Abstract
Sickle cell anemia is common in the Middle East and India where the HbS gene is sometimes associated with the Arab-Indian (AI) β-globin gene (HBB) cluster haplotype. In this haplotype of sickle cell anemia, fetal hemoglobin (HbF) levels are 3-4 fold higher than those found in patients with HbS haplotypes of African origin. Little is known about the genetic elements that modulate HbF in AI haplotype patients. We therefor studied Saudi HbS homozygotes with the AI haplotype (mean HbF 19.2±7.0%, range 3.6 to 39.6%) and known genotyped cis- and trans-acting elements associated with HbF expression. All cases, regardless of HbF concentration, were homozygous for AI haplotype-specific elements cis to HBB. SNPs in BCL11A and HBS1L-MYB that were associated with HbF in other populations explained only 8.8% of the variation of HbF. KLF1 polymorphisms associated previously with high HbF were not present In the 44 patients tested. The SNPs and genetic loci we have chosen for this study do not explain the high HbF in sickle cell patients with AI haplotype or its variation among patients with this haplotype. The dispersion of HbF levels among AI haplotype patients suggests that other genetic elements modulate the effects of the known cis- and trans-acting regulators. These regulatory elements, which remain to be discovered, might be specific in the Saudi and some other populations where HbF levels are especially high.
Introduction
Fetal hemoglobin (HbF) protects against many of the hematologic and clinical complications of sickle cell anemia [[homozygosity for the sickle hemoglobin (HbS) gene; HBB glu6val; reviewed in(1)]. This is dependent on the ability of HbF to hinder deoxyHbS polymerization. HbF level is variable among patients and populations with sickle cell anemia and is highly heritable.(2, 3) HbF expression is regulated by elements linked to the β-globin gene (HBB) gene cluster (11p15.5) and other quantitative trait loci (QTL) in trans to HBB, two of which are the HBS1L-MYB intergenic region on chromosome 6q22-23 and BCL11A on chromosome 2p16.1. Together, these QTL accounted for 15 to 30% of HbF variation in sickle cell anemia patients with African origins of the sickle β-globin gene.(2, 4-8)
The HbS gene is also autochthonous to the Middle East and India where it is sometimes on an indigenous Arab-Indian (AI) HBB globin gene cluster haplotype.(9-11) This haplotype is marked by an Xmn1 restriction site polymorphism (C>T 158 bp 5′ to HBG2; rs7482144) and other single nucleotide polymorphisms (SNPs) and insertion-deletion polymorphisms that distinguish it from all African-origin haplotypes, including the Senegal haplotype that also has the Xmn1 restriction site polymorphism. Individuals with sickle cell anemia and the AI haplotype had higher mean HbF levels [average 17%, range 4-32%, reviewed in(11)] than patients with HbS haplotypes of African origin.(12) For example, untreated African Americans had mean HbF levels of 5.8% in males and 7.3% in females with mean ages of about 17 years.(6) Because of their higher HbF, AI haplotype sickle cell anemia patients had milder, albeit not asymptomatic disease when compared with carriers of African HBB haplotypes.(13, 14) As the QTL modifying HbF levels in sickle cell anemia patients with the AI haplotype have not been comprehensively studied, we genotyped the major known HbF-modulating QTL in 137 individuals and additional known cis- and trans-acting elements in subsets of these patients to study their association with HbF.
Methods
Patients
Subjects with sickle cell anemia who attended clinics at King Fahd Hospital, Al-Ahsa and King Saud University, Riyadh, Saudi Arabia were selected on the basis of homozygosity for the HbS gene and the AI haplotype, age of at least 10 years; they were not taking hydroxyurea at the time HbF was measured. HbF was measured by high performance liquid chromatography (HPLC).
HbS and the HBB haplotype
Homozygosity for the HbS mutation was confirmed using amplification refractory mutation system analysis (Table S1).(15) The AI haplotype was ascertained by genotyping the HBG2 Xmn1 C>T restriction site (rs7482144) and a Hinc2 site 5′ to HBE1 (rs3834466), and confirmed by the presence of a C>T polymorphism 68 bp 5′ to HBD.(16)
Sanger sequencing/TaqMan assay of selected cis- and trans- regions to βS-globin gene
Polymorphisms in the β-globin gene cluster region were detected by Sanger sequencing or TaqMan assays in HbS homozygote with the Arab Indian haplotype. For many of these assays we selected a smaller group for testing. Selection was based on availability of samples at time of testing or at random from the subjects we had confirmed as homozygous for HbS and the AI haplotype.
HBB gene cluster regulatory regions (11p15): (Table S1)
Regions selected for sequencing were based on their potential functional role in globin gene regulation and included the KLF1 binding site in the HBB promoter region, an AT motif 530 bp 5′ to HBB, TTTTA repeats 1412 bp 5′ to HBB, the HBD-HBG1 intergenic region, promoters of HBG2 and HBG1, and the cores of hypersensitive sites 2, 3 and 4 (HS-2, 3, 4) of the β-globin gene cluster locus control region (LCR). A −68 HBD C>T SNP in the promoter of HBD was detected using a custom designed TaqMan assay.
BCL11A, HBS1L-MYB, KLF1, DLX4
We genotyped SNPs in BCL11A and HBS1L-MYB using either pre-made or custom TaqMan Assays (Applied Biosystems). As KLF1 is a known regulator of BCL11A and globin switching, and has been associated with the phenotype of hereditary persistence of HbF, and as BP1 (DLX4) binds to AT motif 530 bp 5′ to HBB and has a down-regulatory effect on HBB expression, we sequenced KLF1 (n=44) and BP1 (DLX4) (n=23) in randomly selected cases, to exclude polymorphisms in these genes that might be associated with HbF levels.(17)
Statistical analysis
Linear regression was performed on HbF for each genetic locus, adjusting for gender of the subjects. No transformation of the HbF values was necessary as the HbF values of these patients were approximately normally distributed. The analysis was performed using an additive genetic model whereby the total number of minor alleles present was counted for each subject. A 2-sample Kolmogorov-Smirnov test was used to compare the distribution of HbF in patients enrolled in Cooperative Study of Sickle Cell Disease (CSSCD) and in patients with the AI haplotype from Saudi Arabia.(18)
Results
HbF
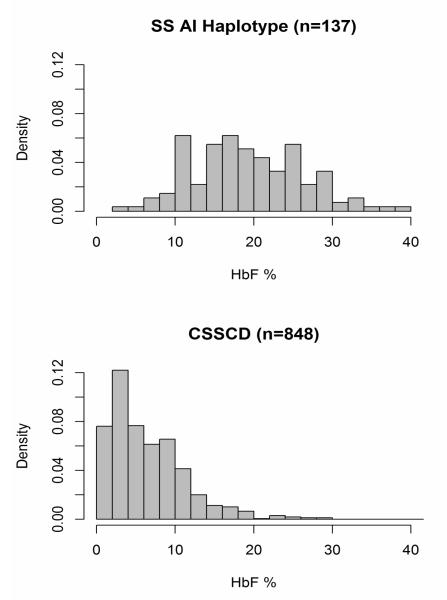
One hundred and thirty-seven sickle cell anemia patients who met our selection criteria were initially examined (Table 1) and their distribution of HbF concentrations is shown in Figure 1. Mean HbF was 19.2±7.0%. For comparison, African Americans with sickle cell anemia had a mean HbF of 6.6±5.5%. The HbF distribution for African Americans with sickle cell anemia was right skewed, whereas the AI haplotype subjects had a Gaussian or normal distribution. The distributions of HbF in these 2 cohorts were significantly different (p-value 2.2e-16).
Table 1.
A comparison of age and HbF in patients with sickle cell anemia (HbSS) and the Arab-Indian (AI) haplotype.
| HbSS AI Haplotype n = 137 |
CSSCD n = 848 |
||||||
|---|---|---|---|---|---|---|---|
| Category | Mean | SD | Range | Mean | SD | Range | P-value |
| Age (yrs) | 26.6 | 10.6 | 11 - 65 | 17.2 | 10.7 | 5 - 63 | <0.001 |
| HbF (%) | 19.2 | 7.0 | 3.6 – 39.6 | 6.6 | 5.5 | 0.1 – 60 | <0.001 |
Patients with sickle cell anemia from the Cooperative Study of Sickle Cell Database (CSSCD) served as reference in the statistical analysis. Although the CSSCD cohort was slightly younger on average, the HbF difference remains significant as age has an inverse relationship with HbF. Only subjects age 10 or older were included in this analysis as HbF changes are minimal after that age cutoff.
Figure 1. Density plots showing the distribution of HbF in sickle cell anemia with the AI haplotype (SS AI Haplotype).
For comparison, results from the Cooperative Study of Sickle Cell Disease (CSSCD), an African American sickle cell anemia population are replotted.(6) All CSSCD subjects were aged ≥5 years and none were taking hydroxyurea. These distributions are significantly different (p=2.2e-16).
Sanger sequencing/TaqMan assays
BCL11A and HBS1L-MYB
Table 2 shows the associations of polymorphisms in BCL11A and the HBS1L-MYB intergenic region and HbF in 137 patients. The 2.9 kb region in the 2nd intron of BCL11A bounded by rs1427407 and rs4671393 was associated with HbF as suggested previously (4-8), but accounted for only 7.5% of the variation in HbF. Together, BCL11A and HBS1L-MYB accounted for 8.8% of HbF variance. The remaining polymorphisms we examined were done in subsets of these patients.
Table 2.
Quantitative trait loci (QTLs) and association with HbF in subjects homozygous for HbS with Arab-Indian haplotype.
|
SS AI Haplotype (n=137)
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Chr. Position |
Major Allele |
Minor Allele |
MAF | Beta | SE | P |
| BCL11A | rs6732518 | 60708847 | C | T | 0.463 | 1.280 | 0.827 | 0.124 |
| BCL11A | rs1427407 | 60718293 | G | T | 0.283 | 2.451 | 0.891 | 0.007* |
| BCL11A | rs766432 | 60719970 | A | C | 0.300 | 2.769 | 0.864 | 0.002* |
| BCL11A | rs11886868 | 60720496 | T | C | 0.455 | 1.773 | 0.807 | 0.030* |
| BCL11A | rs4671393 | 60721201 | G | A | 0.301 | 2.697 | 0.859 | 0.002* |
| BCL11A | rs6729815 | 60723922 | T | C | 0.471 | −0.161 | 0.843 | 0.849 |
| HMIP | rs7775698 | 135418385 | G | A | 0.056 | −0.991 | 1.811 | 0.585 |
| HMIP | rs9399137 | 135418768 | T | C | 0.073 | 1.358 | 1.621 | 0.404 |
| HMIP | rs4895441 | 135426323 | A | G | 0.090 | 0.703 | 1.593 | 0.660 |
| HMIP | rs9402686 | 135427567 | G | A | 0.091 | 0.699 | 1.489 | 0.640 |
| HMIP | rs73555746 | 135521239 | A | C | 0.004 | 5.583 | 7.085 | 0.432 |
QTLs found to be significantly associated with HbF are denoted with *. MAF = minor allele frequency; Beta = effect size on HbF levels; SE=standard error; P= p-value.
HBB-HBD intergenic region
Mutations within HBB-HBD region have been linked to elevated HbF.(17, 19) We investigated polymorphisms within this region to further characterize the AI haplotype and to identify candidates that may be associated with elevated HbF. An AT motif, (AC)2(AT)9(T)5, 530 bp 5′ to HBB is found with the AI haplotype and postulated to play a role in ameliorating the sickle phenotype due to suppression of HBB by binding the repressor protein BP1.(17) In 6 AI haplotype homozygotes that we examined, all had the previously described AI haplotype AT repeat motif. (17)
In 35 patients, the (TTTTA)6 insertion 1412 bp 5′ of HBB was present . In previous studies, Benin haplotype cases were reported to have a (TTTTA)5 insertion (rs61168339; Table 3).
Table 3.
Polymorphisms in globin gene cluster region detected by Sanger sequencing or TaqMan assays in homozygous sickle cell anemia patients with the Arab Indian haplotype.
|
Location or Gene (# screened) |
Polymorphism or Sequence |
|---|---|
| HBB-HBD intergenic region | |
| −530 5′ to HBB (6) | (AC)2(AT)9T5 |
| −1412 5′ to HBB (35) | G(TTTTA)6TTTTG |
| HBD-HBG1 intergenic region | |
| −68 5′ to HBD (137) | C>T |
| HBG1 promoter | |
| −222 5′ to HBG1 (30) | AAGC insertion |
| HBG2 promoter | |
| −158 5′ to HBG2 (137) | C>T |
| LCR | |
| HS-2 Core (30) | (TA)10(CA)2(TA)2CG(TA)12 |
In 6 cases with a mean HbF level of 24.5%, the sequence of the KLF1 binding site in the HBB promoter (CCACACCCT) was identical to the reference sequence (GenBank U01317) suggesting a lack of novel polymorphisms that could account for the uniquely elevated HbF found in the AI haplotype.
A C>T SNP 68 bp 5′ to HBD that we previously found to be a specific marker for the AI haplotype was homozygous in all 137 cases.(16)
HBG1 and HBG2 promoters
Thirty patients were screened for promoter mutations. One polymorphism, a C>G 369 bp 5′ to HBG1 (rs2855040), was identified in the HBG1 promoter region and a 4 bp (AAGC) insertion at 222 bp 5′ to HBG1 was found. These findings are not unique to sickle patients with the AI haplotype and are therefore unlikely to have a significant role in modulating the elevated HbF that distinguishes the AI haplotypes from other HBB haplotypes. The HBG2 promoter contained the Xmn1 polymorphism, C>T SNP 158 bp 5′ to HBG2 (rs7482144), which is a marker for the AI haplotype.
LCR core regions of HS-2, 3, and 4
Thirty patients were studied. The core regions of HS-3 and HS-4 were identical to the GenBank U01317 reference sequences. In the core of HS-2, the unique TA)10(CA)2(TA)2CG(TA)12 motif known to be typical of the AI haplotype, was present.
KLF1
Forty-four cases with HbF levels between 9 and 30% were studied . Table 4 shows the HbF levels and the SNPs detected in KLF1 exons. None of the SNPs in KLF1 we studied showed an association with elevated HbF in AI haplotype patients and the polymorphisms we detected were “neutral.” SNPs previously associated with increased HbF were not present.(20-22) There was no significant difference in HbF level between the 12 patients, who were heterozygous for the promoter nt −188 C>G polymorphism (rs3817621) and had HbF of 13.2±6.8%, compared with the 10 cases without the same promoter polymorphism with HbF of 16.4±4.5%.
Table 4.
KLF1 polymorphisms in sickle cell anemia.
| M39L | S102P | F182L | Number | Gender (M/F) |
Age (yrs.) (Mean±SD) |
HbF (Mean±SD) |
|||
|---|---|---|---|---|---|---|---|---|---|
| homo | hetero | homo | hetero | homo | hetero | ||||
| − | − | + | − | − | − | 2 | 1/1 | 17.5±2.1 | 15.3±5.2 |
| − | − | − | + | − | − | 17 | 11/6 | 26.2±10.4 | 16.3±7.2 |
| − | + | − | + | − | − | 3 | 1/2 | 30±10.5 | 9.3±0.6 |
| − | − | − | + | − | + | 2 | 0/2 | 37±24 | 26.1±4.0 |
| − | − | + | − | − | + | 1 | 0/1 | 25 | 23 |
| − | − | − | − | − | − | 18 | 11/7 | 24.8±7.3 | 15.6±5.1 |
S102P: T>C (rs2072597); M39L: A>C (rs112631212); F182L: T>C (rs2072596)
DLX4 (BP1)
Twenty-three patients were studied: 4 were heterozygous for SNP rs58769681 (G>A) in the 5′UTR of DLX4; 2 were heterozygous for rs61749026 (A>G; N44S); no mutation was present in the remaining 17 patients.
Discussion
The distribution of HbF concentrations in AI haplotype sickle cell anemia was significantly different than that of patients with African-derived HBB haplotypes suggesting differential regulation of HBG2 and HBG1 in these genetically distinct populations (Figure 1). We genotyped the known cis- and trans-acting elements associated with HbF regulation to try and explain these differences.
Cis-acting elements
Unique polymorphisms cis to HBB and associated with the HbS gene distinguish the AI from other haplotypes. Every patient was homozygous for the cis-acting elements we examined suggesting that although these elements are linked to the sequences, or are the sequences causing high levels of expression of the γ-globin genes, they alone cannot account for the heterogeneity of HbF among AI haplotype carriers.
We confirmed the presence of the (AC)2(AT)9(T)5 motif in the HBB-HBD intergenic region. This is a binding site for BP1 (DLX4), which binds more tightly to this region in the AI haplotype compared with other haplotypes, and reduces βS-globin gene expression.(17, 23) This particular AT repeat motif was associated with the lowest HBB mRNA expression in differentiated MEL cells.(24) A similar AT motif was associated with β thalassemia in carriers without a detectable mutation in HBB.(25, 26) HBB suppression might play a role in the high level of γ-globin gene expression found with the AI haplotype. The (TTTTA)6 insertion 1412 bp upstream of HBB is also seen with the AI haplotype but the functional significance of this insertion/deletion polymorphism is unknown.(27) Functional studies may help to clarify whether the differences in the copy number of this motif can explain the HbF difference between these two HBB haplotypes.
The discovery of the Corfu δβ thalassemia deletion first suggested that the HBD-HBG1 intergenic region had a potential role in activating the γ-globin genes and repressing the downstream β-globin gene.(19) A 3.5 kb region near the 5′ portion of HBD binds the HBG silencer BCL11A, GATA-1 and HDAC1. Its deletion delayed HBG to HBB switching.(28) A functional C>T SNP 68 bp 5′ to HBD that is a specific marker for the AI haplotype was previously reported; whether this affects HBG expression is unknown.(16)
The −158 C>T SNP (rs7482144) is present in the AI haplotype and is associated with high HBG2 expression in this haplotype and the African-derived Senegal haplotype. However, the functional role of this polymorphism is ill-defined.(29, 30) Variation in HS-2 of the LCR has been reported to be associated with HbF levels in sickle cell anemia.(31) Further studies are needed to clarify whether or not the HS-2 signature sequence found in the AI haplotype plays a role in γ-globin gene regulation.
Trans-acting elements
QTL on chr6q22-23 (HBS1L-MYB)(32, 33) and chr2p16.1 (BCL11A) (2, 4, 6, 34) modulate HBG expression in all populations studied, albeit with different effect sizes. SNPs in these loci accounted for 15 to 30% of the HbF phenotypic variation in African American sickle cell anemia.(2, 4-7, 35) Among AI haplotype homozygotes, variants in BCL11A accounted for 7.5% of HbF variance. SNPs in HBS1L-MYB that have been associated with HbF level in other populations were not associated with HbF in the AI haplotype. However the minor allele frequencies of these HBS1L-MYB SNPs were about 1% so that a SNP of importance is likely to be rare and have a small effect in this population. It remains possible that AI haplotype patients with high HbF have novel functional in BCL11A and HBS1L-MYB that are not tracked by the SNPs we genotyped. Alternatively, SNPs in these genes play a lesser role in HbF variance in the AI haplotype than in African haplotypes.
KLF1, a regulator of BCL11A, also mediates hemoglobin switching.(22, 36) Its polymorphisms have been associated with the phenotype of hereditary persistence of HbF.(20, 22, 37) We did not find mutations in the KLF1 zinc finger domains that were previously associated with increased HbF in the 44 cases we studied for these polymorphisms. The SNPs that were present did not show a correlation with HbF in the AI haplotype patients we studied, and were likely to be “neutral.” Although a few SNPs were found in BP1 (DLX4) in some patients, their role in HbF regulation is unclear.
Rarely, African American HbS homozygotes can have a baseline HbF similar to that associated with the AI haplotype. These individuals more commonly have the minor alleles of BCL11A and the HBS1L-MYB intergenic region compared with counterparts with low HbF.(38) In some instances, rare missense mutations in MYB have been associated with higher HbF in African Americans with sickle cell anemia.(2) We did not test these specific variants in AI haplotype patients.
Conclusions
To increase our understanding of HbF regulation in AI haplotype HbS homozygotes we compared polymorphisms in selected regions that define this haplotype and also examined known trans acting elements associated with HbF expression. Although all polymorphisms were not studied in all patients, previous studies of the AI haplotype and our results suggest that any variants in these sequences that might be detected if a larger patient sample were to be examined are apt to be rare.
HbF levels are determined by interactions of cis and trans acting elements and are likely to be, modulated by stress erythropoiesis.(39, 40) Carriers of the AI haplotype and sickle cell trait have nearly normal HbF levels(11) suggesting that homozygosity for the AI haplotype and/or the presence of hemolytic anemia is required for the clinical expression of the high HbF phenotype. Some elements cis to HBB in the AI haplotype, like the −530 AT motif, down-regulate HBB; others, such as the −68 C>T HBD promoter SNP might down regulate HBD expression.(16, 17) Coupled with continued expression of HBG2, this might reduce post-transcriptional competition and enhance the stability of γ-globin RNA.(41, 42) Known trans-acting QTL accounted for only 8.8% of HbF variance in this population and SNPs in BCL11A explained only 7.5% of the variance, or about half that found in the largest reported study of African-origin patients. (4) Additional QTL could have evolved in the Arab population and might be discovered by sufficiently powered genome-wide association studies or by whole genome sequencing of highly selected patients. Determining transcription factor binding patterns and histone modification in the HBB gene cluster region in AI haplotype cases could provide a better understanding of the influence of different cis acting elements on HbF globin gene expression.
The variation in HbF among patients with the AI haplotype suggests that other genetic elements modulate the effects of the known cis- and trans-acting regulators. These regulatory elements, which remain to be discovered, might be specific for the Saudi and some other populations where HbF levels are especially high.
Supplementary Material
Acknowledgments
Supported by R01 HL 068970, RC2 HL 101212 and R01 87681 and King Abdulaziz City for Science and Technology (KACST ARP-30-367)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict-of Interest Disclosure: The authors declare no competing financial interests.
References
- 1.Steinberg MH, Sebastiani P. Genetic modifiers of sickle cell disease. Am. J. Hematol. 2012;87:795–803. doi: 10.1002/ajh.23232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galarneau G, Palmer CD, Sankaran VG, Orkin SH, Hirschhorn JN, Lettre G. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat. Genet. 2010;42:1049–51. doi: 10.1038/ng.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dover GJ, Boyer SH, Charache S, Heintzelman K. Individual variation in the production and survival of F cells in sickle-cell disease. N. Engl. J. Med. 1978;299:1428–35. doi: 10.1056/NEJM197812282992603. [DOI] [PubMed] [Google Scholar]
- 4.Lettre G, Sankaran VG, Bezerra MA, Araujo AS, Uda M, Sanna S, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11869–74. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae HT, Baldwin CT, Sebastiani P, Telen MJ, Ashley-Koch A, Garrett M, et al. Meta-analysis of 2040 sickle cell anemia patients: BCL11A and HBS1L-MYB are the major modifiers of HbF in African Americans. Blood. 2012;120:1961–2. doi: 10.1182/blood-2012-06-432849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solovieff N, Milton JN, Hartley SW, Sherva R, Sebastiani P, Dworkis DA, et al. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5′ olfactory receptor gene cluster. Blood. 2010;115:1815–22. doi: 10.1182/blood-2009-08-239517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sedgewick A, Timofeev N, Sebastiani P, So JCC, Ma ESK, Chan LC, et al. BCL11A (2p16) is a major HbF quantitative trait locus in three different populations. Blood Cells, Mol. Dis. 2008;41:255–8. doi: 10.1016/j.bcmd.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makani J, Menzel S, Nkya S, Cox SE, Drasar E, Soka D, et al. Genetics of fetal hemoglobin in Tanzanian and British patients with sickle cell anemia. Blood. 2011;117:1390–2. doi: 10.1182/blood-2010-08-302703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labie D, Srinivas R, Dunda O, Dode C, Lapoumeroulie C, Devi V, et al. Haplotypes in tribal Indians bearing the sickle gene: evidence for the unicentric origin of the beta S mutation and the unicentric origin of the tribal populations of India. Hum. Biol. 1989;61:479–91. [PubMed] [Google Scholar]
- 10.Kulozik AE, Wainscoat JS, Serjeant GR, Kar BC, Al-Awamy B, Essan GJF, et al. Geographical survey of the ás-globin gene haplotypes: Evidence for an independent Asian origin of the sickle-cell mutation. Am. J. Hum. Genet. 1986;39:239–44. [PMC free article] [PubMed] [Google Scholar]
- 11.Miller BA, Salameh M, Ahmed M, Wainscoat J, Antognetti G, Orkin S, et al. High fetal hemoglobin production in sickle cell anemia in the eastern province of Saudi Arabia is genetically determined. Blood. 1986;67:1404–10. [PubMed] [Google Scholar]
- 12.Akinsheye I, Alsultan A, Solovieff N, Ngo D, Baldwin CT, Sebastiani P, et al. Fetal hemoglobin in sickle cell anemia. Blood. 2011;118:19–27. doi: 10.1182/blood-2011-03-325258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adekile AD. Limitations of Hb F as a phenotypic modifier in sickle cell disease: study of Kuwaiti Arab patients. Hemoglobin. 2011;35:607–17. doi: 10.3109/03630269.2011.617230. [DOI] [PubMed] [Google Scholar]
- 14.El-Hazmi MAF. Clinical and haematological diversity of sickle cell disease in Saudi children. J. Trop. Pediatr. 1992;38:106–12. doi: 10.1093/tropej/38.3.106. [DOI] [PubMed] [Google Scholar]
- 15.Little S. Amplification-refractory mutation system (ARMS) analysis of point mutations. Curr. Protoc. Hum. Genet. 2001 May; doi: 10.1002/0471142905.hg0908s07. Chapter 9:Unit 9 8. [DOI] [PubMed] [Google Scholar]
- 16.Alsultan A, Ngo DA, Farrell JJ, Akinsheye I, Solovieff N, Ghabbour HA, et al. A functional promoter polymorphism of the delta-globin gene is a specific marker of the Arab-Indian haplotype. Am. J. Hematol. 2012;87:824–6. doi: 10.1002/ajh.23239. [DOI] [PubMed] [Google Scholar]
- 17.Elion J, Berg PE, Lapoumeroulie C, Trabuchet G, Mittelman M, Krishnamoorthy R, et al. DNA sequence variation in a negative control region 5′ to the beta-globin gene correlates with the phenotypic expression of the beta s mutation. Blood. 1992;79:787–92. [PubMed] [Google Scholar]
- 18.Massey FJJ. The Kolmogorov-Smirnov test for goodness of fit. J. Am. Stat. Assoc. 1951;46:68–78. [Google Scholar]
- 19.Chakalova L, Osborne CS, Dai YF, Goyenechea B, Metaxotou-Mavromati A, Kattamis A, et al. The Corfu deltabeta thalassemia deletion disrupts gamma-globin gene silencing and reveals post-transcriptional regulation of HbF expression. Blood. 2005;105:2154–60. doi: 10.1182/blood-2003-11-4069. [DOI] [PubMed] [Google Scholar]
- 20.Borg J, Papadopoulos P, Georgitsi M, Gutierrez L, Grech G, Fanis P, et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat. Genet. 2010;42:801–5. doi: 10.1038/ng.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat. Genet. 2010;42:742–4. doi: 10.1038/ng.637. [DOI] [PubMed] [Google Scholar]
- 22.Borg J, Patrinos GP, Felice AE, Philipsen S. Erythroid phenotypes associated with KLF1 mutations. Haematologica. 2011;96:635–8. doi: 10.3324/haematol.2011.043265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mpollo MS, Beaudoin M, Berg PE, Beauchemin H, D’Agati V, Trudel M. BP1 is a negative modulator of definitive erythropoiesis. Nucl. Acids Res. 2006;34:5232–7. doi: 10.1093/nar/gkl680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan PK, Ma ES, Philipsen S, Tan-Un KC. The study of sequence configuration and functional impact of the (AC)n(AT)xTy motif in human beta-globin gene promoter. Am. J. Hematol. 2007;82:342–8. doi: 10.1002/ajh.20836. [DOI] [PubMed] [Google Scholar]
- 25.Semenza GL, Delgrosso K, Poncz M, Malladi P, Schwartz E, Surrey S. The silent carrier allele: beta thalassemia without a mutation in the beta-globin gene or its immediate flanking regions. Cell. 1984;39:123–8. doi: 10.1016/0092-8674(84)90197-1. [DOI] [PubMed] [Google Scholar]
- 26.Murru S, Loudianos G, Cao A, Vaccargiu S, Pirastu M, Sciarratta GV, et al. A beta-thalassemia carrier with normal sequence within the beta-globin gene. Blood. 1990;76:2164–5. [PubMed] [Google Scholar]
- 27.Spritz RS. Duplication/deletion polymorphism 5′ - to the human beta globin gene. Nucl. Acids Res. 1981;9:5037–47. doi: 10.1093/nar/9.19.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sankaran VG, Xu J, Byron R, Greisman HA, Fisher C, Weatherall DJ, et al. A functional element necessary for fetal hemoglobin silencing. N. Engl. J. Med. 2011;365:807–14. doi: 10.1056/NEJMoa1103070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller BA, Salameh M, Ahmed M, Olivieri N, Antognetti G, Orkin SH, et al. Analysis of hemoglobin F production in Saudi Arabian families with sickle cell anemia. 1987;70:716–20. [PubMed] [Google Scholar]
- 30.Miller BA, Olivieri N, Salameh M, Ahmed M, Antognetti G, Huisman TH, et al. Molecular analysis of the high-hemoglobin-F phenotype in Saudi Arabian sickle cell anemia. N. Engl. J. Med. 1987;316:244–50. doi: 10.1056/NEJM198701293160504. [DOI] [PubMed] [Google Scholar]
- 31.Oner C, Dimovski AJ, Altay C, Gurgey A, Gu YC, Huisman TH, et al. Sequence variations in the 5′ hypersensitive site-2 of the locus control region of beta S chromosomes are associated with different levels of fetal globin in hemoglobin S homozygotes. 1992;79:813–9. [PubMed] [Google Scholar]
- 32.Creary LE, Ulug P, Menzel S, McKenzie CA, Hanchard NA, Taylor V, et al. Genetic variation on chromosome 6 influences F cell levels in healthy individuals of African descent and HbF levels in sickle cell patients. PLoS One. 2009;4:e4218. doi: 10.1371/journal.pone.0004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thein SL, Menzel S, Peng X, Best S, Jiang J, Close J, et al. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11346–51. doi: 10.1073/pnas.0611393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1620–5. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thein SL, Menzel S, Lathrop M, Garner C. Control of fetal hemoglobin: new insights emerging from genomics and clinical implications. Hum, Mol, Genet. 2009;18:R216–R23. doi: 10.1093/hmg/ddp401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011;118:2044–54. doi: 10.1182/blood-2011-03-331371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallienne AE, Dreau HM, Schuh A, Old JM, Henderson S. Ten novel mutations in the erythroid transcription factor KLF1 gene associated with increased fetal hemoglobin levels in adults. Haematologica. 2012;97:340–3. doi: 10.3324/haematol.2011.055442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akinsheye I, Solovieff N, Ngo D, Malek A, Sebastiani P, Steinberg MH, et al. Fetal hemoglobin in sickle cell anemia: Molecular characterization of the unusually high fetal hemoglobin phenotype in African Americans. Am. J. Hematol. 2012;87:217–219. doi: 10.1002/ajh.22221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005;33:259–71. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mabaera R, West RJ, Conine SJ, Macari ER, Boyd CD, Engman CA, et al. A cell stress signaling model of fetal hemoglobin induction: what doesn’t kill red blood cells may make them stronger. Exp. Hematol. 2008;36:1057–72. doi: 10.1016/j.exphem.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 41.Russell JE. A post-transcriptional process contributes to efficient gamma-globin gene silencing in definitive erythroid cells. Eur. J. Haematol. 2007;79:516–25. doi: 10.1111/j.1600-0609.2007.00970.x. [DOI] [PubMed] [Google Scholar]
- 42.Dover GJ, Boyer SH. Fetal hemoglobin-containing cells have the same mean corpuscular hemoglobin as cells without fetal hemoglobin: a reciprocal relationship between gamma- and beta-globin gene expression in normal subjects and in those with high fetal hemoglobin production. Blood. 1987;69:1109–13. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.