Abstract
Marginal periodontitis is not a homogeneous disease but is rather influenced by an intricate set of host susceptibility differences as well as diversities in virulence among the harbored organisms. It is likely that clonal heterogeneity of subpopulations with both high and low levels of pathogenicity exists among organisms harbored by individuals with negligible, slight, or even severe periodontal destruction. Therefore, specific virulent clones of periodontal pathogens may cause advanced and/or aggressive periodontitis. Porphyromonas gingivalis is a predominant periodontal pathogen that expresses a number of potential virulence factors involved in the pathogenesis of periodontitis, and accumulated evidence shows that its expression of heterogenic virulence properties is dependent on clonal diversity. Fimbriae are considered to be critical factors that mediate bacterial interactions with and invasion of host tissues, with P. gingivalis shown to express two distinct fimbria-molecules, long and short fimbriae, on the cell surface, both of which seem to be involved in development of periodontitis. Long fimbriae are classified into six types (I to V and Ib) based on the diversity of fimA genes encoding FimA (a subunit of long fimbriae). Studies of clones with type II fimA have revealed their significantly greater adhesive and invasive capabilities as compared to other fimA type clones. Long and short fimbriae induce various cytokine expressions such as IL-1α, IL-β, IL-6, and TNF-α, which result in alveolar bone resorption. Although the clonal diversity of short fimbriae is unclear, distinct short fimbria-molecules have been found in different strains. These fimbriae variations likely influence the development of periodontal disease.
Keywords: P. gingivalis, long fimbriae, short fimbriae, FimA, genotype, mfa1
Porphyromonas gingivalis, a Gram-negative black-pigmented anaerobic rod residing in subgingival biofilms, is widely recognized as a contributor to development of periodontal infections together with other oral pathogens (1–5). The species has also been reported to cause extraoral infections (6–9) and is suggested to play a role in the development of coronary heart disease, stroke, and diabetes mellitus, as well as preterm delivery of low birth-weight infants (7–13).
Although the central cause of periodontitis is loss of a healthy balance between microbial virulent agents and host immunity in host–parasite interactions, there are marked differences in progression rate and severity, as well as response to therapy in individuals affected by this infectious disorder (3, 5). Thus, periodontitis is not considered to be a homogeneous disease but is rather influenced by an intricate set of host susceptibility differences along with diversities in virulence among the harbored organisms. Indeed, P. gingivalis is present in periodontal pockets undergoing destruction as well as in healthy gingival margins (14, 15). Furthermore, clonal heterogeneity of subpopulations with both high and low levels of pathogenicity has been suggested to exist among periodontal pathogens harbored by individuals with negligible, slight, or even severe periodontal destruction. Therefore, specific virulent clones of the pathogens may be the cause of advanced and/or aggressive periodontitis.
P. gingivalis harbors an arsenal of virulence factors, including fimbriae, cysteine proteinases, hemagglutinins, and lipopolysaccharide (LPS), which along with its many interactions with the host immune system strongly support its potency as a pathogen (2, 16–18). Among those, fimbriae are a critical factor for colonization of P. gingivalis in subgingival regions, as they promote both bacterial adhesion to and invasion of targeted sites. Fimbriae variations and their effects on bacterial virulence are discussed below.
Pili/fimbriae
Bacteria commonly express proteinaceous appendages on their outer surfaces. One class of extracellular polymers, known as pili or fimbriae (non-flagellar appendages), is used in attachment to and invasion of host cells, biofilm formation, cell motility, and transport of proteins and DNA across cell membranes. Pili and fimbriae are synonymous terms, with both commonly used, and are derived from Latin; pili for ‘hair’ or ‘fur’ and fimbriae for ‘fringe’. Since the first observations of these non-flagellar peritrichous appendages in the early 1950s, several distinct types of structures have been identified and characterized in Gram-negative bacteria, and later in Gram-positive bacteria (19–21).
Aggressive bacterial virulence factors that promote adherence and colonization of host organisms also include the well-studied protein adhesins, toxins, and translocated effector proteins. In all life forms, the general secretory pathway (GSP) (19, 20) provides a generic mechanism of protein transport across the cytoplasmic membrane. For secretion of proteins in Gram-positive species, the GSP may be sufficient, while Gram-negative bacteria have a more complex cell membrane and face the specific problem of management of protein transport across the outer membrane (OM) of their bacterial cells. The OM functions as a protective barrier against various antimicrobial host defenses, as well as a structure that enables the organisms to effectively colonize host cells and tissues. Therefore, Gram-negative bacteria have evolved special systems (20) for secretion and transport of proteins across the cell envelope.
Adhesins are a group of extracytoplasmic proteins found in pathogenic bacteria as well as environmental species, and they can be divided into two major classes. Fimbrial adhesins are composed of heteropolymers of pili subunits, while non-fimbrial adhesins consist of homotrimers or a single protein. Classification of the different types of pili/fimbriae has been established based on the different biosynthetic pathways of protein secretion and assembly (21).
The most studied fimbrial adhesins are type I pili [chaperone-usher (CU) pili], expressed by the Pap pili of uropathogenic Escherichia coli, and commensal E. coli isolates; type IV pili, expressed by the enteropathogenic E. coli, Pseudomonas, and Neisseria species, and curli pili, which are expressed by some strains of E. coli. In electron microscopic observations, type I pili have a peritrichous appearance on the cell surface, with rigid and rod-like structures 1–2 µm in length, while type IV pili are similar in length, though they appear to be more flexible (22), and curli pili have a coiled structure. All three types are formed by a non-covalent association of pilin subunits that comprise regular polymeric structures (20, 21). In addition, two other types have been described, type III secretion needle and type IV secretion pili (23).
P. gingivalis fimbriae
P. gingivalis fimbriae seem to participate in nearly all interactions between the bacterium and the host, as well as with other bacteria (24). This pathogen expresses two distinct fimbria-molecules on its cell surface, one of which is composed of a subunit protein (named FimA or fimbrillin) encoded by the fimA gene, and termed long or long fimbriae, while the other consists of a subunit Mfa protein encoded by the mfa1 gene and termed short, minor, or Mfa fimbriae (henceforth referred to as simply long and short fimbriae, respectively, in this review) (25). Short fimbriae are homopolymers of a subunit protein encoded by mfa1, with an apparent molecular mass of 75 kDa and antigenicity distinct from long fimbriae (26). Short fimbriae are shorter than long fimbriae and can only be visualized when the latter are absent. Both are apparently involved in the development of periodontitis (27).
Long fimbriae
The fimbriae of P. gingivalis have been a focus of research in periodontal microbiology and pathogenesis for many years since Slots and Gibbons (28) first published their paper ‘Bacteroides melaninogenicus subsp. asaccharolyticus attachment to oral surfaces, and its possible role in colonization of the mouth and periodontal pockets’ in 1978. Earlier, bacterial surface components of other species that mediated attachment to host tissues were recognized as important pathogenic determinants (29). Electron microscopic observations of several black-pigmented anaerobic rods revealed that the organisms exhibited fine fibrillar appendages arranged in a peritrichous manner on the cell surface (30, 31). Some years later, Okuda and co-workers (31) noted that cell surface morphology and adherence to erythrocytes and human buccal epithelial cells vary among Bacteroides strains, including B. gingivalis. In negatively stained preparations, a dense network of fibers radiating from all dimensions of the cell surface of all strains examined was regularly observed. However, differential expression of fimbriae among strains of P. gingivalis has been reported (32).
Ultrastructural examinations of P. gingivalis strains have shown that peritrichous fimbriae vary in length from 0.3 to 3 µm and are 5 nm wide (2, 33). These structures have been classified as major or long fimbriae (FimA), based on their fimbrillin monomer composition, and range in size from 41 to 49 kDa (34–36). Even though ultrastructural studies were introduced 30 years ago, such examinations are still often used in combination with others for revealing additional information about this species (37). Various investigators have used purified fimbriae, recombinant fimbrillin, and antibodies to show that P. gingivalis fimbriae mediate bacterial adherence to a wide variety of molecules and oral substrates. These include salivary molecules, such as proline-rich proteins, proline-rich glycoproteins, statherins, oral epithelial cells, fibrinogen, fibronectin, and lactoferrin, and bacteria, such as oral streptococci and actinomyces species, which will be detailed (later in the article). Thus, long fimbriae are considered to be directly responsible for many of the adhesive properties of the organism, binding specifically to and activating various host cells, such as human epithelial, endothelial, and spleen cells, as well as peripheral blood monocytes, resulting in the release of inflammatory cytokines and several different adhesion molecules (4, 17, 27, 38).
fimA genotypes and virulence
FimA is encoded by the fimA gene and occurs as a single copy in the chromosome of P. gingivalis (34, 39, 40). Based on its nucleotide sequence variation, the gene has been classified into six types (I, Ib, II, III, IV, V) (18, 39, 41, 42). That variation led to development of a PCR-based fimA genotyping method that is used for detection of possible relationships among the different genotypes, and virulence and disease. This method has been used in both experimental and clinical studies.
Nakagawa et al. (43) demonstrated that recombinant FimA protein corresponding to fimA genotype II has a greater ability to adhere to and invade human epithelial cells than FimA corresponding to the protein from other genotypes. The pathogenicity of the various fimA genotypes has also been evaluated in animal models, with fimA genotypes II, Ib, and IV shown to cause stronger infectious symptoms and inflammatory changes as compared to strains harboring fimA genotypes I and III (27, 44–46). On the other hand, Umeda et al. (47) found no significant difference between strains with different fimA genotypes in regard to adhesion to and invasion of epithelial KB cells. In addition, mutants in which the fimA type I gene was substituted with type II showed enhanced bacterial adhesion/invasion. In contrast, substitution of type II with type I resulted in diminished efficiency, supporting the notion that type II fimbriae are a critical determinant of virulence (48).
Results of several clinical studies also support findings that nucleotide variation of the gene is likely related to virulence. In chronic marginal periodontitis, P. gingivalis isolates with fimA genotypes II, IV, and Ib have been shown to be significantly more prevalent than isolates with other genotypes (27, 41, 42, 49–53). Also, studies of the pathogenic potential of distinct fimA genotypes in patients suffering from aggressive periodontitis have indicated that genotype II strains are more prevalent (54). In contrast, isolates with fimA genotype I are the most prevalent among P. gingivalis-positive healthy adults, followed by genotype V (52). In addition, fimA genotyping of cultured clinical strains of P. gingivalis sampled from individuals with periodontitis support previous findings that genotypes II, IV, and Ib are related to virulence (50).
A common observation among studies utilizing the fimA genotyping method is positive PCR cross reactivity to several different primer sets. Various explanations have been proposed, such as the presence of several different genotypes colonizing the same site (49, 51) and possible existence of new unidentified genotypes shown by the existence of fimA non-typable strains in some studies (41, 42, 49, 51, 53). However, genotyping of cultured P. gingivalis strains has detected only typable isolates and revealed similar findings as presented in clinical studies (50). Also, in endodontic infections, a fimA genotyping method resulted in similar findings as with strains from periodontal infections (55). To explain the observation of strains with multiple positive PCR reactions to several of the fimA primer sets, Enersen and co-workers (50) sequenced a selected number of isolates with new primers and then focused mainly on those showing multiple positive PCR reactions in a genotyping study. Their analysis verified a conserved fimA gene with only minor variations among the examined strains.
Multiple sequence alignments were presented by Enersen and co-workers (50), indicating that the genotyping method should be reevaluated partly due to the scant genetic variation between isolates within each genotype and between the different groups, in combination with the small differences in design of the primer sets. This was most important for detection of fimA I, Ib, and II. Their findings led to further development of new primers for detection of genotype II that was claimed to increase the accuracy of detection of the most prevalent genotype in diseased periodontal sites (56). A few studies suggested that other characteristics besides fimA gene variation may be responsible for the adhesion and invasion abilities (27, 47). That was supported by Inaba and co-workers (57), who reported heterogenic virulence among fimA genotype II strains, indicating that the variations in pathogenic potential and invasive efficiency were related to extracellular secreted gingipains, which will be discussed later.
In spite of the discrepancies related to the fimA genotyping method, a large number of experimental and clinical studies have indicated that some fimA genotypes may be important determinants of virulence for P. gingivalis. In addition, they may have a possible role in initiation and progression of cardiovascular diseases (see ‘Long fimbriae and cardiovascular diseases’ section).
Protein structure of long fimbriae
The primary protein sequences of FimA share no significant homology with other described fimbrial proteins, indicating that P. gingivalis may possess a unique class of fimbria subunits (2, 36, 37, 39, 50).
The presence of an extremely long signal peptide, and requirements for Arg- and Lys-specific proteases (gingipains) for extracellular maturation, indicates that FimA is a novel group of fimbriae different from the type I and IV families (20). Shoji and co-workers (37) demonstrated that the major component proteins of the two cell surface structures, FimA and the 75-kDa protein related to short fimbriae, which are detailed later, seem to utilize the lipoprotein transport system with signal peptidase II, indicating a novel transport and assembly machinery with extracellular proteolytic polymerization.
In contrast, the major protein component of cell surface filaments of type I pili makes use of signal peptidase I for translocation across the cytoplasmic membrane, while type IV pili are dependent upon type IV-specific signal peptidases for their biogenesis.
The hypothesis and presented data supporting the variable virulence potential among different fimA genotypes of P. gingivalis imply a possible role for the tertiary structure in the function of FimA. However, the level of transcription of the gene is also an important factor (32).
Translation of fimA nucleotide sequences performed in a study by Enersen et al. (50) resulted in the same number of primary protein structures as sequence variants. Although the fimA gene was conserved, there were some minor variations between isolates belonging to the same genotype, resulting in corresponding variations in the primary protein sequence of the FimA monomer, which was also shown by Fujiwara and co-workers (39). Whether these mutations result in a FimA monomer with a changed structure that influences the pathogenicity of the isolate may partly depend on how the secondary structure of the molecule folds into a tertiary structure.
Presently, the tertiary structure of FimA is unknown, and an experimental structure resembling a protein with high homology to FimA has not been found (50, 57). Advanced bioinformatic data yet unpublished (58), based on results presented by Shoji and co-workers (37) as well as other bioinformatic sources, indicate that the structure of FimA may resemble the structure of protein NP_809975 of Bacteroides thetaiotaomicron found in the RCSB Protein Data Bank (structure 3GF8). From this structure, it is possible to speculate that fimbrillin generates multimers employing a ‘donor strand exchange’ in a manner resembling E. coli type I pili (59). FimA also appears to be hypervariable, which is consistent with its importance as a virulence factor for the species. Furthermore, the FimA sequence and subunits are changing rapidly over short evolutionary distances, making an experimental structure less useful, since the bacterium may be under positive selective pressure.
Based on alignments with similar sequences, it seems that conservation is highest in the signal sequence of fimA in P. gingivalis (58). This is in contrast to other bacteria in which the signal sequences have been found to be less conserved than the remaining part of the primary protein (58, 60), also indicating positive selection.
Although it has been claimed that FimA is unique due to lack of sequence homology between FimA and other fimbrial proteins, corresponding sequence alignments of fimbriae from other Bacteroides and parabacteroides species show a similarity to FimA, but the sequence homology may quickly become lost in this protein family due to extensive positive selection.
Involvement of fimbriae in biofilm formation
Biofilm formation is a complex process involving reversible and irreversible bacterial attachment, microcolony formation, formation of a stable three-dimensional structure, and dispersion (61). Interspecies interactions help to develop the complex bacterial consortia in the gingival crevice. In the early phase, the initial bacterial colonizers (early colonizers), including streptococci, attach to available oral surfaces such as a salivary pellicle-coated tooth surface. Thereafter, later colonizers attach to the antecedent organisms and assemble into polymicrobial biofilm, which is mediated by co-adhesion (co-aggregation) with other bacterial species. P. gingivalis is able to aggregate with various oral Gram-positive and -negative species (62). Long fimbriae extend a significant distance from the bacterial cell wall, which suggests that they are the first bacterial components to interact with other bacteria as well as host cells. Indeed, P. gingivalis long fimbriae have been reported to mediate coadhesion with Actinomyces viscosus (63), Treponema denticola (64), Streptococcus gordonii (65), and Streptococcus oralis (66) via specific interactions with their receptors, including dentilisin of T. denticola, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of S. gordonii and S. oralis (25). GAPDH is a well-characterized glycolytic protein involved in energy production and has been suggested to be a multi-functional house-keeping protein that is conserved by eubacteria and eukaryotic cells. The interaction between long fimbriae and GAPDH is the initial contact event that allows for localization of P. gingivalis on the streptococcal surface (67). The binding domains of the subunit protein FimA that mediate attachment to streptococci are localized to a C-terminal region spanning amino acid residues 266–337 (68). Human GAPDH has also been shown to bind to long fimbriae (69).
Short fimbriae reportedly mediate co-adhesion between P. gingivalis and S. gordonii via adhesin-receptor interactions with streptococcal SspA and SspB surface proteins (antigen I/II family) (61). SspA and B bind to short fimbriae, which increases binding avidity with a higher affinity than that of GAPDH to long fimbriae. Interestingly, following the development of a P. gingivalis–S. gordonii community, mfa1 expression is down-regulated, presumably to alter the adhesin requirements of the antecedent organisms, as the streptococcal substrate becomes unavailable to later arriving P. gingivalis (70). The resulting phenotypic adaptation of P. gingivalis along with production of signaling molecules promote community development by recruitment of additional P. gingivalis cells from the planktonic phase (61).
The roles of long and short fimbriae in biofilm formation are likely different. The effects of a set of fimbriae as well as gingipains (Rgp and Kgp) were examined with regard to homotypic biofilm formation using deficient mutants (71). Those results suggested that long fimbriae promote initial biofilm formation and then exert a restraining regulation on biofilm maturation, whereas short fimbriae and Kgp have suppressive and regulatory roles during biofilm development. Furthermore, Rgp likely controls microcolony morphology and biovolume. Collectively, these molecules seem to act in a coordinated manner to regulate the development of mature biofilm.
Streptococcus cristatus is a later colonizer of tooth surfaces and attaches to P. gingivalis long fimbriae. However, arginine deiminase on the surface of S. cristatus initiates a signal transduction cascade in P. gingivalis that down-regulates fimA expression, resulting in fewer long fimbriae present on the P. gingivalis surface and no community formation between these microbes (72). Similarly, a secreted/excreted arginine deiminase from Streptococcus intermedius reportedly represses the expressions of fimA and mfa1 (73), while it was also shown that this arginine deiminase prevents mono-species biofilm development by P. gingivalis, because P. gingivalis auto-aggregation is attributable to FimA protein (74, 75).
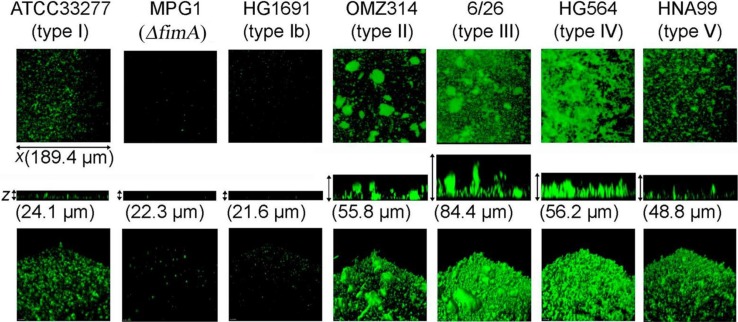
Long fimbria variations interfere with the structures of biofilms formed by P. gingivalis. The effects of such variations on homotypic biofilm formation were examined using representative strains of each long fimbria type (I to V and Ib) (76). Biofilm structures formed by the six representative long fimbria type strains were apparently different (Fig. 1), and their characteristic features were confirmed to be closely related to long fimbria type in assays that utilized mutants with fimA substituted from type I to II and from type II to I.
Fig. 1.
Homotypic biofilm formation by P. gingivalis strains with different types of long fimbriae and FimA (type I)-deficient mutant. P. gingivalis strains shown in the figure were stained with fluorescein (green). Biofilms developed on cover-glasses were observed with a confocal laser scanning microscope. Optical sections were obtained along the z-axis at 0.7-µm intervals, and three-dimensional images were reconstructed with imaging software. Data presented here were reproduced from Ref. 71, with permission.
Induction of inflammatory responses by fimbriae
Toll-like receptors (TLRs) are a class of proteins that play a key role in the innate immune system. A previous study that employed a P. gingivalis mutant lacking long fimbriae identified reduced alveolar bone loss in a gnotobiotic rat model as compared with the wild type, while the inflammatory role of long fimbriae in atherosclerosis was also shown (77, 78). Since presentation of those findings, the effects of long fimbriae on immune responses have become well established (79).
TLRs co-cluster with CD14, CD36, CD55 (decay accelerating factor), complement receptor 3 (CR3; CD11b/CD18), CXC-chemokine receptor 4 (CXCR4), and growth differentiation factor 5 (GDF5) (80, 81). Long fimbriae have been shown to stimulate nuclear factor-κB (NF-κB) via TLR 2 and CD14, which results in induction of cytokines involved in bone resorption, such as tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), IL-8, and IL-6 (80–84). Although CD14 is an essential co-receptor for activation of epithelial cells by long fimbriae (80), it was found that the cytokine responses (IL-6, IL-8, granulocyte-macrophage colony-stimulating factor, and TNF-α) of primary gingival epithelial cells to P. gingivalis are modest because of a lack of membrane-associated CD14 (85). In contrast, monocytes with CD14 respond vigorously to long fimbriae and secrete substantial quantities of IL-6, IL-8, and TNF-α (85). Long fimbriae also induce IL-1β, IL-8, MCP-1 (define), ICAM-1(vascular cell adhesion molecule-1), and E-selectin in human aortic endothelial cells (86). Although long fimbriae are only one of several inflammatory molecules of P. gingivalis, long fimbriae-deficient mutants showed significantly weaker cytokine responses as compared to wild-type strains (48). This is possibly due to the fact that long fimbriae are the most exterior components and likely the first to interact with host cell receptors, which is followed by initiation of intracellular signaling cascades.
Other studies have reported that short fimbriae strongly interacted with TLR2 and CD14, but not TLR4, and induced the expressions of cytokines, including IL-1α, IL-β, IL-6, and TNF-α, in both human monocyte cell lines and mouse macrophages (87, 88). In another study, P. gingivalis ATCC33277T significantly induced periodontal bone loss in a mouse model, which was clearly suppressed by mfa1 deletion in contrast to fimA deletion (74).
Immune subversion mediated by long fimbriae
TLR signaling pathways crosstalk with the complement system, which is now recognized to exert functions above and beyond simple pathogen tagging and elimination (89). Recent evidence shows that P. gingivalis can suppress cell-mediated immunity by reducing the levels of interferon (IFN)-γ, another activator of cell-mediated immunity (90). P. gingivalis long fimbriae interact with CR3 to activate extracellular signal-regulated kinase 1/2 signaling, which inhibits IL-12 production mediated by TLR 2 signaling (91). In addition, the interaction of P. gingivalis or its purified fimbriae with CR3 inhibits the immune ability of LPS from other bacteria, such as A. actinomycetemcomitans, to induce IL-12 and IFN-γ in mouse macrophages or human monocytes. IL-12 is a key cytokine involved in pathogen clearance via its regulatory effects on the production of IFN-γ, a potent activator of macrophage microbicidal activity (92). Thus, CR3 activation, which signals cross talking with TLR 2 pathways and IL-12 inhibition, promotes the survival of P. gingivalis in vitro and in vivo (93).
Long fimbriae interact with CXCR4, a TLR2-associated receptor, which limits TLR2 activation in human monocytes and mouse macrophages. Furthermore, long fimbriae induce CXCR4-mediated activation of cAMP-dependent protein kinase A, which in turn inhibits TLR2-induced NF-κB activation in response to P. gingivalis (94). These results suggest that long fimbriae enable P. gingivalis to resist clearance in vitro and in vivo, promoting its adaptive fitness. In addition, Type II fimbriae from strain OMZ314 were shown to significantly induce cytokine expressions (48), thus the immune subversion potential may vary among fimA types, though definitive findings have not been presented.
Long fimbriae and cardiovascular diseases
Long fimbria-deficient mutants were found to be relatively avirulent as compared to wild-type strains of P. gingivalis with regard to accelerating atherosclerotic plaque formation (79). Recently, oral-hematogenous spreading of P. gingivalis has received special attention for its possible association with several types of cardiovascular diseases. Several studies have reported detection of P. gingivalis in specimens collected from patients with cardiovascular diseases. In one of those, 42% of endarterectomy specimens showed a histological association of P. gingivalis with ulcer and thrombus formation (95), while another utilized a PCR assay method and found that 26% of carotid endarterectomy specimens were positive for the 16S rRNA fragment of P. gingivalis (96). Additional PCR analyses demonstrated that approximately 10% of studied heart valves and atheromatous plaque specimens were positive for P. gingivalis (97–99). On the other hand, Kozarov et al. (100) reported that the detection rate of P. gingivalis in atheromatous plaque specimens was approximately 90%, though they also noted that the rate for specimens taken from young patients was approximately 20%. Together, these results indicate localization of P. gingivalis in cardiovascular lesions, suggesting its association with development of cardiac diseases.
Long fimbriae were also shown to be associated with a necessary initial event in the development of atherogenesis by stimulating endothelial cell activation (101), while fimbria-mediated invasion was found to up-regulate the expressions of genes related to inflammation in aortic endothelial cells, leading to accelerated inflammatory responses directly in the aorta (102). The association of fimA genotype with virulence for cardiovascular diseases has been studied by several researchers. Pérez-Chaparro and co-workers (103) investigated seven blood isolates of P. gingivalis from dental patients who underwent scaling and root planing treatments, and showed that the fimA gentotypes of those isolated strains were composed of type IV (n=4), type II (n=2), and type III (n=1). As for the fimA genotypes of P. gingivalis-positive cardiovascular specimens, type IV was most frequently detected (45%), followed by type II (30%) (98). In addition, the abovementioned study demonstrated that approximately 50% of the dental biofilm specimens collected from cardiovascular patients were positive for P. gingivalis, with fimA type II found in 36%, type I in 29%, and type IV in 21%. These findings suggest that strains with specific fimA types, such as type II and IV, are associated with development of cardiovascular diseases. In fact, fimA type II, IV, and Ib strains were shown to cause more severe systemic inflammation in a mouse abscess model following subcutaneous injection as compared to those with other types (46). However, comparisons of the pathogenicity of strains with various fimA types to cardiovascular diseases in humans remain to be performed.
Minor components of long fimbriae
Prototypical type I fimbriae of uropathogenic E. coli are structurally composed of a fim gene cluster encoding proteins, including the pilus shaft, adhesive tip components on the shaft, and proteins involved in the translocation of subunits across the cell envelope (25). After examining a typical model, the flanking region of the P. gingivalis fimA gene was comprehensively analyzed (104). The downstream ORFs, designated ORF 1, 2, 3, and 4, were found to encode 15-, 50-, 80-, and 19-kDa proteins, respectively. Among the specific antibodies against each of those proteins, two against the 50- and 80-kDa products reacted with purified fimbriae and were proposed to be minor components associated with fimbriae (105). Very recently, three ORFs (fimC, fimD, and fimE, named ORF 2, 3, and 4, respectively) were reported to encode minor components associated with FimA protein, and long fimbriae were suggested to comprise polymerized FimA and accessory proteins (FimCDE) encoded by genes of the fimbrial operon (105). FimE is known to be required for assembly of FimC and FimD onto FimA fibers (106), while the two genes upstream of fimA are involved in regulation of fimA expression under the control of the FimS–FimR two-component system (107, 108). In addition, fimA expression is controlled by expression levels of the FimA protein itself, as well as by Rgp and Kgp gingipains (109).
It was also reported that fimC-, fimD-, and fimE-deficient mutants lost their auto-aggregation ability, and long fimbriae purified from those mutants showed diminished efficiencies to bind to GAPDH of S. oralis as well as fibronectin and type I collagen (106). Thus, FimC, FimD, and FimE are adhesive tip components likely associated with long fimbriae, whereas recombinant FimA protein was demonstrated to express various binding activities, while it is also known to be an adhesive molecule (38). Another study showed that fimbriae of the fimCDE mutant lost their ability to down-regulate IL-12, a key cytokine involved in intracellular bacterial clearance, and induced a significantly lower level of alveolar bone loss as compared with the wild type (110). Furthermore, the mutant failed to exploit CXCR4 in vivo for immune subversion. Indeed, purified FimC and FimD (but not FimE) were shown to interact with CXCR4. In addition, FimC and FimD bound to fibronectin and type 1 collagen, whereas FimE failed to interact with these matrix proteins. Together, these reports indicate the importance of FimCDE in the virulence of P. gingivalis and assembly of fully functional fimbriae.
Conclusions
Development of a useful genotyping testing tool for periodontal pathogens is necessary for therapeutic use. Future dentistry-related research will certainly produce such bacterial testing techniques for periodontal diagnosis, as well as medication and treatment strategies for affected individuals. However, additional efforts are required to investigate the exact relationship between genotypic variation and bacterial pathogenicity in periodontitis. Genomic variations of the long fimbria structures of P. gingivalis seem to be related to periodontitis initiation and progression. Furthermore, pan-genome analysis of P. gingivalis is expected to clarify the differences of virulence among strains. Future developments will be vital to identify the virulence/pathogenicity-related genes of P. gingivalis, while they will also be necessary for advancements in periodontal therapy and assessment of prognosis, by elucidating periodontal-related bacterial clones that contribute to disease.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors.
References
- 1.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 2.Lamont RJ, Jenkinson H. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis . Microbiol Mol Biol Rev. 1998;62:1244–63. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–87. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 4.Holt S, Ebersole J. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 5.Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics N, Chalmers NI, Diaz P. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 6.Iida Y, Honda K, Suzuki T, Matsukawa S, Kawai T, Shimahara T, et al. Brain abscess in which Porphyromonas gingivalis was detected in cerebrospinal fluid. Br J Oral Maxillofac Surg. 2004;42:180. doi: 10.1016/S0266-4356(03)00190-6. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Kolltveit KM, Tronstad L, Olsen I. Systemic diseases caused by oral infection. Clin Microbiol Rev. 2000;13:547–58. doi: 10.1128/cmr.13.4.547-558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scannapieco FA, Bush RB, Paju S. Association between periodontal disease and risk for nosocomial bacterial pneumonia and chronic obstructive pulmonary disease. A systematic review. Ann Periodontol. 2003;8:54–69. doi: 10.1902/annals.2003.8.1.54. [DOI] [PubMed] [Google Scholar]
- 9.Scannapieco FA. Systemic effects of periodontal diseases. Dent Clin North Am. 2005;49:533–50. doi: 10.1016/j.cden.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Chun YH, Chun KR, Olguin D, Wang HL. Biological foundation for periodontitis as a potential risk factor for atherosclerosis. J Periodontal Res. 2005;40:87–95. doi: 10.1111/j.1600-0765.2004.00771.x. [DOI] [PubMed] [Google Scholar]
- 11.Demmer RT, Desvarieux M. Periodontal infections and cardiovascular disease. J Am Dental Assoc. 2006;137:14–20. doi: 10.14219/jada.archive.2006.0402. [DOI] [PubMed] [Google Scholar]
- 12.Leishman SJ, Do HL, Ford PJ. Cardiovascular disease and the role of oral bacteria. J Oral Microbiol. 2010;2 doi: 10.3402/jom.v2i0.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pussinen PJ, Alfthan G, Jousilahti P, Paju S, Tuomilehto J. Systemic exposure to Porphyromonas gingivalis predicts incident stroke. Atherosclerosis. 2007;193:222–8. doi: 10.1016/j.atherosclerosis.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Dzink JL, Socransky SS, Haffajee AD. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 1988;15:316–23. doi: 10.1111/j.1600-051x.1988.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 15.Frandsen EV, Poulsen K, Curtis MA, Kilian M. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect Immun. 2001;9:4479–85. doi: 10.1128/IAI.69.7.4479-4485.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis . Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 17.Lamont RJ, Jenkinson HF. Subgingival colonization by Porphyromonas gingivalis . Oral Microbiol Immunol. 2000;15:341–9. doi: 10.1034/j.1399-302x.2000.150601.x. [DOI] [PubMed] [Google Scholar]
- 18.O-Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Prot Pept Sci. 2003;4:409–26. doi: 10.2174/1389203033487009. [DOI] [PubMed] [Google Scholar]
- 19.Bendtsen JD, Binnewies TT, Hallin PF, Sicheritz-Ponten T, Ussery DW. Genome update: prediction of secreted proteins in 225 bacterial genomes. Microbiology. 2005;151:1725–7. doi: 10.1099/mic.0.28029-0. [DOI] [PubMed] [Google Scholar]
- 20.Gerlach RG, Hensel M. Protein secretion systems and adhesins: the molecular armory of gram-negative pathogens. Int J Med Microbiol. 2007;297:401–15. doi: 10.1016/j.ijmm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Telford JL, Barocchi M, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nature Rev Microbiol. 2006;4:509–19. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 22.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nature Rev Microbiol. 2004;2:363–78. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 23.Kimberley AK, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18:224–32. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamada S, Amano A, Kimura S, Nakagawa I, Kawabata S, Morisaki I. The importance of fimbriae in the virulence and ecology of some oral bacteria. Oral Microbiol Immunol. 1998;13:129–38. doi: 10.1111/j.1399-302x.1998.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 25.Amano A. Bacterial adhesins to host components in periodontitis. Periodontol 2000. 2010;52:12–37. doi: 10.1111/j.1600-0757.2009.00307.x. [DOI] [PubMed] [Google Scholar]
- 26.Hamada N, Sojar HT, Cho MI, Genco RJ. Isolation and characterization of a minor fimbria from Porphyromonas gingivalis . Infect Immun. 1996;64:4788–94. doi: 10.1128/iai.64.11.4788-4794.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amano A, Nakagawa I, Okahashi N, Hamada N. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res. 2004;39:136–42. doi: 10.1111/j.1600-0765.2004.00719.x. [DOI] [PubMed] [Google Scholar]
- 28.Slots J, Gibbons RJ. Attachment of Bacteriodes melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and periodontal pockets. Infect Immun. 1978;19:254–64. doi: 10.1128/iai.19.1.254-264.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith H. Microbial surfaces in relation to pathogenicity. Bacteriol Rev. 1977;41:475–500. doi: 10.1128/br.41.2.475-500.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Listgarten MA, Lai CH. Comparative ultrastructure of Bacteroides melaninogenicus subspecies. J Periodontal Res. 1979;14:332–40. doi: 10.1111/j.1600-0765.1979.tb00797.x. [DOI] [PubMed] [Google Scholar]
- 31.Okuda K, Slots J, Genco RJ. Bacteroides gingivalis, Bacteroides asaccharolyticus and Bacteroides melaninogenicus subspecies: cell surface morphology and adherence to erythrocytes and human buccal epithelial cells. Curr Microbiol. 1981;6:7–12. [Google Scholar]
- 32.Zheng C, Wu J, Xie H. Differential expression and adherence of Porphyromonas gingivalis fimA genotypes. Mol Oral Microbiol. 2011;26:388–95. doi: 10.1111/j.2041-1014.2011.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handley PS, Tipler LS. An electron microscopic survey of the surface structures and hydrophobicity of oral and non-oral species of the bacterial genus Bacteroides . Arch Oral Biol. 1986;31:325–35. doi: 10.1016/0003-9969(86)90047-6. [DOI] [PubMed] [Google Scholar]
- 34.Dickinson DP, Kubiniec MA, Yoshimura F, Genco RJ. Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis . J Bacteriol. 1988;170:1658–65. doi: 10.1128/jb.170.4.1658-1665.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JY, Sojar HT, Bedi GS, Genco RJ. Porphyromonas (Bacteroides) gingivalis fimbrillin: size, amino-terminal sequence, and antigenic heterogeneity. Infect Immun. 1991;59:383–9. doi: 10.1128/iai.59.1.383-389.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H, Fives-Taylor P. Molecular strategies for fimbrial expression and assembly. Crit Rev Oral Biol Med. 2001;12:101–115. doi: 10.1177/10454411010120020101. [DOI] [PubMed] [Google Scholar]
- 37.Shoji M, Naito M, Yukitake H, Sato K, Sakai E, Ohara N, et al. The major structural components of two cell surface filaments of Porphyromonas gingivalis are matured through lipoprotein precursors. Mol Microbiol. 2004;52:1513–25. doi: 10.1111/j.1365-2958.2004.04105.x. [DOI] [PubMed] [Google Scholar]
- 38.Amano A. Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J Periodontol. 2003;74:90–6. doi: 10.1902/jop.2003.74.1.90. [DOI] [PubMed] [Google Scholar]
- 39.Fujiwara T, Morishima S, Takahashi I, Hamada S. Molecular cloning and sequencing of the fimbrillin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem Biophys Res Commun. 1993;197:241–7. doi: 10.1006/bbrc.1993.2467. [DOI] [PubMed] [Google Scholar]
- 40.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalisstrain W83. J Bacteriol. 2003;185:5591–601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa I, Amano A, Kimura RK, Nakamura T, Kawabata S, Hamada S. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of fimA gene. J Clin Microbiol. 2000;38:1909–14. doi: 10.1128/jcm.38.5.1909-1914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa I, Amano A, Ohara-Nemoto Y, Endoh N, Morisaki I, Kimura S, et al. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J Periodontal Res. 2002;37:425–32. doi: 10.1034/j.1600-0765.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa I, Amano A, Kubinowa M, Nakamura T, Kawabata S, Hamada S. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect Immun. 2002;70:277–85. doi: 10.1128/IAI.70.1.277-285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neiders ME, Chen PB, Suido H, Reynolds HS, Zambon JJ, Shlossman M, et al. Heterogeneity of virulence among strains of Bacteroides gingivalis . J Periodontal Res. 1989;24:192–8. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 45.Genco CA, Van Dyke T, Amar S. Animal models for Porphyromonas gingivalis-mediated periodontal disease. Trends Microbiol. 1998;6:444–9. doi: 10.1016/s0966-842x(98)01363-8. [DOI] [PubMed] [Google Scholar]
- 46.Nakano K, Kubinowa M, Nakagawa I, Yamamura T, Nomura R, Okahashi N, et al. Comparison of inflammatory changes caused by Porphyromonas gingivalis with distinct fimA genotypes in a mouse abscess model. Oral Microbiol Immunol. 2004;19:205–9. doi: 10.1111/j.0902-0055.2004.00133.x. [DOI] [PubMed] [Google Scholar]
- 47.Umeda JE, Missailidis C, Longo PL, Anzai D, Wikstrom M, Mayer MPA. Adhesion and invasion to epithelial cells by fimA genotypes of Porphyromonas gingivalis . Oral Microbiol Immunol. 2006;21:415–9. doi: 10.1111/j.1399-302X.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- 48.Kato T, Kawai S, Nakano K, Inaba H, Kubinowa M, Nakagawa I, et al. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell Microbiol. 2007;9:753–65. doi: 10.1111/j.1462-5822.2006.00825.x. [DOI] [PubMed] [Google Scholar]
- 49.Amano A, Nakagawa I, Kataoka K, Morisaki I, Hamada S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol. 1999;37:1426–30. doi: 10.1128/jcm.37.5.1426-1430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Enersen M, Olsen I, Kvalheim Ø, Caugant DA. FimA genotypes and multilocus sequence types of Porphyromonas gingivalis from periodontitis. J Clin Microbiol. 2008;46:31–42. doi: 10.1128/JCM.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Missailidis CG, Emeda JE, Ota-Tsuzuki C, Anzai D, Mayer MPA. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various forms of periodontal conditions. Oral Microbiol Immunol. 2004;19:224–9. doi: 10.1111/j.1399-302X.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 52.Amano A, Kubinowa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79:1664–8. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 53.Beikler T, Peters U, Prajaneh S, Prior K, Ehmke B, Fleming TF. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur J Oral Sci. 2003;111:390–4. doi: 10.1034/j.1600-0722.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- 54.Miura M, Hamachi T, Fujise O, Maeda K. The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J Periodontal Res. 2005;40:147–52. doi: 10.1111/j.1600-0765.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 55.Rocas IN, Siquiera JF., Jr Distribution of Porphyromonas gingivalis fimA genotypes in primary endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;109:474–8. doi: 10.1016/j.tripleo.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 56.Moon JH, Shin SI, Chung JH, Lee SW, Amano A, Lee JY. Development and evaluation of new primers for PCR-based identification of type II fimA of Porphyromonas gingivalis . FEMS Immunol Med Microbiol. 2012;64:1–4. doi: 10.1111/j.1574-695X.2011.00889.x. [DOI] [PubMed] [Google Scholar]
- 57.Inaba H, Nakano K, Kato T, Nomura R, Kawai S, Kubinowa M, et al. Heterogenic virulence and related factors among clinical isolates of Porphyromonas gingivalis with type II fimbriae. Oral Microbiol Immunol. 2008;23:29–35. doi: 10.1111/j.1399-302X.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 58.Laerdal J, Enersen M. Preliminary study of Porphyromonas gingivalis major fimbriae (FimA) Unpublished data, University of Oslo; 2010. [Google Scholar]
- 59.Nishiyama M, Ishikawa T, Rechsteiner H, Glockshuber R. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science. 2008;320:376–9. doi: 10.1126/science.1154994. [DOI] [PubMed] [Google Scholar]
- 60.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–8. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuboniwa M, Lamont RJ. Subgingival biofilm formation. Periodontol 2000. 2010;52:38–52. doi: 10.1111/j.1600-0757.2009.00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whittaker CJ, Klier CM, Kolenbrander PE. Mechanisms of adhesion by oral bacteria. Ann Rev Microbiol. 1996;50:513–52. doi: 10.1146/annurev.micro.50.1.513. [DOI] [PubMed] [Google Scholar]
- 63.Goulbourne PA, Ellen RP. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus . J Bacteriol. 1991;173:5266–74. doi: 10.1128/jb.173.17.5266-5274.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hashimoto M, Ogawa S, Asai Y, Takai Y, Ogawa T. Binding of Porphyromonas gingivalis fimbriae to Treponema denticola dentilisin. FEMS Microbiol Lett. 2003;226:267–71. doi: 10.1016/S0378-1097(03)00615-3. [DOI] [PubMed] [Google Scholar]
- 65.Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W, et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii . Infect Immun. 2005;73:3983–9. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maeda K, Nagata H, Yamamoto Y, Tanaka M, Tanaka J, Minamino N, et al. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect Immun. 2004;72:1341–8. doi: 10.1128/IAI.72.3.1341-1348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology. 2002;148:1627–36. doi: 10.1099/00221287-148-6-1627. [DOI] [PubMed] [Google Scholar]
- 68.Amano A, Fujiwara T, Nagata H, Kuboniwa M, Sharma A, Sojar HT, et al. Prophyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dent Res. 1997;76:852–7. doi: 10.1177/00220345970760040601. [DOI] [PubMed] [Google Scholar]
- 69.Sojar HT, Genco RJ. Identification of glyceraldehyde-3-phosphate dehydrogenase of epithelial cells as a second molecule that binds to Porphyromonas gingivalis fimbriae. FEMS Immunol Med Microbiol. 2005;45:25–30. doi: 10.1016/j.femsim.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 70.Park Y, James CE, Yoshimura F, Lamont RJ. Expression of the short fimbriae of Porphyromonas gingivalis is regulated in oral bacterial consortia. FEMS Microbiol Lett. 2006;262:65–71. doi: 10.1111/j.1574-6968.2006.00357.x. [DOI] [PubMed] [Google Scholar]
- 71.Kuboniwa M, Amano A, Inaba H, Hashino E, Shizukuishi S. Homotypic biofilm structure of Porphyromonas gingivalis is affected by FimA type variations. Oral Microbiol Immunol. 2009;24:260–3. doi: 10.1111/j.1399-302X.2009.00511.x. [DOI] [PubMed] [Google Scholar]
- 72.Xie H, Cook GS, Costerton JW, Bruce G, Rose TM, Lamont RJ. Intergeneric communication in dental plaque biofilms. J Bacteriol. 2000;182:7067–9. doi: 10.1128/jb.182.24.7067-7069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Christopher AB, Arndt A, Cugini C, Davey ME. A streptococcal effector protein that inhibits Porphyromonas gingivalis biofilm development. Microbiology. 2010;156:3469–77. doi: 10.1099/mic.0.042671-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Umemoto T, Hamada N. Characterization of biologically active cell surface components of a periodontal pathogen. The roles of major and minor fimbriae of Porphyromonas gingivalis . J Periodontol. 2003;74:119–22. doi: 10.1902/jop.2003.74.1.119. [DOI] [PubMed] [Google Scholar]
- 75.Kuramitsu H, Tokuda M, Yoneda M, Duncan M, Cho MI. Multiple colonization defects in a cysteine protease mutant of Porphyromonas gingivalis . J Periodontal Res. 1997;32:140–2. doi: 10.1111/j.1600-0765.1997.tb01395.x. [DOI] [PubMed] [Google Scholar]
- 76.Kuboniwa M, Amano A, Hashino E, Yamamoto Y, Inaba H, Hamada N, et al. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis . BMC Microbiol. 2009;9:e105. doi: 10.1186/1471-2180-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malek R, Fisher JG, Caleca A, Stinson M, van Oss CJ, Lee JY, et al. Inactivation of Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994;176:1052–9. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gibson FC, Hong C, 3rd, Chou HH, Yumoto H, Chen J, Lien E, et al. Innate immune recognition of invasive bacteria accelerates atherosclerosis in apolipoprotein E-deficient mice. Circulation. 2004;109:2801–6. doi: 10.1161/01.CIR.0000129769.17895.F0. [DOI] [PubMed] [Google Scholar]
- 79.Pathirana RD, O'Brien-Simpson NM, Reynolds EC. Host immune responses to Porphyromonas gingivalis antigens. Periodontol. 2010;52:218–37. doi: 10.1111/j.1600-0757.2009.00330.x. [DOI] [PubMed] [Google Scholar]
- 80.Hajishengallis G, Tapping RI, Harokopakis E, Nishiyama S, Ratti P, Schifferle RE, et al. Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell Microbiol. 2006;8:1557–70. doi: 10.1111/j.1462-5822.2006.00730.x. [DOI] [PubMed] [Google Scholar]
- 81.Asai Y, Ohyama Y, Gen K, Ogawa T. Bacterial fimbriae and their peptides activate human gingival epithelial cells through Toll-like receptor 2. Infect Immun. 2001;69:7387–95. doi: 10.1128/IAI.69.12.7387-7395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davey M, Liu X, Ukai T, Jain V, Gudino C, Gibson FC, 3rd, et al. Bacterial fimbriae stimulate proinflammatory activation in the endothelium through distinct TLRs. J Immunol. 2008;180:2187–95. doi: 10.4049/jimmunol.180.4.2187. [DOI] [PubMed] [Google Scholar]
- 83.Harokopakis E, Albzreh MH, Martin MH, Hajishengallis G. TLR2 transmodulates monocyte adhesion and transmigration via Rac1- and PI3K-mediated inside-out signaling in response to Porphyromonas gingivalis fimbriae. J Immunol. 2006;176:7645–56. doi: 10.4049/jimmunol.176.12.7645. [DOI] [PubMed] [Google Scholar]
- 84.Ogawa T, Asai Y, Hashimoto M, Uchida H. Bacterial fimbriae activate human peripheral blood monocytes utilizing TLR2, CD14 and CD11a/CD18 as cellular receptors. Eur J Immunol. 2002;32:2543–50. doi: 10.1002/1521-4141(200209)32:9<2543::AID-IMMU2543>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 85.Eskan MA, Hajishengallis G, Kinane DF. Differential activation of human gingival epithelial cells and monocytes by Porphyromonas gingivalis fimbriae. Infect Immun. 2007;75:892–8. doi: 10.1128/IAI.01604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takahashi Y, Davey M, Yumoto H, Gibson FC, III, Genco CA. Fimbria-dependent activation of pro-inflammatory molecules in Porphyromonas gingivalis infected human aortic endothelial cells. Cell Microbiol. 2006;8:738–57. doi: 10.1111/j.1462-5822.2005.00661.x. [DOI] [PubMed] [Google Scholar]
- 87.Hiramine H, Watanabe K, Hamada N, Umemoto T. Porphyromonas gingivalis 67-kDa fimbriae induced cytokine production and osteoclast differentiation utilizing TLR2. FEMS Microbiol Lett. 2003;5:49–55. doi: 10.1016/S0378-1097(03)00788-2. [DOI] [PubMed] [Google Scholar]
- 88.Hamada N, Watanabe K, Arai M, Hiramine H, Umemoto T. Cytokine production induced by a 67-kDa fimbrial protein from Porphyromonas gingivalis . Oral Microbiol Immunol. 2002;17:197–200. doi: 10.1034/j.1399-302x.2002.170311.x. [DOI] [PubMed] [Google Scholar]
- 89.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–97. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hajishengallis G. Immune evasion strategies of Porphyromonas gingivalis . J Oral Biosci. 2011;53:233–40. doi: 10.2330/joralbiosci.53.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hajishengallis G, Shakhatreh MAK, Wang M, Liang S. Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J Immunol. 2007;179:2359–67. doi: 10.4049/jimmunol.179.4.2359. [DOI] [PubMed] [Google Scholar]
- 92.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 93.Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol. 2011;11:187–200. doi: 10.1038/nri2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hajishengallis G, Wang M, Liang S, Triantafilou M, Triantafilou K. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc Natl Acad Sci USA. 2008;105:13532–7. doi: 10.1073/pnas.0803852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chiu B. Multiple infections in carotid atherosclerotic plaques. Am Heart J. 1999;138:S534–6. doi: 10.1016/s0002-8703(99)70294-2. [DOI] [PubMed] [Google Scholar]
- 96.Haraszthey VI, Zambon JJ, Trevisan M, Zeid M, Genco RJ. Identification of periodontal pathogens in atheromatous plaques. J Periodontol. 2000;71:1554–60. doi: 10.1902/jop.2000.71.10.1554. [DOI] [PubMed] [Google Scholar]
- 97.Nakano K, Nemoto H, Nomura R, Inaba H, Yoshioka H, Taniguchi K, et al. Detection of oral bacteria in cardiovascular specimens. Oral Microbiol Immunol. 2009;24:64–8. doi: 10.1111/j.1399-302X.2008.00479.x. [DOI] [PubMed] [Google Scholar]
- 98.Nakano K, Inaba H, Nomura R, Nemoto H, Takeuchi H, Yoshioka H, et al. Distribution of Porphyromonas gingivalis fimA genotypes in cardiovascular specimens from Japanese patients. Oral Microbiol Immunol. 2008;23:170–2. doi: 10.1111/j.1399-302X.2007.00406.x. [DOI] [PubMed] [Google Scholar]
- 99.Nakano K, Wada K, Nomura R, Nemoto H, Inaba H, Kojima A, et al. Characterization of aortic aneurysms in cardiovascular disease patients harboring Porphyromonas gingivalis . Oral Dis. 2011;17:370–8. doi: 10.1111/j.1601-0825.2010.01759.x. [DOI] [PubMed] [Google Scholar]
- 100.Kozarov E, Sweier D, Shelburne C, Progulske-Fox A, Lopatin D. Detection of bacterial DNA in atheromatous plaques by quantitative PCR. Microbes Infect. 2006;8:687–93. doi: 10.1016/j.micinf.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 101.Takahashi Y, Davey M, Yumoto H, Gibson FC, 3rd, Genco CA. Fimbria-dependent activation of pro-inflammatory molecules in Porphyromonas gingivalis infected human aortic endothelial cells. Cell Microbiol. 2006;8:738–57. doi: 10.1111/j.1462-5822.2005.00661.x. [DOI] [PubMed] [Google Scholar]
- 102.Chou HH, Yumoto H, Davey M, Takahashi Y, Miyamoto T, Gibson FC, 3rd, et al. Porphyromonas gingivalis fimbria-dependent activation of inflammatory genes in human aortic endothelial cells. Infect Immun. 2005;73:5367–78. doi: 10.1128/IAI.73.9.5367-5378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pérez-Chaparro PJ, Lafaurie GI, Gracieux P, Meuric V, Tamanai-Shacoori Z, Castellanos JE, et al. Distribution of Porphyromonas gingivalis fimA genotypes in isolates from subgingival plaque and blood sample during bacteremia. Biomedica. 2009;29:298–306. [PubMed] [Google Scholar]
- 104.Yoshimura F, Murakami Y, Nishikawa K, Hasegawa Y, Kawaminami S. Surface components of Porphyromonas gingivalis . J Periodontal Res. 2009;44:1–12. doi: 10.1111/j.1600-0765.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- 105.Watanabe K, Onoe T, Ozeki M, Shimizu Y, Sakayori T, Nakamura H, et al. Sequence and product analyses of the four genes downstream from the fimbrilin gene (fimA) of the oral anaerobe Porphyromonas gingivalis . Microbiol Immunol. 1996;40:725–34. doi: 10.1111/j.1348-0421.1996.tb01133.x. [DOI] [PubMed] [Google Scholar]
- 106.Nishiyama S, Murakami Y, Nagata H, Shizukuishi S, Kawagishi I, Yoshimura F. Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology. 2007;153:1916–25. doi: 10.1099/mic.0.2006/005561-0. [DOI] [PubMed] [Google Scholar]
- 107.Hayashi J, Nishikawa K, Hirano R, Noguchi T, Yoshimura F. Identification of a two-component signal transduction system involved in fimbriation of Porphyromonas gingivalis . Microbiol Immunol. 2000;44:279–82. doi: 10.1111/j.1348-0421.2000.tb02496.x. [DOI] [PubMed] [Google Scholar]
- 108.Nishikawa K, Yoshimura F, Duncan MJ. A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Mol Microbiol. 2004;54:546–60. doi: 10.1111/j.1365-2958.2004.04291.x. [DOI] [PubMed] [Google Scholar]
- 109.Xie H, Chung WO, Park Y, Lamont RJ. Regulation of the Porphyromonas gingivalis fimA (fimbrillin) gene. Infect Immun. 2000;68:6574–9. doi: 10.1128/iai.68.12.6574-6579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang M, Shakhatreh MA, James D, Liang S, Nishiyama S, Yoshimura F, et al. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J Immunol. 2007;179:2349–58. doi: 10.4049/jimmunol.179.4.2349. [DOI] [PubMed] [Google Scholar]