Abstract
Bacteria are highly social organisms that communicate via signaling molecules, move collectively over surfaces and make biofilm communities. Nonetheless, our main line of defense against pathogenic bacteria consists of antibiotics – drugs that target individual-level traits of bacterial cells and thus, regrettably, select for resistance against their own action. A possible solution lies in targeting the mechanisms by which bacteria interact with each other within biofilms. The emerging field of microbial social evolution combines molecular microbiology with evolutionary theory to dissect the molecular mechanisms and the evolutionary pressures underpinning bacterial sociality. This exciting new research can ultimately lead to new therapies against biofilm infections that exploit evolutionary cheating or the trade-off between biofilm formation and dispersal.
Introduction
The past few decades have witnessed a major change in the way microbiologists view bacteria. Rather than being solitary organisms, many bacteria live in biofilm communities, share nutrient scavenging molecules, communicate by cell-cell signaling, form fruiting bodies and migrate collectively by swarming motility (Fig. 1). Biofilms, in particular, may be the norm rather than the exception: an often cited number is that 60% of all human bacterial infections involve biofilms [1]. Even though our view of bacteria has undergone a dramatic change [2], the concepts of microbial sociality have only in rare instances been translated into therapies. Antibiotics, which are nonetheless our main line of defense against infectious bacteria, target individual-level traits such as cell wall assembly or DNA replication and therefore select for resistance against their own action [3] (Fig. 2a). In contrast, innovative therapeutics that disrupt population-level traits such as quorum sensing [4] or phage-based therapies that disperse biofilms [5, 6] can be potentially be used to reduce virulence while avoiding selection for resistance. Here we discuss recent results in the field of microbial social evolution and how this emerging field can open up new therapeutic avenues against biofilm-related infections. We focus on the opportunistic pathogen Pseudomonas aeruginosa, a well-known biofilm pathogen. The principles of social evolution are general and they should be applicable to all biofilm-forming microbes as well as other microbial social traits [7–9].
Fig. 1.
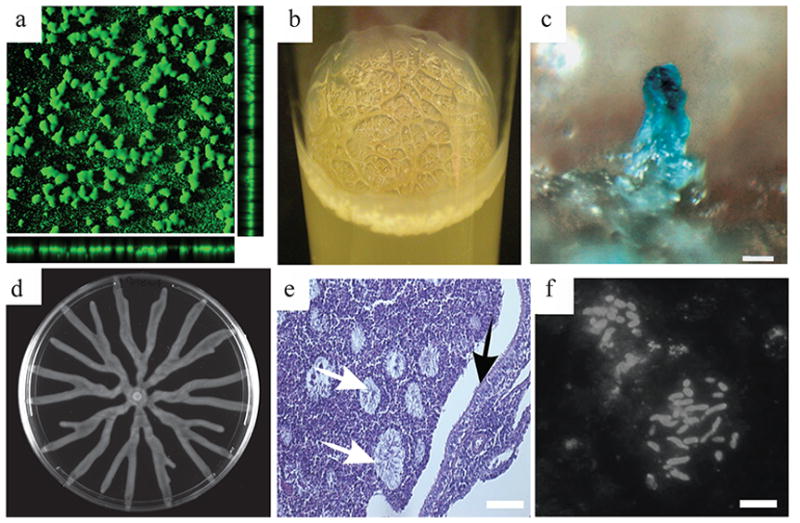
Microbial social traits. a) Vibrio cholera wild type rugose biofilm (10x magnification). b) V. cholera wild type rugose pellicle (a and b images contributed by Yildiz laboratory, UC Santa Cruz). c) A fruiting body in Bacillus subtilis (reprinted with permission from [37]). d) A Pseudomonas aeruginosa swarming colony (9 cm wide). e) P. aeruginosa macrocolonies in obstructed cystic fibrosis bronchus (reprinted with permission from [38]). f) cystic fibrosis lung P. aeruginosa macrocolonies stained with antibodies against P. aeruginosa (reprinted with permission from [38]) (scale: c-50 μm, e-100 μm, f-10 μm).
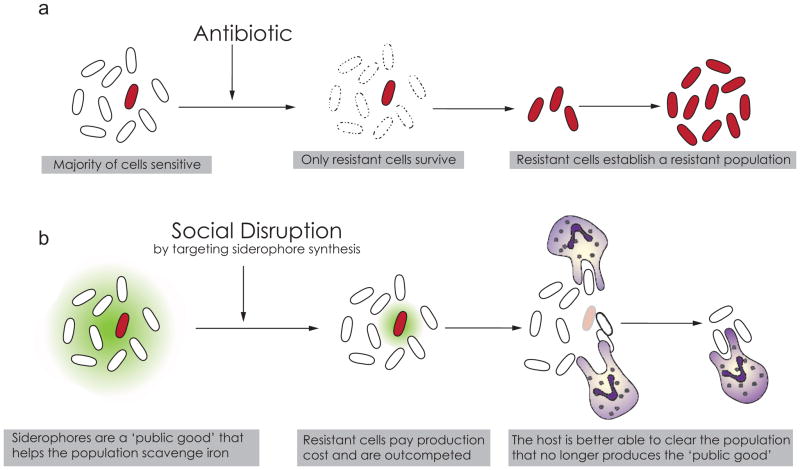
Fig. 2.
Antibiotics versus social disruption. Traditional antibiotic approaches (a) are prone to emergence of resistance. Strategies based on social evolutionary theory (b) can shift selection away from resistance allowing the immune system to clear the weakened infection.
Public good cooperation
P. aeruginosa is a gram-negative bacterium notorious for causing diverse infections in multiple anatomic niches, including wounds, chronic lung infections in cystic fibrosis, septicemia, bacterial keratitis and urinary tract infections. This opportunistic pathogen is also a highly social organism. P. aeruginosa lives in close interaction with microbial strains of the same and other species and is becoming a model system for microbial social evolution.
Much of the social activity of P. aeruginosa involves cooperation through the secretion of public goods. ‘Public good’ is an umbrella term used in the microbial social evolution literature to refer to a resource, such as a secondary metabolite, that is secreted by bacteria and becomes publically available to other cells within a population [8]. Many public goods are costly to produce, making them susceptible to exploitation by cheater strains that benefit from the public good without producing it themselves. For example, the siderophores of P. aeruginosa are iron-scavenging molecules that are costly to produce but are necessary for colonization of iron-limited environments such as eukaryotic tissues [10, 11]. The potential for cheater exploitation makes public good production an attractive target for therapies that select against resistance. The prediction based on social evolutionary theory is that in contrast to antibiotic-resistant mutants, mutants that resist a drug to prevent public good production would not be favored by natural selection. Because drug-sensitive bacteria would get a small initial growth advantage due to not producing costly public goods when the drug is present, drug-sensitive cheaters would eventually outcompete the drug-resistant producers. In the case of siderophores, a cheater population would be weakened without iron and more easily cleared by the host (Fig. 2b).
This conceptual example illustrates how a drug targeting a microbial social trait may succeed where antibiotics fail by enabling cheaters to outcompete more virulent strains. However, the reality can be more complex. For example, social evolutionary experiments have shown that less virulent quorum sensing signal-blind mutants, which do not produce a range of public goods, outcompete virulent P. aeruginosa strains in vitro [12–14]. The selective advantage may explain why signal-blind mutants are often found in long-term chronic cystic fibrosis infections [15] and suggests that quorum sensing inhibitors would be a suitable therapy targeting population-level traits [13]. Nevertheless, another recent study tested a quorum sensing inhibitor in patients and the results were quite the opposite: instead of attenuating the P. aeruginosa infection the quorum sensing inhibitor aggravated infection by decreasing the relative advantage of cheaters [16]. The exact selective pressures involved in social disruptions should be thoroughly investigated.
Social evolution in biofilms
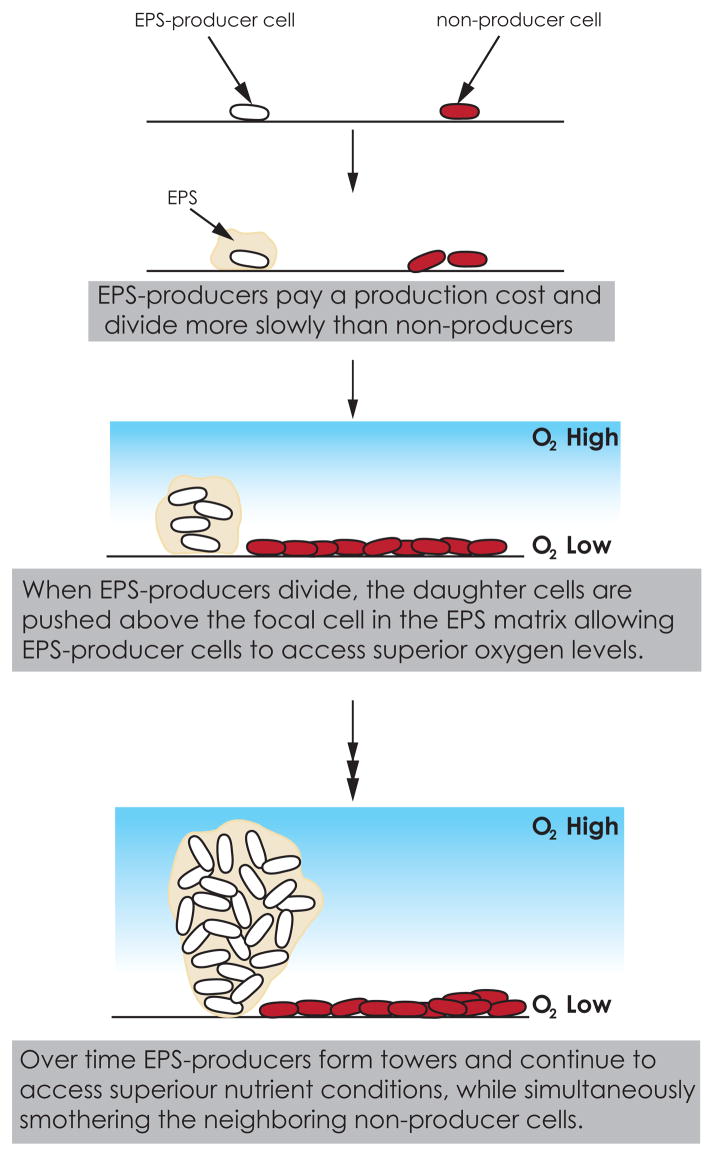
Biofilms are a continuing problem in the clinic because biofilm bacteria are often more robust against antibiotic and metabolic stresses than planktonic bacteria [17]. Biofilm formation is itself is a social trait that requires the production and secretion of shared substances. For example, extracellular polymeric substances (EPS) that encase bacteria in biofilms [18] are shared products, which, like siderophores, are potentially available to neighboring cells after they have been secreted. Unlike siderophores EPS production seems not to be exploited by cheaters. A mechanism for this protection was recently proposed based on computer simulations that modeled the dynamics of EPS production and nutrient diffusion in biofilms [19]. The model showed that even when EPS-producers pay a large cost of EPS production, EPS-producers are capable of outcompeting EPS non-producers in the same biofilm. EPS-producers are able to overcome their growth disadvantage because daughter cells of EPS-producers (unlike daughter cells of non-producers) are pushed up above the focal cell in the EPS matrix, allowing the EPS-producer cells to access superior nutrient conditions such as higher oxygen levels. As the cells grow, divide and secrete EPS, EPS-producers form high tower-like structures within the biofilm, smothering the neighboring non-producer cells, which remain close to the substrate (Fig 3).
Fig. 3.
EPS-producers in biofilm competition with non-producers. Even when EPS-producers have a significantly slower growth rate than non-producers because of costly EPS production, they are able to win in direct competitions with non-producers due to their ability to access superior nutrient conditions [19, 21].
Nutrient limitation is key to the EPS-producer advantage. In simulations where there is no nutrient gradient, the non-producers win due to their higher growth rate. However, conditions without nutrient gradients are unrealistic as nutrients are transported into the biofilm by diffusion, which can be a slow process compared with the fast rates of nutrient uptake by bacteria [20]. As a consequence, biofilm bacteria rarely grow in conditions where nutrients or oxygen levels are non-limiting. In realistic conditions, EPS-producers are able to overcome the cost of production and outcompete cheaters by their ability to take a more advantageous location within the biofilm [19].
While the computer simulations were originally conducted with P. aeruginosa in mind [19], the proposed role of EPS in the competition between cell lineages within a biofilm was confirmed by experiments in Vibrio cholerae [21]. In these experiments, the EPS-producers (ΔflaAΔhapR) were more competitive and increased in population fraction during the course of co-culture competitions with non-producers (ΔflaAΔhapRΔvpsL) even though EPS-producers pay a substantial production cost and have a slower maximum growth rate compared to non-producers. Similar to the in silico study, V. cholerae EPS-producers formed high towers in the biofilm while the non-producers remained in a flat layer [21]. Taken together, these studies demonstrate the importance of social interaction for the success of bacterial strains in a community and illustrate that rather than being the result of a purely cooperative process, the formation of complex biofilms can involve a balance between cooperative and competitive interactions [22].
Another recent study investigated the role of quorum sensing cheaters on P. aeruginosa biofilm stability. The experimentalists demonstrated that cheaters reduce the overall productivity of a biofilm and render biofilms more susceptible to antibiotics [23]. Importantly, the effects of cheaters were more severe in biofilms than in planktonic populations, suggesting that biofilm infections are particularly susceptible to social disruption strategies. With both competitive and cooperative traits playing important roles for biofilm stability, the next question is which social traits are appropriate targets for biofilm disruption therapies.
Inducing biofilm dispersal
Inducing dispersal of unwanted biofilms is an appealing strategy [17, 24]. However, the use of extrinsic detachment promoting agents can be limited by a slow diffusion of the agent into the biofilm [25]. An alternative is to manipulate the bacterial regulatory mechanisms for different modes of growth to make bacteria less successful at forming biofilms or even disperse established biofilms. In P. aeruginosa biofilm formation and motility behaviors are inversely regulated [26, 27]. The sad genes, chemotaxis genes and intracellular c-di-GMP levels have been shown to play roles in this inverse relationship between biofilms and motility [27]. As more molecular mechanisms are uncovered (Table 1) such systems could be manipulated to direct cells away from forming a stable biofilm.
Table 1.
Examples of intrinsic mechanisms for dispersal of bacterial biofilms.
| Bacillus subtilis - D-Amino Acids | D-Amino acids naturally produced by B. subtillis induce biofilm dispersal by inducing release of amyloid fibers from cells within the biofilm [39]. |
| Pseudomonas aeruginosa - Rhamnolipids | Rhamnolipid biosurfactants naturally produced by P. aeruginosa induce detachment of P. aeruginosa cells from the biofilm and disperse biofilms of other species [28, 29]. |
| Staphylococcus aureus - extracellular proteases | Extracellular proteases regulated by the S. aureus agr quorum sensing system mediate detachment of mature biofilms. Dispersed cells have increased sensitivity to antibiotic treatment [40]. |
| Bacteriophage engineered delivery of dispersal enzymes | Biofilms of E. coli can be dispersed by expression of the active biofilm-degrading enzyme, dispersin B, introduced by an engineered bacteriophage [6]. |
P. aeruginosa produces rhamnolipid biosurfactants that promote detachment of its own biofilms [28]. The ability to secrete the right amount of rhamnolipids is crucial for proper biofilm architecture and stability. Therefore, strains that overproduce rhamnolipids are deficient in biofilm formation, forming thin, flat biofilms compared to the wild type’s voluminous towers. Artificially-induced production of rhamnolipids, on the other hand, leads to detachment of cells within the biofilm [28]. Rhamnolipid-induced dispersal is not limited to P. aeruginosa making these biosurfactants appealing candidates for dispersal of multi-species biofilms in both medical and industrial settings [29]. Rhamnolipid secretion has the potential to induce biofilm dispersal in vivo and make bacteria more vulnerable to clearing by the host or by traditional antibiotics
Here, again, proper caution must be taken. The rhamnolipids of P. aeruginosa have detrimental effects on eukaryotic cells, lysing red blood cells and causing necrotic cell death in polymorphonuclear leukocytes (PMNs) and macrophages [30]. This function may have an important role in protecting biofilms from the host immune cells such as PMNs, which are critical to clearing P. aeruginosa in vivo [31]. Biofilms of rhamnolipid-deficient strains cause significantly less necrosis in PMNs [30]. Additionally, experiments using implants colonized by P. aeruginosa in mice showed that robust implant colonization requires biosurfactant production [31, 32]. Rhamnolipid-deficient strains can be phagocytosed and cleared by predating PMNs, thereby failing to form biofilms comparable to those of wild type rhamnolipid-producing strains. The role of P. aeruginosa rhamnolipids is therefore multifactorial and their use as a therapeutic target requires further investigation.
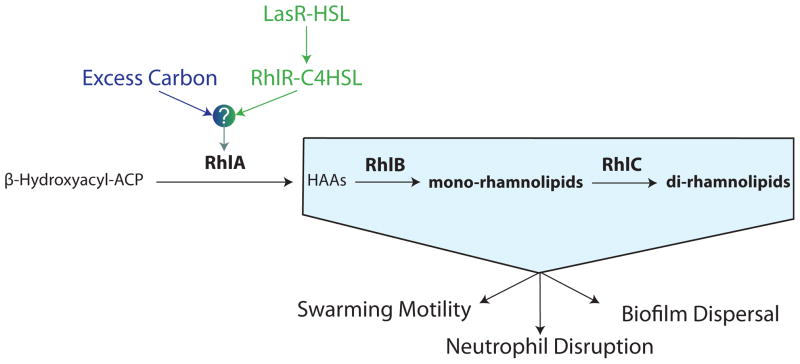
Rhamnolipid-targeting drugs may take advantage of a complex transcriptional regulation of rhamnolipid synthesis that integrates metabolic and quorum sensing signals. Rhamnolipids are produced by the action of three sequentially functioning enzymes: RhlA, RhlB and RhlC [33]. RhlA is required for any rhamnolipid synthesis in the cell and catalyzes the initial conversion of β-hydroxyacyl-ACP to 3-(3-hydroxyalkanoyloxy) alconoic acids (HAAs) [34]. After this conversion, RhlB and RhlC are sequentially required to add rhamnose units, producing mono- and di-rhamnolipids, respectively. Quorum sensing signals are necessary but not sufficient for the expression of the rhlAB operon, the rate-controlling step for rhamnolipid production. New studies suggest that bacteria require excess carbon in addition to quorum sensing signals to trigger the synthesis of rhamnolipids [35]. Thus, the bacterial cells carry out an integration of metabolic and quorum sensing signals in order to initiate rhamnolipid production (Fig 4). The mechanism of metabolic and quorum sensing integration is called metabolic prudence, since it prevents wild type strains from being outcompeted by cheaters even though rhamnolipid production requires significant metabolic resources [35, 36]. Through metabolic prudence, rhamnolipid synthesis is delayed until other nutrients such as nitrogen have been depleted and excess carbon used in rhamnolipid synthesis is free to be diverted away from the production of biomass.
Fig. 4.
The pathway for synthesis of rhamnolipid biosurfactants in P. aeruginosa. Expression of the enzyme RhlA is the rate-limiting step for rhamnolipid synthesis [34] and implements a molecular decision-making process by which bacteria start producing rhamnolipids. The process requires the integration of quorum sensing (Las and Rhl systems) and metabolic cues [35]. The molecular details of the integration, represented here by a question mark, remain unknown.
In addition to biofilm dispersal, targeting rhamnolipid production with social disruption strategies could have substantial impacts on the ability of bacteria to compete successfully in a biofilm or infection. Engineered rhamnolipid-producer strains, which have rhamnolipid synthesis under an inducible promoter and therefore lack metabolic prudence, are highly susceptible to cheaters and are quickly outcompeted in co-culture swarming colonies [35]. Manipulating rhamnolipid secretion will require a more complete understanding of the system, as key molecular details of the integration between quorum sensing and metabolic sensing are still unknown. Further study into the molecular basis of metabolic prudence may reveal new avenues to exploit the intrinsic mechanism of biofilm dispersal of P. aeruginosa.
Conclusion
There is a pressing need for alternatives to antibiotics, our main defense against bacterial pathogens that is increasingly threatened by the emergence of resistance. The solution may come from targeting population-level traits such as biofilm formation and quorum sensing. Microbial social evolution can help identify novel therapeutic targets and assist in the rational design of therapies that avoid selection for resistance. The coming years are sure to bring more insights from the fascinating interface between molecular microbiology and social evolution theory.
Highlights.
Instead of isolated cells bacteria are now acknowledged to lead highly social lives
Biofilm formation and dispersal are favorable targets for new therapeutic avenues
The selective pressures involved must be understood thoroughly before translation
Acknowledgments
We thank Karina Xavier and Carlos Carmona-Fontaine for comments on the manuscript. We acknowledge support from the National Institutes of Health (grant DP2OD008440 to J. B. X.) and a James S. McDonnell postdoctoral fellowship to S.H..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Costerton JW. Bacterial Biofilms: A Common Cause of Persistent Infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg EP. Microbe Magazine. ASC; 2010. Sociomicrobiology: A personal perspective on an emerging research area. [Google Scholar]
- 3.Bérdy J. Thoughts and facts about antibiotics- Where we are now and where we are heading.pdf. The Journal of Antibiotics. 2012;65:441. doi: 10.1038/ja.2012.54. [DOI] [PubMed] [Google Scholar]
- 4.Jimenez PN, et al. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76(1):46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nature Reviews Genetics. 2010;11(5):367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007;104(27):11197–202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster KR. The sociobiology of molecular systems. Nat Rev Genet. 2011;12(3):193–203. doi: 10.1038/nrg2903. [DOI] [PubMed] [Google Scholar]
- 8.Xavier JB. Social interaction in synthetic and natural microbial communities. Mol Syst Biol. 2011:7. doi: 10.1038/msb.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33(1):206–24. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 10.Jiricny N, et al. Fitness correlates with the extent of cheating in a bacterium. Journal of Evolutionary Biology. 2010;23(4):738–747. doi: 10.1111/j.1420-9101.2010.01939.x. [DOI] [PubMed] [Google Scholar]
- 11.Griffin Ashleigh S, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430(7003):1024–7. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 12.Diggle SP, et al. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450(7168):411–4. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 13.Mellbye B, Schuster M. The Sociomicrobiology of Antivirulence Drug Resistance: a Proof of Concept. mBio. 2011;2(5) doi: 10.1128/mBio.00131-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proceedings of the National Academy of Sciences. 2007;104(40):15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bjarnsholt T, et al. Quorum Sensing and Virulence of Pseudomonas aeruginosa during Lung Infection of Cystic Fibrosis Patients. PLoS ONE. 2010;5(4):e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Kohler T, et al. Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog. 2010;6(5):e1000883. doi: 10.1371/journal.ppat.1000883. This study tested the quorum sensing inhibiting drug, azxithromycin, to reduce pathogenesis of infecting P. aeruginosa. Instead, the drug decreased the advantage quorum sensing cheaters giving an advantage to the virulent wild type bacteria. The paper highlights the importance of thoroughly understanding the selective pressures involved. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoiby N, et al. Antibiotic resistance of bacterial biofilms. International Journal of Antimicrobial Agents. 2010;35(4):322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Flemming HC, Wingender J. The biofilm matrix. Nature Reviews Microbiology. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 19.Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci U S A. 2007;104(3):876–81. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadell CD, Foster KR, Xavier JB. Emergence of Spatial Structure in Cell Groups and the Evolution of Cooperation. PLoS Comput Biol. 2010;6(3):e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Nadell CD, Bassler BL. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci U S A. 2011;108(34):14181–5. doi: 10.1073/pnas.1111147108. This paper demonstrates that, similar to in silico models, V. Cholera EPS-producers outcompete non-producers in biofilms. The paper goes on to show that the advantage has a trade-off: EPS-producers depart from the biofilm and seed new areas at a lower rate than they increase in the biofilm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dandekar AA, Chugani S, Greenberg EP. Bacterial Quorum Sensing and Metabolic Incentives to Cooperate. Science. 2012;338(6104):264–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Popat R, et al. Quorum-sensing and cheating in bacterial biofilms. Proc Biol Sci. 2012;279(1748):4765–71. doi: 10.1098/rspb.2012.1976. This paper shows that P. aeruginosa quorum sensing cheaters reduce growth of a population and that this reduction is more pronounced in biofilms. Biofilms are also more susceptible to antibiotic treatment when cheaters are present. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brindle ER, Miller DA, Stewart PS. Hydrodynamic deformation and removal of Staphylococcus epidermidis biofilms treated with urea, chlorhexidine, iron chloride, or DispersinB. Biotechnol Bioeng. 2011;108(12):2968–77. doi: 10.1002/bit.23245. [DOI] [PubMed] [Google Scholar]
- 25.Xavier JB, et al. Biofilm-control strategies based on enzymic disruption of the extracellular polymeric substance matrix--a modelling study. Microbiology. 2005;151(Pt 12):3817–32. doi: 10.1099/mic.0.28165-0. [DOI] [PubMed] [Google Scholar]
- 26.Caiazza NC, et al. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189(9):3603–12. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Toole GA. At surfaces, these bacteria either form biofilms or swarm, a regulated behavior with important consequences for pathogenesis. ASM Microbe Magazine. 2008;3(2):65–71. [Google Scholar]
- 28.Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol. 2005;57(5):1210–23. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- 29.Zeraik AE, Nitschke M. Biosurfactants as Agents to Reduce Adhesion of Pathogenic Bacteria to Polystyrene Surfaces: Effect of Temperature and Hydrophobicity. Current Microbiology. 2010;61(6):554–559. doi: 10.1007/s00284-010-9652-z. [DOI] [PubMed] [Google Scholar]
- 30.Jensen PO, et al. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology. 2007;153(Pt 5):1329–38. doi: 10.1099/mic.0.2006/003863-0. [DOI] [PubMed] [Google Scholar]
- 31.Van Gennip M, et al. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS. 2009;117(7):537–46. doi: 10.1111/j.1600-0463.2009.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Gennip M, et al. Interactions between Polymorphonuclear Leukocytes and Pseudomonas aeruginosa Biofilms on Silicone Implants In Vivo. Infection and Immunity. 2012;80(8):2601–2607. doi: 10.1128/IAI.06215-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdel-Mawgoud AM, Lepine F, Deziel E. Rhamnolipids: diversity of structures, microbial origins and roles. Applied Microbiology and Biotechnology. 2010;86(5):1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu K, Rock CO. RhlA converts beta-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the beta-hydroxydecanoyl-beta-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J Bacteriol. 2008;190(9):3147–54. doi: 10.1128/JB.00080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35*.Xavier JB, Kim W, Foster KR. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol Microbiol. 2011;79(1):166–79. doi: 10.1111/j.1365-2958.2010.07436.x. This paper shows that P. aeruginosa uses a mechanism of metabolic prudence to delay production of rhamnolipid biosurfactants, a public good required for swarming migration, until nitrogen is depleted and growth has slowed. Metabolic prudence reduces the cost of producing the public good, making the wild type more competitive against cheaters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Ditmarsch D, Xavier J. High-resolution time series of Pseudomonas aeruginosa gene expression and rhamnolipid secretion through growth curve synchronization. BMC Microbiology. 2011;11(1):140. doi: 10.1186/1471-2180-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Branda SS, et al. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98(20):11621–6. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Worlitzsch D, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. Journal of Clinical Investigation. 2002;109(3):317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolodkin-Gal I, et al. D-amino acids trigger biofilm disassembly. Science. 2010;328(5978):627–9. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blaise R, Boles ARH. agr-Mediated Dispersal of Staphylococcus aureus Biofilms. PloS Pathogens. 2008;4(4):e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]