Abstract
Mutations of the epidermal growth factor receptor (EGFR) are the strongest predictive factor for response to EGFR tyrosine kinase inhibitors (TKIs), such as gefitinib and erlotinib. EGFR TKIs are approved in Korea as a first-line treatment for lung cancer patients with mutated EGFR. Rapid and accurate EGFR mutation testing is essential for patient selection and establishing targeted therapies with EGFR TKIs. Thus, a standard set of guideline recommendations for EGFR mutation testing suitable for the Korean medical community is necessary. In this article, we propose a set of guideline recommendations for EGFR mutation testing that was discussed and approved by the Cardiopulmonary Pathology Study Group of the Korean Society of Pathologists.
Keywords: Mutation; Receptor, epidermal growth factor; Guideline
Recent advances in molecular pathology and targeted therapies have opened a new era of personalized medicine for lung cancer treatment. Driver genetic alterations such as epidermal growth factor receptor (EGFR) mutations, as well as Kirsten rat sarcoma viral oncogene homolog (KRAS) and anaplastic lymphoma kinase (ALK) rearrangements, have been identified and are currently used as predictive biomarkers for targeted therapies.1 Activating somatic mutations in the EGFR gene are known to be major driver mutations in that they exhibit a high incidence in lung cancers and have played an important role in the development of targeted molecular therapies for lung cancer.2
EGFR tyrosine kinase inhibitors (TKIs), such as gefitinib and erlotinib, are associated with anti-tumor activity, inhibiting multiple downstream signaling processes that activate cell proliferation and other cell responses, including cell migration and angiogenesis.3 EGFR TKIs are approved in Korea as a first-line treatment for advanced non-small cell lung cancer (NSCLC) with mutated EGFR (Fig. 1). In the Iressa Pan-Asia Study (IPASS) trial, tumors with mutated EGFR exhibited a 71.2% clinical response to first-line gefitinib treatment, while only 1.1% of tumors with wild-type EGFR responded to the treatment.4 Therefore, patient selection is critical for the clinical use of EGFR TKIs as a first-line treatment. Clinical characteristics such as female gender, never-smoker status, and Asian ethnicity were also found to be associated with the response to EGFR TKIs; however, the results of the IPASS study confirmed that molecular selection-based EGFR mutation testing is the strongest predictive factor for EGFR TKI treatment response.4,5
Fig. 1.
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) are approved as a first-line treatment for advanced non-small cell lung cancer harboring EGFR mutation.
Thus, EGFR mutation testing is very important for lung cancer therapy. Likewise, rapid and accurate EGFR mutation testing is essential for proper patient selection when considering targeted therapy with EGFR TKIs. In addition, a standard set of guidelines suitable for the Korean medical community is necessary. In this article, we propose guideline recommendations for EGFR mutation testing that were discussed and approved by the Cardiopulmonary Pathology Study Group of the Korean Society of Pathologists (Table 1).
Table 1.
Recommendation summary for EGFR mutation testing

EGFR, epidermal growth factor receptor.
aIn this regard, poorly differentiated non-small cell carcinoma should be further classified into a more specific type whenever possible. A minimum immunohistochemical panel (such as thyroid transcription factor 1/napsin A/p63 or p40) is recommended in small specimens to preserve as much tissue as possible for molecular testing.
PATIENT SELECTION
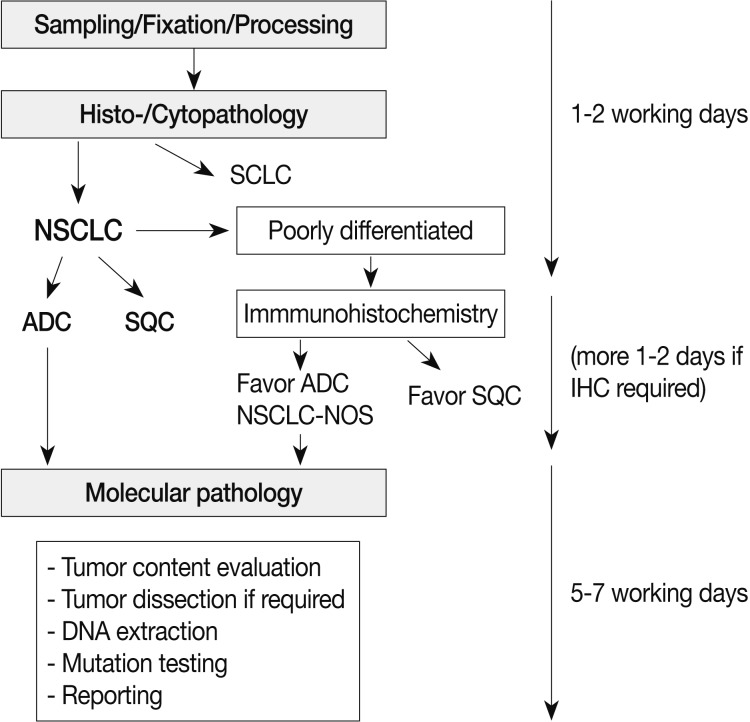
The most important reason for EGFR mutation testing is to select patients who might benefit from EGFR TKI therapy. Patients that receive EGFR mutation testing are primarily those with advanced stage disease. EGFR mutations are more prevalent in female patients, never-smokers, and patients of Asian ethnicity. However, clinical features alone cannot entirely predict EGFR mutation status.6,7 Most of the guidelines published thus far recommend histologic type as the most important factor for determining whether EGFR mutational testing should be performed.8-10 Specifically, when patients are diagnosed with NSCLC including an adenocarcinoma component or NSCLC-not-otherwise-specified after immunohistochemistry, EGFR mutation testing is routinely recommended.9 Thus, pathologists should try to further classify poorly differentiated NSCLC into more specific types, such as adenocarcinoma or squamous cell carcinoma, whenever possible (Fig. 2). In addition, to preserve as much tissue as possible for molecular testing in small specimens, a minimum immunohistochemical panel such as thyroid transcription factor 1/napsin A/p63 or p40 is recommended.9,11,12
Fig. 2.
Overall process for pathologic diagnosis and molecular analysis with recommended turnaround times. SCLC, small cell lung carcinoma; NSCLC, non-small cell lung carcinoma; ADC, adenocarcinoma; SQC, squamous cell carcinoma; NSCLC-NOS, non-small cell carcinoma-not otherwise specified; IHC, immunohistochemistry.
EGFR mutations are detected in approximately 40% of Korean NSCLC patients with adenocarcinoma histology.13 Furthermore, it has been reported that EGFR mutations are more prevalent in specific subtypes of adenocarcinomas such as lepidic, papillary, or micropapillary, although it should be noted that these subtypes are not fully predictive of EGFR mutation status.14,15 Although previous studies have reported that a small fraction of squamous cell carcinomas or small cell carcinomas harbor EGFR mutations,16-19 routine examination is not recommended because the incidence in pure types is very low. However, in cases of female never-smokers, those with a combined tumor type, or when otherwise clinically indicated, mutation testing can be performed.
SAMPLE SOURCES
Various small biopsy and cytology specimens can be used as samples for mutation testing. More specifically, acceptable tissue specimens include transbronchial biopsy, gun biopsy, computed tomography-guided needle aspiration, endobronchial ultrasound-guided transbronchial needle aspiration, bronchial brushing/washing, and pleural fluid sampling.8,20,21 Many studies have shown that cytology specimens are suitable for assessing EGFR mutations, and that the results are highly concordant with those of corresponding histological specimens, especially when using more sensitive methods.20-24
There have been several reports on the heterogeneous distribution of EGFR mutations and discordance of EGFR mutation status between primary tumors and corresponding metastatic tumors.25-27 In contrast, Yatabe et al.28 reported that a heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. Although there is an ongoing debate with respect to these reports, and further studies are needed,29,30 samples from a small portion of primary or metastatic tumor can be used equally.
SAMPLE PROCESSING
Routinely prepared samples are mostly formalin-fixed, paraffin-embedded (FFPE) tissues. Although there has been a report of fixation-related artifacts,31 routinely prepared FFPE tissues are the most practical and standard resource for EGFR mutation analysis. There is consensus that 10% neutral-buffered formalin is the optimum fixative for preparing FFPE samples,8,31 while the optimal fixation time ranges from 6 to 24 hours to avoid underfixation or overfixation, respectively.8,31
Routinely prepared cytology specimens, such as alcohol-fixed smears or ThinPrep slides prepared by transferring cells in suspension20,23 and cell block specimens,32 are also suitable materials for EGFR mutation analysis.
ESTABLISHING ADEQUATE TUMOR CONTENT FOR MUTATION TESTING
Before mutation testing, the presence of tumor cells in the sample must be assessed by a pathologist. The ratio of tumor cells to normal cells is crucial for adequate mutation testing. For direct sequencing, the percentage of tumor cells in the sample should ideally be at least 50%, although reliable results can be influenced by a variety of factors. Thus, determination of the percentage of tumor cells in a given tissue or cell specimen is recommended. Macro- or microdissection may be used to increase the ratio of tumor to normal tissues if required. A study performed by Sun et al. showed that the following parameters correlate with the most reliable EGFR mutation results when using cytology samples: DNA concentration >25 µg/µL, content of >30 tumor cells, and tumor percentage >30%.23 The minimum number of tumor cells required for adequate testing and the minimum ratio of tumor to normal cells is influenced by the testing method (see below).
METHODS FOR EGFR MUTATION TESTING
Various methods can be used for detecting EGFR mutations.33-35 Pathologists should consider the available facilities and the pros and cons of each method, including the sensitivity and turnaround time. In addition, new techniques must be approved by the Korean government.
Direct sequencing is considered to be the gold standard for EGFR mutation analysis. In Korea, most pathology laboratories perform direct sequencing for the detection of EGFR mutations using FFPE tissue samples. However, for directing DNA sequencing, a high ratio of tumor tissue to normal tissue content is required (more than 50% tumor content). In contrast, real time polymerase chain reaction (PCR)-based methods exhibit high sensitivity, requiring a mutant DNA content of only 1%.33 However, these methods can only detect previously known mutations or targeted sites. The peptide nucleic acid (PNA)-mediated PCR clamping method was recently developed and approved in Korea. The PNA clamping method exhibits high sensitivity compared with direct sequencing, and clinical outcomes are not significantly different between groups harboring EGFR mutations detected by direct sequencing or PNA-mediated PCR clamping.36-38 Highly sensitive methods can also be useful for detection of EGFR mutations associated with acquired resistance, such as T790M.39
TURNAROUND TIME
Gefitinib is approved as a first-line treatment for advanced NSCLC-harboring EGFR mutations. Thus, the results of mutation analysis should be made available to physicians as soon as possible. Pathologic diagnosis and molecular testing are a combined and continuous process and should be supervised by a pathologist (Fig. 2). It is recommended that testing be completed within five to seven working days after ordering EGFR mutation testing.
Several factors may influence turnaround time. In general, pathologic diagnoses and EGFR mutation testing performed within the same department rather than at separate laboratories would have a shorter turnaround time. When mutation testing is performed by an outside laboratory, communication and coordination between the pathology department and the external laboratory are encouraged.
REPEATED EXAMINATION
Several criteria for repeated examination of EGFR mutation status have been recommended. The test should be repeated in cases of poor sequencing data, a cycle threshold too close to the defined cut-off limit (with pyrosequencing or PNA clamp kit), and/or mutation results that are not matched with previously well-defined clinical-pathologic characteristics. Specifically, pathologists should carefully interpret the results of EGFR mutations found in heavy smokers, solid or mucinous cancer types, or when EGFR mutations are concurrent with other exclusive driver mutations.
REPORTING FORMAT
Molecular testing reports should contain the following information: pathologic number, age, sex, hospital unit number, biopsy site, sample source, requesting physician, requesting department, adequacy for testing (estimated tumor cell content), receipt day, report day, methodology used, exons tested and associated range of detectable mutations, mutation status, comments, testing technician, and corresponding pathologist.
PATHOLOGIST'S ROLES
The pathologist plays an essential role in EGFR mutation testing.40 The pathologist can either perform the test at the home institution or transfer the tissue to a reference laboratory for external examination. In both situations, the pathologist is responsible through the procedures. First, the pathologist should choose the most appropriate tissue to be tested.7,8,23 Second, the pathologist should verify that the selected tissue block for EGFR mutation testing contains sufficient tumor cells required for analysis. The proportion of the tumor cells in the tissue or cytology samples is very important to prevent contamination with non-tumor cells.7,23 Lastly, the pathologist is responsible for accurate and prompt reporting, which should include results from routing diagnostic information (such as histologic diagnosis), as well as from EGFR mutation testing. If the test is performed by an external reference laboratory, the pathologist integrates the test results into the pathology report of his/her institute.40 We recommend all patients with NSCLC having an adenocarcinoma component or NSCLC-not-otherwise-specified after immunohistochemistry should be tested for EGFR mutation. In the cases of squamous cell carcinomas or small cell carcinomas arising from never-smokers, mutation testing can be performed. As for the small biopsy or cytology specimens, macro- or microdissection may enhance mutation testing sensitivity.
PERSPECTIVES AND ADDITIONAL RECOMMENDATIONS
EGFR mutations and ALK rearrangements are currently used as predictive biomarkers for targeted lung cancer therapy. In addition, other driver mutations are now receiving attention, including ROS1 rearrangement,41 BRAF mutation,42,43 HER2 mutation,44 and RET rearrangement.45 In terms of molecular diagnostics, these other targetable mutations have developed the need for multiplex mutational testing. However, because most patients with lung cancer present with advanced-stage disease at the time of diagnosis, the diagnosis of lung cancer is often based on small specimens from a biopsy or cytology alone. Thus, each pathology department must develop a strategy to manage clinical samples and collaborate with clinicians.9 As mentioned above, these strategies include minimization of diagnostic stains in order to maximize the available tissue for molecular studies9,12 and reduction of the number of trimmings for slide sections.
CONCLUSION
As targetable mutations are discovered and corresponding targeted agents are developed, molecular diagnostics using clinical samples has become increasingly important. EGFR mutations are the most robust predictive factors for response to EGFR TKIs. Thus, each pathology department should maintain an optimal organization for the entire workflow of EGFR mutation testing, from sample collection to the final report. Lastly, pathologists should keep in mind that personalized medicine is driven by pathology and molecular diagnostics.
Acknowledgments
The authors appreciate all members of the Korean Cardio-Pulmonary Pathology Study Group, their support, and their excellent opinions. This research was conducted with support from an Investigator Sponsored Study Programme of AstraZeneca; Partly supported by a grant from the Korea Healthcare Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (A111405, to JH Chung).
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Cagle PT, Chirieac LR. Advances in treatment of lung cancer with targeted therapy. Arch Pathol Lab Med. 2012;136:504–509. doi: 10.5858/arpa.2011-0618-RA. [DOI] [PubMed] [Google Scholar]
- 2.Janku F, Stewart DJ, Kurzrock R. Targeted therapy in non-small-cell lung cancer: is it becoming a reality? Nat Rev Clin Oncol. 2010;7:401–414. doi: 10.1038/nrclinonc.2010.64. [DOI] [PubMed] [Google Scholar]
- 3.Linardou H, Dahabreh IJ, Bafaloukos D, Kosmidis P, Murray S. Somatic EGFR mutations and efficacy of tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol. 2009;6:352–366. doi: 10.1038/nrclinonc.2009.62. [DOI] [PubMed] [Google Scholar]
- 4.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J Clin Oncol. 2011;29:2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 6.D'Angelo SP, Pietanza MC, Johnson ML, et al. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. J Clin Oncol. 2011;29:2066–2070. doi: 10.1200/JCO.2010.32.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun PL, Seol H, Lee HJ, et al. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol. 2012;7:323–330. doi: 10.1097/JTO.0b013e3182381515. [DOI] [PubMed] [Google Scholar]
- 8.Pirker R, Herth FJ, Kerr KM, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol. 2010;5:1706–1713. doi: 10.1097/JTO.0b013e3181f1c8de. [DOI] [PubMed] [Google Scholar]
- 9.Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salto-Tellez M, Tsao MS, Shih JY, et al. Clinical and testing protocols for the analysis of epidermal growth factor receptor mutations in East Asian patients with non-small cell lung cancer: a combined clinical-molecular pathological approach. J Thorac Oncol. 2011;6:1663–1669. doi: 10.1097/JTO.0b013e318227816a. [DOI] [PubMed] [Google Scholar]
- 11.Rekhtman N, Ang DC, Sima CS, Travis WD, Moreira AL. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod Pathol. 2011;24:1348–1359. doi: 10.1038/modpathol.2011.92. [DOI] [PubMed] [Google Scholar]
- 12.Noh S, Shim H. Optimal combination of immunohistochemical markers for subclassification of non-small cell lung carcinomas: a tissue microarray study of poorly differentiated areas. Lung Cancer. 2012;76:51–55. doi: 10.1016/j.lungcan.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Yatabe Y. EGFR mutations and the terminal respiratory unit. Cancer Metastasis Rev. 2010;29:23–36. doi: 10.1007/s10555-010-9205-8. [DOI] [PubMed] [Google Scholar]
- 14.Zakowski MF, Hussain S, Pao W, et al. Morphologic features of adenocarcinoma of the lung predictive of response to the epidermal growth factor receptor kinase inhibitors erlotinib and gefitinib. Arch Pathol Lab Med. 2009;133:470–477. doi: 10.1043/1543-2165-133.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shim HS, Lee da H, Park EJ, Kim SH. Histopathologic characteristics of lung adenocarcinomas with epidermal growth factor receptor mutations in the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society lung adenocarcinoma classification. Arch Pathol Lab Med. 2011;135:1329–1334. doi: 10.5858/arpa.2010-0493-OA. [DOI] [PubMed] [Google Scholar]
- 16.Park SH, Ha SY, Lee JI, et al. Epidermal growth factor receptor mutations and the clinical outcome in male smokers with squamous cell carcinoma of lung. J Korean Med Sci. 2009;24:448–452. doi: 10.3346/jkms.2009.24.3.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyamae Y, Shimizu K, Hirato J, et al. Significance of epidermal growth factor receptor gene mutations in squamous cell lung carcinoma. Oncol Rep. 2011;25:921–928. doi: 10.3892/or.2011.1182. [DOI] [PubMed] [Google Scholar]
- 18.Tatematsu A, Shimizu J, Murakami Y, et al. Epidermal growth factor receptor mutations in small cell lung cancer. Clin Cancer Res. 2008;14:6092–6096. doi: 10.1158/1078-0432.CCR-08-0332. [DOI] [PubMed] [Google Scholar]
- 19.Shiao TH, Chang YL, Yu CJ, et al. Epidermal growth factor receptor mutations in small cell lung cancer: a brief report. J Thorac Oncol. 2011;6:195–198. doi: 10.1097/JTO.0b013e3181f94abb. [DOI] [PubMed] [Google Scholar]
- 20.Rekhtman N, Brandt SM, Sigel CS, et al. Suitability of thoracic cytology for new therapeutic paradigms in non-small cell lung carcinoma: high accuracy of tumor subtyping and feasibility of EGFR and KRAS molecular testing. J Thorac Oncol. 2011;6:451–458. doi: 10.1097/JTO.0b013e31820517a3. [DOI] [PubMed] [Google Scholar]
- 21.Navani N, Brown JM, Nankivell M, et al. Suitability of endobronchial ultrasound-guided transbronchial needle aspiration specimens for subtyping and genotyping of non-small cell lung cancer: a multicenter study of 774 patients. Am J Respir Crit Care Med. 2012;185:1316–1322. doi: 10.1164/rccm.201202-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allegrini S, Antona J, Mezzapelle R, et al. Epidermal growth factor receptor gene analysis with a highly sensitive molecular assay in routine cytologic specimens of lung adenocarcinoma. Am J Clin Pathol. 2012;138:377–381. doi: 10.1309/AJCPVAGIUC1AHC3Y. [DOI] [PubMed] [Google Scholar]
- 23.Sun PL, Jin Y, Kim H, Lee CT, Jheon S, Chung JH. High concordance of EGFR mutation status between histologic and corresponding cytologic specimens of lung adenocarcinomas. Cancer Cytopathol. 2012 Dec 05; doi: 10.1002/cncy.21260. [Epub]. http://dx.doi.org/10.1002/cncy.21260. [DOI] [PubMed] [Google Scholar]
- 24.da Cunha Santos G, Saieg MA, Geddie W, Leighl N. EGFR gene status in cytological samples of nonsmall cell lung carcinoma: controversies and opportunities. Cancer Cytopathol. 2011;119:80–91. doi: 10.1002/cncy.20150. [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi K, Okami J, Kodama K, Higashiyama M, Kato K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008;99:929–935. doi: 10.1111/j.1349-7006.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 27.Gow CH, Chang YL, Hsu YC, et al. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol. 2009;20:696–702. doi: 10.1093/annonc/mdn679. [DOI] [PubMed] [Google Scholar]
- 28.Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29:2972–2977. doi: 10.1200/JCO.2010.33.3906. [DOI] [PubMed] [Google Scholar]
- 29.Chen ZY, Zhong WZ, Zhang XC, et al. EGFR mutation heterogeneity and the mixed response to EGFR tyrosine kinase inhibitors of lung adenocarcinomas. Oncologist. 2012;17:978–985. doi: 10.1634/theoncologist.2011-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai H, Wang Z, Chen K, et al. Influence of chemotherapy on EGFR mutation status among patients with non-small-cell lung cancer. J Clin Oncol. 2012;30:3077–3083. doi: 10.1200/JCO.2011.39.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eberhard DA, Giaccone G, Johnson BE Non-Small-Cell Lung Cancer Working Group. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983–994. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- 32.Nicholson AG, Gonzalez D, Shah P, et al. Refining the diagnosis and EGFR status of non-small cell lung carcinoma in biopsy and cytologic material, using a panel of mucin staining, TTF-1, cytokeratin 5/6, and p63, and EGFR mutation analysis. J Thorac Oncol. 2010;5:436–441. doi: 10.1097/JTO.0b013e3181c6ed9b. [DOI] [PubMed] [Google Scholar]
- 33.Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007;13:4954–4955. doi: 10.1158/1078-0432.CCR-07-1387. [DOI] [PubMed] [Google Scholar]
- 34.Ellison G, Zhu G, Moulis A, Dearden S, Speake G, McCormack R. EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol. 2013;66:79–89. doi: 10.1136/jclinpath-2012-201194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HJ, Xu X, Kim H, et al. Comparison of direct sequencing, PNA clamping-real time polymerase chain reaction, and pyrosequencing methods for the detection of EGFR mutations in non-small cell lung carcinoma and the correlation with clinical responses to EGFR tyrosine kinase inhibitor treatment. Korean J Pathol. 2013;47:52–60. doi: 10.4132/KoreanJPathol.2013.47.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HJ, Kim WS, Shin KC, et al. Comparative analysis of peptide nucleic acid (PNA)-mediated real-time PCR clamping and DNA direct sequencing for EGFR mutation detection. Tuberc Respir Dis. 2011;70:21–27. [Google Scholar]
- 37.Kim HJ, Lee KY, Kim YC, et al. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer. 2012;75:321–325. doi: 10.1016/j.lungcan.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Han HS, Lim SN, An JY, et al. Detection of EGFR mutation status in lung adenocarcinoma specimens with different proportions of tumor cells using two methods of differential sensitivity. J Thorac Oncol. 2012;7:355–364. doi: 10.1097/JTO.0b013e31823c4c1b. [DOI] [PubMed] [Google Scholar]
- 39.Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17:1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Krieken JH, Jung A, Kirchner T, et al. KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: proposal for an European quality assurance program. Virchows Arch. 2008;453:417–431. doi: 10.1007/s00428-008-0665-y. [DOI] [PubMed] [Google Scholar]
- 41.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29:3574–3579. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 43.Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29:2046–2051. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res. 2012;18:4910–4918. doi: 10.1158/1078-0432.CCR-12-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30:4352–4359. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]