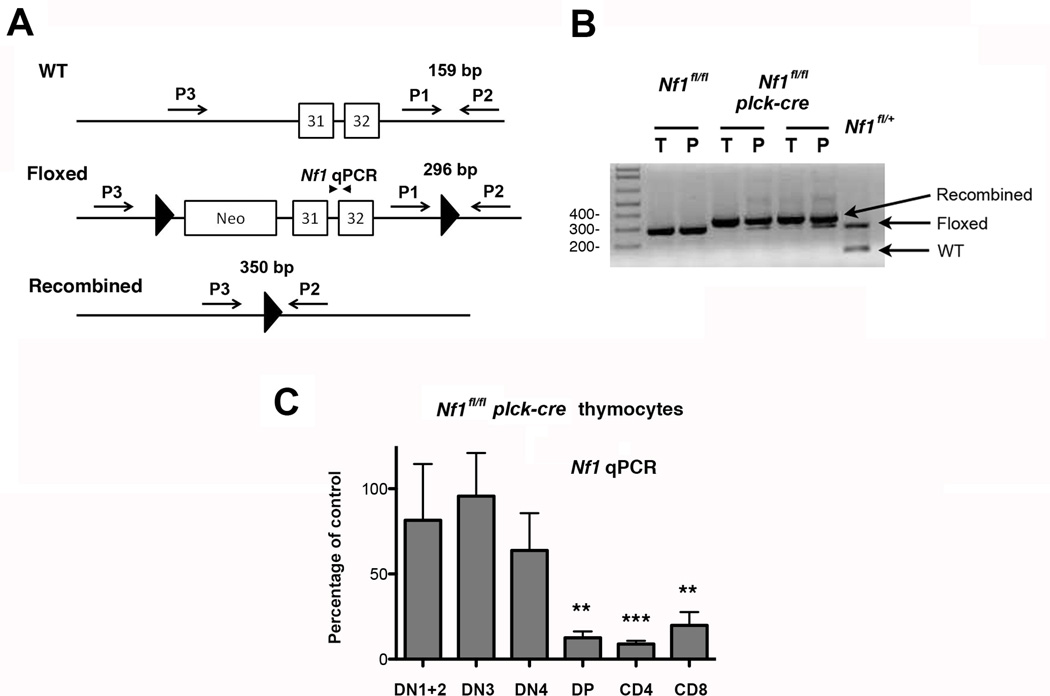
To generate T cell-specific NF1-deficient mice we crossed Nf1fl/fl mice with Tg mice that express the cre recombinase under control of the proximal lck promoter (plck-cre). In these mice, cre is expressed from the DN3 (CD44lo, CD25+) stage of T cell development onward, i.e. is first expressed prior to the TCRβ selection checkpoint (Hennet et al., 1995). PCR analysis of genomic DNA using previously published primer sets indicated that recombination at the Nf1 locus, which results in a null Nf1 allele, occurred in the vast majority of thymocytes and peripheral T cells from Nf1fl/fl plck-cre mice (Fig. 1A and B) (Zhu et al., 2001). To confirm this finding and to determine the extent of NF1 deletion in different thymocyte subsets, we performed qPCR upon genomic DNA prepared from FACS-purified subpopulations using a primer/probe set located at the Nf1 intron 31-exon 32 boundary that is deleted upon cre-mediated recombination (Fig. 1A and C). These analyses revealed little deletion of Nf1 in DN1 (CD44hi, CD25−), DN2 (CD44hi, CD25−) or DN3 thymocytes from Nf1fl/fl plck-cre mice compared to Nf1fl/fl mice. Some deletion was observed in DN4 (CD44−, CD25−) thymocytes, albeit this was not statistically significant over repeat experiments. By contrast, substantial deletion of Nf1 (80–90%) was observed in DP and SP thymocytes from Nf1fl/fl plck-cre mice (Fig. 1C).
Fig. 1.
T cell-specific deletion of NF1. A, Shown is part of the wild type (WT), floxed and recombined Nf1 locus and position of primers P1-P3 (arrows) and qPCR primers (arrowheads) used to determine the extent of NF1 deletion in Nf1fl/fl plck-cre mice. B, Genomic DNA was extracted from whole thymus (T) and purified peripheral splenic and LN T cells (P) from Nf1fl/fl and Nf1fl/fl plck-cre mice and analyzed by PCR using primers P1-P3 in a single reaction. Shown are the results of a representative analysis performed with two Nf1fl/fl plck-cre mice and a littermate Nf1fl/fl control. For comparison, the same PCR reaction was performed upon tail genomic DNA from a Nf1fl/+ mouse. C, Genomic DNA was prepared from the indicated sorted thymocyte subpopulations from littermate Nf1fl/fl and Nf1fl/fl plck-cre mice and relative Nf1 gene amounts were determined by qPCR using primers shown in A. For each subpopulation, the percentage amount of Nf1 in Nf1fl/fl plck-cre mice compared to Nf1fl/fl mice was calculated. Experiments were performed with three independent pairs of mice. Shown is the mean percentage of Nf1 expression + 1 SEM in Nf1fl/fl plck-cre mice compared to Nf1fl/fl mice. Statistical significance was determined using a Student one-sample t test. **p <0.01, ***p <0.001.
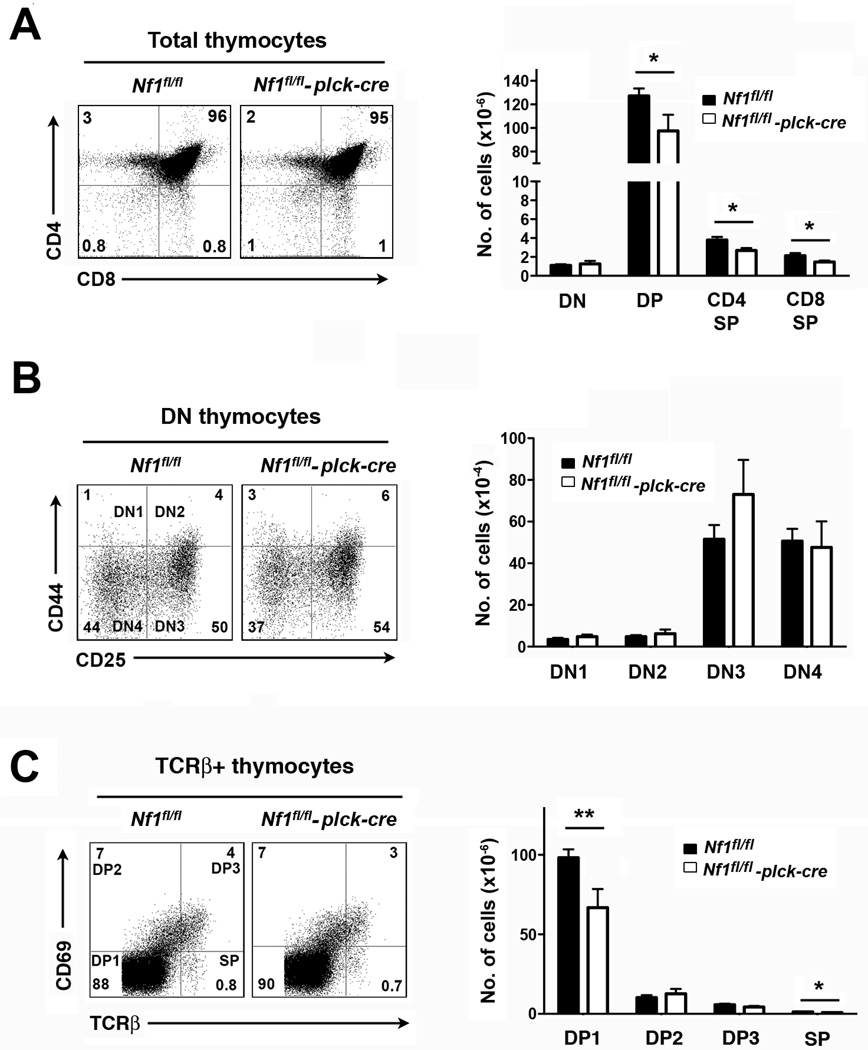
Contrary to previous findings in bone marrow chimeras (Ingram et al., 2002), thymic cellularity was not increased in the absence of NF1 in this model. In fact, numbers of DP and SP thymocytes were moderately decreased in Nf1fl/fl plck-cre mice compared to littermate controls (Fig. 2A). Numbers of DN thymocytes were not affected in Nf1fl/fl plck-cre mice, consistent with the poor deletion of the Nf1, gene in DN3 and DN4 subsets (Fig. 2B). Ratios of CD4 SP or CD8 SP to DP thymocytes in Nf1fl/fl plck-cre thymi were not altered compared to controls suggesting that positive selection is unaffected by loss of NF1 (Fig. 2A). To explore this further, we performed TCRβ versus CD69 staining to determine numbers of pre-positive selection DP thymocytes (DP1; TCRβ +CD69−) and immediate post-positive selection DP thymocytes (DP2; TCRβ +CD69+) (Fig. 2C) (Hernandez-Hoyos et al., 2003; Lauritsen et al., 2008). The DP2/DP1 ratio was slightly higher in Nf1fl/fl plck-cre thymi suggestive of increased positive selection. However, this increase could be explained entirely by a decrease in the number of DP1 cells and no increase in the number of DP2 cells was evident. This finding, therefore, argues against increased positive selection in this model.
Fig. 2.
T cell development in T cell-specific NF1-deficient mice. All experiments were performed with littermate Nf1fl/fl and Nf1fl/fl plck-cre mice. A, At left are shown representative two-color flow cytometry plots of CD4 versus CD8 staining of whole thymi (CD90.2+ gate). Numbers indicate the percent of cells in each quadrant. The bar graph at right shows the mean number plus 1 SEM of the indicated thymocyte populations (Nf1fl/fl, n=10; Nf1fl/fl plck-cre n=7). B, At left are shown representative plots of CD44 versus CD25 staining of gated DN thymocytes (CD90.2+ and CD24hi gates). The bar graph at right shows the mean number plus 1 SEM of DN1 (CD44+, CD25−), DN2 (CD44+, CD25+), DN3 (CD44lo, CD25+) and DN4 (CD44−, CD25−) thymocytes (Nf1fl/fl, n=10; Nf1fl/fl plck-cre n=7). C, At left are shown representative plots of CD69 versus TCRβ staining of thymocytes (TCRβ gate). The bar graph at right shows the mean number plus 1 SEM of DP1 (TCRβ +CD69−), DP2 (TCRβ +CD69+), DP3 (TCRβ hi, CD69+) and SP (TCRβ hi, CD69−) thymocytes (Nf1fl/fl, n=10; Nf1fl/fl plck-cre n=7). Statistical significance was determined using a Student two-sample t test. *p < 0.05, **p <0.01.
3.2 A role for NF1 in positive selection in HY TCR Tg mice
We also examined a role for NF1 in positive selection in TCR Tg mice. The rationale was that any alterations in positive selection might be more readily revealed in a TCR monoclonal setting. To this end, we generated AND TCR and HY TCR Tg Nf1fl/fl plck-cre mice and Nf1fl/fl controls. The AND TCR recognizes PCC peptide 88–104 in the context of the MHC class II molecule, I-Ek. In C57BL/6 × 129 S/v H-2b mice, DP thymocytes that express the AND TCR are positively selected on I-Ab and mature into CD4 SP cells (Kaye et al., 1989). The HY TCR recognizes the male specific HY peptide in the context of the MHC class I molecule, H-2Db. In female C57BL/6×129 S/v H-2b mice, DP thymocytes that express the HY TCR are positively selected on H-2Db and mature into CD8 SP cells (Kisielow et al., 1988). Therefore, use of these two models affords the opportunity to examine positive selection of MHC class II and I-restricted T cells at a clonal level.
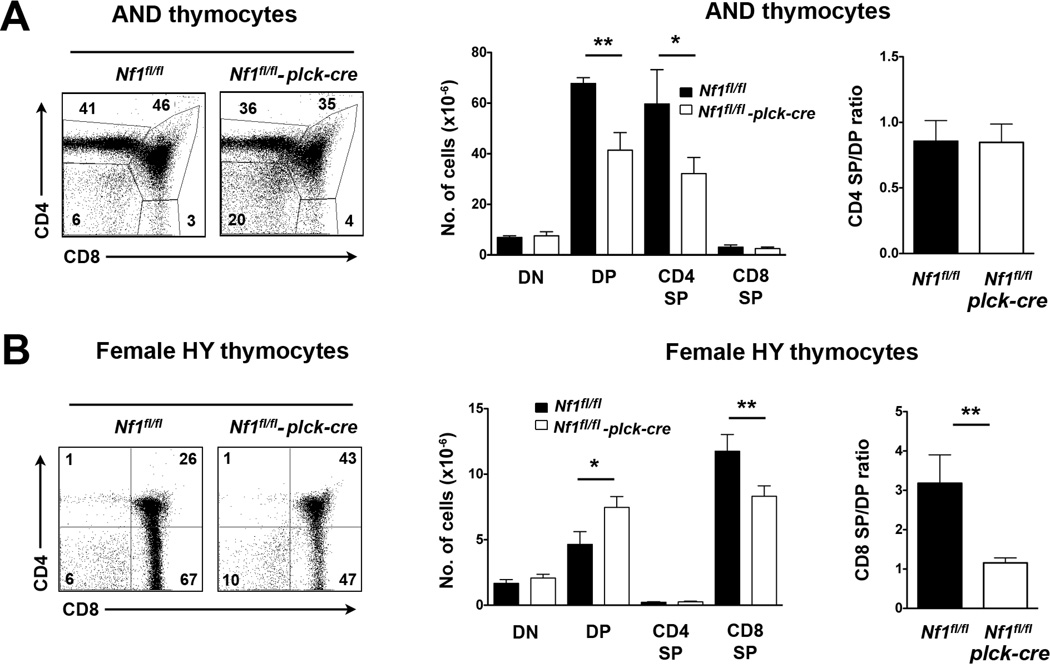
In AND TCR Tg mice, absence of NF1 resulted in substantially reduced numbers of DP and CD4 SP cells as observed in non-TCR Tg mice (Fig. 3A). Loss of RASA1 similarly resulted in a substantial decrease in the number of DP cells in AND TCR Tg thymi and was attributed to increased death of DP cells as evidenced by increased annexin V cell surface staining (Lapinski et al., 2011). Likewise, DP and SP cells in AND TCR Tg NF1-deficient thymi showed increased annexin V cell surface staining (data not shown). However, despite the decreased survival of DP and SP cells, the CD4 SP/DP ratio was not changed in NF1-deficient AND TCR Tg thymi (Fig. 3A). This indicates that NF1 does not participate in the positive selection of AND TCR Tg T cells.
Fig. 3.
Positive selection in T cell-specific NF1-deficient TCR Tg mice. A, All experiments were performed with littermate Nf1fl/fl and Nf1fl/fl plck-cre AND TCR Tg mice. At left are shown representative two-color flow cytometry plots of CD4 versus CD8 staining of AND TCR+ thymocytes (TCR Vα 11+, Vα 3+) in whole thymi. The bar graph at middle shows the mean number plus 1 SEM of the indicated thymocyte populations (Nf1fl/fl, n=5; Nf1fl/fl plck-cre n=10). For each mouse the CD4 SP/DP ratio was calculated. The bar graph at right shows the mean CD4 SP/DP ratio plus 1 SEM. B, All experiments were performed with littermate female Nf1fl/fl and Nf1fl/fl plck-cre HY TCR Tg mice. At left are shown representative two-color flow cytometry plots of CD4 versus CD8 staining of HY TCR+ thymocytes (reactive with clonotypic HY TCR antibody) in whole thymi. Bar graphs show the mean number plus 1 SEM of the indicated thymocyte populations and the mean CD8 SP/DP ratio plus 1 SEM calculated as in A (n=6 mice each genotype). Statistical significance was determined using a Student two-sample t test. *p < 0.05, **p <0.01.
In HY TCR Tg mice a different effect of NF1 deletion was observed. Loss of NF1 in these Tg mice did not affect overall thymic cellularity. However, a significant increase in the number of DP cells was observed together with a corresponding decrease in the number of CD8 SP cells (Fig. 3B). Consequently, the CD8 SP/DP ratio decreased three-fold in HY TCR Tg Nf1fl/fl plck-cre mice thymi compared to Nf1fl/fl control thymi (Fig. 3B). This change cannot be explained by a non-specific effect of the plck-cre transgene upon positive selection in the HY TCR Tg model since no alteration in the efficiency of positive selection was observed in Rasa1fl/fl plck-cre mice previously (Lapinski et al., 2011). Thus, NF1 is required for efficient positive selection of HY TCR Tg T cells.
3.3 NF1 is not required for negative selection
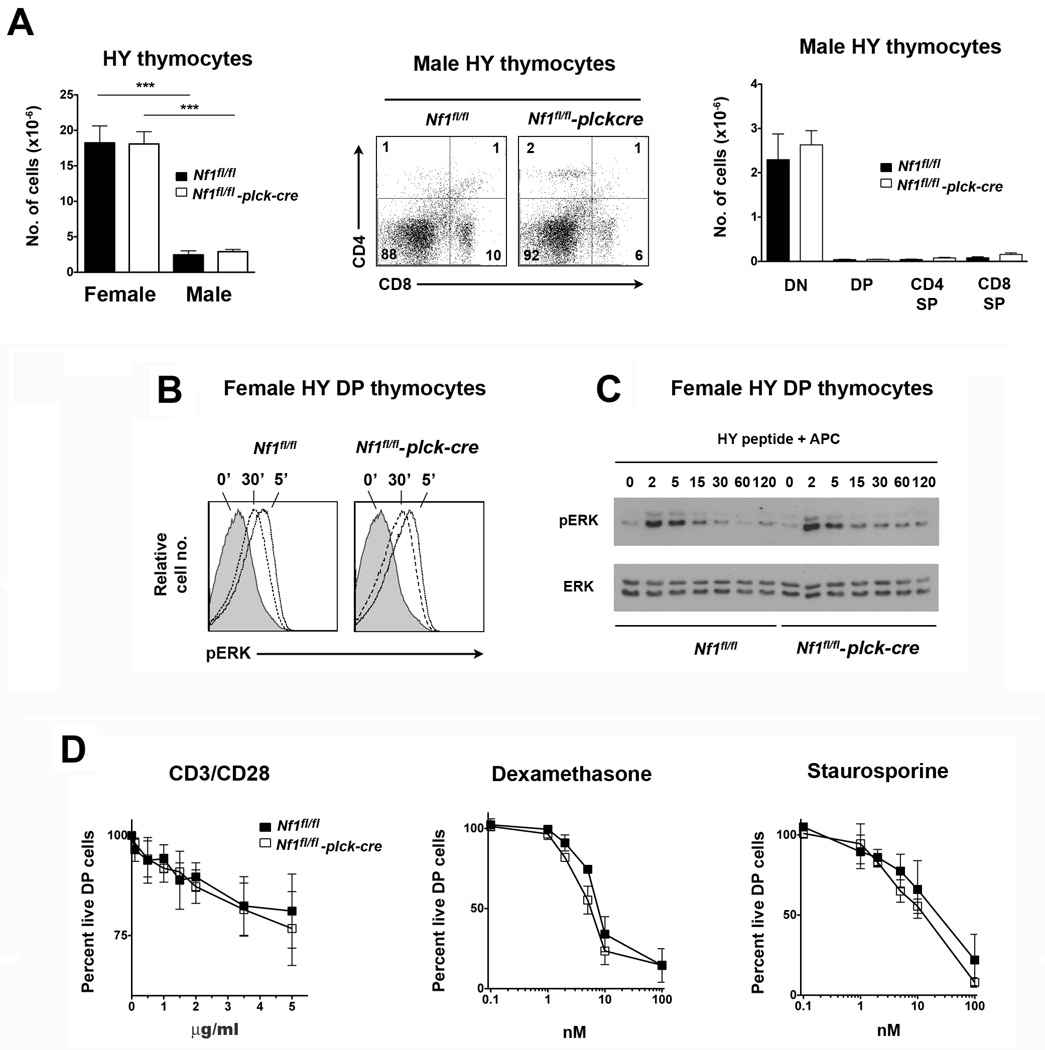
In male HY TCR Tg mice expression of the HY antigen on thymic stromal cells results in negative selection of HY TCR Tg thymocytes (Kisielow et al., 1988). Therefore, we examined thymocyte development in male Nf1fl/fl plck-cre HY TCR Tg mice to determine if NF1 is also required for negative selection in this model. As expected, much fewer HY TCR Tg T cells were found in control Nf1fl/fl male compared to Nf1fl/fl female HY TCR Tg thymi, which is explained by negative selection of HY TCR Tg T cells at the DP stage (Fig. 4A). A similarly small number of total and DP HY TCR Tg T cells were found in thymi of male NF1fl/fl plck-cre HY TCR Tg mice. Thus, NF1 is not required for negative selection of HY TCR Tg thymocytes.
Fig. 4.
Negative selection in T cell-specific NF1-deficient mice. A, Experiments were performed with littermate Nf1fl/fl and Nf1fl/fl plck-cre HY TCR Tg mice. Bar graph at left shows the mean number of HY TCR+ thymocytes plus 1 SEM in female compared to male mice (n=6 females and 5 males of each genotype). In middle is shown a representative CD4 versus CD8 flow cytometry plot of male thymi gated upon HY TCR+ cells. Bar graph at right shows the mean number plus 1 SEM of the indicated HY TCR+ populations in male thymi (n=5 mice of each genotype). B, Whole thymocytes from female Nf1fl/fl and Nf1fl/fl plck-cre HY TCR Tg mice were stimulated with HY peptide-pulsed H-2b APC in vitro for the indicated times. Ras-MAPK activation within gated HY TCR+ DP thymocytes was determined by flow cytometry using a phospho-ERK mAb. C, DP thymocytes were stimulated with HY peptide-pulsed APC as in B. ERK activation was determined by Western blotting using a phospho-ERK antibody. For B and C, similar results were obtained in two repeat experiments. D, Thymocytes from littermate Nf1fl/fl and Nf1fl/fl plck-cre non-TCR Tg mice were stimulated with CD3 and CD28 mAb, dexamthasone or staurosporine at the indicated concentrations. After 20 h the percentage of live DP cells was determined by flow cytometry. Results were normalized to the percentage of live DP cells that were cultured in the absence of stimuli. Shown is the mean percentage of live DP cells + 1 SEM (n=3 mice of each genotype). Statistical significance was determined using a Student two-sample t test. ***p <0.001.
In contrast to positive selection, negative selection is independent of MAPK (McGargill et al., 2009). Therefore, lack of a role for NF1 in negative selection of HY TCR Tg thymocytes might not necessarily reflect an inability of NF1 to regulate Ras-MAPK activation in DP cells in response to strong agonist stimuli. Furthermore, although negative selection does not require MAPK, excessive MAPK activation could potentially augment negative selection. However, any increased negative selection resulting from loss of an inhibitory function of NF1 toward Ras would be obscured in the male HY TCR Tg model since the negative selection that occurs already causes almost complete depletion of DP cells. Therefore, we examined directly if NF1 regulates the Ras-MAPK pathway in HY TCR Tg DP thymocytes in response to a negatively selecting MHC-HY peptide stimulus. For this purpose, whole thymocytes from female HY TCR Tg Nf1fl/fl plck-cre and Nf1fl/fl mice were stimulated in vitro with HY peptide-pulsed H-2b APC and the extent of activation of the Ras-MAPK pathway in DP cells was assessed by flow cytometry using a phospho-ERK mAb (Fig. 4B). As shown, the Ras-MAPK pathway was activated to the same degree in NF1-deficient and control DP cells by HY peptide. To confirm these findings, we also examined activation of the Ras-MAPK pathway in HY TCR Tg DP cells by Western blotting. In these experiments, HY TCR Tg thymi were first depleted of CD8 SP T cells and then stimulated with HY peptide-pulsed H-2b APC before lysis and examination of ERK activation by Western blotting (Fig. 4C). In agreement with data from flow cytometric analyses, ERK was activated to the same degree and with similar kinetics in NF1-deficient DP thymocytes compared to control DP thymocytes in these experiments. Thus, NF1 does not regulate activation of the Ras-MAPK pathway in HY TCR Tg DP cells stimulated with full MHC-HY peptide agonist.
We also examined apoptosis of non-TCR Tg DP cells in vitro in response to CD3 mAb (directed to the TCR complex) and mAb against the CD28 costimulatory receptor (Fig. 4D). Apoptotic responses were the same for NF1-sufficient and NF1-deficient DP cells over a range of CD3/CD28 mAb concentrations. Likewise, apoptosis in response to the recognized apoptosis-inducing agents dexamethasone and staurosporine was not impaired in NF1-deficient DP cells. Therefore, loss of NF1 in thymocytes neither inhibits nor promotes negative selection.
3.4 Diminished numbers of naïve T cells in T cell specific NF1-deficient mice
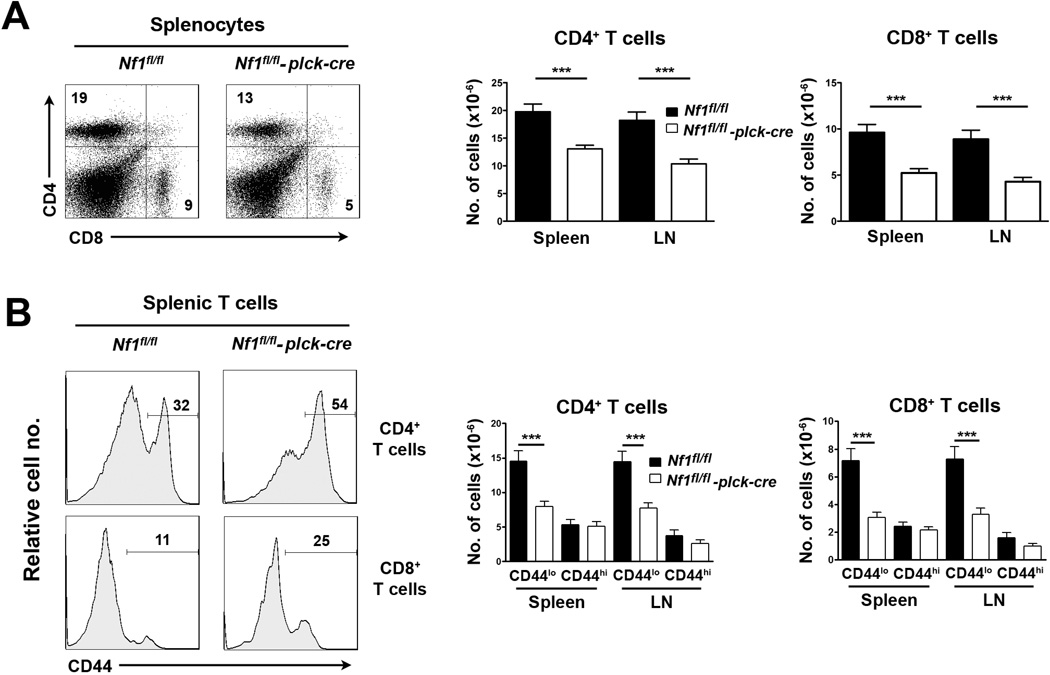
In NF1-deficient bone marrow chimeras increased numbers of CD4+ and CD8+ T cells were observed in spleen as well as thymus (Ingram et al., 2002). Therefore, we examined carefully numbers of T cells in peripheral lymphoid organs of T cell-specific NF1-deficient mice. In contrast to findings in bone marrow chimeras, total numbers of CD4+ and CD8+ T cells in spleen and LN were significantly reduced in T cell-specific NF1-deficient mice (Fig. 5A).
Fig. 5.
Peripheral T cells in T cell-specific NF1-deficient mice. All experiments were performed with littermate Nf1fl/fl and Nf1fl/fl plck-cre mice. A, At left are shown representative two-color flow cytometry plots of CD4 versus CD8 staining of splenocytes. The bar graph at right shows the mean number plus 1 SEM of CD4+ and CD8+ cells in spleen and LN (Nf1fl/fl, n=23; Nf1fl/fl plck-cre, n=24). B, At left are representative histograms of CD44 staining upon gated CD4+ and CD8+ splenic T cells. Bar graphs at right show the mean number plus 1 SEM of the indicated CD44lo and CD44hi populations in spleen and LN (Nf1fl/fl, n=23; Nf1fl/fl plck-cre, n=24). Statistical significance was determined using a Student two-sample t test. ***p <0.001.
We also examined numbers of naïve and memory T cells by staining for the CD44 memory marker. The percentage representation of CD44+ cells amongst CD4+ or CD8+ T cells was consistently higher in T cell-specific NF1-deficient mice compared to controls (Fig. 5B). For both T cell subsets this was attributed to a specific diminution in the number of naïve T cells. By contrast, numbers of memory T cells were unaffected by NF1 loss (Fig. 5B). Numbers of TCRβ T cells, NKT cells and Treg were normal in T cell-specific NF1-deficient mice (data not shown).
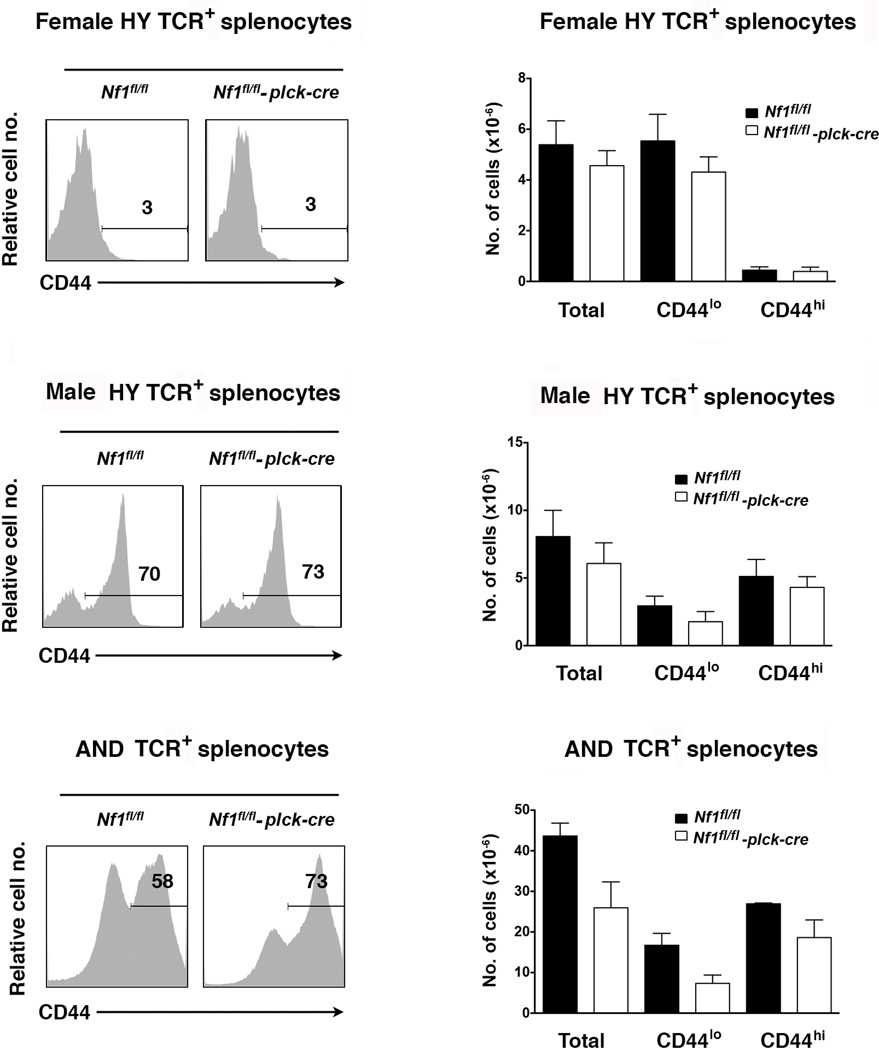
We also examined numbers of T cells in the periphery of TCR Tg T cell-specific NF1-deficient mice (Fig. 6). In female HY TCR Tg mice, numbers of naïve and memory HY TCR+ CD8+ T cells were not affected by the loss of NF1. It has been shown previously that peripheral T cells in female HY Tg mice are unable to undergo homeostatic proliferation, which is consistent with the fact that few CD44+ HY TCR Tg T cells are present in these animals (Ernst et al., 1999; Rocha and von Boehmer, 1991). Therefore, that loss of NF1 did not affect peripheral T cell numbers in the female HY TCR Tg model suggests that impaired homeostatic proliferation of peripheral T cells may be the cause of reduced T cell numbers in NF1-deficient non-TCR Tg mice. This is supported by the finding that loss of NF1 did result in reduced numbers of total and naïve T cells in AND TCR Tg mice (Fig. 6).
Fig. 6.
Peripheral T cells in T cell-specific NF1-deficient TCR Tg mice. All experiments were performed with littermate Nf1flfl and Nf1fl/fl plck-cre HY TCR Tg or AND TCR Tg mice. Histograms show representative CD44 staining upon gated HY or AND TCR+ T cells in spleens. Bar graphs show the mean number plus 1 SEM of total, CD44lo and CD44hi HY or AND TCR+ splenocytes. HY Tg female (Nf1fl/fl, n=9; Nf1fl/fl plck-cre, n=11), HY Tg male (Nf1fl/fl, n=9; Nf1fl/fl plck-cre, n=10), AND Tg (Nf1fl/fl, n=3; Nf1fl/fl plck-cre, n=3).
In male HY TCR Tg mice, some HY TCR+ T cells escape negative selection and enter the periphery where they are able to recognize and respond to HY peptide presented by H-2b APC (Suzuki et al., 2001). Therefore, a much higher percentage of memory HY TCR+ CD8+ T cells are observed in the periphery of male compared to female HY TCR Tg NF1-sufficient mice, which is presumably reflective of full agonist MHC-peptide stimulation in vivo (Fig. 6). Notably, loss of NF1 did not affect numbers of naïve or memory HY TCR+ T cells in male mice (Fig. 6). This indicates that NF1 does regulate T cell activation in response to full agonist MHC-peptide stimulation (see below).
3.5 Normal peripheral T cell function in T cell-specific NF1-deficient mice
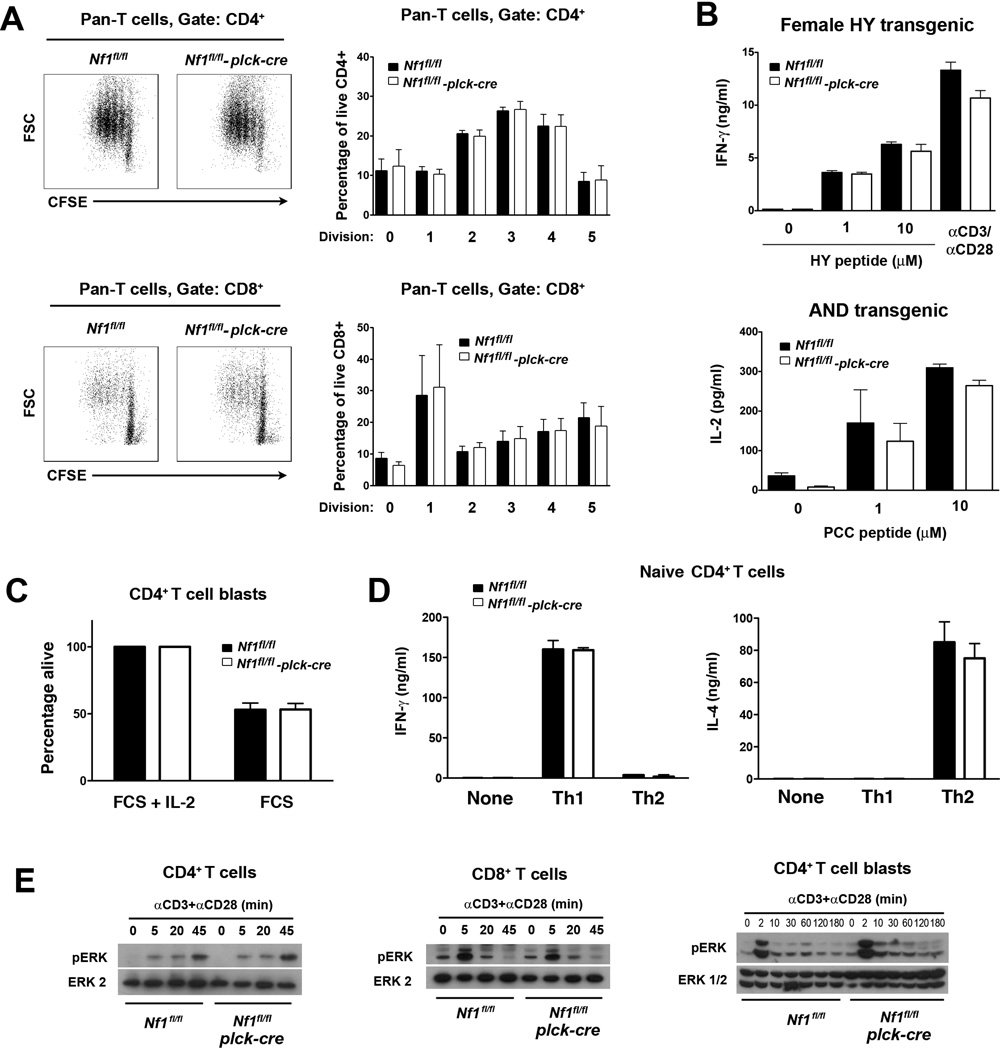
T cells from NF1-deficient BM chimeras showed reduced proliferation in response to CD3 and CD28 mAb stimulation in vitro (Ingram et al., 2002). Therefore, we examined if T cells from T cell-specific NF1-deficient mice also showed reduced proliferation in response to the same stimulus. Proliferation of non-TCR Tg T cells was assessed by dilution of CFSE fluorescence (Fig. 7A). No significant differences in the proliferative capacity of CD4+ or CD8+ T cells between NF1-deficient and control mice was observed. In parallel, we examined T cell AICD (Fig. 7C). For this purpose, CD4+ T cell blasts were generated and cultured in the presence or absence of IL-2, which provides anti-apoptotic survival signals to T cell blasts under these conditions (Lapinski et al., 2008). However, the extent of T cell apoptosis in response to IL-2 withdrawal was not affected by the loss of NF1.
Fig. 7.
Function of NF1-deficient T cells. All experiments were performed with littermate Nf1flfl and Nf1fl/fl plck-cre non-TCR Tg, AND TCR Tg or HY TCR Tg mice. A, Purified pan T cells (CD4+ and CD8+) from non-TCR Tg mice were labeled with CFSE and stimulated with CD3 and CD28 mAb. Flow cytometry plots at left show representative CFSE fluoresence versus forward scatter upon gated CD4+ and CD8+ T cells after 72 h culture. Bar graphs at right show the mean percentage plus 1 SEM of CD4+ or CD8+ T cells at the indicated number of divisions at 72 h (n=4 mice of each genotype). B, Purified CD8+ HY TCR Tg T cells from spleens of female HY TCR Tg mice (all naïve CD44lo) and purified naïve CD44lo T cells from spleens of AND TCR Tg mice were stimulated with HY peptide-pulsed H-2b APC or PCC peptide-pulsed H-2k APC respectively. Concentrations of IFN-γ or IL-2 in culture supernatants were determined by ELISA after 48 h. Bar graphs show mean concentration of cytokines plus 1 SEM (n=3 mice of each genotype). C, CD4+ T cell blasts from non-TCR Tg mice were cultured in complete medium with or without IL-2 for 48 h at which point the percentage of viable cells was by determined by flow cytometry. Viability in the absence of IL-2 was normalized to viability in the presence of IL-2. Shown is mean percent viability + 1 SEM (n=3 mice of each genotype). D, Purified CD44− naïve CD4+ T cells from non-TCR Tg mice were stimulated for 5 d under Th1 or Th2 polarizing conditions. Cells were harvested and restimulated with CD3 mAb for 24 h. Concentrations of IFN-γ and IL-4 in culture supernatants were determined by ELISA. Shown are mean concentrations of cytokines plus 1 SEM for each polarizing condition (n=3 mice each genotype). E, CD4+, CD8+ T cells or CD4+ T cell blasts were stimulated with CD3 and CD28 mAb for the indicated times. Activation of the Ras-MAPK pathway was determined by Western blotting using a phospho-ERK antibody. Results are representative of three repeat experiments.
We also examined cytokine synthesis by naïve NF1-deficient T cells (Fig. 7B). No significant differences in the ability of female HY TCR Tg T cells to secrete IFN-γ in response to HY peptide-pulsed H-2b APC or in the ability of AND TCR Tg T cells to secrete IL-2 in response to PCC peptide-pulsed H-2k APC were apparent. Similarly, no significant differences in the ability of non-TCR Tg naïve or memory CD4+ or CD8+ T cells to secrete cytokines in response to CD3 or CD28 mAb stimulation were noted (data not shown).
To examine T helper cell differentiation, naïve CD4+ T cells from non-TCR Tg mice were stimulated under Th1 or Th2 polarizing conditions. T cells were then restimulated with CD3 mAb and synthesis of IFN-γ or IL-4 was examined (Fig. 7D). No difference in the amounts of IFN-γ or IL-4 secreted under Th1 or Th2 polarizing conditions respectively were noted between NF1-deficient and control T cells. Therefore, NF1 is not required for Th1 or Th2 cell differentiation.
Consistent with the lack of a role for NF1 in T cell proliferation, survival, cytokine synthesis and differentiation, CD3/CD28 mAb-induced activation of ERK in non-TCR Tg CD4+ and CD8+ T cells as well as in CD4+ T cell blasts was normal in the absence of NF1 (Fig. 7E). Similarly, activation of ERK in HY and AND TCR Tg T cells in response to peptide-pulsed APC was unaffected by the loss of NF1 (data not shown).
4. Discussion
Knowledge of which RasGAPs regulate Ras in the T cell lineage is incomplete. Our previous studies of T cell-specific RASA1-deficient mice indicated that this RasGAP functions as a negative regulator of Ras during positive selection but neither regulates pre-TCR signaling nor controls Ras activation in peripheral T cells activated by full agonist MHC-peptide complexes (Lapinski et al., 2011). Therefore, other RasGAPs are likely to participate in the control of Ras activation in T cells. One a priori candidate is NF1. Thymocytes in mice transplanted with NF1-deficient BM showed elevated basal levels of Ras-GTP and both thymocyte and peripheral T cell numbers were increased in these animals (Ingram et al., 2002). In addition, and paradoxically, NF1-deficient peripheral T cells showed reduced proliferation in response to CD3 plus CD28 mAb-induced stimulation (Ingram et al., 2002). However, since other hematopoietic cells would also lack NF1 expression in these BM chimeras, it is unclear from these studies whether the apparent influence of NF1 deficiency on T cell function is T cell intrinsic or not. Moreover, the influence of NF1 loss on events such as thymocyte positive and negative selection and peripheral T cell differentiation was not examined in these studies. Therefore, in the current studies, we used T cell-specific NF1-deficient mice to address definitively the function of NF1 in T cells.
Findings in T cell-specific NF1-deficient mice were strikingly different from those reported in NF1-deficient BM chimeras. First, thymocyte numbers were not elevated in T cell specific NF1-deficient mice and, in fact, on non-TCR Tg and AND TCR Tg backgrounds thymocyte number was significantly reduced. Second, the number of peripheral T cells in T cell-specific NF1-deficient mice was diminished and this was accounted for by a specific decrease in the number of naive CD4+ and CD8+ T cells. Third, no evidence was obtained from the study of T cell-specific NF1-deficient mice that NF1 was a significant regulator of peripheral T cell proliferation, survival, cytokine secretion, differentiation or Ras activation triggered in response to full agonists. These differences, therefore, are most likely explained by additional loss of NF1 in hematopoietic cells other than T cells in NF1-deficient BM chimeras. A cell extrinsic role for NF1 as a contributing factor in the development of NF1-deficiency related phenotypes has been demonstrated beforehand. For example, plexiform neurofibroma formation resulting from biallelic loss of Nf1 in Schwann cells additionally requires at least heterozygous loss of Nf1 in BM-derived mast cells (Yang et al., 2008; Zhu et al., 2002). An alternative explanation for the differences in phenotype relates to the stage of T cell development at which NF1 loss first occurs. In BM chimeras, NF1 would be absent throughout T cell development whereas in T cell-specific NF1-deficient mice, NF1 would not be lost until later stages of DN development. Theoretically, therefore, earlier deletion of NF1 in BM chimeras could account for the observed differences.
In non-TCR Tg and AND TCR Tg mice loss of NF1 had little influence upon thymocyte positive selection. By contrast, in female HY TCR Tg mice, loss of NF1 resulted in reduced positive selection indicating that NF1 is required for efficient positive selection in this model. As a potential negative-regulator of Ras this finding was unexpected since increased TCR-mediated Ras activation would be predicted to promote positive selection. Whether NF1 regulates Ras activation during positive selection in female HY TCR Tg mice cannot be readily determined at present since the identity of the peptide that mediates positive selection in this model is not known. Notably, however, NF1 does not function as a negative regulator of Ras during negative selection or, as noted above, during activation of peripheral T cells with CD3 and CD28 mAb or full agonist MHC-peptide complexes. Furthermore, it is important to point out that if NF1 did regulate Ras during positive selection then, based upon the observed phenotype, it is predicted that it would promote rather than inhibit Ras activation, which would be unprecedented. For this reason, we favor the idea that NF1 performs a Ras independent role during positive selection. The possibility that NF1 has Ras independent functions in cellular signal transduction has been raised beforehand. A large number of disease-causing mutations in the human NF1 gene have been identified that are not predicted to affect GAP function or NF1 protein stability (Abernathy et al., 1997; Fahsold et al., 2000). Furthermore, whilst the isolated GAP domain of NF1 was able to rescue cardiovascular development in non-conditional NF1-deficient mice, this domain did not rescue overgrowth of neural crest-derived tissues in these mice (Ismat et al., 2006). By whichever mechanism NF1 does promote positive selection, presumably this mechanism is either not operative or is not required for thymocyte negative selection which is not decreased or increased by the loss of NF1.
In TCR Tg models, T cell-specific loss of RASA1 had the opposite effect upon positive selection as loss of NF1 (Lapinski et al., 2011). Thus, loss of RASA1 had no influence upon positive selection of HY TCR Tg cells but resulted in increased positive selection of AND TCR Tg cells, the latter of which was associated with increased Ras-MAPK activation in DP thymocytes. Together, these findings point to the complexity of RasGAP-mediated control of thymocyte positive selection. It is possible that the participation of different RasGAPs in the two studied TCR Tg models applies generally to the positive selection of MHC class I versus MHC class II-restricted T cells. However, additional TCR Tg models would need to be studied in order to reach this conclusion.
Reduced numbers of naïve CD4+ and CD8+ T cells were observed in T cell-specific non-TCR Tg NF1-deficient mice. A similar phenotype was observed in T cell-specific RASA1-deficient mice (Lapinski et al., 2011). In both situations, the reduced number of naive T cells cannot be accounted for by altered T cell proliferation or AICD responses induced by full agonist stimulation which are unchanged compared to control T cells. The basis for the reduced naïve T cell numbers in both types of mice remains unclear. However, reduced numbers of naïve T cells were not observed in the periphery of female HY TCR Tg T cell-specific NF1-deficient mice. HY TCR Tg T cells are peculiarly unable to undergo homeostatic proliferation in the periphery of females which suggests that impaired homeostatic proliferation in non-TCR Tg NF1-deficient mice is responsible for their reduced numbers (Ernst et al., 1999; Rocha and von Boehmer, 1991). Whether the reduced number of T cells is caused by dysregulated Ras activation is also uncertain. Notably, however, mice that are Tg for a constitutively active form of ERK-2 known as sevenmaker also contain substantially reduced numbers of peripheral naïve T cells (Sharp et al., 1997). This suggests that over-stimulation of the Ras-MAPK pathway in vivo may be the cause of the naïve T cell lymphopenia.
In summary, we demonstrate here a role for NF1 at the positive selection stage of thymocyte development. In addition, we show that NF1 functions as a regulator of naïve peripheral T cell numbers. Which RasGAP(s) regulate the activation of peripheral T cells stimulated with full agonist MHC-peptide complexes has yet to be determined.
Highlights.
-
! !
The NF1 RasGAP is necessary for thymocyte positive selection in TCR transgenic mice
-
! !
NF1 is not required for thymocyte negative selection
-
! !
Loss of NF1 results in reduced numbers of naïve T cells but is dispensable for T cell activation
Acknowledgements
This work was supported by National Institutes of Health grants AI050699 and HL096498 to PDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: BM, bone marrow; DN, double negative; DP, double positive; LN, lymph node; NF1, neurofibromin 1; PCC, pigeon cytochrome c peptide; RASA1, p120 RasGAP; RasGAP, Ras GTPase-activating protein; SP, single positive; Tg, transgenic.
Conflict of interest
The authors have no financial conflicts of interest
References
- Abernathy CR, Rasmussen SA, Stalker HJ, Zori R, Driscoll DJ, Williams CA, Kousseff BG, Wallace MR. NF1 mutation analysis using a combined heteroduplex/SSCP approach. Hum Mutat. 1997;9:548–554. doi: 10.1002/(SICI)1098-1004(1997)9:6<548::AID-HUMU8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Alberola-Ila J, Forbush KA, Seger R, Krebs EG, Perlmutter RM. Selective requirement for MAP kinase activation in thymocyte differentiation. Nature. 1995;373:620–623. doi: 10.1038/373620a0. [DOI] [PubMed] [Google Scholar]
- Alberola-Ila J, Hernandez-Hoyos G. The Ras/MAPK cascade and the control of positive selection. Immunol Rev. 2003;191:79–96. doi: 10.1034/j.1600-065x.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- Bauler TJ, Hendriks WJ, King PD. The FERM and PDZ domaincontaining protein tyrosine phosphatases, PTPN4 and PTPN3, are both dispensable for T cell receptor signal transduction. PLoS One. 2008;3:e4014. doi: 10.1371/journal.pone.0004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- Bollag G, Clapp DW, Shih S, Adler F, Zhang YY, Thompson P, Lange BJ, Freedman MH, McCormick F, Jacks T, Shannon K. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12:144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- Cawthon RM, Weiss R, Xu GF, Viskochil D, Culver M, Stevens J, Robertson M, Dunn D, Gesteland R, O'Connell P, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Crompton T, Gilmour KC, Owen MJ. The MAP kinase pathway controls differentiation from double-negative to double-positive thymocyte. Cell. 1996;86:243–251. doi: 10.1016/s0092-8674(00)80096-3. [DOI] [PubMed] [Google Scholar]
- Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- Donovan S, Shannon KM, Bollag G. GTPase activating proteins: critical regulators of intracellular signaling. Biochim Biophys Acta. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- Ebinu JO, Stang SL, Teixeira C, Bottorff DA, Hooton J, Blumberg PM, Barry M, Bleakley RC, Ostergaard HL, Stone JC. RasGRP links T-cell receptor signaling to Ras. Blood. 2000;95:3199–3203. [PubMed] [Google Scholar]
- Ernst B, Lee DS, Chang JM, Sprent J, Surh CD. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity. 1999;11:173–181. doi: 10.1016/s1074-7613(00)80092-8. [DOI] [PubMed] [Google Scholar]
- Fahsold R, Hoffmeyer S, Mischung C, Gille C, Ehlers C, Kucukceylan N, Abdel-Nour M, Gewies A, Peters H, Kaufmann D, Buske A, Tinschert S, Nurnberg P. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am J Hum Genet. 2000;66:790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AM, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 2005;23:431–443. doi: 10.1016/j.immuni.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Genot E, Cantrell DA. Ras regulation and function in lymphocytes. Curr Opin Immunol. 2000;12:289–294. doi: 10.1016/s0952-7915(00)00089-3. [DOI] [PubMed] [Google Scholar]
- Hennet T, Hagen FK, Tabak LA, Marth JD. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc Natl Acad Sci U S A. 1995;92:12070–12074. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- Hope DG, Mulvihill JJ. Malignancy in neurofibromatosis. Adv Neurol. 1981;29:33–56. [PubMed] [Google Scholar]
- Ingram DA, Zhang L, McCarthy J, Wenning MJ, Fisher L, Yang FC, Clapp DW, Kapur R. Lymphoproliferative defects in mice lacking the expression of neurofibromin: functional and biochemical consequences of Nf1 deficiency in T-cell development and function. Blood. 2002;100:3656–3662. doi: 10.1182/blood-2002-03-0734. [DOI] [PubMed] [Google Scholar]
- Ismat FA, Xu J, Lu MM, Epstein JA. The neurofibromin GAP-related domain rescues endothelial but not neural crest development in Nf1 mice. J Clin Invest. 2006;116:2378–2384. doi: 10.1172/JCI28341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- Kaye J, Hsu ML, Sauron ME, Jameson SC, Gascoigne NR, Hedrick SM. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 1989;341:746–749. doi: 10.1038/341746a0. [DOI] [PubMed] [Google Scholar]
- King PD, Lubeck BA, Lapinski PE. Non-redundant functions for Ras GTPase-activating proteins in tissue homeostatsis. Science Signaling. 2013 doi: 10.1126/scisignal.2003669. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- Kortum RL, Sommers CL, Pinski JM, Alexander CP, Merrill RK, Li W, Love PE, Samelson LE. Deconstructing Ras signaling in the thymus. Mol Cell Biol. 2012 doi: 10.1128/MCB.00317-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinski PE, King PD. Regulation of Ras signal transduction during T cell development and activation. Am J Clin Exp Immunol. 2012;1:147–153. [PMC free article] [PubMed] [Google Scholar]
- Lapinski PE, Oliver JA, Kamen LA, Hughes ED, Saunders TL, King PD. Genetic analysis of SH2D4A, a novel adapter protein related to T cell-specific adapter and adapter protein in lymphocytes of unknown function, reveals a redundant function in T cells. J Immunol. 2008;181:2019–2027. doi: 10.4049/jimmunol.181.3.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinski PE, Qiao Y, Chang CH, King PD. A role for p120 RasGAP in thymocyte positive selection and survival of naive T cells. J Immunol. 2011;187:151–163. doi: 10.4049/jimmunol.1100178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Largaespada DA, Brannan CI, Jenkins NA, Copeland NG. Nf1 deficiency causes Ras-mediated granulocyte/macrophage colony stimulating factor hypersensitivity and chronic myeloid leukaemia. Nat Genet. 1996;12:137–143. doi: 10.1038/ng0296-137. [DOI] [PubMed] [Google Scholar]
- Lauritsen JP, Kurella S, Lee SY, Lefebvre JM, Rhodes M, Alberola-Ila J, Wiest DL. Egr2 is required for Bcl-2 induction during positive selection. J Immunol. 2008;181:7778–7785. doi: 10.4049/jimmunol.181.11.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGargill MA, Ch'en IL, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. Cutting edge: Extracellular signal-related kinase is not required for negative selection of developing T cells. J Immunol. 2009;183:4838–4842. doi: 10.4049/jimmunol.0902208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JA, Stolberg VR, Chensue SW, King PD. IL-4 acts as a potent stimulator of IFN-gamma expression in CD8+ T cells through STAT6-dependent and independent induction of Eomesodermin and T-bet. Cytokine. 2012;57:191–199. doi: 10.1016/j.cyto.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priatel JJ, Teh SJ, Dower NA, Stone JC, Teh HS. RasGRP1 transduces low-grade TCR signals which are critical for T cell development, homeostasis, and differentiation. Immunity. 2002;17:617–627. doi: 10.1016/s1074-7613(02)00451-x. [DOI] [PubMed] [Google Scholar]
- Rincon M, Flavell RA, Davis RJ. Signal transduction by MAP kinases in T lymphocytes. Oncogene. 2001;20:2490–2497. doi: 10.1038/sj.onc.1204382. [DOI] [PubMed] [Google Scholar]
- Rocha B, von Boehmer H. Peripheral selection of the T cell repertoire. Science. 1991;251:1225–1228. doi: 10.1126/science.1900951. [DOI] [PubMed] [Google Scholar]
- Roose JP, Mollenauer M, Gupta VA, Stone J, Weiss A. A diacylglycerol-protein kinase C-RasGRP1 pathway directs Ras activation upon antigen receptor stimulation of T cells. Mol Cell Biol. 2005;25:4426–4441. doi: 10.1128/MCB.25.11.4426-4441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JP, Mollenauer M, Ho M, Kurosaki T, Weiss A. Unusual interplay of two types of Ras activators, RasGRP and SOS, establishes sensitive and robust Ras activation in lymphocytes. Mol Cell Biol. 2007;27:2732–2745. doi: 10.1128/MCB.01882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sharp LL, Schwarz DA, Bott CM, Marshall CJ, Hedrick SM. The influence of the MAPK pathway on T cell lineage commitment. Immunity. 1997;7:609–618. doi: 10.1016/s1074-7613(00)80382-9. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yamaguchi MT, Ohteki T, Sasaki T, Kaisho T, Kimura Y, Yoshida R, Wakeham A, Higuchi T, Fukumoto M, Tsubata T, Ohashi PS, Koyasu S, Penninger JM, Nakano T, Mak TW. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 2001;14:523–534. doi: 10.1016/s1074-7613(01)00134-0. [DOI] [PubMed] [Google Scholar]
- Swan KA, Alberola-Ila J, Gross JA, Appleby MW, Forbush KA, Thomas JF, Perlmutter RM. Involvement of p21ras distinguishes positive and negative selection in thymocytes. Embo J. 1995;14:276–285. doi: 10.1002/j.1460-2075.1995.tb07001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, Culver M, Carey JC, Copeland NG, Jenkins NA, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, Fountain JW, Brereton A, Nicholson J, Mitchell AL, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249:181–186. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- Yang FC, Ingram DA, Chen S, Zhu Y, Yuan J, Li X, Yang X, Knowles S, Horn W, Li Y, Zhang S, Yang Y, Vakili ST, Yu M, Burns D, Robertson K, Hutchins G, Parada LF, Clapp DW. Nf1-dependent tumors require a microenvironment containing Nf1+/–- and c-kit-dependent bone marrow. Cell. 2008;135:437–448. doi: 10.1016/j.cell.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]