Abstract
Autophagy plays a critical role in multiple aspects of the immune system, including the development and function of T lymphocytes. In mammalian cells, the class III phosphoinositide 3-kinase Vps34 is thought to play a critical role in autophagy. However, recent studies have cast doubt on the role of Vps34 in autophagy, at least in certain cell types. In order to study the effects of Vps34 on autophagy in T lymphocytes, we generated mice that selectively lack Vps34 in the T cell lineage. Vps34 ablation in T cells caused profound defects in autophagic flux, resulting in accumulation of cellular organelles and apoptosis. These animals exhibited normal intrathymic development of conventional T cells, but were profoundly impaired in the intrathymic development of invariant natural killer T cells. In peripheral organs, T cell-specific ablation of Vps34 had a profound impact on T cell homeostasis and function. Furthermore, aged animals developed an inflammatory wasting syndrome characterized by weight loss, intestinal inflammation and anemia. Consistent with this phenotype, Vps34 was required for the peripheral maintenance and function of CD4+FoxP3+ regulatory T cells. Collectively, our study reveals a critical role for Vps34 in autophagy and for the peripheral homeostasis and function of T lymphocytes.
Introduction
Autophagy is a ‘self-eating’ catabolic process used to break down and recycle long-lived proteins and organelles in order to maintain a homeostatic environment within the cell (1). This process is usually functional at a low level, but is upregulated in response to nutrient starvation, stress or cellular damage (2, 3). Autophagy is initiated by the formation of a cup-shaped membrane structure that extends around the cellular organelles, forming double membrane vesicles called autophagosomes (4–6). Autophagosomes fuse with lysosomes and late endosomes for degradation of the inner autophagosomal membrane and their cargo (4).
Genetic analyses in yeast have identified a large number of evolutionary conserved genes, termed autophagy-related (Atg) genes, that are required for autophagy (7–9). The class III phosphoinositide 3 (PI3)-kinase vacuolar protein sorting 34 (Vps34; also called PIK3C3) and its binding partner Atg6 (also called Beclin 1), have been reported to play important roles for the initiation of autophagy, including formation of the cup-shaped ‘omegasome’ or isolation membrane (10, 11). Elongation of the isolation membrane is regulated by two ubiquitin-like systems, the Atg12 and the Atg8 (or its mammalian homolog LC3) conjugation pathways that are required for the generation of LC3-bound phosphatidylethanolamine as a building block to form double-membraned autophagosomes (12, 13). The class III PI3-kinase Vps34 converts phosphatidylinositol (PtdIns) to PtdIns-3-phosphate (PtdIns3P), which recruits FYVE domain-containing proteins and members of the WD-repeat domain PtdIns-interacting (WIPI) family of proteins to the site of autophagosome formation. This, in turn, provides a scaffold for Atg proteins to initiate autophagy (11, 14). In yeast, Vps34 activity is critical for the recruitment of Atg proteins to the pre-autophagosomal structure and for autophagy initiation (15). However, in higher eukaryotes, the role of Vps34 and its product PtdIns3P in autophagy is less well understood. Recent studies have described the deletion of Vps34 in embryonic fibroblasts, heart, liver (16), sensory neurons (17), or T cells (18) in mice. While the study with embryonic fibroblasts, heart and liver argued for a critical role of Vps34 in regulating functional autophagy (16), the study in sensory neurons favored a predominant role of Vps34 in endocytosis but not autophagy (17), and the study in T cells concluded that Vps34 is critical for IL-7Rα chain expression (18).
Here, we have generated mice with a T cell-specific deletion in Vps34 to study the role of Vps34 in T cell homeostasis and function. We found that deletion of Vps34 in T cells results in severe defects in autophagic flux and accumulation of cellular organelles. This phenotype correlated with enhanced apoptosis in Vps34-deficient T cells. Mice with T cell-specific deletion of Vps34 exhibited normal intrathymic development of conventional CD4+ and CD8+ T cells but impaired intrathymic development of invariant natural killer T (iNKT) cells. In peripheral organs, T cell-specific ablation of Vps34 resulted in a profound loss of T cells. Furthermore, we found that Vps34 was required for the peripheral homeostasis and function of CD4+FoxP3+ regulatory T (Treg) cells. Consequently, aged animals developed an inflammatory wasting syndrome characterized by weight loss, intestinal inflammation and anemia.
Methods
Mice
Vps34flox/flox (Vps34f/f) mice have been described (16). T cell-specific deletion of Vps34 was achieved by crossing the Vps34f/f mice with CD4-Cre transgenic mice (Taconic). Mice were genotyped as described previously (16). Six- to 8-week-old animals were used in this study. An inflammatory wasting phenotype in 18- to 25-week-old Vps34f/f;CD4-Cre mice was observed. These mice consistently developed a rectal prolapse and were carefully monitored. Mice were treated with an antibiotic ointment three times per week. Rag2−/− mice were obtained from the Jackson Laboratories. All breeder and experimental mice were housed in specific pathogen-free conditions in compliance with guidelines from the Institutional Animal Care and Use Committee at Vanderbilt University.
Activation of T cells
T cells were purified from Vps34f/+;CD4-Cre or Vps34f/f;CD4-Cre splenocytes by negative selection using magnetic sorting (Miltenyi Biotec). Cell purity was routinely >96% as determined by flow cytometry. T cells were activated with plate-bound anti-CD3ε and anti-CD28 antibodies in complete RPMI 1640 medium for 36 hrs and were utilized for electron microscopy, Western blot analyses, and measurements of glycolytic rate and β-oxidation.
Transmission electron microscopy
Activated T cells were fixed with buffer (50 mM cacodylate [pH 7.2], 50 mM KCl, and 2.5 mM MgCl2) containing 2.5% glutaraldehyde at 4°C overnight. For pre-embed immunolabeling, 2% paraformaldehyde-fixed cells were stained with anti-LC3B antibodies, followed by goat anti-rabbit IgG conjugated to 10 nm gold particles (BBinternational). The cells were washed and fixed as above (19). The cells were washed 3 times (5 min) in 0.1 M cacodylate buffer followed by incubation for 1 hr in 1% osmium tetraoxide at RT. The cells were then washed three times with 0.1 M cacodylate buffer and were dehydrated through a series of increasing ethanol washes followed by propylene oxide incubations then infiltration with Poly/Bed® 812 (Polysciences, Inc.) resin and finally embedded in Poly/Bed® 812 resin. Sections (70 nm thick) were collected on copper grids and stained with uranyl acetate and lead citrate and subsequently analyzed with a Philips/FEI Tecnai T-12 or Philips CM-12 electron microscope equipped with a side-mounted 2k X 2k AMT CCD camera. Thirty to 35 individual cells in each group were imaged to measure the size of mitochondria. The size of mitochondria was determined using the Image J program by manually marking single mitochondria, measuring their area, and normalization against the scale bar as previously described (20). The data presented are the total size of all the mitochondria normalized against the size of the cell. A similar analysis was carried out with the nuclear size as a negative control.
Measurement of LC3 punctae
Purified T cells were washed with PBS and adhered to polylysin-coated coverslips for 5 min and fixed with 2% paraformaldehyde. Cells were then treated with 0.1% saponin (Sigma-Aldrich) in PBS containing 0.5% BSA for 10 min on ice and intracellularly stained with an anti-LC3B antibody (Enzo) overnight in a humified chamber at 4°C. After washing, anti-rabbit–Cy3 Ab (Jackson ImmunoResearch Laboratories) was added and incubated on ice for 30 min. Z-stack images were acquired at 1 μm spacing using 60x oil objective by a Zeiss LSM 510 Meta Confocal Microscope. Three-dimensional deconvolution of the images was performed and the LC3 punctae were quantified by MetaMorph 7.6 software (Universal Imaging) as described previously (21). The threshold was set to >4 pixels for LC3 puncta count and a total of 30–35 cells from each group were analyzed.
Western blotting
Activated T cells or CD4+ thymocytes purified by magnetic sorting were washed 3x with PBS before preparation of cellular proteins for Western blotting. Some T cells were treated with 3-methyladenine (3-MA) (5 mM; Sigma) for the last 5 hrs before cell lysate preparation for Western blot analysis. Cells were lysed with lysis buffer (Cell Signaling Technology) containing protease inhibitor cocktail (Sigma-Aldrich) for 2 hrs at 4°C. The protein samples were separated by 12% SDS-PAGE (Bio-Rad Laboratories) and transferred to nitrocellulose membranes overnight. The membranes were blocked with 5% nonfat milk and incubated with primary antibody overnight at 4°C, followed by HRP-conjugated secondary antibody for 2 hrs at room temperature. The blots were developed with enhanced chemiluminescence method (GE Healthcare). The protein bands were quantified by densitometric analysis using Image J software (NIH) and the presented data are normalized results against respective β-actin controls. LC3 antibody (5F10) was obtained from Enzo Life Sciences, p62 antibody from Cell Signaling Technology, Bim and Bax antibodies from eBiosciences, and β-actin, Atg5, Atg7, Bad and Vps34 antibodies from Sigma-Aldrich. All secondary antibodies were obtained from Promega.
Autophagic flux assays
Autophagic flux assay was performed by culture of CD4+ thymocytes or activated T cells in the presence of E64D (10 μg/ml; Sigma-Aldrich) plus pepstatin A (10 μg/ml; Sigma-Aldrich) for 90 min. After three washes with PBS, cell lysates were prepared for immunoblot analysis with anti-LC3B antibody.
Flow cytometry
Single-cell suspensions of the spleen, thymus, lymph nodes and liver were prepared and stained with fluorescently labeled monoclonal antibodies in fluorescence-activated cell sorter (FACS) buffer (PBS containing 2% FBS and 0.05% sodium azide) as described previously (22). In all experiments, dead cells were excluded from the analysis by electronic gating. Fluorescently labeled antibodies against mouse TCRβ, CD11b, CD11c, TCRγδ, CD4, CD8, Ki67, IL-2, IFN-γ, IL-13, IL-17A, CD24, CD25, Bcl-2, and isotype control antibodies were obtained from BD Biosciences. Anti-human Bcl-xL antibody and its isotype control were obtained from SouthernBiotech and FoxP3 staining sets were obtained from eBioscience. Apoptosis was measured using Annexin V apoptosis detection kit from BD Biosciences. CD1d/α-GalCer tetramers(CD1d-tetramers) were obtained from the NIH tetramer core facility (Emory University, Atlanta, GA). The iNKT cell population was identified as B220−TCR-β+CD1d-tetramer+ cells as described (22). For analysis of intracellular cytokines produced by CD4 and CD8 T cells, 18- to 25-week-old mice with rectal prolapse were sacrificed and splenocytes and mesenteric lymph node cells were activated with or without ionomycin (ION, 1 μM) and phorbol myristate acetate (PMA, 100 ng/ml) in the presence of GolgiPlug™ (all from BD Biosciences-Pharmingen) for 5 hrs. Intracellular cytokine staining was performed with Cytofix/Cytoperm reagents (BD Biosciences-Pharmingen) according to the manufacturer’s protocol. For measurement of endoplasmic reticulum and mitochondrial content in T cells by flow cytometry, splenocytes and lymph node cells were prepared and stained with MitoTracker® Red (50 nM) or ER-Tracker™ red (1 μM; Invitrogen) at 37°C for 30 min in complete medium. The cells were then washed and stained with anti-CD4 and -CD8 antibodies at 4°C in FACS buffer. Flow cytometric analyses were performed using a FACSCalibur™ instrument (BD Biosciences) and studies with ER-Tracker™ red were analyzed on a LSRFortessa™ instrument (BD Biosciences). The acquired data were analyzed using FlowJo software (Tree Star Inc.).
Quantitative real-time PCR
Total RNA as purified using Trizol® reagent (Invitrogen) according to the manufacturer’s instructions, including a DNase I digestion step. cDNA was prepared from 1–2 μg RNA using ThermoScript™ Reverse Transcriptase system (Invitrogen). Real-time PCR was performed using Sybr green master mix (Biorad) and 20 ng of cDNA per well in duplicates for each sample on a CFX96 real-time PCR machine (Biorad). The CT values were collected for the housekeeping gene β-actin and the genes of interest during the log phase of the cycle. The level of the gene of interest was normalized to that of β-actin for each sample and compared with the values obtained for the test sample. Each gene was compared with every normalizer in succession and the ΔCt was calculated (ΔCT= CT Gene of interest − CT Normalizer). The normalized expression (ΔΔC(t)) of the gene of interest was calculated using the CFX manager software (BioRad). The primers used for PCR were as follows: Vps34 forward, 5′-CAGCCCTGGATGAGATGTTT-3′ and reverse, 5′-GGATGGGTGACAGAACCAAG-3′; β-actin forward, 5′-TACAGCTTCACCACCACAGC-3′ and reverse, 5′-AAGGAAGGCTGGAAAAGAGC-3′.
T regulatory (Treg) cell suppression assay
Treg cells were purified on the basis of CD25 staining from negatively selected CD4+ T cells by magnetic sorting according to the manufacturer’s protocol (Miltenyi Biotec). Two × 105 CD4+CD25− T effector (Teff) cells were activated with anti-CD3ε antibody (1 μg/ml) and 2×104 dendritic cells (DCs) in the presence of Treg cells purified from splenic CD4+ T cells derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice at various Teff to Treg cell ratios for 60 hrs with the addition of 1 μCi of [3H]thymidine (PerkinElmer Life Sciences) for the last 12 hrs of culture.
Adoptive transfer of T cells and evaluation of colitis
CD4+ T cells were enriched from pooled splenic and mesenteric lymph node cells by magnetic sorting using negative selection, followed by enrichment of CD25− T cells and CD25+ Treg cells by positive selection. CD4+CD25− cells (2×106 cells) derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice were adoptively transferred into Rag2−/− mice to induce colitis as described (23). Groups of recipient mice also received 5×105 CD4+CD25+ Treg cells derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice. The mice were followed for signs of colitis twice a week for up to 12 weeks. Animals with severe clinical signs of disease were sacrificed according to the guidelines from the Institutional Animal Care and Use Committee at Vanderbilt University. Mice were monitored for weight change and signs of colitis, including diarrhea, rectal bleeding and scruffiness, each given a clinical score of 1. Histological sections of the colons were graded for signs of lymphocyte infiltration, loss of globlet cells and ulcers as described (24). Each of these histological parameters was given a score ranging from 0–3 according to severity. The clinical and the histological scores were pooled to obtain the final disease score.
Measurement of glycolytic rate
The glycolytic rate was measured as described (25). In brief, one million activated cells were suspended in 0.5 ml of RPMI that was previously equilibrated at 37°C under 5% CO2. Ten μCi of 5-[3H] glucose was added to each well and samples were incubated for 1 hr at 37°C in a humified chamber. The reaction was terminated by mixing 50 μl of cells with an equal volume of 0.2 N HCl in a PCR vial and samples were then placed upright in 4 ml scintillation vials containing 0.5 ml water. The vials were sealed with parafilm and [3H]2O generated during glycolysis in the PCR tubes was allowed to equilibrate with the water in the scintillation vials for 2 days. The contents of the PCR tubes were transferred into a new scintillation vial and scintillation fluid was added to the original vials for diffused counts and to the new vials for undiffused counts. The fraction of [3H]2O that diffused in 2 days was determined with control PCR tubes containing 1 μCi of [3H]2O. The background diffusion rate was determined using tubes containing cell-free medium. The glycolytic rate was calculated as sample diffusion ratio minus the background diffusion ratio divided by the diffusion fraction from the [3H]2O control. This number was multiplied by 5500 (the nmoles of glucose in 0.5 ml of RPMI) to obtain the glycolytic rate as nmols of glucose consumed/million cells/hr.
β-oxidation assay
β-oxidation of lipids was measured in activated T cells as described (26, 27). In brief, T cells were activated as described above, washed twice with PBS and cultured in 0.4 ml of RPMI containing 2% fatty acid free BSA (Sigma) for half an hour. After two washes with PBS, 4×106 cells were cultured in 0.4 ml of RPMI containing 2 μCi 9,10-[3H] palmitate (MP), 2% of BSA (fatty acid free) and 0.25 mM L-carnitine for 4 hrs under 5% CO2 in an incubator. The oxidation of 9,10-[3H] palmitate was measured by release of [3H]2O in the supernatant. The excess of radioactive lipid from the supernatant was removed twice by precipitation of lipids with an equal volume of 10% trichloroacetic acid followed by incubation at RT for 15 min and centrifugation at 13,000 g for 10 min. The resultant supernatant was treated with 750 μl of chloroform:methanol mixture (2:1) and 300 μl of 2 M KCl:HCl, and then centrifuged at 3,000 g for 5 min. 500 μl of the supernatant was collected and counted after the addition of 5 ml of scintillation fluid (Ecolite™, MP). The rate of β-oxidation was determined by subtracting counts per minute (CPM) of cell-free supernatant (background) from [3H]2O released in the sample supernatant and was expressed as CPM per 4×106 cells.
Measurement of antigen-specific T cell responses
Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice were immunized with 200 μg of ovalbumin (OVA) emulsified in CFA at day 0, and were boosted with the same amount of antigen emulsified in IFA at day 7. The mice were sacrificed at day 15 and splenic and draining lymph node cells were stimulated in vitro with 25–100 μg/ml of OVA. The supernatants were collected at 72 hrs of culture and assayed for IL-17A and IFN-γ by ELISA.
CFSE dilution analysis
CFSE dilution analysis was performed as described in our previous study (22). In brief, splenocytes derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice were labeled with 5 mM CFSE (Invitrogen) for 10 min at 37°C in PBS containing 0.1% BSA and washed twice with complete RPMI 1640 medium. Labeled cells (3×105 cells/well) were then stimulated with anti-CD3ε antibody (1 μg/ml) with or without the apoptosis inhibitor z-VAD-fmk (50 μM, Sigma) for 65 hrs in complete RPMI 1640 medium. At the end of the culture, cells were harvested, stained with anti-CD4 and anti-CD8 antibodies and analyzed by flow cytometry.
Hybridoma activation assay
DN32.D3 iNKT cell hybridoma cells (28) (obtained from Dr. A. Bendelac, University of Chicago, Chicago, IL) were used as responder cells in cultures with thymocytes as described (29). Briefly, 3×104 DN32.D3 cells were cultured with 5×105 or 106 total thymocytes from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice per well for 20 hrs. Supernatants were collected and IL-2 was measured by ELISA.
ELISA
A standard sandwich ELISA was performed to measure mouse IL-2, IL-17A and IFN-γ in the culture supernatants using BD Biosciences BD OptEIA™ kit. For detection, streptavidin-HRP conjugate (Zymed Laboratories Inc.) was used, and the color was developed with the substrate 3,3′,5,5′-tetramethylbenzidine (Dako Corp.) in the presence of H2O2. Total IgG antibodies in the serum were measured by anti-mouse IgG (H+L) capture antibody and biotin labeled anti-mouse IgG reagents (SouthernBiotec) according to manufacturer’s instructions.
Histological and blood analyses
Complete small and large intestinal rolls were prepared and fixed in formalin. Sections (8 μm thick) from paraffin-embedded tissues were stained with H&E. Total blood counts were carried out in an automated analyzer at the Translational Pathology Shared Resource core facility at Vanderbilt Medical Center.
Statistical analyses
Statistical significance between the groups was determined by application of an unpaired 2-tailed Student’s t test using Graphpad Prism software. A P value less than 0.05 was considered significant.
Results
Defective autophagy in Vps34-deficient T cells
Vps34f/f mice (16) were crossed with transgenic mice expressing Cre recombinase driven by the CD4 promoter, resulting in T cell-specific deletion of the Vps34 gene starting at the CD4+CD8+ (double-positive, DP) thymocyte stage (Supplemental Fig. 1A). Such deletion resulted in complete loss of Vps34 mRNA and protein expression in thymocytes and splenic CD4+ and CD8+ T cells (Supplemental Fig. 1 B,C).
We assessed the steady-state levels of LC3 in Vps34-deficient T cells. In purified CD4+ thymocytes, we observed increased levels of both LC3B-I and LC3B-II in mutant T cells (Fig. 1 A,B). In purified splenic T cells activated with plate-bound anti-CD3 and anti-CD28 antibodies for 36 hrs, we detected little LC3B in wild-type cells, but increased levels of both forms of LC3B were observed in Vps34-deficient T cells (Fig. 1A,B).
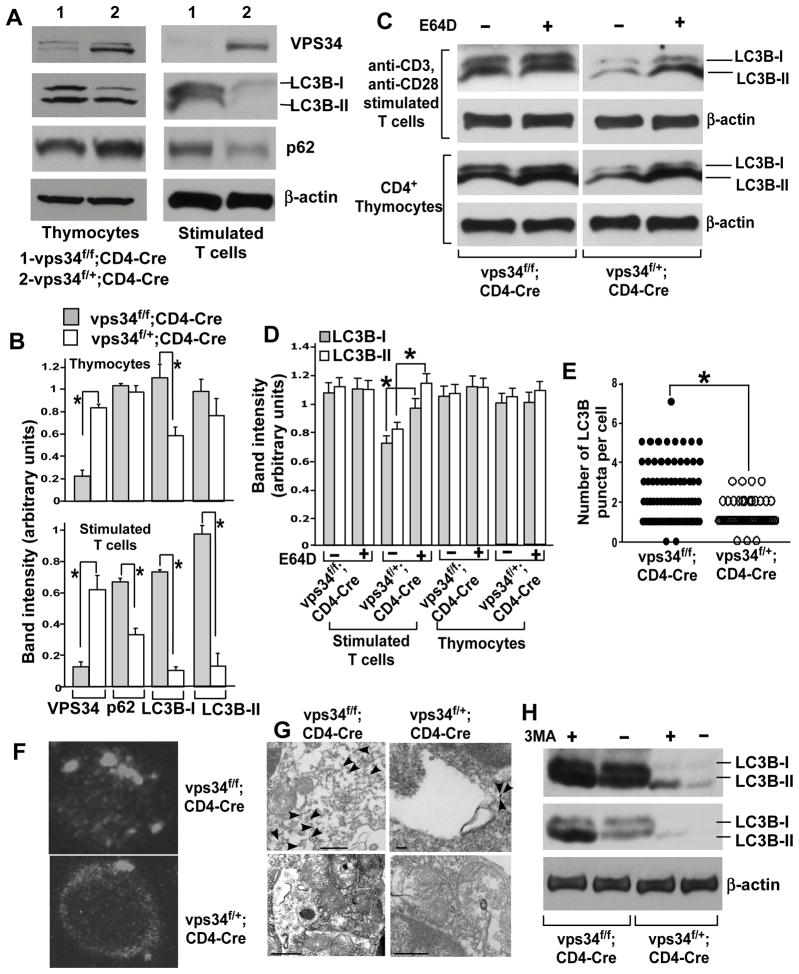
FIGURE 1.
Vps34-deficient T cells display defective autophagy. (A) CD4+ thymocytes or total T cells were purified from 6- to 8-week-old Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice using magnetic sorting. Proteins were prepared from CD4+ thymocytes or from anti-CD3/CD28 stimulated T cells for immunoblot analysis with antibodies against Vps34, p62, LC3B or β-actin. Representative data from three independent experiments are shown. (B) Densitometric analysis of protein bands obtained in (A). Data are from an average of 4 mice from 3 independent experiments. *p<0.05. (C) Autophagic flux assay was performed by culture of CD4+ thymocytes or activated T cells in the absence or presence of E64D (10 μg/ml) plus pepstatin A (10 μg/ml) for 90 min, followed by immunoblot analysis with the anti-LC3B antibody. (D) Densitometric analysis of protein bands obtained in (C). Data are from an average of 4 mice from 3 independent experiments. *p<0.05. (E) Purified total T cells were stained with the anti-LC3B and Cy3-labeled donkey anti-mouse antibodies. Z-stack images were captured and spaced at 1 μm using confocal microscopy and a total of 30–35 cells in each group were analyzed. The number of LC3 punctae was quantified at a threshold set at pixels >4. *p<0.05. (F) A representative image from (E) is shown. (G) Activated T cells were stained with anti-LC3B antibodies followed by anti-rabbit antibodies labeled with 10 nm gold particles. The cells were embedded, sectioned and stained with uranyl acetate and lead citrate followed by analysis with a Philips/FEI T-12 transmission electron microscope. Note the predominant staining of LC3B associated with autophagosomes (staining indicated with arrows) in Vps34f/+;CD4-Cre T cells (scale bar=100nm) (upper panel), while increased cytosolic staining is observed in Vps34f/f;CD4-Cre T cells (scale bar=500nm) (upper panel). Controls with secondary antibody alone rarely showed gold staining in T cells from either Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice (scale bar=500nm) (lower panel). Note in the control image of T cells derived from Vps34f/f;CD4-Cre mice that cellular organelles accumulate and are clumped together due to defective autophagy. (H) T cells were enriched and activated as in (A). The cells were treated with 3-MA (5 mM) for the last 5 hours of culture, followed by immunoblot analysis with anti-LC3B antibody. Blots with high (upper panel) or low (lower panel) exposure time are shown. The data are representative of 3 independent experiments with similar results.
p62, a protein that binds polyubiquitinated proteins and is degraded in lysosomes during autophagy (30), serves as an important indicator of defective autophagy (31). While no significant change in p62 levels was observed in CD4+ thymocytes, profound accumulation of p62 was observed in activated Vps34-deficient T cells as compared with wild-type T cells (Fig. 1A,B), suggesting a defect in autophagic protein degradation.
Abnormally high levels of LC3B and accumulation of p62 in Vps34-deficient T cells suggested that these cells have a defect in autophagic flux. To test this possibility, we performed autophagic flux assays by using the lysosomal protease inhibitor E64D. In wild-type thymocytes or activated splenic T cells, such treatment increased the protein levels of LC3B-II as compared with cells treated with medium alone, whereas in Vps34-deficient cells, high levels of LC3B remained unchanged with E64D treatment (Fig. 1C). These results indicated a failure of autophagic protein turnover in Vps34-deficient T cells. Consistent with decreased LC3B turnover in Vps34-deficient T cells, an increased amount of large-sized LC3-positive structures was observed in these cells as compared with wild-type T cells (Fig. 1D,E). Immunostaining of LC3B on electron micrographs of activated T cells derived from Vps34f/+;CD4-Cre mice revealed that LC3B gold staining was predominantly localized to autophagic membrane structures and partly to the cytoplasm. However, in activated T cells from Vps34f/f;CD4-Cre mice, LC3B staining failed to associate with membranes and, instead, increased staining was observed in the cytoplasm (Fig. 1G). Since Vps34 and autophagy are inhibited by 3-MA, we next treated activated T cells derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice with 3-MA for 5 hrs and observed the accumulation of LC3B by Western bot analysis. The results showed that 3-MA treatment caused accumulation of LC3B in both Vps34-sufficient and -deficient T cells, suggesting that Vps34 is not the sole target of 3-MA (Fig. 1H). Collectively, these results indicated that functional autophagy is severely blocked in Vps34-deficient T cells.
Normal intrathymic development but reduced numbers of peripheral TCRαβ+ T cells in mice lacking Vps34 in the T cell lineage
Analysis of TCRαβ+ T cells in the peripheral lymphoid organs revealed that the percentage of both CD4+ and CD8+ T cells was significantly reduced in the spleen, lymph nodes and peripheral blood of Vps34f/f;CD4-Cre mice as compared with Vps34f/+;CD4-Cre controls (Fig. 2A,B). In the liver, the percentage of CD8+ T cells in Vps34f/f;CD4-Cre mice was slightly increased, whereas the percentage of CD4+ T cells was decreased. A similar trend was observed when absolute numbers of T cells were calculated based on total organ lymphoid cellularity (Fig. 2B and Supplemental Fig. 2A).
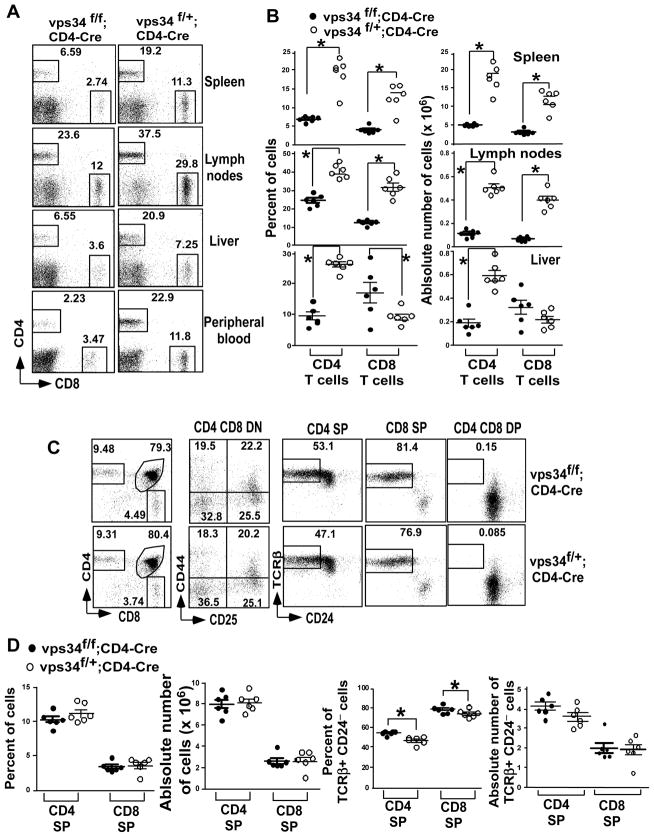
FIGURE 2.
Frequency of T cells in T cell-specific Vps34-deficient mice. (A) The frequency of CD4+ T cells and CD8+ T cells was analyzed in spleen, lymph nodes, liver and peripheral blood after live cell gating by flow cytometry. (B) Summary of the frequency of CD4+ and CD8+ T cells as shown in (A) and their absolute numbers calculated based on the organ cellularity with 5–6 mice analyzed in each group. *p<0.05. (C) Thymocytes were stained with anti-CD4 and -CD8 antibodies and double negative (DN) cells were analyzed using anti-CD44 and -CD25 antibodies. Positively selected cells were identified by the expression of TCRβ and loss of CD24 on T cells gated on CD4 single-positive (SP), CD8 SP and CD4 CD8 double-positive (DP) cells. (D) Summary of the frequency and absolute numbers from (C) is shown with 5–6 mice analyzed per group and from at least 3 independent experiments. *p<0.05.
To understand the mechanism for the loss of peripheral T cells in Vps34-deficient organs, we analyzed thymic T cell development. We found that the percentages of CD4 single-positive (SP), CD8 SP and CD4 CD8 DP cells were comparable between Vps34f/f;CD4-Cre and Vps34f/+;CD4-Cre mice (Fig. 2C,D). We found that the thymic cellularity in Vps34f/f;CD4-Cre mice was slightly higher as compared to Vps34f/+;CD4-Cre mice (Supplemental Fig. 2A). However, differences in the absolute numbers of CD4 and CD8 SP cells did not reach statistical significance (Fig. 2D). Analysis of DN cells, based on expression of CD44 and CD25 during various stages of thymocyte development revealed that Vps34f/f;CD4-Cre mice had no defect in transition from the DN1 to DN4 stages as compared with Vps34f/+;CD4-Cre mice (Fig. 2C,D), which is consistent with initiation of CD4 transgene expression at the DP thymocyte stage (32). We further analyzed the mature T cell subset that was positively selected, based on the acquisition of TCRβ expression and loss of CD24 expression (33). Surprisingly, we found that Vps34f/f;CD4-Cre mice had a higher percentage of TCRβ+CD24− cells among the CD4 and CD8 SP fractions as compared with the controls (Fig. 2D). The DP population was used as a negative control, as they hardly displayed any positively selected cells (Fig. 2C). Such an increase in positively selected cells in the thymus may represent a compensatory mechanism for peripheral T cell loss. These results further supported the notion that thymic development of conventional T cells in T cell-specific Vps34-deficient mice was largely unperturbed.
Vps34 is required for invariant natural killer T cell development
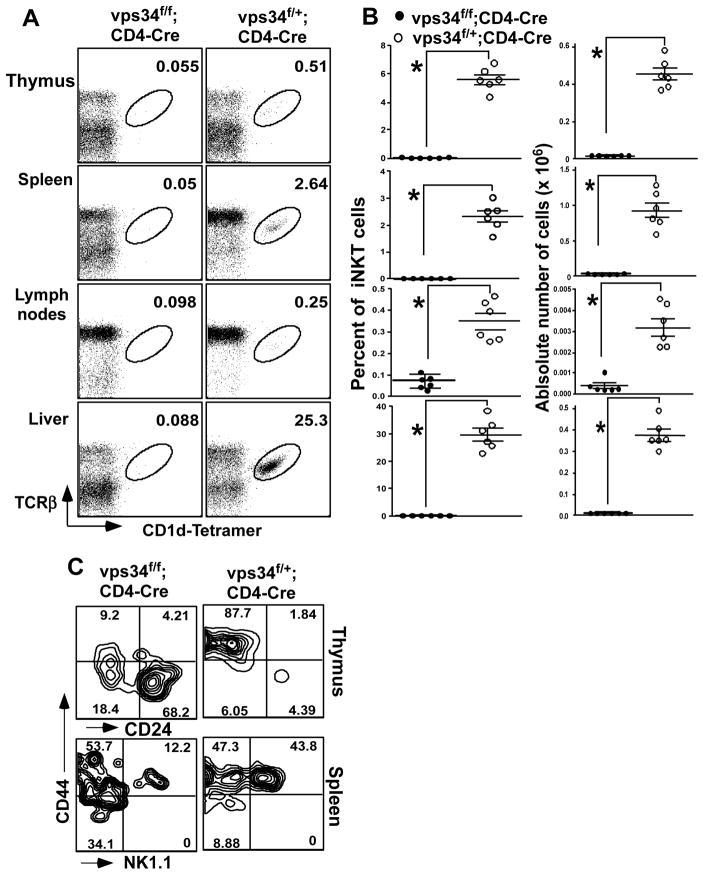
Invariant natural killer T cells (iNKT) cells are a small population of T cells that recognize glycolipid antigens bound with the MHC class I-related protein CD1d (34–37). iNKT cell development in the thymus requires expression of the Vα14-Jα18 invariant TCR and its recognition of endogenous glycolipid ligands presented by CD1d expressed on thymocytes (28, 29, 38). The positively selected iNKT cells then arise from the DP stage to mostly become CD4+ iNKT cells and a smaller population of CD4−CD8− iNKT cells (39, 40). iNKT cell development progresses through various stages, based on their early expression of CD24 and CD44 and late expression of NK1.1 (41–43). After their development, these cells populate peripheral lymphoid organs, mainly liver and spleen. We found that Vps34f/f;CD4-Cre mice exhibited a profound defect in intrathymic iNKT cell development, as hardly any TCRβ+CD1d-tetramer+ cells were observed as compared with the heterozygote controls (Fig. 3A,B). Consistent with these findings, the iNKT cell population was lacking in the spleen, liver and lymph nodes (Fig. 3A,B). We next assessed the precise stage of iNKT cell developmental blockade. The developmental stages of iNKT cells are defined as CD1d-tetramer+ cells that are CD24+CD44−NK1.1− (stage 0), CD24−CD44−NK1.1− (stage 1), CD24−CD44+NK1.1− (stage 2) and CD24−CD44+NK1.1+ (stage 3). We found that most iNKT cells in the thymus of Vps34f/f;CD4-Cre mice were CD44−CD24+, with few cells of the other stages (Fig. 3C), suggesting that iNKT cells in these mice exhibited a developmental blockade at stage 0. Since loss of CD24 expression on thymocytes is a marker for positive selection, we considered the possibility that iNKT cells in these animals had a defect in positive selection due to defective expression of endogenous glycolipid ligands that mediate this process. We therefore measured activation of DN32.D3 iNKT cell hybridoma responder cells, which react with endogenous glycolipids displayed by CD1d (28), by thymocytes derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice (29, 38). The results showed comparable DN32.D3 cell activation (data not shown), suggesting that Vps34-deficient thymocytes express normal levels of endogenous iNKT cell glycolipid ligands. Consistent with these findings, surface expression of CD1d on thymocytes derived from both groups of animals was comparable (data not shown). These results suggested that the blockade in iNKT cell development at stage 0 in Vps34f/f;CD4-Cre mice was most likely due to a cell-intrinsic defect rather than a defect in the expression of endogenous glycolipid ligands on the selecting thymocytes.
FIGURE 3.
iNKT cells in T cell-specific Vps34-deficient mice. (A) Splenic, lymph node, thymic and liver mononuclear cells were prepared from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice and were stained with anti-TCR-β, CD1d-tetramer and anti-B220 antibodies. (B) Frequency of iNKT cells in various lymphoid organs with 5–6 mice analyzed in each group from at least 3 independent experiments. *p<0.05. (C) Thymic iNKT cells were stained with CD1d-tetramer, and anti-CD8, -CD44 and -CD24 antibodies. Cells gated on tetramer+CD8α− cells are shown. For splenic iNKT cell analysis, B220+ cells were excluded from the analysis. A representative experiment of 3 independent experiments is shown.
Consistent with previous findings demonstrating that iNKT cells and NK cells share common requirements for homeostatic cytokines such as IL-15 and IL-7 (44), Vps34f/f;CD4-Cre mice exhibited a reciprocal increase in NK cells in the spleen, liver and lymph nodes (Supplemental Fig. 3A). Due to partial loss of T cells, the overall percentage of other cells of the immune system, including γδ T cells, DCs and B cells (Supplemental Fig. 3B–D) were increased in spleen, liver and lymph nodes of mice with a T cell-specific deficit in Vps34, but the absolute numbers of these cells remained largely unaffected.
Loss of peripheral T cells due to accumulation of cellular organelles and apoptosis
To understand the mechanism of peripheral T cell loss in Vps34f/f;CD4-Cre mice, we first tested if these cells were dying because of apoptosis. Staining of splenic and lymph node T cells with annexin V showed that Vps34-deficient T cells consistently exhibited a higher percentage of early apoptotic cells (annexin-V+7AAD−) (Fig. 4A and Supplemental Fig. 2B). We also observed a higher percentage of late apoptotic cells (annexin-V+7AAD+) in Vps34f/f;CD4-Cre mice as compared with the heterozygote controls. However, this was not observed in the thymus of Vps34f/f;CD4-Cre mice (Fig. 4B and Supplemental Fig. 2B), which is consistent with the normal cellularity of thymic SP T cells (Supplemental Fig. 2A).
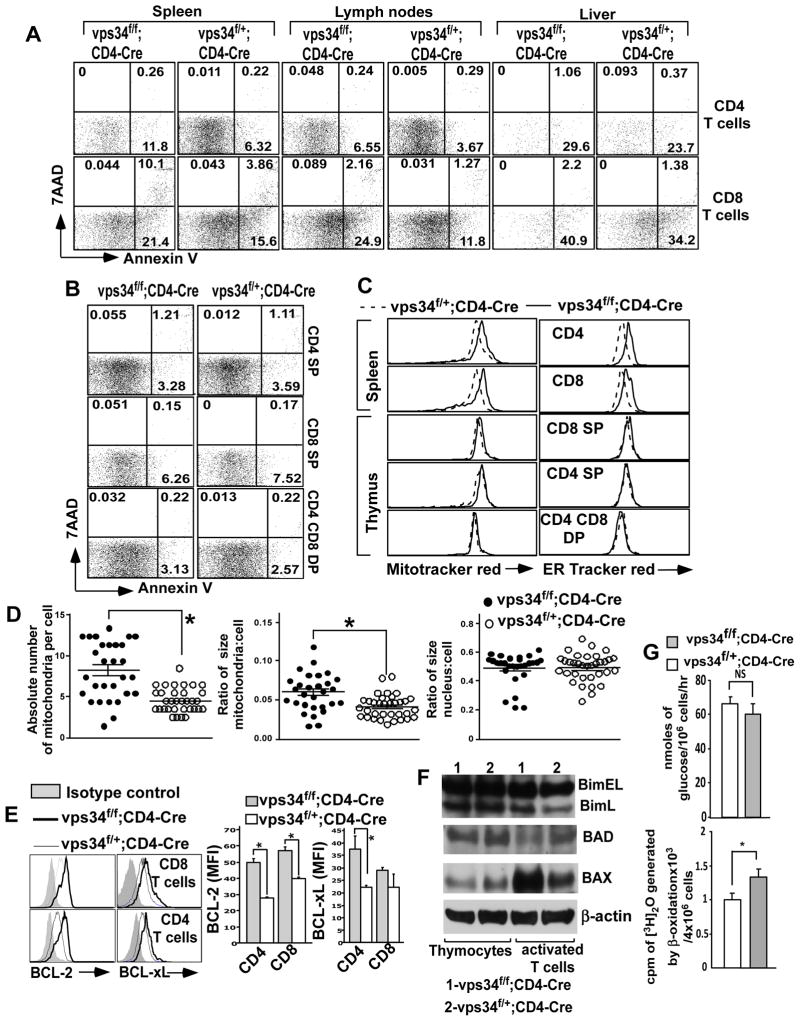
FIGURE 4.
Apoptosis and cellular organelles in Vps34-deficient T cells. (A,B) Splenic (A), lymph node (A), liver (A) or thymic (B) mononuclear cells were prepared from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice and were stained with anti-CD4 and -CD8 antibodies, 7-AAD and Annexin V. Annexin V+7-AAD− cells were considered early apoptotic cells and annexin V+7-AAD+ cells were considered late apoptotic cells. A representative of 4 independent experiments is shown. The average ± SEM of 6 mice is shown. A summary of these results is shown in Supplemental Fig. 2B. (C) Splenocytes or thymocytes were stained with MitoTracker® Red (50 nM) or ER-Tracker™ red (1 μM) at 37°C for 30 min in complete medium followed by surface staining with anti-CD4 and -CD8 antibodies at 4°C. The data shown are gated on splenic CD4+ or CD8+ T cells or thymic CD4 SP, CD8 SP or CD4 CD8 DP cells. A representative of 3 independent experiments is shown. (D) Absolute number of mitochondria and the total size of the mitochondria were determined in activated T cells using transmission electron microscopy. The Image J software was used to manually mark single mitochondria, measuring their surface area, and normalization against the scale bar from transmission electron microscopy images. The data presented are the total size of all the mitochondria or nucleus (as negative control) normalized against the size of the cell. A representative from 30–35 individually imaged cells in each group is shown to the bottom. Scale bar = 500 nm. *p<0.05. (E) Splenocytes derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice were stained with anti-CD4 and -CD8 antibodies followed by intracellular staining with anti-Bcl-2 or -Bcl-xL antibodies or their respective isotype controls. The data shown are gated on CD4+ or CD8+ T cells. A representative experiment of 3 individual experiments is shown to the left and the average ± SEM of 4 mice is summarized. (F) T cells were activated as described in the legend to Fig. 1 and subjected to immunoblot analysis with anti-Bim, -Bad or -Bax antibodies. (G) The rate of glycolysis was measured in activated T cells by incubating 106 cells with ten μCi of 5-[3H] glucose for 1 hr in RPMI medium. The rate of β-oxidation was measured by culture of 4×106 activated T cells in 0.4 ml of RPMI containing 2 μCi 9,10-[3H] palmitate (MP), 2% of BSA (fatty acid free) and 0.25 mM L-carnitine for 4 hrs.
Previous studies with autophagy-related genes such as Atg3 (21) and Atg5 (45) in T cells have shown that defective autophagy causes accumulation of cellular organelles, ultimately leading to apoptosis of T cells. Consistent with these findings, we similarly found, using organelle-specific dyes and flow cytometry, that Vps34f/f;CD4-Cre mice have an increase in mitochondrial and endoplasmic reticulum size in their splenic and lymph node T cells but not in thymic T cells (Fig. 4C). Further, electron microscopic images showed increased numbers and sizes of mitochondria in splenic Vps34-deficient T cells (Fig. 4D).
Next, we measured expression of the anti-apoptotic factors Bcl-2 and Bcl-xL in T cells. We found that expression of Bcl-2 was significantly increased and expression of Bcl-xL was modestly increased in Vps34-deficient CD4+ and CD8+ T cells as compared with the controls (Fig. 4E), suggesting that the increase in apoptosis in Vps34-deficient T cells was not due to lack of induction of anti-apoptotic factors in response to impaired growth factor signaling. We also measured the expression of pro-apoptotic molecules such as Bcl-2–interacting mediator of cell death (Bim), Bcl-2–associated X protein (Bax) and Bcl-2–associated death protein (Bad) in control and Vps34-deficient T cells by Western blot analysis. The results showed that expression of Bim and Bad remained unchanged in thymocytes as well as in activated T cells derived from Vps34f/+;CD4-Cre mice or Vps34f/f;CD4-Cre mice. Interestingly, the expression of Bax was significantly elevated in activated T cells derived from Vps34f/f;CD4-Cre mice as compared with Vps34f/+;CD4-Cre mice, while its expression in thymocytes remained unchanged (Fig. 4F).
To further understand the mechanism for increased cellular apoptosis in Vps34-deficient T cells, we next measured parameters related to energy metabolism. We found that the rate of glycolysis in activated T cells derived from control and Vps34-deficient mice was similar. Interestingly, however, the rate of β-oxidation of fatty acids was significantly higher in Vps34-deficient than wild-type T cells (Fig. 4G), suggesting dysregulated energy metabolism in autophagy-defective T cells.
Collectively these results suggested that the defect in autophagy in Vps34-deficient T cells caused an accumulation of cellular organelles, leading to apoptosis, possibly due to dysregulated energy metabolism.
Loss of quiescence and defective response to TCR engagement in Vps34-deficient T cells
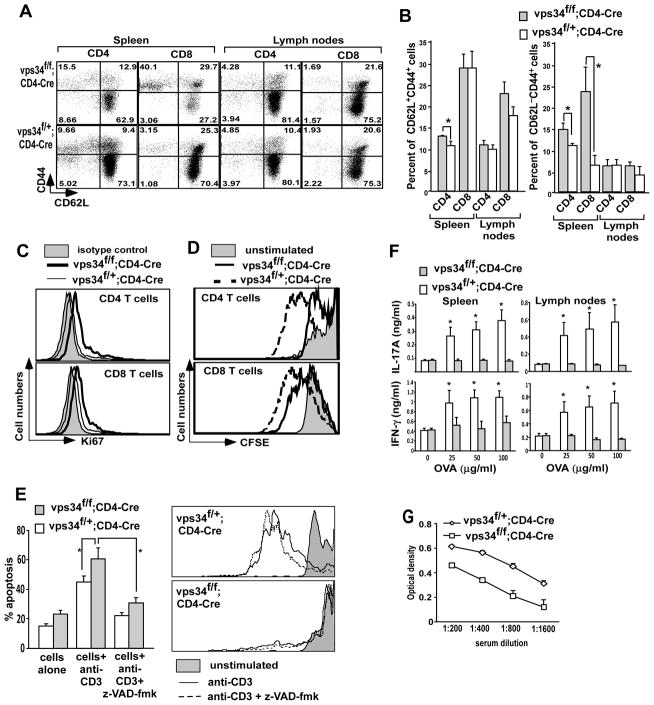
Previous studies with Atg3 conditional knockout mice have described lymphopenia-mediated T cell proliferation with acquisition of the CD44 activation marker and loss of the naïve T cell marker CD62L (21). Phenotyping of T cells using antibodies against markers such as CD44, CD69, CD25 and CD62L showed that Vps34f/f;CD4-Cre mice have an increased frequency of CD44+CD62L− T cells as compared with Vps34f/+;CD4-Cre mice, predominantly reflected among splenic T cells (Fig. 5A,B). While none of the T cells derived from Vps34 conditional knockout or control mice expressed CD69 under steady-state conditions, expression of CD25 was mostly confined to FoxP3+ Treg cells (see Fig. 7 below). This phenotype correlated well with the increased expression of intracellular Ki67 in Vps34-deficient T cells as compared with control T cells (Fig. 5C), suggesting a higher level of homeostatic proliferation among Vps34-deficient T cells, possibly due to their gradual loss of quiescence. Next, we tested how Vps34-deficient T cells respond to TCR-mediated stimulation. For this purpose, we stimulated CFSE-labeled splenocytes with anti-CD3 antibodies and measured proliferation of CD4+ and CD8+ T cells using CFSE-dilution assay. We found that Vps34-deficient T cells proliferated less efficiently as compared with Vps34-sufficient T cells, suggesting a defect in the response to TCR-mediated stimulation (Fig. 5D). Such a defect was T cell intrinsic and was not due to higher levels of apoptosis observed in Vps34f/f;CD4-Cre mice as compared to Vps34f/+;CD4-Cre mice, as inclusion of the apoptosis inhibitor zVAD-fmk in these cultures failed to restore the T cell proliferative defect observed in Vps34-deficient T cells (Fig. 5E).
FIGURE 5.
Homeostatic markers and proliferation of T cells in response to TCR engagement. (A) Splenocytes and lymph node cells derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice were stained with anti-CD44, -CD62L, -CD4 and -CD8 antibodies. The data shown are gated on CD4+ or CD8+ T cells. (B) Summary of results obtained in (A). The average ± SEM of 6 mice is shown. (C) Splenocytes were stained with anti-CD4-PerCP and -CD8-APC antibodies followed by intracellular staining with anti-Ki67 antibody. The data shown are gated on CD4+ or CD8+ T cells. A representative experiment of 3 individual experiments is shown. (D) Splenocytes from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cremice were labeled with the CFSE dye and 3×105 cells/well were activated with anti-CD3 antibody (1 μg/ml) for 60 hrs. Cells were stained with anti-CD4 and -CD8 antibodies and CFSE dilution analysis was performed on the respective T cell subset. (E) CFSE-labeled cells were cultured as in (D) with or without z-VAD-fmk (20 μM). The percentage total apoptotic cells was measured (left) as described in the legend of Fig. 4 and CFSE dilution analysis was performed on CD4+ T cells. A representative experiment of 3 individual experiments is shown. (F) Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice were immunized with ovalbumin (OVA). The mice were sacrificed at day 15 and splenic and draining lymph node cells were stimulated in vitro with 100 μg/ml of OVA. The supernatants were collected at 72 hrs of culture and assayed for IL-17A and IFN-γ by ELISA. (G) Total serum IgG derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice was measured by ELISA. The average ± SEM of 4 mice is shown.
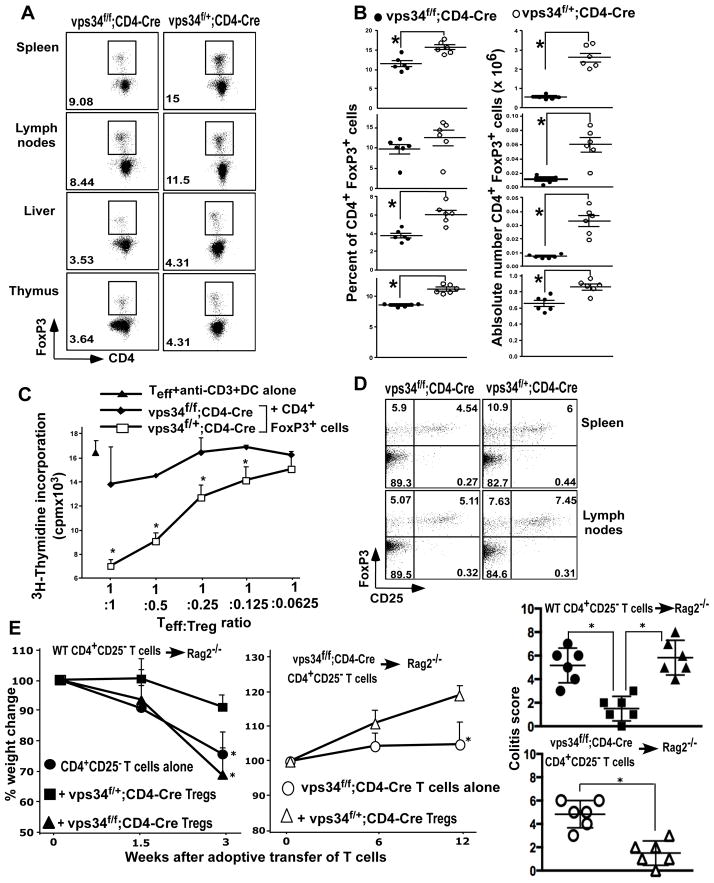
FIGURE 7.
CD4+FoxP3+ Treg cells in T cell-specific Vps34-deficient mice. (A) Splenic, lymph node, thymic and liver mononuclear cells were prepared from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice and were stained with anti-CD4, -CD8 and -CD25 antibodies followed by intracellular staining with anti-FoxP3 antibody. Data shown are for cells gated on CD4+ T cells or CD4 SP thymocytes. (B) Summary of the frequency of CD4+FoxP3+ T cells and their absolute numbers calculated based on the organ cellularity with 5–6 mice analyzed in each group. *p<0.05. (C) Two × 105 CD4+CD25− T effector (Teff) cells were activated with anti-CD3ε antibody (1 μg/ml) and 2×104 DCs in the presence of Treg cells purified from splenic CD4+ T cells derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice at various Teff to Treg cell ratios for 60 hrs with the addition of 1μCi of [3H]thymidine for the last 12 hrs of culture. *p<0.05. (D) Splenic and lymph node cells were stained as in (A) and data are shown for cells gated on CD4+ T cells. Results shown are representative of 3 individual experiments and 6 mice in each group. (E, F) CD4+CD25− cells (2×106 cells) derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice were adoptively transferred into Rag2−/− mice to induce colitis. Groups of mice were co-transferred with 5×105 CD4+CD25+ Treg cells derived from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice. The mice were followed for 12 weeks for weight loss and signs of colitis. The data shown are the average ± SEM of 6 mice pooled from two experiments. Representative histology pictures from each group are shown in Supplemental Fig. 4B.
We next tested the capacity of Vps34-deficient T cells to induce an antigen-specific response in vivo. For this purpose Vps34f/f;CD4-Cre mice and Vps34f/+;CD4-Cre mice were immunized with ovalbumin (OVA) antigen emulsified in CFA and boosted at day 7. At day 15 after immunization, splenocytes and draining lymph node cells were restimulated with OVA in vitro and IFN-γ and IL-17A responses were measured in the culture supernatant. The results showed that Vps34f/f;CD4-Cre mice displayed a profound defect in the generation of OVA-specific IFN-γ and IL-17A responses (Fig. 5F). Consistent these findings, we observed reduced levels of total IgG antibodies in the serum of Vps34f/f;CD4-Cre mice (Fig. 5G). Collectively, these results suggested a defect in TCR-mediated stimulation, resulting in defective antigen-specific immune responses in vivo.
Development of a wasting syndrome in aged T cell-specific Vps34 knockout mice
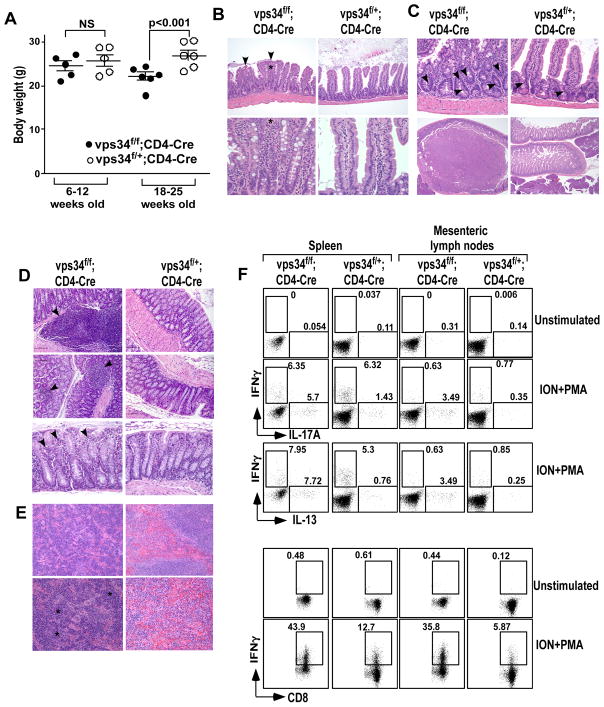
We observed that aged (>18 weeks old) Vps34f/f;CD4-Cre mice developed a wasting syndrome with significant weight loss (Fig. 6A). This was often accompanied by a rectal prolapse, which suggested intestinal inflammation leading to the development of colitis. Histological examination of tissue sections showed significant but mild inflammation within both the small and large intestines (Fig. 6B–D). We found an increased frequency of lymphoid follicles that had a larger size, located in the mucosa of the colons of Vps34f/f;CD4-Cre mice. In two of the Vps34f/f;CD4-Cre mice we found unusually large lymphoid follicles in the small intestine, and this was never observed in control mice (Supplemental Fig. 4). The small and large intestinal lamina propria displayed infiltration of inflammatory cells such as neutrophils, macrophages and plasma cells, as determined by their nuclear morphology and cytoplasmic stains (Fig. 6B,D). Other significant observations included fusion of the crypts (Fig. 6B), hyperplasia of epithelial cells as determined by nuclear crowding in the crypts, increased numbers of mitotic cells, and polyp formation in the small intestine (Fig. 6C). We further observed inflammatory lesions ranging from hyperplasia to adenoma in the proximal duodenum of Vps34f/f;CD4-Cre mice (Fig. 6C). We next carried out full body necropsy to determine if other organs of the body were affected. The results showed that inflammation was mostly confined to the intestine, whereas all the vital organs of the body such as heart, liver, stomach, kidney, lungs and the brain appeared normal in Vps34f/f;CD4-Cre mice as compared with the controls. Interestingly, pockets of erythropoiesis (but not myelopoiesis) were observed in the splenic sections (Fig. 6E), which could be suggestive of extramedullary erythropoiesis due to anemia. Consistent with this possibility, complete blood counts showed low hematocrit and a compensatory increase in the numbers of reticulocytes in the blood of Vps34f/f;CD4-Cre mice (Table I).
FIGURE 6.
Development of a wasting disease in T cell-specific Vps34-deficient mice. (A) Absolute weights of mice in 6- to 12-week-old or 18- to 25-week-old mice. (B) H&E staining of small intestinal sections from Vps34f/f;CD4-Cre or Vps34f/+;CD4-Cre mice depicting intestinal villus blunting and fusion (arrows), and inflammatory infiltrates (*) in the lamina propria consisting of neutrophils, lymphocytes, plasma cells, and macrophages in Vps34f/f;CD4-Cre mice (magnification: upper panels, 20x; lower panels, 60x). (C) Top panels depict H&E staining of small intestinal sections (magnification: 40x) showing hyperplastic crypts in the Vps34f/f;CD4-Cre mice with crowded nuclei and increased mitosis (arrows) while few scattered mitotic cells were observed in the Vps34f/+;CD4-Cre mice. Lower panels (magnification: 4x) depict adenomas in the anterior duodenum of Vps34f/f;CD4-Cre mice. (D) H&E staining of colon sections showing increased lymphoid follicles in the mucosa of Vps34f/f;CD4-Cre mice (upper and middle panels: 20x magnification, bottom panels: 40x magnification) with mild infiltrates of neutrophils, lymphocytes and plasma cells (arrows) in the lamina propria of the distal colon. (E) H&E staining of splenic sections showing expanded red pulp area due to extramedullary erythropoiesis (*) in Vps34f/f;CD4-Cre mice (magnification: upper panels: 20x, lower panels, 40x). (F) Splenic and mesenteric lymph node mononuclear cells from 18- to 25-week-old mice were prepared and 2×105 cells were activated with PMA and ionomycin (ION+PMA) in the presence of GolgiPlug™ to allow intracellular accumulation of cytokines. Cells were then harvested and surface-stained with anti-CD4 and -CD8 antibodies, followed by intracellular staining with anti-IL-17A and anti-IFN-γ antibodies, or with anti-IL-13 and anti-IFN-γ antibodies. The data shown are gated on CD4+ or CD8+ T cells. Results shown are representative of 3 individual experiments and 6 mice in each group.
Table I.
Complete blood counts of aged Vps34f/f;CD4-Cre and Vps34f/+;CD4-Cre mice.a
| Blood test (unit) | Mouse genotype
|
|
|---|---|---|
| Vps34f/f;CD4-Cre | Vps34f/+;CD4-Cre | |
| White blood cells (thousands/μl) | 6.28 ± 0.374 | 4.88 ± 0.906 |
| Neutrophils (thousands/μl) | 1.81 ± 0.272 | 1.42 ± 0.517 |
| Lymphocytes (thousands/μl) | 4.19 ± 0.123 | 3.116 ± 0.310* |
| Monocytes (thousands/μl) | 0.243 ± 0.029 | 0.343 ± 0.0876 |
| Eosinophils (thousands/μl) | 0.0233 ± 0.0133 | 0 |
| Basophils (thousands/μl) | 0.0066 ± 0.0066 | 0.0033 ± 0.0033 |
| Nucleated red blood cells (thousands/μl) | 0 | 0 |
| Neutrophils (%) | 28.56 ± 2.780 | 26.88 ± 6.146 |
| Lymphocytes (%) | 67 ± 2.486 | 66.19 ± 6.782 |
| Macrophages (%) | 3.92 ± 0.399 | 6.83 ± 0.676* |
| Eosinophils (%) | 0.383 ± 0.217 | 0.0466 ± 0.024 |
| Basophils (%) | 0.116 ± 0.088 | 0.046 ± 0.032 |
| Nucleated red blood cells (%) | 0 | 0 |
| Hematocrit (%) | 31.83 ± 2.142 | 39.93 ± 1.36* |
| Red blood cells (millions/μl) | 7.88 ± 0.458 | 9.65 ± 0.362* |
| Hemoglobin (g/dL) | 9.7 ± 0.838 | 12.6 ± 0.351* |
| Mean corpuscular erythrocyte volume (fL) | 40.36 ± 0.536 | 41.4 ± 0.305 |
| Mean corpuscular erythrocyte hemoglobin (pg) | 12.26 ± 0.392 | 13.06 ± 0.120 |
| Mean corpuscular erythrocyte hemoglobin concentration (g/dL) | 30.4 ± 0.608 | 31.56 ± 0.284 |
| Red cell volume distribution width (%) | 21.16 ± 3.112 | 13.73 ± 0.272* |
| Red cell standard deviation (%) | 8.53 ± 1.156 | 5.7 ± 0.115* |
| Reticulocytes (thousands/μl) | 175.26 ± 43.74 | 16.66 ± 4.605* |
| Reticulocytes (%) | 2.27 ± 0.615 | 0.176 ± 0.055* |
| Platelet count (thousands/μl) | 1381 ± 154.77 | 968 ± 82.52* |
| Mean platelet volume (fL) | 5.56 ± 0.120 | 5.466 ± 0.166 |
| Platelet volume distribution width (%) | 42.2 ± 0.115 | 41.93 ± 0.95 |
| Plateletcrit (%) | 0.7686 ± 0.085 | 0.531 ± 0.057* |
Blood was collected from 18- to 25-week-old Vps34f/f;CD4-Cre and Vps34f/+;CD4-Cre mice in heparinized tubes. Complete blood counts were carried out in an automated analyzer. The data shown are the mean ± SEM of 3 mice in each group.
p<0.05
We next measured cytokine responses of T cells in aged mice. The results showed that Vps34f/f;CD4-Cre mice had an increased percentage of CD4+ T cells in spleen and lymph nodes that produced IL-17A and IL-13 cytokines, whereas CD8+ T cells expressed higher levels of IFN-γ as compared with cells from the heterozygous controls (Fig. 6F). These results suggested the development of T cell-mediated autoinflammatory disease in Vps34f/f;CD4-Cre mice.
The combination of intestinal inflammation and anemia may provide an explanation for the wasting syndrome observed in Vps34f/f;CD4-Cre mice.
Vps34 is required for normal homeostatic maintenance and function of CD4+FoxP3+ Treg cells
To provide a possible explanation for the development of the inflammatory wasting syndrome in Vps34f/f;CD4-Cre mice, we next assessed the development of the CD4+FoxP3+ T regulatory (Treg) cell population of these animals. We found that Treg cell development within the thymus was evident among the CD4 SP cells in Vps34f/f;CD4-Cre mice, but at reduced levels as compared with Vps34f/+;CD4-Cre mice (Fig. 7A,B). In the peripheral lymphoid organs such as spleen and lymph nodes and in the liver, the percentage of Treg cells among CD4+ T cells was reduced by 20–30% in Vps34f/f;CD4-Cre mice as compared with the heterozygote controls. We also found that, due to reduced numbers of T cells and a decrease in overall cellularity of all peripheral lymphoid organs (Supplemental Fig. 2A), the absolute numbers of Treg cells were significantly reduced in Vps34f/f;CD4-Cre mice (Fig. 7B). Next, we tested the capacity of these Treg cells to function as suppressor cells using an ex vivo Treg cell functional assay. For this purpose we purified CD4+CD25+ Treg cells from Vps34f/f;CD4-Cre and Vps34f/+;CD4-Cre mice on the basis of their CD25 expression and cultured these cells with CD4+CD25− T effector (Teff) cells from wild-type animals at various Teff to Treg cell ratios in the presence of DCs and anti-CD3 antibodies. The results clearly showed that Treg cells derived from Vps34f/f;CD4-Cre mice, compared with Treg cells derived from Vps34f/+;CD4-Cre mice, had severely reduced functional capacity to suppress the Teff cell response (Fig. 7C). This reduced suppressive capacity of Tregs derived from Vps34f/f;CD4-Cre mice was not due to increased contamination with CD4+FoxP3−CD25+ T cells, as these cells were equally represented in both groups of mice (Fig. 7D).
We next tested the capacity of Treg cells derived from Vps34f/+;CD4-Cre or Vps34f/f;CD4-Cre mice to prevent the development of colitis induced by adoptive transfer of wild-type CD4+CD25− T cells into Rag2−/− mice as described (23). Adoptive transfer of wild-type CD4+CD25− T cells into Rag2−/− mice resulted in the development of colitis characterized by weight loss, moderate diarrhea, hunched posture, anal inflammation, infiltration of cells in the lamina propria, loss of goblet cells and occasional ulceration between 2–3 weeks after transfer. These clinical signs of colitis were significantly reversed by co-transfer of CD4+CD25+ Treg cells derived from Vps34f/+;CD4-Cre mice. However, co-transfer of Treg cells derived from Vps34f/f;CD4-Cre mice failed to prevent the development of colitis in recipient animals (Fig. 7E). These findings provide further evidence for a functional defect of Treg cells in Vps34f/f;CD4-Cre mice. Next, we determined the effect of adoptive transfer of CD4+CD25− T cells derived from Vps34f/f;CD4-Cre mice into Rag2−/− mice and found that these cells failed to cause a wasting disease between 2–3 weeks after transfer. Instead, development of a wasting syndrome was observed 10 weeks after adoptive transfer (Fig. 7E). This disease was characterized by loss of weight gain, diarrhea, infiltration of cells in the colonic lamina propria, loss of goblet cells, anal bleeding and colonic ulcers, without signs of severe weight loss or hunched posture (Fig. 7E). These disease signs were prevented by co-transfer of Treg cells derived from Vps34f/+;CD4-Cre mice. These results therefore suggested a cell intrinsic capacity of T cells from Vps34f/f;CD4-Cre mice to cause wasting disease, which could be counteracted by functional Treg cells from wild-type mice.
Collectively, our findings showed that intrathymic Treg cell development was evident in Vps34f/f;CD4-Cre mice but that peripheral Treg cell homeostasis and acquisition of functional capacity were profoundly defective.
Discussion
Autophagy has emerged as a key process that regulates many aspects of T cell function, including their development, survival and homeostasis (6, 21, 45, 46). Here, we describe the role of the autophagy-related class III PI3-kinase Vps34 in the survival, peripheral homeostatic maintenance and TCR-stimulated proliferation of conventional T cells. While we were unable to identify a defect in the development of conventional T lymphocytes in the thymus of Vps34f/f;CD4-Cre mice, the development of iNKT cells was profoundly blocked. Vps34-deficient thymocytes and peripheral T cells were defective in autophagic flux, making these cells susceptible to apoptosis, as reflected by their reduced numbers in the periphery. Our findings further suggested that the remaining T cells gradually lost quiescence due to the mild lymphopenic environment or/and due to reduced and ineffective Treg cells. While these T cells had defects in TCR-induced proliferation, they retained the capacity to produce inflammatory cytokines, which might explain the development of an inflammatory wasting disease in these animals.
Vps34 has been suggested to play a critical role in the initiation of the autophagic process by generating PtdIns3P at the site of autophagosome formation, which recruits PtdIns3P-binding proteins (15). In yeast, the absence of Vps34 results in a severe blockade in autophagosome formation, but the role of Vps34 in autophagy in mammalian cells has been controversial. Recent studies have described the deletion of Vps34 in sensory neurons (17), embryonic fibroblasts, heart, liver (16) or T cells (18) from mice, indicating an essential role of Vps34 in cellular function and survival. While the studies with neuron- or T cell-specific ablation of Vps34 failed to identify a defect in autophagy (17, 18), a study using the same Vps34f/f mice as in the present study to ablate Vps34 in embryonic fibroblasts, liver, and heart showed that Vps34 is required for autophagy (16). Our results presented here provide evidence that ablation of Vps34 in T cells results in defective autophagic flux, and impaired turnover of autophagic cargo. In turn, these abnormalities in autophagy result in the accumulation of damaged proteins and subcellular organelles and subsequent T cell death. Immunostaining of LC3B in electron microscopic sections revealed that LC3B staining in Vps34-defcient T cells failed to localize to membrane structures but instead localized to the cytoplasm. This might be indicative of accumulating LC3B protein aggregates due to defective autophagic flux. The Vps34-independent conversion of LC3BI to LC3BII found in our study (Fig. 1A,C,H) is in accordance with previous reports describing the function of Vps34 in heart, liver, T cells and neurons (16–18, 47). Consistent with these findings, we found that Vps34 deletion led to enhanced T cell apoptosis, similar to that caused by T cell-specific deletion of Atg3 (21), Atg5 (45), or Atg7 (21, 45, 48). Our findings reported here are consistent with a recent study reporting a critical role of Vps34 in autophagy in T cells (49).
The precise mechanism for the induction of apoptosis in Vps34-deficient T cells and the factors that mediate this process remain incompletely understood. A recent study with T cell-specific Vps34-deficient mice suggested a role for defective IL-7Rα expression, resulting in reduced induction of Bcl-2 and T cell loss (18). While we similarly found decreased expression of IL-7Rα by T cells in our line of Vps34 conditional knockout mice (data not shown), we observed an increase in anti-apoptotic factors such as Bcl-2 and Bcl-xL. The reasons for such discrepancies could be due to differences in the Cre transgenes employed (Lck-Cre employed by McLeod et al. (18) vs CD4-Cre employed by us and by Willenger and Flavell (49) to selectively ablate Vps34 in T cells or due to differences in specific targeting of Vps34 exons (exon 17/18 employed by McLeod et al (18) vs exon 4 employed by us (16) and by Willenger and Flavell (49). Our results, nevertheless, are in line with high Bcl-2 expression found in T cells from T cell-specific Atg7 knockout mice (48), suggesting that defective cytokine signaling and lack of induction of anti-apoptotic factors may not be the primary cause of T cell loss induced by Vps34-deficiency. Based on the similarities between the T cell phenotype observed in mice with a T cell-specific deletion of Vps34 and mice with a T cell-specific deletion in Atg3, Atg5 or Atg7 (50), and our results demonstrating an increase in fatty acid oxidation in Vps34-null T cells, we speculate that nutrient starvation in the face of reduced autophagy results in apoptotic death of Vps34-deficient T cells.
The most striking phenotype we observed in T cell-specific Vps34 mutant mice was the development of a wasting syndrome in aged animals. This was often accompanied by a rectal prolapse and anemia. Histological analysis showed mild inflammation throughout the intestine and this was typically characterized by the presence of large lymphoid follicles in the mucosa, infiltration of inflammatory cells such as neutrophils, macrophages and plasma cells in the lamina propria, crypt hyperplasia in the small intestine, and occasional polyp formation and lateral crypt fusion. Interestingly, crypt hyperplasia has often been observed during inflammatory diseases in the intestine and is typically induced by inflammatory cytokines such as IFN-γ and -α (44, 45), whereas lateral crypt fusion has been observed during prolonged low-grade inflammation in the intestine of patients with inflamed appendices (51). Vps34-deficient T cells exhibited a proliferative defect in response to TCR engagement but retained the capacity to induce inflammatory cytokines. This capacity to produce cytokines was not dependent on the age of the animals, as young (6- to 8-week-old) Vps34-deficient mice also displayed high cytokine production (Supplemental Fig. 4C). Further, adoptive transfer of enriched Vps34-null CD4+CD25+ T cells was sufficient to cause colonic disease in recipient Rag2−/− mice. This disease resembled the wasting syndrome observed in aged Vps34f/f;CD4-Cre mice in terms of its timing. Although colitis induced by adoptive transfer of CD4+CD25− T cells derived from either Vps34-sufficient or -deficient mice differed in its kinetics, disease in both groups of animals was ameliorated by co-transfer with Treg cells derived from wild-type mice (Fig. 7E). However, Treg cells derived from Vps34f/f;CD4-Cre mice were defective in suppressing disease induced by adoptive transfer of CD4+CD25− T cells derived from Vps34f/+;CD4-Cre mice. Therefore, we conclude that the inflammatory disease observed in the intestine of T cell-specific Vps34 mutant mice is mediated by T cells in an environment that is devoid of fully competent Treg cells.
Another interesting phenotype observed in T cell-specific Vps34-deficient mice was anemia. These mice displayed a low hematocrit, low numbers of RBCs and reduced hemoglobin in the blood (Table 1). Thus, a combination of inflammation in the intestine and anemia may explain the wasting syndrome observed in these animals. Such a phenotype was not observed in conditional Vps34-deficient mice where the Cre transgene with driven by the Lck promoter (18) and we have confirmed the lack of such a wasting disease in an independently generated line of Vps34f/f;Lck-Cre mice (unpublished observations). Reasons for such divergent effects are unclear but might be due to the differential timing of Vps34 deletion induced by the distinct Cre transgenes during thymic development. Nevertheless, the wasting disease observed in Vps34f/f;CD4-Cre mice resembles the phenotype of IL-2-deficient mice, which develop intestinal autoinflammatory disease and anemia (52–54). In these animals, IL-2-deficiency results in the generation of functionally defective Treg cells, which in turn, causes colitis and the generation of hemolytic autoantibodies against RBCs. However, we did not observe any evidence of autoantibodies against RBCs in Vps34-null mice (data not shown). Consistent with reduced numbers of T cells and defective TCR stimulation, the total IgG antibody levels in the serum of Vps34-null mice were significantly reduced as compared with control mice (Fig. 5G). While the phenotype observed in T cell-specific Vps34-deficient mice was milder and required aging of the animals, it was interesting that both of these strains developed intestinal inflammation and anemia that could be attributed to defective Treg cell function. We did not find any defect in IL-2 production by Vps34-deficient T cells, suggesting that the functional defect in Treg cells due to Vps34-deficiency is likely due to an intrinsic defect in autophagy in these cells. Further experiments will be required to precisely delineate the mechanism by which Vps34 and autophagy affect Treg cell function.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI070305 (to L.V.K.), HL089667 (to L.V.K.), DK081536 (to L.W. and L.V.K.), CA129536 (to W.X.Z), GM97355 (to W.X.Z.), and NS064090 (to J.Z.), a VA merit award (to J.Z.), the Vanderbilt Digestive Diseases Research Center (supported by NIH grant P30DK058404), and a postdoctoral fellowship from the National Multiple Sclerosis Society of America (to V.V.P.).
We thank the NIH tetramer facility for providing CD1d-tetramers and Dr. Albert Bendelac (University of Chicago, Chicago, IL) for providing the DN32.D3 hybridoma. We thank Drs. Sung Hoon Cho, Mark Boothby and Masakazu Shiota for providing reagents related to glycolysis and β-oxidation assays. We thank the VUMC cell imaging shared resource core (CISR) for confocal microscopy and transmission electron microscopy.
Abbreviations used in this article
- Atg
autophagy-related
- DN
double-negative
- DP
double-positive
- iNKT
invariant natural killer T
- PI3
phosphoinositide
- PtdIns
phosphatidylinositol
- PtdIns3P
PtdIns-3-phosphate
- SP
single-positive
- vps
vacuolar protein sorting
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xue L, Fletcher GC, Tolkovsky AM. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol Cell Neurosci. 1999;14:180–198. doi: 10.1006/mcne.1999.0780. [DOI] [PubMed] [Google Scholar]
- 3.Bursch W. The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ. 2001;8:569–581. doi: 10.1038/sj.cdd.4400852. [DOI] [PubMed] [Google Scholar]
- 4.Fengsrud M, Erichsen ES, Berg TO, Raiborg C, Seglen PO. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur J Cell Biol. 2000;79:871–882. doi: 10.1078/0171-9335-00125. [DOI] [PubMed] [Google Scholar]
- 5.Munz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- 6.Crotzer VL, Blum JS. Autophagy and intracellular surveillance: Modulating MHC class II antigen presentation with stress. Proc Natl Acad Sci U S A. 2005;102:7779–7780. doi: 10.1073/pnas.0503088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 8.Khalfan WA, Klionsky DJ. Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae. Curr Opin Cell Biol. 2002;14:468–475. doi: 10.1016/s0955-0674(02)00343-5. [DOI] [PubMed] [Google Scholar]
- 9.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 10.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 13.Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 14.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 6 doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 16.Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, Zong WX. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X, Wang L, Hasegawa H, Amin P, Han BX, Kaneko S, He Y, Wang F. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci U S A. 2010;107:9424–9429. doi: 10.1073/pnas.0914725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLeod IX, Zhou X, Li QJ, Wang F, He YW. The class III kinase Vps34 promotes T lymphocyte survival through regulating IL-7Ralpha surface expression. J Immunol. 2011;187:5051–5061. doi: 10.4049/jimmunol.1100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanner VA, Ploug T, Tao-Cheng JH. Subcellular localization of SV2 and other secretory vesicle components in PC12 cells by an efficient method of preembedding EM immunocytochemistry for cell cultures. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1996;44:1481–1488. doi: 10.1177/44.12.8985140. [DOI] [PubMed] [Google Scholar]
- 20.Ott C, Ross K, Straub S, Thiede B, Gotz M, Goosmann C, Krischke M, Mueller MJ, Krohne G, Rudel T, Kozjak-Pavlovic V. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol Cell Biol. 2012;32:1173–1188. doi: 10.1128/MCB.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immunol. 2011;186:5313–5322. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- 22.Parekh VV, Wilson MT, Olivares-Villagomez D, Singh AK, Wu L, Wang CR, Joyce S, Van Kaer L. Glycolipid antigen induces long-term natural killer T cell anergy in mice. J Clin Invest. 2005;115:2572–2583. doi: 10.1172/JCI24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mottet C, Uhlig HH, Powrie F. Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. J Immunol. 2003;170:3939–3943. doi: 10.4049/jimmunol.170.8.3939. [DOI] [PubMed] [Google Scholar]
- 24.Olivares-Villagomez D, Algood HM, Singh K, Parekh VV, Ryan KE, Piazuelo MB, Wilson KT, Van Kaer L. Intestinal epithelial cells modulate CD4 T cell responses via the thymus leukemia antigen. J Immunol. 2011;187:4051–4060. doi: 10.4049/jimmunol.1101052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edinger AL, Linardic CM, Chiang GG, Thompson CB, Abraham RT. Differential effects of rapamycin on mammalian target of rapamycin signaling functions in mammalian cells. Cancer research. 2003;63:8451–8460. [PubMed] [Google Scholar]
- 26.Dunning KR, Cashman K, Russell DL, Thompson JG, Norman RJ, Robker RL. Beta-oxidation is essential for mouse oocyte developmental competence and early embryo development. Biology of reproduction. 2010;83:909–918. doi: 10.1095/biolreprod.110.084145. [DOI] [PubMed] [Google Scholar]
- 27.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, Brutkiewicz RR. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 29.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 33.Crispe IN, Bevan MJ. Expression and functional significance of the J11d marker on mouse thymocytes. J Immunol. 1987;138:2013–2018. [PubMed] [Google Scholar]
- 34.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr Opin Immunol. 2008;20:358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 36.Taniguchi M, Harada M, Kojo S, Nakayama T, Wakao H. The regulatory role of Valpha14 NKT cells in innate and acquired immune response. Annu Rev Immunol. 2003;21:483–513. doi: 10.1146/annurev.immunol.21.120601.141057. [DOI] [PubMed] [Google Scholar]
- 37.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 38.Honey K, Benlagha K, Beers C, Forbush K, Teyton L, Kleijmeer MJ, Rudensky AY, Bendelac A. Thymocyte expression of cathepsin L is essential for NKT cell development. Nat Immunol. 2002;3:1069–1074. doi: 10.1038/ni844. [DOI] [PubMed] [Google Scholar]
- 39.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 40.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 41.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 43.Yue X, Izcue A, Borggrefe T. Essential role of Mediator subunit Med1 in invariant natural killer T-cell development. Proc Natl Acad Sci U S A. 108:17105–17110. doi: 10.1073/pnas.1109095108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuda JL, Gapin L, Sidobre S, Kieper WC, Tan JT, Ceredig R, Surh CD, Kronenberg M. Homeostasis of V alpha 14i NKT cells. Nat Immunol. 2002;3:966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 45.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, Lu B. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 47.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1205305109. Ahead of publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186:1564–1574. doi: 10.4049/jimmunol.1001822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A. 2012;109:8670–8675. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Millikin PD. On the possible origin of extra-epithelial enterochromaffin cells by budding from the crypts of Lieberkuhn. J Pathol. 1984;143:51–56. doi: 10.1002/path.1711430108. [DOI] [PubMed] [Google Scholar]
- 52.Sadlack B, Lohler J, Schorle H, Klebb G, Haber H, Sickel E, Noelle RJ, Horak I. Generalized autoimmune disease in interleukin-2-deficient mice is triggered by an uncontrolled activation and proliferation of CD4+ T cells. Eur J Immunol. 1995;25:3053–3059. doi: 10.1002/eji.1830251111. [DOI] [PubMed] [Google Scholar]
- 53.Wolf M, Schimpl A, Hunig T. Control of T cell hyperactivation in IL-2-deficient mice by CD4(+)CD25(-) and CD4(+)CD25(+) T cells: evidence for two distinct regulatory mechanisms. Eur J Immunol. 2001;31:1637–1645. doi: 10.1002/1521-4141(200106)31:6<1637::aid-immu1637>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 54.Ma A, Datta M, Margosian E, Chen J, Horak I. T cells, but not B cells, are required for bowel inflammation in interleukin 2-deficient mice. J Exp Med. 1995;182:1567–1572. doi: 10.1084/jem.182.5.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.