Abstract
The OmpR/PhoB family of response regulators (RRs) is the largest class of two-component system (TCS) signal transduction proteins. Extensive biochemical and structural characterization of these transcription factors has provided insights into their activation and DNA-binding mechanisms. For the most part, OmpR/PhoB family proteins are thought to become activated through phosphorylation from their cognate histidine kinase (HK) partners, which in turn facilitates an allosteric change in the RR enabling homodimerization and subsequently enhanced DNA binding. Incongruently, it has been suggested that OmpR, the eponymous member of this RR family, becomes activated via different mechanisms, whereby DNA binding plays a central role in facilitating dimerization and phosphorylation. Characterization of the rate and extent of the phosphorylation of OmpR and OmpR DNA-binding mutants following activation of the EnvZ/OmpR TCS shows that DNA binding is not essential for phosphorylation of OmpR in vivo. In addition, detailed analyses of the energetics of DNA binding and dimerization of OmpR in both its unphosphorylated and phosphorylated state indicate that phosphorylation enhances OmpR dimerization and that this dimerization enhancement is the energetic driving force for phosphorylation-mediated regulation of OmpR-DNA binding. These findings suggest that OmpR phosphorylation mediated activation follows the same paradigm as the other members of the OmpR/PhoB family of RRs in contrast to previously proposed models of OmpR activation.
Keywords: Bacterial two-component systems, Bacterial response regulators, Thermodynamics of DNA binding, Phosphorylation-mediated dimerization, Allosteric regulation
Introduction
The Escherichia coli EnvZ/OmpR two-component system (TCS) is an extensively studied signal transduction system that has been implicated in the regulation of over 100 genes in response to changes in the osmotic milieu of the cell. In this TCS, the sensory histidine kinase (HK), EnvZ, autophosphorylates a conserved histidine residue and the phosphoryl group is subsequently transferred to a specific aspartic acid in the N-terminal domain of the response regulator (RR), OmpR.3–5 Regulation of the transcriptional activity of OmpR is mediated by strict control of the cellular level of phosphorylated OmpR (OmpR~P) through the kinase and potentially the OmpR-phosphatase activities of EnvZ.
For other proteins of the OmpR/PhoB winged-helix transcription factor RR family of which OmpR is an eponymous member, phosphorylation has been shown to enhance homodimerization affinity and thus increase DNA-binding affinity and specificity.8–10 This mechanism is summarized pictorially in Scheme A of Figure 1. In contrast, evidence for the dimerization of OmpR or OmpR~P in solution has been lacking11–13 except for qualitative assessment in a systematic study of Escherichia coli RR dimerization.14 The absence of data demonstrating dimerization, coupled with observations that interactions of OmpR with DNA at equilibrium require an obligatory OmpR dimer,12 and that interactions of OmpR with DNA enhance the phosphorylation of OmpR,15 and conversely, that OmpR phosphorylation enhances interactions with DNA, have been used to infer stepwise mechanisms for OmpR activation. In one proposed mechanism, the interaction of one molecule of OmpR with its DNA recognition sequence is followed by a conformational change in OmpR that then enhances OmpR phosphorylation activity and thus OmpR dimerization affinity, culminating in a 2OmpR~P•DNA complex13 (Fig. 1, Scheme B). An alternative mechanism culminating in the same end product, supposes that phosphorylation of OmpR is followed by a conformational change in OmpR that enables it to form an OmpR~P•DNA complex that then provides a high-affinity site for binding of the second OmpR~P protomer18 (Fig. 1, Scheme C).
Fig. 1.
Potential mechanisms for the phosphorylation-mediated activation of OmpR-regulated transcription. Scheme A) Standard mechanism of RR transcriptional activation.8–10 Schemes B and C) Mechanisms of OmpR transcriptional activation proposed previously.13,18 Blue ovals and starbursts represent OmpR receiver domains in inactive and active conformational states, respectively. Red arrows represent OmpR DNA-binding domains and DNA is depicted in green. Pink circles and yellow circles represent phosphoryl group donors and phosphoryl groups, respectively.
Each of the above models of OmpR-mediated transcriptional activation and the standard model of OmpR/PhoB family RR-mediated transcriptional regulation yield the same final active DNA-bound complex, 2RR~P•DNA. What is distinct about each of the individual models is that the rate-limiting steps controlling the formation of the transcriptionally active complex would be inherently different. Furthermore, a deviation of the regulatory mechanism of OmpR activation from the standard mechanism of OmpR/PhoB family RR activation would imply that the structural similarities of OmpR/PhoB family members could not be associated with functional similarities in DNA recognition and transcriptional regulation for this class of proteins.
The goal of this study was to determine the model that best describes the mechanism of transcriptional regulation by OmpR. To this end, we monitored in vitro and in vivo phosphorylation kinetics of OmpR and DNA binding-deficient OmpR mutant proteins to elucidate the rate-limiting step in the transcription activation pathway. Although the ability of OmpR to bind DNA can influence its phosphorylation in vitro, it was found to have no significant effect on OmpR phosphorylation in vivo. This finding suggests that DNA binding is not a rate-limiting step in the formation of the OmpR transcriptional activation complex. Analytical centrifugation and calorimetric analyses have defined the energetics of OmpR and OmpR~P dimerization and DNA binding in vitro. The phosphorylation-induced change in the free energy of OmpR dimerization yields a greater negative value than the phosphorylation-induced change in the free energy of DNA binding, suggesting that dimerization is the energetic driving force for the formation of the interaction of OmpR with DNA. Together these findings were used to generate a model for the thermodynamic linkage of phosphotransfer to OmpR dimerization and DNA-binding equilibria. This model is consistent with published data on DNA binding and transcriptional regulation, not only for OmpR, but also for other OmpR/PhoB family RRs and therefore likely represents a common mechanism for phosphorylation-mediated control of RR-regulated transcription.
Results
Procaine treatment produces a quantifiable increase in OmpR phosphorylation in vivo
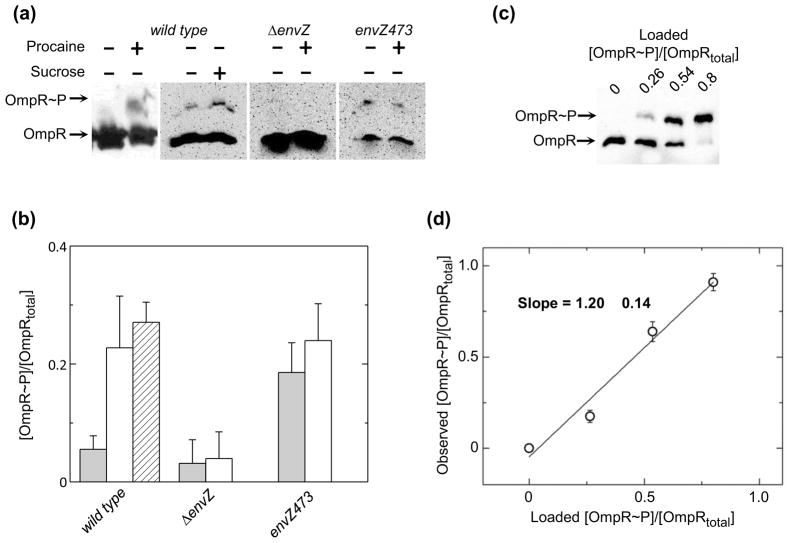
Despite the difficulty in monitoring RR phosphorylation due to the lability of the acyl-phosphate moiety of the phospho-Asp residue, previous studies have demonstrated that the steady-state level of OmpR phosphorylation (OmpR~P) is increased following treatments that perturb the cell membrane.19 Two drawbacks to the previous analyses are (1) only relative values for the fraction of OmpR~P can be determined, and (2) the rapid kinetics of the change in OmpR~P cannot be directly followed. The recently described method of phosphoprotein-affinity gel electrophoresis using Phos-tag™ acrylamide was used to address the shortcomings of the previous studies and to glean new information regarding the regulation of the cellular response to membrane stress. To this end, the change in the fraction of OmpR~P relative to total OmpR was compared in strain BW25114 expressing the wild-type ompR and envZ genes (henceforth referred to as wild type)grown in low osmolarity medium A in the absence and presence of procaine, an analgesic compound that alters membrane fluidity22 and sucrose, a commonly used osmolyte (Figs. 2a and b). This strain demonstrates little OmpR phosphorylation (5 ± 2 %) in the absence of procaine or sucrose stimuli. As expected, following treatment with 20 mM procaine or 20 % sucrose, the fraction of OmpR that is phosphorylated is significantly enhanced (23 ± 9 % and 27 ± 3 %, respectively). The approximately five-fold increase in the fraction of phosphorylated OmpR following cell membrane perturbation is entirely consistent with previously reported measurements of in vivo OmpR phosphorylation.19 Also note, the effect of procaine treatment is similar to the effect of sucrose mediated osmotic stress on the phosphorylation of OmpR in the cell. The two conditions also have similar effects on porin protein levels in these strains (Fig. S1). For subsequent studies procaine is used to minimize the impact of carbon source mediated repression of inducible plasmid encoded genes. In a ΔenvZ strain, OmpR remains in a constant state of low phosphorylation (3 ± 4 %) whether procaine is present or not, indicating that the effects of stimuli on OmpR phosphorylation are directly related to the sensory histidine kinase EnvZ. The low level of OmpR phosphorylation observed in the absence of a cognate histidine kinase is likely due to nonspecific phosphorylation from small molecule phosphodonors or from noncognate histidine kinases.25
Fig. 2.
Characterization of OmpR phosphorylation in E. coli cells. (a) Representative western blots of phosphoprotein affinity gel electrophoretic separation of wild type, ΔenvZ and envZ473 mutant cell lysates in the presence and absence of procaine or sucrose, as indicated. Representative blots are taken from separate phosphoaffinity SDS-PAGE gels. Therefore, differences in signal intensity do not reflect differences in cellular OmpR concentration. (b) Quantitation of the extent of OmpR phosphorylation in wild type, ΔenvZ and envZ473 cell lysates, untreated (clear bars), following treatment with procaine (shaded bars), or following addition of sucrose to the media (hashed bar). Values reflect the mean values of 3 separate experiments with error bars reflecting the standard deviation from the mean. (c) Representative western blot of known fractions of [OmpR~P]/[OmpRtotal] mixed with ΔompR cells and subsequently lysed. (d) Quantitative efficiency of western blots of phosphoprotein affinity gel electrophoretic separations of [OmpR~P]/[OmpRtotal]. Points represent the average values from 2 separate experiments with error bars reflecting the standard deviation from the mean. Solid line indicates the best-fit linear regression analysis of the plotted data.
One intriguing observation from these data is that phosphorylation of OmpR appears to be significantly less than complete under conditions that are thought to maximally activate the OmpR/EnvZ TCS. A strain that contains a mutant of envZ, envZ473, which is deficient in OmpR phosphatase activity, also exhibits less than complete phosphorylation of OmpR whether or not procaine is present in the media, 18 ± 5 % and 24 ± 6 % phosphorylation of OmpR, respectively. Not only does OmpR phosphorylation in envZ473 appear less than stoichiometric, but in envZ473 the fraction of OmpR that is phosphorylated is essentially equivalent to the maximum fraction of phosphorylated OmpR in wild type, a finding that has been previously reported for another OmpR-phosphatase deficient strain envZ11.19 This finding suggests that either the assay is causing hydrolysis of OmpR~P, resulting in a smaller amount of OmpR~P being observed than is present in the cell, or that EnvZ-mediated phosphatase activity is not the only mechanism regulating OmpR phosphorylation in the cell. To further test the quantitative accuracy of the phosphoprotein affinity gel electrophoresis assay, purified protein with known fractions of [OmpR~P]/[OmpRtotal] were added to cultures of a ΔompR strain which were subsequently lysed and characterized by the phosphoprotein affinity gel electrophoresis assay (Fig. 2c). The densitometrically quantified observed fraction of [OmpR~P]/[OmpRtotal] plotted versus the fraction of [OmpR~P]/[OmpRtotal] loaded onto the gel is shown in Figure 2d. The data were well fit with a line having a slope of approximately 1, indicating that phosphoprotein affinity gel electrophoresis analysis does not cause significant hydrolysis of OmpR~P. Thus, even under a state of constant stimulation of the EnvZ/OmpR TCS, the majority of the OmpR in the cell is not phosphorylated even when the phosphatase activity of the HK EnvZ is diminished.
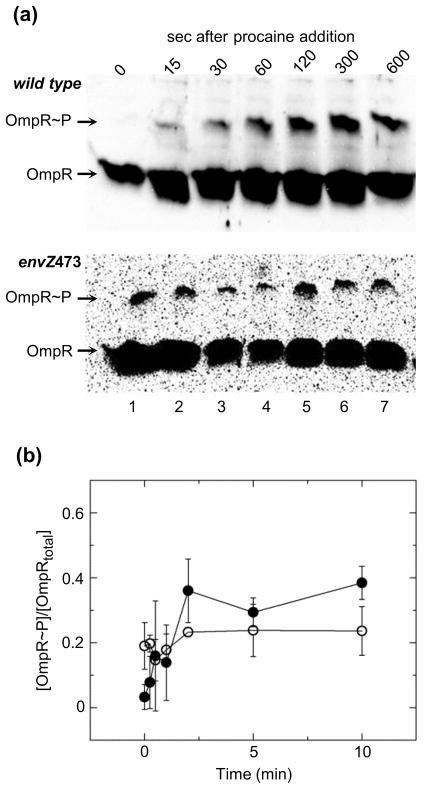
In some TCSs RR phosphorylation is tempered to a submaximal level following an initial surge in phosphorylation following the initial stimulation.26 In systems that follow this type of multistate activation process the observed fraction of activated protein would appear to reach a peak concentration followed by a decrease to a steady state level. Phosphoprotein affinity gel electrophoresis allows assessment of phosphorylation over relatively short time scales and was used to monitor the increase in [OmpR~P]/[OmpRtotal] following treatment with procaine for wild type and envZ473 (Fig. 3). Lane 1 of both blots contains lysate of untreated cells while lanes 2–7 contain cell lysates following treatment with procaine for the amount of time indicated. The fraction [OmpR~P]/[OmpRtotal] in wild-type lysates increases following procaine addition, reaching a maximum after ~2 min post-treatment and remaining at that maximum level thereafter. [OmpR~P]/[OmpRtotal] in envZ473 lysates is maintained at a constant level nearly equivalent to the maximum level of phosphorylation observed in wild type. Also note that the maximum fraction [OmpR~P]/[OmpRtotal] is equivalent to that observed in the steady state experiment, demonstrating that activation of OmpR follows a simple model of induction.
Fig. 3.
Time-dependent change in OmpR phosphorylation in E. coli cells following treatment with procaine. (a) Representative western blots of phosphoprotein affinity gel electrophoretic separation of wild type and envZ473 following treatment with procaine. Lane 1 in both blots contains untreated cells lysates and lanes 2–7 contain cell lysates at indicated times following treatment with procaine. (b) Quantitation of the extent of OmpR phosphorylation in wild type and envZ473 cell lysates following treatment with procaine. Closed and open circles reflect the mean values of 3 separate experiments, for wild type and envZ473 cell lysates, respectively, with error bars reflecting the standard deviation from the mean.
OmpR-DNA binding is not required for phosphorylation in vivo
DNA binding has been shown to enhance phosphorylation of OmpR in vitro15 and it has been suggested that DNA binding plays a role in phosphorylation of OmpR in vivo.13 Recent NMR13 and crystal structures (B. Benoff & A. Stock unpublished data) have identified important contacts between OmpR and the specific DNA sequences to which it binds. Mutations to residues of OmpR that make specific contacts to DNA should disrupt OmpR-mediated transcriptional regulation. If DNA binding substantially enhances OmpR phosphorylation, then these mutations should diminish EnvZ-mediated OmpR phosphorylation. Arg-207 to Ala (R207A) and Trp-226 to Ala (W226A) mutations disrupt DNA binding by different mechanisms. R207A removes a critical hydrogen bond between the recognition helix and guanine base in the DNA major groove. W226A disrupts extensive van der Waals interactions between the Wing 1 loop and the minor groove of DNA. Previous studies using reporter genes have demonstrated that these mutations eliminate OmpR-mediated transcriptional regulation of the porin genes ompF and ompC.13 In addition, ITC experiments reveal that each point mutation reduces DNA-binding affinity of OmpR by at least 100 fold (Fig. S2 and Table S1).
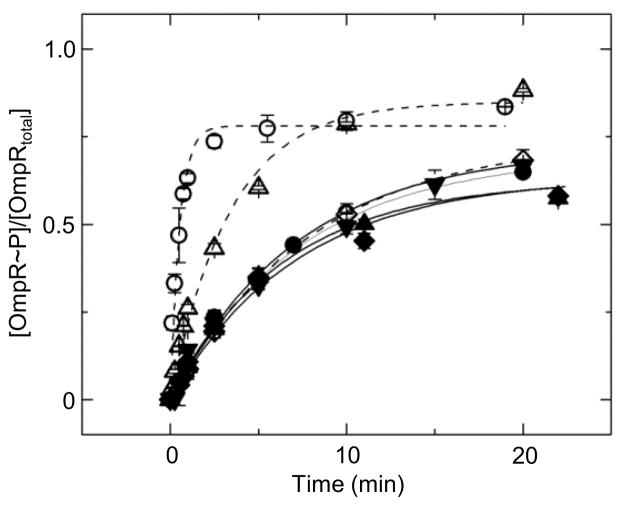
Autophosphorylation activity of wild-type OmpR, R207A, and W226A with the small molecule phosphodonor phosphoramidate was monitored (Fig. 4 and Table 1). The autophosphorylation rates were similar for all proteins, indicating that the substitutions did not affect autophosphorylation capability. The enhancement of autophosphorylation by DNA binding was also monitored (Table 1). Note that the presence of a saturating concentration of C1 DNA increases the rate of phosphorylation of wild-type OmpR 14 fold. A finding that is similar to what has been previously described using wild-type OmpR in the presence of oligonucleotides containing specific OmpR biding sites.15 One potential cause for this DNA-induced enhancement in phosphorylation is that DNA binding removes an interaction between the N- and C-terminal domains that stabilizes an unphosphorylatable conformation of the receiver domain. While such inhibitory interactions have been observed in some other RRs of the OmpR/PhoB family, the isolated receiver domain of OmpR (OmpRN)does not have increased autophosphorylation relative to that of the full-length protein.27 This suggests that there are weak contacts, if any, between the two domains and therefore that DNA binding enhances OmpR autophosphorylation through another mechanism. Furthermore, DNA binding appears to be required for the observed enhancement of OmpR autophosphorylation because the autophosphorylation rates of DNA-binding domain mutants of OmpR, in the presence of C1 at a concentration that saturates the wild-type protein, are affected to a much lesser extent. The R207A mutant, which binds to C1 with a higher affinity than the W226A mutant, has a slight increase in autophosphorylation rate in the presence of C1 DNA, while W226A displays no detectable increase. The increase in autophosphorylation for R207A and W226A is enhanced when the reactions are performed at lower salt concentrations, i.e. conditions that would increase ionic interactions between the protein and DNA (data not shown). These findings strongly suggest that the extent of increase in autophosphorylation of OmpR in the presence of specific DNA sequences is dependent on the extent of occupancy of the DNA-binding domain of OmpR.
Fig. 4.
Autophosphorylation activity of wild-type and mutant OmpR proteins. Rates of autophosphorylation of OmpR, R207A, and W226A in the absence and presence of the C1-DNA duplex (closed and open circles, triangles, and diamonds, respectively), as well as OmpRN (inverted triangles). Data were each fit with a first-order exponential decay (solid lines for proteins alone, dashed lines for mixtures of protein and C1 DNA).
Table 1.
Rates of autophosphorylation of wild-type OmpR, OmpRN and R207A and W226A mutants in the presence and absence of C1 DNA.
| Protein | DNA | kobsa (min−1) | F∞a |
|---|---|---|---|
| OmpR | NA | 0.069 ± .004 | 0.72 ± .01 |
| OmpR | 20 μM | 0.961 ± .078 | 0.78 ± .02 |
| OmpRN | NA | 0.067 ± .006 | 0.70 ± .02 |
| R207A | NA | 0.076 ± .005 | 0.63 ± .01 |
| R207A | 20 μM | 0.147 ± .014 | 0.85 ± .03 |
| W226A | NA | 0.070 ± .006 | 0.64 ± .02 |
| W226A | 20 μM | 0.062 ± .006 | 0.75 ± .01 |
Observed rates of autophosphorylation are calculated using the single exponential decay, F = F∞(1 − exp(kobs×t)), where F is the fraction of protein phosphorylated, F∞ is the maximum fraction of protein phosphorylated, kobs is the observed rate of phosphorylation, and t represents time following addition of the phosphoramidate.
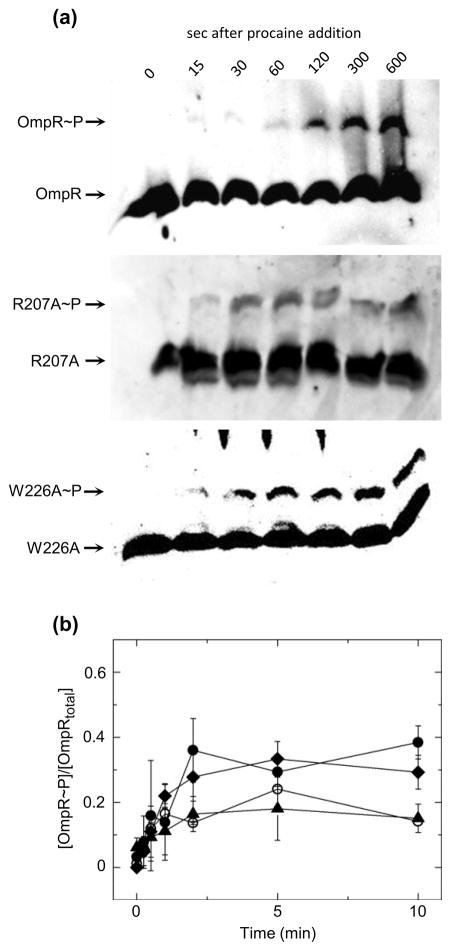
To test whether phosphorylation of OmpR in the cell is impaired in DNA-binding mutants, wild-type OmpR, R207A, and W226A were expressed at near wild-type levels in a ΔompR strain (Fig. S1) and phosphorylation of each was monitored(Fig. 5). Note that for each OmpR protein, regardless of its ability to bind DNA, phosphorylation is enhanced upon exposure of the culture to procaine. A comparison of the data in Figure 3a and Figure 5 shows that wild-type OmpR, expressed chromosomally or from a plasmid, undergoes similar phosphorylation kinetics suggesting that EnvZ acts to phosphorylate OmpR whether the two proteins are expressed in the same operon or separately. Furthermore, the extent of phosphorylation and rate of change in phosphorylation are similar for both the wild-type and mutant proteins. Note that enhanced phosphorylation of OmpR, or its associated DNA-binding domain mutants, is initiated within seconds following addition of procaine and the phosphorylation extent reaches a maximum within approximately 2 minutes. These data suggest that DNA binding is neither the rate-limiting step in the OmpR phosphorylation reaction nor is it a substantial inhibitor of OmpR dephosphorylation in vivo. The in vivo measurements of OmpR phosphorylation indicate that this RR does not likely regulate transcription via a mechanism where phosphorylation is predicated on DNA binding13 (Fig. 1, Scheme B). What remains to be determined, however, is whether dimerization in the absence of DNA binding is a necessary component of OmpR-mediated regulation of transcription. Such data would differentiate between the model that requires DNA binding by OmpR to facilitate dimerization (Fig. 1, Scheme C) and the standard model for RR transcription activation where phosphorylation mediated enhancement in dimerization facilitates DNA binding (Fig. 1, Scheme A).
Fig. 5.
Time-dependent change in plasmid-expressed OmpR, R207A and W226A phosphorylation in ΔompR cells following treatment with procaine. (a) Representative western blots of phosphoprotein affinity gel electrophoretic separation of ΔompR cells expressing either OmpR, R207A or W226A following treatment with procaine. Lane 1 in all blots contains untreated cells lysates and lanes 2–7 contain cell lysates at indicated times following treatment with procaine. (b)Quantitation of the extent of phosphorylation of OmpR (circles), R207A(triangles) or W226A (diamonds) following treatment with procaine. Points reflect the mean values of 3 separate experiments, with error bars reflecting the standard deviation from the mean.
OmpR does not bind to its DNA target sequence as a monomer
To distinguish between the two remaining models for the OmpR-DNA interaction the stoichiometry of the OmpR-DNA interaction must be accurately defined under conditions where potential intermediate states of the binding reaction would be populated. The ITC-derived data (Fig. S2 and Table S1) indicate that binding of OmpR to DNA occurs with a 2:1 stoichiometry under equilibrium conditions, yet yields little information regarding the mechanism of this binding interaction. Previous studies have shown that OmpRC can interact with a DNA half-site as a monomer,13 but monomeric binding of full-length OmpR has not been observed.15,16,18 To ascertain whether a monomeric state in the binding reaction is populated under conditions that should favor such an intermediate, sedimentation velocity experiments were performed using a fluorescein-labeled oligonucleotide containing the OmpR F1 binding site (henceforth referred to as F1-FAM). The label on this oligonucleotide allows for absorbance analysis of sedimentation of the oligonucleotide without interference from the absorbance of protein. The data were analyzed using the Sedfit program to determine the sedimentation coefficients of the various species in the solution (Fig. 6a). Note that each plot contains either one peak, as in the case of F1-FAM alone, or two peaks, as in the mixtures of OmpR and F1-FAM. No intermediate states corresponding to one OmpR bound to F1-FAM were observed. The findings indicate that dimerization of OmpR is essential for the OmpR-DNA interaction and, based on the limits of detection, the OmpR monomer has >200-fold weaker affinity for DNA than the dimer of OmpR.
Fig. 6.
Characterization of stoichiometry of OmpR DNA interaction. (a) SV-AUC analyses of F1-FAM DNA in the presence and absence of various concentrations of OmpR. Each plot contains 10 μM F1-FAM with 0, 1, 2.5, 5, 10 or 20 μM OmpR (plots 1–6 respectively). (b) Integrated ITC analyses of the interactions of OmpR (circles) and BeF3−-treated OmpR (diamonds) with DNA containing sequences accommodating either 1 (open symbols) or 2 (closed symbols) OmpR molecules, F1b and F1, respectively. Data sets were each fit with a model for one set of binding sites (solid lines).
A high positive cooperativity for the OmpR-DNA interaction could account for the inability to observe monomeric OmpR bound to DNA in the above experiment and in ITC analysis (Figs. 6b and S2). To remove the effects of cooperativity on DNA binding by OmpR, a duplex oligonucleotide, F1b, was created, containing the strongest OmpR-binding DNA consensus sequence18 with a limited number of bps to accommodate the binding of only a single OmpR molecule. The ability of OmpR to bind to F1b was monitored using ITC (Fig. 6b, open circles). To analyze weak binding to this site, 1-μl aliquots of 2 mM F1b-half site were injected into 10 μM OmpR to yield a final concentration of 400 μM F1b. No interaction was observed, indicating that the dissociation constant for DNA binding (KD) is greater than 1 × 10−3 M. This finding strongly suggests that an interaction between an OmpR monomer and DNA is not an essential step in forming the stable 2OmpR•DNA complex. Therefore, these findings provide additional evidence arguing against the model described in Scheme B of Figure 1 and suggest that both in vivo and in vitro, unphosphorylated OmpR does not interact with DNA as a monomer.
The remaining models for OmpR-dependent transcriptional regulation, depicted in Figure 1 Schemes A and C, infer that phosphorylation is important for the OmpR-DNA interaction. In Scheme C, monomeric OmpR~P is suggested to bind to DNA and subsequently facilitates the binding of another OmpR~P molecule, while in Scheme A, the OmpR-DNA interaction is driven by OmpR~P dimerization. Previous studies have clearly demonstrated that DNA binding by OmpR is enhanced by phosphorylation. These findings have been replicated using the noncovalent small molecule phosphoryl analog BeF3−,30 which stabilizes the active conformation of OmpR via a non-enzymatically reversible mechanism, thus making it suitable for ITC and other experiments requiring long incubations (Figs. 6b and S2). The data generated using BeF3−, as well as previously reported data, are entirely consistent with the models in Schemes A and C for OmpR-dependent transcriptional regulation. To distinguish between these models, the effect of phosphorylation on the ability of OmpR to bind DNA as a monomer was determined using ITC (Fig. 6b, open diamonds). Note that although phosphorylation greatly enhances DNA binding by OmpR when a full-length OmpR recognition sequence is used (KD = 2.0 × 10−6 M versus 1.5 × 10−7 M in the absence and presence of BeF3−, respectively), when the half site oligonucleotide F1b is used, phosphorylation has essentially no effect on the affinity this DNA sequence for OmpR (KD> 1.0 × 10−3 M regardless of BeF3− in solution). The absence of an observed increase in binding of OmpR to F1b in the presence of a phosphoryl mimic casts doubt on the model described in Scheme C in which phosphorylation-enhanced DNA binding drives dimerization of OmpR on the DNA.
Phosphorylation enhances OmpR dimerization
The above findings argue against previously proposed models for the role of phosphorylation in OmpR-DNA interactions (Fig. 1, Schemes B and C). What remains to be determined, however, is whether data for OmpR support the standard model of phosphorylation-mediated dimerization enhancing DNA binding (Fig. 1, Scheme A). The ITC and SV-AUC studies presented above indicate that there is no detectable binding of monomeric OmpR to DNA. If the model in Scheme C is correct, then OmpR dimers must be able to form in solution and subsequently become stabilized by the presence of a DNA target sequence. Also, dimerization affinity must be enhanced by phosphorylation. The demonstration of phosphorylation-mediated dimerization of OmpR using fluorescent protein tagged constructs14 suggests that this is true. In order to determine the validity of these findings in a non-labeled protein and to quantify the effect of phosphorylation on dimerization affinity, sedimentation equilibrium analytical ultracentrifugation (SE-AUC) was performed on OmpR alone and in the presence of BeF3− (Fig. 7 and Table 3). These data show that in the absence of BeF3− OmpR dimerization is weak (KD = 1.1 ×10−3 M, (95 % CE (0.97 to 1.3) ×10−3 M), while in the presence of BeF3− dimerization is significantly enhanced (KD = 7.0 ×10−6 M, (95 % CE (2.8 to 38) ×10−6 M). These values for KD are entirely consistent with those reported for other unphosphorylated and phosphorylated RR proteins such as E. coli PhoB (KD = 3.8 ×10−4 M and 5.1 ×10−6 M for unphosphorylated and phosphorylated forms, respectively)31 and Bacillus subtilis Spo0A (KD = 1.8 ×10−3 M and 1.3 ×10−5 M for unphosphorylated and phosphorylated forms, respectively)32. The similarity of KD values for phosphorylated forms of OmpR and PhoB is not surprising, since dimerization of these proteins is presumed to involve a highly conserved interface. Furthermore, phosphorylation mediated increase in OmpR dimerization is entirely consistent with observed differences in the thermodynamics of the OmpR-F1 and OmpR-C1 interactions in the absence and presence of BeF3−. The enhancement in binding free energy (ΔΔG = 1.6 kcal/mol) for formation of the 2OmpR•DNA complex appears to be driven by an enhancement in binding enthalpy (ΔΔH = 4.5 or 3.3 kcal/mol for F1 and C1 respectively). When dimerization is coupled to DNA binding the apparent enthalpy of the interaction would have contributions from not only enthalpies associated with OmpR direct interactions with DNA and OmpR dimerization, which are likely to be similar in regardless of the presence of BeF3−, but also the free energy of the dimerization interaction which is enhanced by the presence BeF3− (−4.3 and −7.3 kcal/mol in the absence and presence of BeF3−, respectively).36
Fig. 7.
Comparison of dimerization propensities for untreated and BeF3−-treated OmpR. SE-AUC profiles for 2 μM OmpR and BeF3−-treated OmpR (closed and open circles, respectively) at 23,000 rpm. Lines represent the fits of the data for a monomer-dimer equilibrium.
Together, the above findings favor the standard model for control of transcription by OmpR, in which phosphorylation-activated dimerization drives an increase in DNA binding (Fig. 1, Scheme A). This model contrasts with those previously proposed for OmpR activation, but corresponds to the model accepted for many other well characterized OmpR/PhoB family members. The apparent rationale for proposals of alternate models for activation of OmpR, distinct from those of other OmpR/PhoB family RRs, has been the previously reported inability to detect dimerization of OmpR in the absence of DNA.11–13 Assessment of dimerzation is hindered by the low solubility of OmpR~P. We have observed significant precipitate in solutions of OmpR~P at concentrations ≥10 μM (data not shown). Note that this value is conspicuously close to the dimerization constant of OmpR~P, 7 μM. Low solubility renders many methodologies for monitoring protein oligomerization, such as gel filtration or SV-AUC, untenable because these methods often require protein concentrations greater than 10 μM. However, SE-AUC, at the low loaded sample concentrations used (≤ 2 μM), provides a method for characterizing dimerization of activated OmpR while maintaining most of the protein in a soluble state.
Discussion
The extent of OmpR phosphorylation might be regulated by multiple mechanisms
The studies above are the first to directly and quantitatively describe changes in the percentage of RR phosphorylation in vivo over sub-minute time scales following HK activation. Previous studies have demonstrated that the EnvZ/OmpR TCS produces graded changes in the expression of OmpR-regulated genes in response to cell membrane perturbations such as increases in media osmotic strength or treatment with anesthetics.37–39 Because the intracellular concentration of OmpR does not vary extensively in response to changes in medium osmotic strength or procaine treatment, the control of OmpR-regulated gene expression is thought to be mediated by the concentration of OmpR~P,40 with control of OmpR~P being mediated by the opposing kinase and phosphatase activities of EnvZ. One important finding from these analyses is that complete phosphorylation of OmpR does not appear to occur even under conditions that are thought to fully activate the pathway. Additionally, the incomplete phosphorylation of OmpR upon activation is not entirely due to the phosphatase activity of EnvZ, suggesting that alternate mechanisms, such as OmpR autodephosphorylation, play important roles in maintaining an intracellular pool of unphosphorylated OmpR, as has been proposed previously.41 These findings also demonstrate that a narrow window of OmpR~P concentrations can have a profound effect on gene expression. The intracellular concentration of OmpR is thought to be significantly greater than the number of OmpR binding sites on DNA, thus saturation of most OmpR DNA-binding sites can potentially occur with substoichiometric phoshphorylation of OmpR.40
In the absence of activation by a membrane altering anesthetic or high osmolarity, a small percentage of OmpR exists as OmpR~P (~5 %). This fraction appears to be maintained at a constant level through the phosphatase activity of EnvZ (compare wild type and envZ473 strains in Fig. 2). Although this observed level could result from high levels of OmpR~P existing in a small percentage of the population of cells, it seems more likely that this low level of OmpR~P is distributed throughout the population, reflected in the requirement of OmpR~P for expression of ompF under conditions that do not perturb the cell membrane (Fig. S2). Similarly, low fractions of OmpR~P are observed in cells expressing OmpR using a PBAD expression system activated by derepression through metabolism of media glucose, a mechanism that should not have significant cell-to-cell variation.44
Stabilization of the OmpR dimer thermodynamically enhances DNA-binding affinity
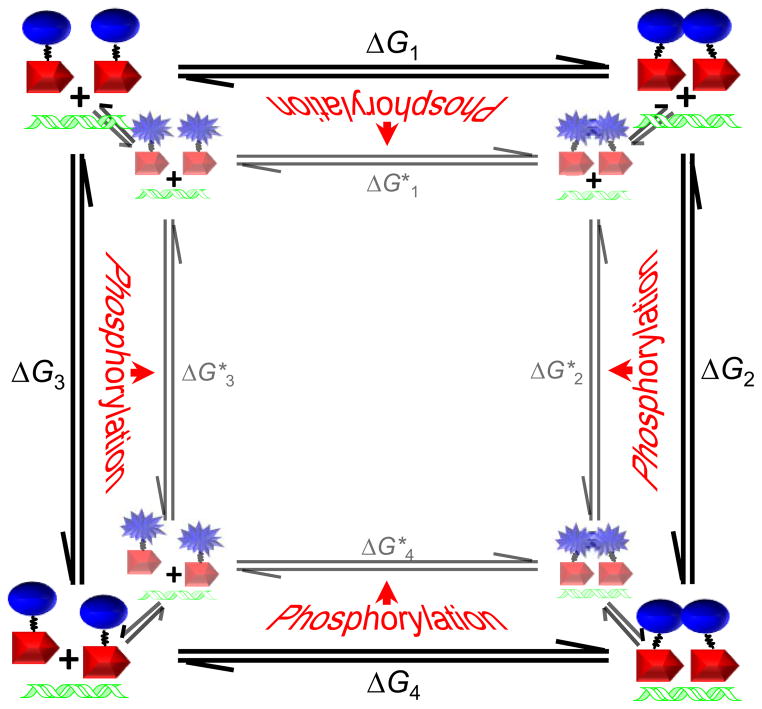
The presented model for phosphorylation-mediated activation of OmpR-regulated transcription represents a departure from previously proposed models, yet the data presented do not contradict previously reported data. The reasoning behind the model presented here as well as the previous models for activation is, for the most part, thermodynamically sound, yet the kinetics of activation implicitly favor the model presented here over other reported models. The coupled thermodynamic cycles that drive the OmpR dimerization and DNA-binding reactions are depicted in Figure 8. Note that there are two distinct cycles depicted for the inactive and active conformational states of OmpR. Previous studies have shown that inactive and active conformations of RRs have different propensities for dimerization, DNA binding, and phosphorylation. Also note that phosphorylation is not indicated to be solely associated with a single state of the RR, reflecting the conformational equilibrium between inactive and active states. Phosphorylation strongly stabilizes the active conformation, but unphosphorylated proteins are also able to adopt the active conformation, albeit with a markedly lower fractional population distribution.47 Due to this distribution of states, our measurements of dimerization and DNA binding each contain energetic contributions from both active and inactive conformational states. In addition, the phosphorylation-dependent differences observed for dimerization and DNA binding do not necessarily reflect different modes of interaction for unphosphorylated versus phosphorylated OmpR, but do reflect different population distributions of the inactive and active protein forms.
Fig. 8.
Coupled thermodynamic cycles linking OmpR dimerization and DNA binding. Blue ovals and starbursts represent OmpR receiver domains in inactive and active conformational states, respectively. Red arrows represent OmpR DNA-binding domains and DNA containing OmpR binding site sequences is depicted in green. Free energies underlying the steps in the cycle are numbered, with free energies associated with OmpR in an active conformational state being indicated by asterisks. Indicated free energies do not correspond directly to any measured values due to the coupling of active to inactive conformational equilibria to each step in the interaction pathways as depicted by the equilibrium arrows between active and inactive OmpR reaction cycles. Phosphorylation is thought to drive the conformational equilibrium toward the active state as indicated by the red arrows pointing toward the active OmpR reaction cycle.
Assays with purified proteins have established that phosphorylation is enhanced when OmpR is bound to DNA. Although this does not appear to be relevant for activation of OmpR in vivo, as has been suggested previously,13 it is consistent with the thermodynamic model presented in Figure 8. It has been suggested that the conformational distribution between inactive and active states influences RR phosphorylation in vitro by establishing a subpopulation of receiver domains that are catalytically competent for autophosphorylation.27 Therefore, interactions that stabilize an active conformation, such as the formation of the 2OmpR•DNA complex, would be likely to enhance the autophosphorylation rate for OmpR. However, in vivo, with EnvZ serving as the phosphodonor, the rate-limiting step for OmpR phosphorylation is likely modulated by the availability EnvZ~P, rather than the rate of phosphotransfer to OmpR~P. Furthermore, within the cell, the concentration of OmpR not specifically bound to DNA is much greater than that of OmpR bound to specific DNA sequences, suggesting that nonspecifically bound OmpR would be preferentially phosphorylated by EnvZ based on simple mass action. Further constraints on phosphorylation of DNA-bound OmpR are presumably imposed by the membrane localization of EnvZ. Our findings suggest that the phosphorylation rate of OmpR within cells is not significantly impacted by DNA binding or dimerization, supporting the hypothesis that OmpR phosphorylation is the first step in the activation pathway.
In correlation with previously published results, the SV-AUC and ITC findings above suggest that DNA binding requires the tandem interaction of two OmpR molecules. These equilibrium studies do not address whether DNA binding stabilizes OmpR dimers already formed in solution or if OmpR only self-associates upon binding to DNA. We have determined the dimerization constant for OmpR alone and in the presence of a phosphoryl analog, providing evidence for a binding mechanism where OmpR dimers form in solution and are stabilized in the presence of DNA. This finding overturns the notion that OmpR dimers form only in the presence of DNA, a conclusion based on the previous failure to observe dimers of OmpR in DNA-free solution and the finding that altering the order of the OmpR-specific DNA half-sites reduces DNA-binding affinity.18 As mentioned above, the low affinity of dimerization of unphosphorylated OmpR and the low solubility of OmpR~P, likely explain the lack of previous detection of OmpR dimers. In light of the data reported here, the lower affinity of OmpR observed for reverse-ordered DNA half-sites is likely explained by alternative mechanisms such as the influence of specific DNA tertiary structure and the partial overlap between DNA half-sites.
The coupling of dimerization and DNA binding allows for any interaction that energetically stabilizes one of these events to enhance the interaction affinity for the other. The studies presented here address the thermodynamics of OmpR binding to DNA and OmpR dimerization both in the presence and absence of a phosphoryl analog. A comparison of the change in ΔG for DNA binding by dimeric OmpR due to the presence of a phosphoryl analog (ΔΔGbinding = 1.6 kcal/mol) whether F1 or C1 DNA is used (compare 0 and 2 mM BeF3− data in Tables 2 and S2) is significantly lower in magnitude than the change in ΔG for OmpR dimerization due to the presence of a phosphoryl analog (ΔΔGdimerization = 3.0 kcal/mol). The larger magnitude phosphorylation-associated change in ΔGdimerization relative to ΔGbinding, and the non-dependence on the DNA sequence used, indicates that dimerization is the sole driving force for the phosphorylation-associated enhancement in DNA binding. The finding that phosphorylation has no observable effect on binding of OmpR to a DNA sequence that accommodates only monomeric OmpR lends further credence to this hypothesis and refutes hypotheses that phosphorylation enhances DNA binding directly.18
Table 2.
ITC-derived parameters for the interactions of OmpR in the presence and absence of BeF3− with F1 and F1b duplex DNA.
| DNA | [BeF3−] (mM) | KD (M) | N | ΔH(kcal/mol) | −TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|---|
| F1 | 0 | (2.0 ± 0.6) × 10−6 | 2.1 ± 0.2 | −3.5 ± 0.2 | −4.3 ± 0.3 | −7.8 ± 0.2 |
| F1 | 2 | (1.5 ± 0.3)×10−7 | 2.1 ± 0.1 | −8.0 ± 0.2 | −1.3 ± 0.3 | −9.3 ± 0.1 |
| F1ba | 0 | > 1 × 10−3 | 1.0 Fixed | > 0 | ND | >−4.0 |
| F1ba | 2 | > 1 × 10−3 | 1.0 Fixed | > −30 | ND | >−4.0 |
The ITC profiles (Fig. 6B) were fit with a model for one set of binding sites. The indicated uncertainties in the fitted values reflect the standard deviation of the experimental data from the fitted curves. Values for ΔG and −TΔS were calculated when possible using the standard formalisms containing the maximum errors as carried through the equations.
Fitted values for OmpR binding to F1b DNA in the absence of BeF3− (a) minimized to KD = (1.2 ± 0.4) × 10−3 M and ΔH= 44 ± 11 kcal/mol, and in the presence of BeF3− (b) minimized to KD = (2.7 ± 7.6) × 10−3 M and ΔH= −29 ± 37 kcal/mol. Due to the concentration of reactants used and the small signal to noise differences in the data it is likely that these values underestimate the actual dissociation constants (KD) and overestimate the magnitudes of the binding enthalpies (ΔH), therefore minimum values are indicated for each parameter.
Conclusions
Dimerization is a common characteristic of transcription factor proteins and phosphorylation is a common mechanism for regulation of dimerization. The thermodynamic cycle depicted above describes the mechanism by which all phosphorylatable OmpR/PhoB family RR proteins allow for phosphorylation-mediated control of gene expression through regulation of dimerization. Similar mechanisms are likely employed by other phosphorylation regulated proteins as well. Although the processes of phosphorylation, dimerization and DNA binding are energetically coupled, kinetic differences in forward and reverse reaction rates for these enzymatic and binding reactions are utilized by the cell to shift the reactions toward the desired outcomes. Because of the kinetic tuning of these processes, certain results obtained in vitro are not applicable to the in vivo activities of the OmpR/EnvZ TCS. Therefore, caution should be used when inferring in vivo mechanisms based on in vitro measurements, as has been done previously for this TCS. As a consequence of dimerization being a primary regulatory event controlled by OmpR/PhoB family signaling pathways, the highly conserved dimerization interface becomes an attractive target for the development of antimicrobial compounds.
Materials and Methods
Plasmid constructs, oligonucleotides, and protein purification
For transcriptional and in vivo phosphorylation analyses, the various OmpR constructs were cloned into the low copy number plasmid pBAD3350 with protein expression driven by the PBAD promoter. For purification, the T7 expression system encoded in pET-21b (Novagen, Madison, WI) was used. Native proteins were used for transcription and in vivo phosphorylation analyses. For purification, OmpR and its associated mutants were expressed with a His6 tag and a thrombin cleavage site. Plasmids used for expression of the various OmpR constructs were generated using previously described cloning strategies.31 All plasmids and strains used in this study are listed in supplemental Table S2.
Oligonucleotides used in these studies were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). For DNA-binding experiments C1 DNA (5′-GGCATTTACATTTTGAAACATCTAGG-3′ and complement), F1 DNA (5′-GCTTTACTTTTGGTTACATATTTCG-3′ and complement), F1-FAM DNA(5′-/56-FAM/CCATTTTACTTTTGGTTACATATTCC-3′ and complement), and F1b DNA (5′-CCTGGTTACATATTCC-3′ and complement) were preheated to 85 °C for 5 min in the buffers indicated for each experiment and passively cooled to room temperature immediately prior to use.
Proteins were expressed in E. coli BL-21(DE3) cells. Cells expressing His-tagged OmpR and mutant forms were grown to mid-log phase at 37 °C in Luria-Bertani medium supplemented with 100 μg/ml ampicillin, then induced with 0.5 mM IPTG and grown for an additional 3 h at 30 °C. All proteins were lysed by sonication in a buffer containing 50 mM Tris-HCl, 0.1 M NaCl, 1 mM EDTA, 0.5 mM PMSF, 2 mM β-mercaptoethanol (β-ME) at pH 8.0. Lysates were applied to a 5-ml His-Trap HP column (GE Healthcare, Piscataway, NJ) equilibrated with 20 mM sodium phosphate, 2 mM β-ME, 75 mM NaCl, 25 mM imidazole at pH 7.5 and eluted using a linear gradient of 20 mM sodium phosphate, 2 mM β-ME, 75 mM NaCl, 500 mM imidazole at pH 7.5. Pooled fractions containing OmpR or mutants were applied to a Superdex 75 26/60 gel-filtration column (GE Healthcare, Piscataway, NJ) pre-equilibrated in 50 mM Tris-HCl, 0.1 M NaCl, 2 mM β-ME at pH 7.5. The purified proteins were stored at −80 °C after rapid freezing in ethanol/solid CO2. The His6 tag was cleaved from the proteins by digestion with 0.4 U/mL biotin-tagged thrombin (EMD biosciences) in a reaction solution containing 50 mM Tris-HCl, 0.1 NaCl, 2 mM β-ME, 2.5 mM CaCl2 at pH 7.5. Following overnight digestion at 20 °C, biotin-tagged thrombin was removed as directed by the manufacturer using streptavidin-agarose beads. Undigested protein and digested His6-tag containing peptides were removed by incubating with 25 μL Ni-NTA beads (Qiagen) for 30 min with constant shaking.
Analysis of autophosphorylation
Autophosphorylation activity of OmpR and its associated mutants was measured at a concentration of 10 μM in a solution containing 10 mM PIPES, 10 mM MgCl2, 2 mM β-ME at pH 7.5, and sufficient NaCl for a final sodium concentration of 150 mM. Ammonium hydrogen phosphoramidate (PA) in the same buffer was added to a final concentration of 20 mM to initiate the reaction. For reactions performed in the presence of C1 DNA, 20 μM C1 was added to the reaction solution. At times ranging from 15 s to 2 h following the addition of PA, 15-μL aliquots were removed and the reaction was stopped by trituration with 5 μL of 4X SDS loading buffer [0.2 M Tris, 8% SDS (w/v), 40% (w/v) glycerol, 4% (v/v) β-ME, 0.08% (w/v) bromphenol blue at pH 6.8]. The fraction of protein phosphorylated for each time point was determined using phosphoprotein affinity gel electrophoresis as described previously. Briefly, Phos-tag™ acrylamide running gels contained 10% (w/v) 29:1 acrylamide/N,N′-methylenebisacrylamide, 375 mM Tris (pH 8.8) and 0.1% (w/v) SDS. Gels were copolymerized with 75 μM Phos-tag™ acrylamide and 150 μM MnCl2. Stacking gels contained 4% (w/v) 29:1 acrylamide/N,N′-methylenebisacrylamide, 125 mM Tris (pH 6.8) and 0.1% (w/v) SDS. All Phos-tag™ acrylamide-containing gels were run with standard denaturing running buffer [0.4% (w/v) SDS, 25 mM Tris, 192 mM glycine] at 4 °C under constant voltage (160 V). The fraction of phosphorylated protein in each gel lane was determined by monitoring the Coomassie Blue staining intensity for the upshifted (phosphorylated) protein band in each lane relative to the total staining intensity for both bands (total protein) in the lane.
Analysis of OmpR and OmpR~P in E. coli cells
E. coli cultures were grown on a shaking incubator at 37 °C in medium A51 until cultures were in mid-log phase growth (0.4–0.6 OD). For steady-state measurements, cultures were either left untreated or were treated with either 10 mM procaine or 20 % (w/v) sucrose. Following an additional 60 min of growth with shaking, 100- to 300-μL samples were removed, cells were pelleted by centrifugation, lysed with 70 μL 1X BugBuster™ (EMD Chemicals, Inc.) and 25 μL of 4X SDS loading buffer was added. For time course measurements, cultures were placed in a 37 °C thermal incubator with shaking and treated with 10 mM procaine. At 15, 30, 60, 120, 300, and 600 s following addition of procaine, 70-μL aliquots were removed from the culture and mixed with 7 μL 10X BugBuster™ using an Elmeco™ vortexer, immediately followed by the addition of 25 μL 4X SDS loading buffer.
All samples were loaded onto gels containing 25 μM Phos-tag™ acrylamide and 50 μM MnCl2 prepared and run as described above. After running, gels were fixed for 10 min in standard transfer buffer, 20% (v/v) methanol, 50 mM Tris, 40 mM glycine, with 1 mM EDTA added to remove Mn2+ from the gel. Gels were incubated for an additional 20 min in transfer buffer without EDTA to remove the chelated metal. Transfer to nitrocellulose membranes was performed using a Bio-Rad semi-dry transfer apparatus under a constant 160 mA for 80 min. Western blotting was performed using standard protocols with rabbit anti-OmpR primary antisera and anti-rabbit secondary antibody coupled to horseradish peroxidase. Blots were imaged on an AlphaInnotech Alphaimager™ using chemiluminescent detection (SuperSignal West Pico Chemiluminescent Substrate; Pierce Biotechnology, Inc., Rockford, IL). The fraction of phosphorylated protein in each gel lane was determined as described above.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) measurements were conducted at 25 °C on a MicroCal iTC200 (MicroCal, Inc., Northampton, MA). The solution conditions for all ITC measurements were 10 mM HEPES and 5 mM MgCl2 at pH 7.5. Experiments on inactivated protein contained 145 mM NaCl and experiments on activated protein contained 2 mM BeCl2, 115 mM NaCl, and 30 mM NaF for final sodium concentrations of 150 mM. In each experiment, either 1- or 2-μL aliquots of solutions of either 100 μM F1 or C1, or 2 mM F1b DNA were sequentially injected from a 40-μL syringe rotating at 1000 rpm into an isothermal sample chamber containing 211 μL of 10 μM protein. The duration for each injection was set to 0.5 μL/s. A 120-s initial delay was used prior to the first injection and a 180-s delay was used between the subsequent injections. Each DNA-protein experiment was accompanied by the corresponding control experiment, in which DNA was injected into a solution of buffer alone. Each injection generated a heat burst curve (μcal/s versus s), the area under which was determined by integration [using Origin version 7.0 software (MicroCal, Inc., Northampton, MA)] to obtain a measure of the heat associated with that injection. The measure of the heat associated with each DNA-buffer injection was subtracted from that of the corresponding heat associated with each DNA-protein injection to yield the heat of DNA binding for that injection. The buffer-corrected ITC profiles for the binding of each protein to DNA were fit with a model for one set of binding sites.
Analytical ultracentrifugation
AUC experiments were carried out in an Optima XL-I ultracentrifuge using an An-50 Ti rotor (Beckman Coulter, Fullerton, CA). Proteins and F1-Fam DNA were equilibrated in a buffer containing 10 mM PIPES disodium salt, 5 mM MgCl2, 1 mM TCEP at pH 7.5. Experiments on inactivated protein contained 130 mM NaCl and experiments on activated protein contained 2 mM BeCl2, 100 mM NaCl and 30 mM NaF for final sodium concentrations of 150 mM. Buffer density, buffer viscosity and the partial specific volumes of the proteins were calculated using the program SEDNTERP.52 All AUC experiments were conducted in epon charcoal-filled double-sector centerpieces and sapphire windows. SV experiments were conducted at 25 °C and the samples were spun at 50,000 RPM. DNA gradients were monitored at 410 nm using the optical absorbance system. Approximately 50 SV scans were used to calculate a c(s) distribution that was generated using the program SEDFIT.53 During the c(s) analysis, the frictional ratio and meniscus position were treated as floating parameters. After optimization of these parameters, the final distribution was calculated using a resolution setting of 200 and a confidence interval of 0.8.
All SE experiments were analyzed at 25 °C at three different speeds 12,000, 19,000 and 23,000 RPM. For the inactivated protein, protein gradients were monitored at 280 nm using 100, 75, and 50 μM OmpR. For the BeF3−-activated protein, protein gradients were monitored at 230 nm using 2, 1, and 0.5μM OmpR. Profiles for active or inactive OmpR were globally fit to a monomer-dimer self-association model using the program SEDPHAT54 with the monomer molecular weight being fixed at 27,354 Da. For data analysis the protein concentration used in each cell was assumed to be constant for all speeds.
Analysis of porin proteins
Wild type, ΔenvZ, ΔompF, ΔompC, ΔompR and ΔompR containing a plasmid expressing either wild-type ompR, or ompR DNA-binding domain mutants were grown to mid log phase in A media. Cultures were then diluted 1:2 into fresh media and incubated for 1 h at 37 °C or fresh media containing either procaine or sucrose such that the final concentrations were 10 mM or 15% (w/v), respectively, and incubated for 2 h. Fractions enriched in membrane proteins were prepared from cells using the following procedure. Cells from 10 mL of each culture were harvested by centrifugation and washed with 30 mM Tris at pH 8.0. Pellets were frozen in a dry ice-ethanol bath, thawed, and were resuspended in 0.2 mL 30 mM Tris, 1 mM MgCl2 at pH 8.0 and containing 20 μg/mL DNase I. Cells were lysed by 30 min incubation with 1 mg/mL lysozyme followed by sonication in an ice bath. Lysed samples were centrifuged at 7000 × g for 10 min at 4 °C to remove unlysed cells; supernatants were centrifuged at 25,000 × g for 30 min to pellet membrane fractions, pellets were washed once with 30 mM Tris at pH 8.0 and resuspended in 1 % SDS, 30 mM Tris at pH 8.0. 60 μL of each sample was mixed with 20 μL 4X SDS loading buffer supplemented with 6 M urea and boiled for 10 min. Proteins were separated by urea-SDS gel electrophoresis using a stacking gel containing 4.5 % (w/v) 37.5:1 acrylamide/N,N′-methylenebisacrylamide and 6 M urea at pH 6.8 and a running gel containing 10% (w/v) 37.5:1 acrylamide/N,N′-methylenebisacrylamide and 6 M urea at pH 8.8. Porin proteins were visualized with Coomassie Blue stain.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the assistance of Rong Gao in assay development and data analysis and thank Jizong Gao for preparation of protein expression plasmids.
Abbreviations used
- AUC
analytical ultracentrifugation
- β-ME
2-mercaptoethanol
- EDTA
ethylenediaminetetraacetic acid
- HK
histidine kinase
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- ITC
isothermal titration calorimetry
- NMR
nuclear magnetic resonance
- RR
response regulator
- RPM
revolutions per minute
- PMSF
phenylmethylsulfonyl fluoride
- PA
phosphoramidate
- SE
sedimentation equilibrium
- SV
sedimentation velocity
- SDS
sodium dodecyl sulfate
- TCEP
tris(2-carboxyethyl)phosphine
- TCS
two-component system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- 2.Kramer R. Bacterial stimulus perception and signal transduction: response to osmotic stress. Chem Rec. 2010;10:217–29. doi: 10.1002/tcr.201000005. [DOI] [PubMed] [Google Scholar]
- 3.Igo MM, Silhavy TJ. EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J Bacteriol. 1988;170:5971–5973. doi: 10.1128/jb.170.12.5971-5973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aiba H, Nakasai F, Mizushima S, Mizuno T. Phosphorylation of a bacterial activator protein, OmpR, by a protein kinase, EnvZ, results in a stimulation of its DNA-binding ability. J Biochem. 1989;106:5–7. doi: 10.1093/oxfordjournals.jbchem.a122817. [DOI] [PubMed] [Google Scholar]
- 5.Forst SA, Delgado J, Inouye M. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattison K, Kenney LJ. Phosphorylation alters the interaction of the response regulator OmpR with its sensor kinase EnvZ. J Biol Chem. 2002;277:11143–11148. doi: 10.1074/jbc.M111128200. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida T, Cai S, Inouye M. Interaction of EnvZ, a sensory histidine kinase, with phosphorylated OmpR, the cognate response regulator. Mol Microbiol. 2002;46:1283–94. doi: 10.1046/j.1365-2958.2002.03240.x. [DOI] [PubMed] [Google Scholar]
- 8.Jeon Y, Lee YS, Han JS, Kim JB, Hwang DS. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J Biol Chem. 2001;276:40873–40879. doi: 10.1074/jbc.M104855200. [DOI] [PubMed] [Google Scholar]
- 9.Toro-Roman A, Mack TR, Stock AM. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the a4-β5-a5 face. J Mol Biol. 2005;349:11–26. doi: 10.1016/j.jmb.2005.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack TR. Characterization of the inter- and intramolecular interactions that mediate the phosphorylation-dependent activation of the response regulator PhoB. University of Medicine and Dentistry; New Jersey: 2008. [Google Scholar]
- 11.Jo YL, Nara F, Ichihara S, Mizuno T, Mizushima S. Purification and characterization of the OmpR protein, a positive regulator involved in osmoregulatory expression of the ompF and ompC genes in Escherichia coli. J Biol Chem. 1986;261:15252–15256. [PubMed] [Google Scholar]
- 12.Harlocker SL, Bergstrom L, Inouye M. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J Biol Chem. 1995;270:26849–26856. doi: 10.1074/jbc.270.45.26849. [DOI] [PubMed] [Google Scholar]
- 13.Rhee JE, Sheng W, Morgan LK, Nolet R, Liao X, Kenney LJ. Amino acids important for DNA recognition by the response regulator OmpR. J Biol Chem. 2008;283:8664–8677. doi: 10.1074/jbc.M705550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao R, Tao Y, Stock AM. System-level mapping of Escherichia coli response regulator dimerization with FRET hybrids. Mol Microbiol. 2008;69:1358–1372. doi: 10.1111/j.1365-2958.2008.06355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ames SK, Frankema N, Kenney LJ. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc Natl Acad Sci USA. 1999;96:11792–11797. doi: 10.1073/pnas.96.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang KJ, Igo MM. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 17.Head CG, Tardy A, Kenney LJ. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol. 1998;281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida T, Qin L, Egger LA, Inouye M. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J Biol Chem. 2006;281:17114–17123. doi: 10.1074/jbc.M602112200. [DOI] [PubMed] [Google Scholar]
- 19.Forst S, Delgado J, Rampersaud A, Inouye M. In vivo phosphorylation of OmpR, the transcription activator of the ompF and ompC genes in Escherichia coli. J Bacteriol. 1990;172:3473–3477. doi: 10.1128/jb.172.6.3473-3477.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbieri CM, Stock AM. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal Biochem. 2008;376:73–82. doi: 10.1016/j.ab.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita-Kikuta E, Aoki Y, Kinoshita E, Koike T. Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol Cell Proteomics. 2007;6:356–366. doi: 10.1074/mcp.T600044-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Taylor RK, Hall MN, Silhavy TJ. Isolation and characterization of mutations altering expression of the major outer membrane porin proteins using the local anaesthetic procaine. J Mol Biol. 1983;166:273–82. doi: 10.1016/s0022-2836(83)80085-0. [DOI] [PubMed] [Google Scholar]
- 23.Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol. 2007;189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCleary WR, Stock JB. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 25.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: A system-level analysis. PLoS Biol. 2005;3:1770–1788. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin D, Lee EJ, Huang H, Groisman EA. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science. 2006;314:1607–1609. doi: 10.1126/science.1134930. [DOI] [PubMed] [Google Scholar]
- 27.Barbieri CM, Mack TR, Robinson VL, Miller MT, Stock AM. Regulation of response regulator autophosphorylation through interdomain contacts. J Biol Chem. 2010;285:32325–32335. doi: 10.1074/jbc.M110.157164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang KE, Lan CY, Igo MM. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc Natl Acad Sci USA. 1997;94:2828–2832. doi: 10.1073/pnas.94.7.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rampersaud A, Harlocker SL, Inouye M. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J Biol Chem. 1994;269:12559–12566. [PubMed] [Google Scholar]
- 30.Wemmer DE, Kern D. Beryllofluoride binding mimics phosphorylation of aspartate in response regulators. J Bacteriol. 2005;187:8229–8230. doi: 10.1128/JB.187.24.8229-8230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack TR, Gao R, Stock AM. Probing the roles of the two different dimers mediated by the receiver domain of the response regulator PhoB. J Mol Biol. 2009;389:349–364. doi: 10.1016/j.jmb.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis RJ, Scott DJ, Brannigan JA, Ladds JC, Cervin MA, Spiegelman GB, Hoggett JG, Barak I, Wilkinson AJ. Dimer formation and transcription activation in the sporulation response regulator Spo0A. J Mol Biol. 2002;316:235–245. doi: 10.1006/jmbi.2001.5331. [DOI] [PubMed] [Google Scholar]
- 33.Bachhawat P, Stock AM. Crystal structures of the receiver domain of the response regulator PhoP from Escherichia coli in the absence and presence of the phosphoryl analog beryllofluoride. J Bacteriol. 2007;189:5987–5995. doi: 10.1128/JB.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachhawat P, Swapna GV, Montelione GT, Stock AM. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure. 2005;13:1353–1363. doi: 10.1016/j.str.2005.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toro-Roman A, Wu T, Stock AM. A common dimerization interface in bacterial response regulators KdpE and TorR. Protein Sci. 2005;14:3077–388. doi: 10.1110/ps.051722805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyman J, Gill SJ. Binding and Linkage: Functional Chemistry of Biological Macromolecules. University Science Books; Mill Valley, CA: 1990. Ligand Control of Aggregation; pp. 203–236. [Google Scholar]
- 37.Batchelor E, Goulian M. Imaging OmpR localization in Escherichia coli. Mol Microbiol. 2006;59:1767–78. doi: 10.1111/j.1365-2958.2006.05048.x. [DOI] [PubMed] [Google Scholar]
- 38.Batchelor E, Silhavy TJ, Goulian M. Continuous control in bacterial regulatory circuits. J Bacteriol. 2004;186:7618–25. doi: 10.1128/JB.186.22.7618-7625.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russo F, Silhavy TJ. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 40.Cai SJ, Inouye M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J Biol Chem. 2002;277:24155–61. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- 41.Kenney LJ. How important is the phosphatase activity of sensor kinases? Curr Opin Microbiol. 2010;13:168–76. doi: 10.1016/j.mib.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longo D, Hasty J. Dynamics of single-cell gene expression. Mol Syst Biol. 2006;2:64. doi: 10.1038/msb4100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lidstrom ME, Konopka MC. The role of physiological heterogeneity in microbial population behavior. Nat Chem Biol. 2010;6:705–12. doi: 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- 44.Megerle JA, Fritz G, Gerland U, Jung K, Radler JO. Timing and dynamics of single cell gene expression in the arabinose utilization system. Biophys J. 2008;95:2103–15. doi: 10.1529/biophysj.107.127191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Da Re S, Schumacher J, Rousseau P, Fourment J, Ebel C, Kahn D. Phosphorylation-induced dimerization of the Fix J receiver domain. Mol Microbiol. 1999;34:504–511. doi: 10.1046/j.1365-2958.1999.01614.x. [DOI] [PubMed] [Google Scholar]
- 46.Kern D, Volkman BF, Luginbuhl P, Nohaile MJ, Kustu S, Wemmer DE. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;40:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- 47.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 48.Qin L, Yoshida T, Inouye M. The critical role of DNA in the equilibrium between OmpR and phosphorylated OmpR mediated by EnvZ in Escherichia coli. Proc Natl Acad Sci USA. 2001;98:908–913. doi: 10.1073/pnas.031383098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison-McMonagle P, Denissova N, Martínez-Hackert E, Ebright RH, Stock AM. Orientation of OmpR monomers within an OmpR:DNA complex determined by DNA affinity cleaving. J Mol Biol. 1999;285:555–566. doi: 10.1006/jmbi.1998.2375. [DOI] [PubMed] [Google Scholar]
- 50.Guzman L, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawaji H, Mizuno T, Mizushima S. Influence of molecular size and osmolarity of sugars and dextrans on the synthesis of outer membrane proteins O-8 and O-9 of Escherichia coli K-12. J Bacteriol. 1979;140:843–7. doi: 10.1128/jb.140.3.843-847.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton J, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 53.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.