Abstract
Single-cell force spectroscopy is a powerful atomic force microscopy modality in which a single living cell is attached to the atomic force microscopy cantilever to quantify the forces that drive cell-cell and cell-substrate interactions. Although various single-cell force spectroscopy protocols are well established for animal cells, application of the method to individual bacterial cells remains challenging, mainly owing to the lack of appropriate methods for the controlled attachment of single live cells on cantilevers. We present a nondestructive protocol for single-bacterial cell force spectroscopy, which combines the use of colloidal probe cantilevers and of a bioinspired polydopamine wet adhesive. Living cells from the probiotic species Lactobacillus plantarum are picked up with a polydopamine-coated colloidal probe, enabling us to quantify the adhesion forces between single bacteria and biotic (lectin monolayer) or abiotic (hydrophobic monolayer) surfaces. These minimally invasive single-cell experiments provide novel, to our knowledge, insight into the specific and nonspecific forces driving the adhesion of L. plantarum, and represent a generic platform for studying the molecular mechanisms of cell adhesion in probiotic and pathogenic bacteria.
Introduction
Studying the molecular mechanisms of bacterial adhesion is critical to our understanding of bacterial-host interactions. Bacterial adhesion results from a complex interplay of physicochemical forces, that can be either specific (receptor-ligand interactions) or nonspecific (hydrophobic and electrostatic interactions) (1). Although various macroscopic assays are available to investigate microbial adhesion, these approaches probe large ensembles of cells and do not provide information on the fundamental forces driving cell adhesion. Consequently, there is a growing need for methods that can quantify bacterial adhesion forces on a single-cell basis (2,3). Such single-cell techniques would be of great help to refine our perception of cellular heterogeneity, and to reveal otherwise invisible adhesive mechanisms (4).
Probiotic bacteria, which mostly belong to the Gram-positive lactic acid bacteria group, offer exciting prospects in medicine owing to their ability to induce various beneficial health effects (5,6). Among these, Lactobacillus plantarum is a promising candidate in view of its ability to survive several days in the human gastrointestinal tract (7), and to adhere to human mucosa in vitro (8). Positive health effects of probiotics are thought to be related to their ability to attach to epithelial cells and mucus. Therefore, the efficient use of probiotics requires a detailed understanding of their adhesive interactions toward inert and living surfaces. These interactions are determined by the main macromolecules that constitute the bacterial cell wall, i.e., peptidoglycan, proteins, and glycopolymers such as polysaccharides and teichoic acids (5,9). Although much is known about the structure and synthesis of these constituents in L. plantarum, the extent to which they contribute to bacterial adhesion, particularly host interactions, is poorly understood. Accordingly, there is much interest in measuring the molecular forces driving the adhesion of L. plantarum to biotic and abiotic surfaces. In probiotic research, such experiments have great promise for the screening of strains exhibiting enhanced adhesive properties and health effects.
In addition to being used as an imaging tool, atomic force microscopy (AFM) is being increasingly applied to quantify the interactions of biological systems, over scales ranging from single molecules to whole cells (10–13). In the past years, there has been considerable progress in the use of single-cell force spectroscopy (SCFS) to quantify cell-cell and cell-solid interactions (10–15). The general idea is to replace the tip of the AFM cantilever by a living cell that is then used to measure interactions toward other cells or substrates (14,15). Several techniques have been developed to attach cells onto cantilevers, including the use of specific receptor-ligand interactions (16), electrostatic (17,18) or hydrophobic (19) interactions, glue (20), or chemical fixation (21). However, none of these methods enable true, reliable single-bacterial cell analysis for at least one of the following reasons: i), the cell-cantilever bond is too weak, leading to cell detachment; ii), the use of chemicals or drying leads to cell surface denaturation and/or cell death; iii), multiple cells are often attached and probed together, meaning reliable single-cell analysis is not accessible. An interesting approach to solve these problems is the use of cantilevers modified with colloids or beads. For instance, Lower and co-workers (22,23) attached bacteria-coated beads to cantilevers to measure the forces between living Shewanella oneidensis bacteria and goethite. In this method, the bead is covered with multiple bacteria, meaning single-cell analysis is difficult to guarantee. Also exciting is the recently developed fluidFM, which uses hollow cantilevers for local liquid dispensing and manipulation of single living cells under physiological conditions (24). FluidFM is currently able to manipulate bacterial cells (25), but its application to bacterial force measurements is not yet fully established.
In this work, we report a noninvasive method for the SCFS analysis of individual bacteria, which combines colloidal probes cantilevers (26) and bioinspired polydopamine adhesive (27). We show that the use of polydopamine-coated colloidal probes is a simple, versatile platform for the controlled attachment of single bacterial cells on AFM cantilevers, and for quantifying their adhesive interactions toward various surfaces (alkanethiol monolayers, lectin monolayers). The results emphasize the important roles of nonspecific (hydrophobic) and specific (glycopolymer-lectin) interactions in mediating the adhesion of L. plantarum. We expect that this SCFS method will be a valuable tool in biomedical research for understanding the molecular interactions between bacteria (probiotics, pathogens) and host cells.
Materials and Methods
Microorganisms and cultures
L. plantarum NZ7100 cells were grown in Mann-Rogosa-Shape broth (Difco) at 30°C without agitation. Overnight cultures were diluted in fresh media to an OD600 nm of 0.1. The cells were harvested in the exponential growth phase (4 h at 30°C), and washed 3 times in acetate buffer. For cell probe preparation, 50 μL of a 100-fold diluted solution were transferred to a glass petri dish.
Preparation of cell probes
Using a Nanoscope VIII Multimode AFM (Bruker, Santa Barbara, CA), triangular shaped tipless cantilevers (NP-O10, Microlevers, Veeco Metrology Group) were slowly immersed in a very thin layer of ultraviolet (UV)-curable glue (NOA 63, Norland Edmund Optics) spread on a glass slide, and slowly brought into contact with a silica microsphere (6.1 μm diameter, Bangs Laboratories). After 3 min of contact, the colloidal probe was cured for 10 min under a UV lamp. The cantilever was then immersed for 1 h in a 10 mM Tris Buffer solution (pH 8.5) containing 4 mg/mL dopamine hydrochloride (99%, Sigma). The probe was then washed and dried under N2. Using a Bioscope Catalyst (Bruker), the colloidal probe was brought into contact with an isolated cell for 3 min, and the obtained cell probe was then transferred without dewetting over a solid substrate for further force measurements.
Viability of attached bacteria was tested using a LIVE/DEAD Baclight Viablility Kit (Invitrogen, kit L7012). Prior attachment, 2 μL of a 1:1 Syto 9/IP mixture at 1.5 mM were added to a 50 μL cell suspension, mixed thoroughly, and incubated for 15 min in the dark. The labeled cells were then attached to polydopamine colloidal probes or substrates, and their viability checked using a Zeiss Axio Observer Z1 equipped with a Hamamatsu camera C10600. Dead bacteria attached onto polydopamine substrates were obtained by immersing them for 1 h in isopropanol.
Substrate preparation
To prepare hydrophobic and hydrophilic substrates, clean glass coverslips coated with a thin layer of gold were immersed overnight in a 1 mM solution of 1-dodecanethiol or 11-mercapto-1-undecanol (Sigma), rinsed with ethanol, and dried under N2. Lectin-coated substrates were obtained by immersing gold-coated coverslips overnight in an ethanol solution containing 1 mM of 10% mercaptododecahexanoic acid/90% 11-mercapto-1-undecanol (Sigma), rinsed with ethanol, and dried with N2. Substrates were then immersed for 30 min into a solution containing 20 g/L N-hydroxysuccinimide (NHS) and 50 g/L 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) (Sigma), rinsed 5 times with MilliQ water (Millipore), incubated with 0.2 mg/mL of Concanavalin A (ConA) for 1 h, rinsed further with acetate, and then immediately used.
AFM
AFM measurements were performed at room temperature (20°C) in sodium acetate buffer—supplemented with Ca2+ and Mn2+ at 1 mM for experiments with ConA lectins—using a Bioscope Catalyst AFM (Bruker AXS, Santa Barbara, CA). Force-distance curves were obtained by recording multiple force curves in different locations, using a maximum applied force of 250 pN and a constant approach and retraction speed of 1000 nm.s−1. For mannoside blocking experiments, a concentrated methyl α-D-mannopyranoside solution (Sigma) was injected into the AFM chamber to reach a final concentration of 200 mM.
Results and Discussion
Cell probe preparation
Marine mussels produce adhesive proteins that strongly bind to solid surfaces in aqueous environments, owing to the presence of the unusual amino acid 3,4-dihydroxy-L-phenylalanine (dopa) (27,28). Mussel-inspired polydopamine films have recently attracted much interest for the design of adhesive interfaces and materials (29,30). In the AFM context, Kang and Elimelech (31) used polydopamine for attaching bacterial cells onto AFM cantilevers for SCFS experiments. Although this approach is an important improvement to earlier methods, it does not offer a precise control of the cell-substrate contact area. Given the small size of bacteria and the tilted orientation of the cantilever, the controlled immobilization of single cells on the same given area of the cantilever ends is difficult; this leads to a lack of control of the interacting area, and sometimes even to a direct contact between the cantilever and the substrate.
We therefore combined the polydopamine method with the use of colloidal probes of large, well-defined geometry (26) (Fig. 1). Colloidal probe cantilevers were produced by attaching silica microspheres (∼6 μm) to tipless cantilevers using a UV-curable glue (Fig. 1, step 1). Colloidal probes were coated with a thin polydopamine film. Using an integrated AFM-inverted optical microscope, polydopamine probes were then approached toward single bacterial cells deposited on a glass petri dish in buffer, kept in contact for 3 min, and withdrawn (Fig. 1, step 2). The obtained cell probes were directly used for SCFS measurements (Fig. 1, step 3), although avoiding dewetting as this may cause cell detachment or cell surface denaturation. Because single cells are precisely attached in the center of the colloids, this method guarantees reliable and reproducible single-cell force measurements.
Figure 1.
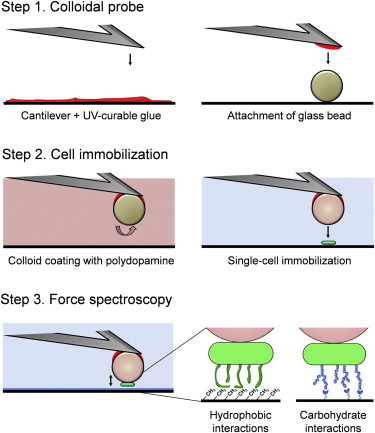
Single-bacterial cell force spectroscopy using colloidal probe cantilevers combined with bioinspired polydopamine polymers. The method involves three main steps, i.e., preparation of the colloidal probe, controlled immobilization of a single cell, and force-distance curve measurements (see text for details).
To confirm that the polydopamine-colloidal probe method is nondestructive, cells were labeled with the Baclight LIVE/DEAD stain, in which living bacteria exhibit green fluorescence, whereas dead bacteria are red. Fig. 2 a shows that most bacteria attached onto a polydopamine-coated substrate were alive (green), whereas bacteria attached onto the same polydopamine substrate but treated with isopropanol were dead (red). Fig. 2 b (left) reveals that single-bacterial cells attached onto polydopamine-colloidal probes were alive even after 60 min measurements, whereas bacteria attached directly onto tipless cantilevers were generally killed (Fig. 2 b, right). Although the reason for this difference is not known yet, a possible explanation is that bacteria in direct contact with the cantilever may be more subject to heating by the laser beam of the AFM optical detection system. Hence, besides affording much better control of the cell-substrate interaction area, the polydopamine-colloidal probe method guarantees that the cells remain alive during the course of the experiment.
Figure 2.

Use of polydopamine-colloidal probe guarantees reliable, single-live cell experiments. (a) Fluorescence images of Lactobacillus plantarum bacteria labeled with the Baclight LIVE/DEAD stain and attached onto polydopamine-coated substrates: comparison of native cells (left) and of cells killed with isopropanol (right). (b) Fluorescence images of bacterial cells labeled with the Baclight LIVE/DEAD stain, attached onto polydopamine-coated cantilevers and imaged either immediately (0 min, top) or after 60 min of force measurements (bottom): comparison of the colloidal probe cantilever method developed here (left) and of the conventional tipless cantilever approach (right).
Quantifying hydrophobic forces
Hydrophobic forces represent one of the driving forces for the adhesion of bacteria to surfaces and tissues (32). To assess whether this indeed applies to L. plantarum, multiple force curves were recorded between bacterial cells and hydrophobic surfaces. Fig. 3, a and b, show the adhesion force histogram with representative force curves, and the rupture length histogram recorded at a short contact time (<100 ms) between single L. plantarum cells and methyl-terminated self-assembled monolayers (SAMs). All curves showed large adhesion forces (from 250 to 2500 pN) with multiple, sequential peaks and extended rupture lengths (from 150 to 600 nm). The general features of the curves did not substantially change when recording consecutive force curves (up to several hundred), or when probing different cells or substrates, supporting the notion that force measurements did not lead to cell surface alteration. These cell-adhesion signatures are reminiscent of those observed on animal cells (15) and indicate that bonds that have been formed between the substrate and the cell break sequentially until the cell has completely separated from the surface. The maximum downward force exerted on the cantilever is referred to as the adhesion force (measured relative to the base line) and used to build the adhesion force histogram, whereas the last rupture peak is used to generate the rupture length histogram.
Figure 3.
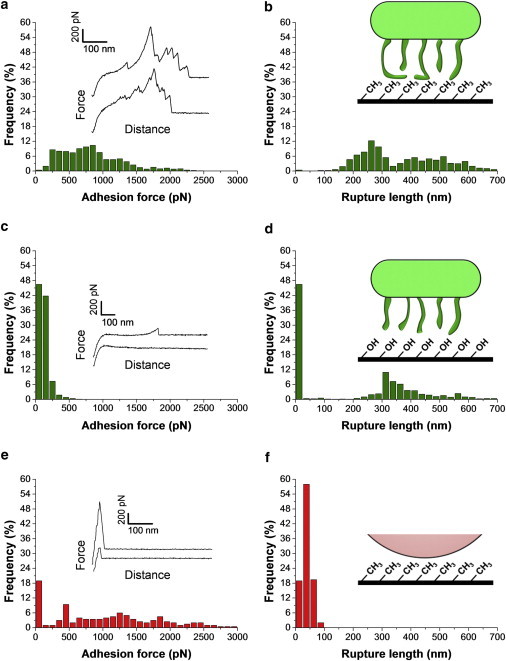
SCFS quantifies hydrophobic interactions between Lactobacillus plantarum and solid surfaces. (a) Adhesion force histogram with representative force curves and (b) histogram of rupture distances recorded in buffer at a short contact time (<100 ms) between single bacterial cells and hydrophobic, methyl-terminated monolayers. The data shown correspond to a total of 1000 force curves obtained from three independent experiments (different cell probes and different substrates). (c) Adhesion force histogram with representative force curves and (d) histogram of rupture distances recorded in buffer between single bacterial cells and hydrophilic, hydroxyl-terminated monolayers. The data shown correspond to a total of 1000 force curves obtained from three independent experiments. (e) Adhesion force histogram with representative force curves and (f) histogram of rupture distances recorded in buffer between polydopamine-coated colloidal probes and Hydrophobic, methyl-terminated monolayers.
We believe that the measured adhesion forces reflect hydrophobic interactions between the substrate and cell surface proteins for the following reasons: i), the extended rupture lengths and multiple force peaks are consistent with the stretching and unfolding of cell surface proteins; by contrast, peptidoglycan that forms fairly compact, stiff structures is not expected to show such extensions (33); ii), unlike proteins, glycopolymers are hydrophilic in nature, thus not expected to strongly bind to hydrophobic surfaces; iii), large, multipeak force signatures were never observed on hydrophilic hydroxyl-terminated SAMs (Fig. 3, c and d); rather, force curves recorded on hydrophilic substrates showed single, well-defined force peaks of ∼200 pN magnitude and 250–500 nm rupture length that we attribute to glycopolymer stretching; iv), control experiments between polydopamine-coated beads and hydrophobic SAMs (Fig. 3, e and f), never exhibited multipeaks typically observed with stretched proteins, but only single, sharp adhesive events with very short extensions. Similar polydopamine signatures were observed when the cells were not well centered on the probe or weakly immobilized. In light of these observations, we suggest that the large adhesion force profiles are associated with the binding and unfolding of hydrophobic domains from cell surface-associated proteins. As observed for the pathogen Candida albicans (34), the force-induced unfolding of protein domains may lead to extended conformations in which hydrophobic groups are freshly exposed and favor hydrophobic interactions toward biotic and abiotic surfaces. In view of the large cell-substrate contact area and of the force magnitude, it is likely that the complex adhesion profiles reflect the simultaneous stretching of multiple proteins.
Measuring glycopolymer interactions
Glycopolymers (teichoic acids, polysaccharides) on the surface of probiotic bacteria play important physiological roles, including controlling cell morphogenesis, and mediating cellular recognition and adhesion, e.g., via lectin binding (3,4). To probe the glycopolymer properties (adhesion, extension) of L. plantarum, we recorded force curves between single bacterial cells and substrates coated with ConA, a lectin that specifically binds the glucose (or mannose) residues contained in glycopolymers (35). As shown in Fig. 4, a and b, most curves (92%) recorded at a short contact time (<100 ms) showed no adhesion. At first sight this seems surprising as i), glucose-rich glycopolymers that decorate the bacterial cell surface are expected to bind to ConA, and ii), SMFS with ConA tips detected substantial amounts of glucose (or mannose) residues on the L. plantarum surface (35). However, this discrepancy can easily be explained by the contact time, which in SCFS experiments dramatically enhances specific interactions (15). For animal cells, the idea is that single receptor-ligand pairs initially anchor the cell to a substrate or another cell, after which these molecular bonds generally increase with time and undergo modifications to greatly increase the total strength of adhesion (15). Consistent with this behavior, Fig. 4, c and d, show that increasing the contact time to 1 s dramatically changed the force profiles: a large fraction (49%) of the curves displayed single or multiple adhesion forces, of 251 ± 146 pN mean magnitude (mean ± SD; total number of force curves = 1000 from three different cell probes and substrates), along with elongation forces and rupture lengths ranging essentially from 25 to 250 nm. The force signatures did not substantially change when recording consecutive force curves, or when probing different cells or substrates. The observed time-dependency suggests that formation of lectin bonds is much slower than hydrophobic bonds, for which full binding was achieved within <100 ms (Fig. 3 a). Earlier SMFS studies have shown that lectin-sugar complexes rupture at forces around 30–60 pN at fairly similar loading rates (36–38). Therefore, we attribute the ∼250 pN forces to the simultaneous detection of multiple ConA-glucose interactions. Finally, we assessed the specificity of the recognition events by blocking the cell surface with free α-methyl mannoside. Fig. 4, e and f, show that treatment of the cells with mannoside led to a much lower frequency of adhesion events (from 49% to 28%), confirming they are associated with specific glucose/mannose binding events.
Figure 4.
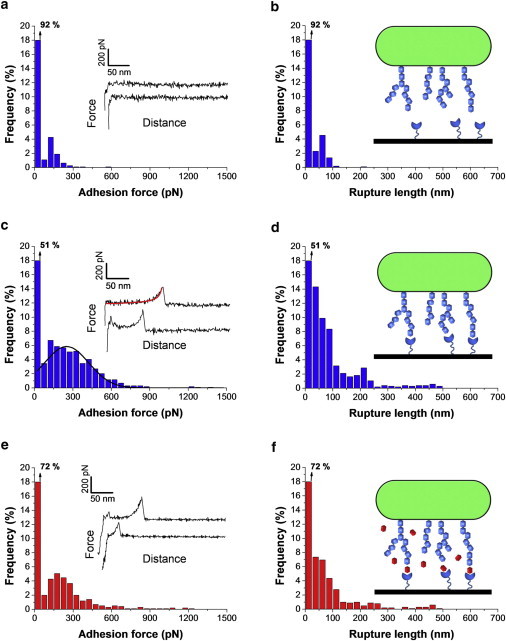
SCFS measures the specific interactions between Lactobacillus plantarum cell surface glycopolymers and lectin surfaces. (a and c) Adhesion force histograms with representative force curves and (b and d) histograms of rupture distances recorded in buffer between single bacterial cells and lectin monolayers, using a contact time of either <100 ms (a and b) or 1 s (c and d). The fit on the top curve of panel c shows that elongation forces were well described by an extended freely jointed chain model with a Kuhn length of 0.4 nm and a segment elasticity of 1 N/m: x(F) = Lc [coth (Flk/kbT)- kbT/Flk] [1 + nF/ksLc], where Lc and lk are the contour length and Kuhn length of the molecule, n and ks the number of segments and their elasticity, kb is the Boltzmann constant, and T the absolute temperature. The line on the histogram of panel c is a Gaussian fit to the data. (e and f) Same measurements performed with a contact time of 1 s, following blocking with 200 mM methyl α-D-mannopyranoside. The data shown for every condition (a and b, c and d, e and f) correspond to a total of 1000 force curves obtained from three independent experiments (different cell probes and different substrates).
The glycopolymers detected here are likely to consist of cell surface polysaccharides because i), elongation forces were well described by an extended freely jointed chain model (Fig. 4 c); ii), the extended rupture lengths are in the range of those observed by SMFS on bacterial cell surface polysaccharides, including those from the probiotic bacterium Lactobacillus rhamnosus GG (37); iii), compared to polysaccharides, teichoic acids give rise to much shorter rupture lengths (35). Our results therefore suggest that glucose-based polysaccharides on the L. plantarum cell surface mediate strong adhesion toward lectin-coated surfaces. Because of their long, flexible nature, polysaccharide chains may strengthen adhesion through long-range polymer bridging interactions. This finding may be of biological significance as lectins on intestinal epithelial and dendritic cells may act as host receptors for probiotic molecules (5).
Conclusions
In conclusion, our experiments have shown that SCFS with polydopamine-coated colloidal probes provide a powerful platform for quantifying bacterial cell adhesion forces on a single-cell basis. Unlike other existing protocols, our methodology is simple, versatile, nondestructive (even after 1 h measurements), and affords much better control of the cell positioning of the cell-substrate contact area. These assets guarantee true and reliable single-bacterial cell analysis. Using this approach, we found that L. plantarum cells show strong adhesive properties toward biotic and abiotic surfaces. Binding to hydrophobic surfaces does not depend on interaction time and gives rise to multiple force peaks and extended rupture lengths that may be attributed to the stretching and unfolding of cell surface proteins. Binding to lectin surfaces is strongly time-dependent and is associated with the stretching of long, flexible glucose (mannose)-based macromolecules, most likely cell-bound polysaccharides. The measured specific and nonspecific adhesive forces are of biological relevance as they are likely to play important roles in mediating L. plantarum adhesive interactions toward inert and living surfaces.
Acknowledgments
Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Y.F.D., P.H., and D.A. are Senior Research Associate, Research Associate, and Postdoctoral Researcher of the FNRS.
References
- 1.Busscher H.J., Norde W., van der Mei H.C. Specific molecular recognition and nonspecific contributions to bacterial interaction forces. Appl. Environ. Microbiol. 2008;74:2559–2564. doi: 10.1128/AEM.02839-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camesano T.A., Liu Y., Datta M. Measuring bacterial adhesion at environmental interfaces with single-cell and single-molecule techniques. Adv. Water Resour. 2007;30:1470–1491. [Google Scholar]
- 3.Busscher H.J., van der Mei H.C. How do bacteria know they are on a surface and regulate their response to an adhering state? PLoS Pathog. 2012;8:e1002440. doi: 10.1371/journal.ppat.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brehm-Stecher B.F., Johnson E.A. Single-cell microbiology: tools, technologies, and applications. Microbiol. Mol. Biol. Rev. 2004;68:538–559. doi: 10.1128/MMBR.68.3.538-559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebeer S., Vanderleyden J., De Keersmaecker S.C.J. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 2010;8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 6.Bron P.A., van Baarlen P., Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 2012;10:66–78. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- 7.Vesa T., Pochart P., Marteau P. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 2000;14:823–828. doi: 10.1046/j.1365-2036.2000.00763.x. [DOI] [PubMed] [Google Scholar]
- 8.Adlerberth I., Ahrne S., Wold A.E. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl. Environ. Microbiol. 1996;62:2244–2251. doi: 10.1128/aem.62.7.2244-2251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delcour J., Ferain T., Hols P. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie van Leeuwenhoek. 1999;76:159–184. [PubMed] [Google Scholar]
- 10.Müller D.J., Dufrêne Y.F. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat. Nanotechnol. 2008;3:261–269. doi: 10.1038/nnano.2008.100. [DOI] [PubMed] [Google Scholar]
- 11.Müller D.J., Dufrêne Y.F. Atomic force microscopy: a nanoscopic window on the cell surface. Trends Cell Biol. 2011;21:461–469. doi: 10.1016/j.tcb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Müller D.J., Dufrêne Y.F. Force nanoscopy of living cells. Curr. Biol. 2011;21:R212–R216. doi: 10.1016/j.cub.2011.01.046. [DOI] [PubMed] [Google Scholar]
- 13.Müller D.J., Helenius J., Dufrêne Y.F. Force probing surfaces of living cells to molecular resolution. Nat. Chem. Biol. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 14.Benoit M., Gaub H.E. Measuring cell adhesion forces with the atomic force microscope at the molecular level. Cells Tissues Organs (Print) 2002;172:174–189. doi: 10.1159/000066964. [DOI] [PubMed] [Google Scholar]
- 15.Helenius J., Heisenberg C.P., Muller D.J. Single-cell force spectroscopy. J. Cell Sci. 2008;121:1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 16.Benoit M., Gabriel D., Gaub H.E. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat. Cell Biol. 2000;2:313–317. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- 17.Le D.T.L., Guérardel Y., Dague E. Measuring kinetic dissociation/association constants between Lactococcus lactis bacteria and mucins using living cell probes. Biophys. J. 2011;101:2843–2853. doi: 10.1016/j.bpj.2011.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ovchinnikova E.S., Krom B.P., Busscher H.J. Force microscopic and thermodynamic analysis of the adhesion between Pseudomonas aeruginosa and Candida albicans. Soft Matter. 2012;8:6454–6461. [Google Scholar]
- 19.Emerson R.J., 4th, Bergstrom T.S., Camesano T.A. Microscale correlation between surface chemistry, texture, and the adhesive strength of Staphylococcus epidermidis. Langmuir. 2006;22:11311–11321. doi: 10.1021/la061984u. [DOI] [PubMed] [Google Scholar]
- 20.Bowen W.R., Lovitt R.W., Wright C.J. Atomic force microscopy study of the adhesion of Saccharomyces cerevisiae. J. Colloid Interface Sci. 2001;237:54–61. doi: 10.1006/jcis.2001.7437. [DOI] [PubMed] [Google Scholar]
- 21.Razatos A., Ong Y.L., Georgiou G. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 1998;95:11059–11064. doi: 10.1073/pnas.95.19.11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lower S.K., Hochella M.F., Jr., Beveridge T.J. Bacterial recognition of mineral surfaces: nanoscale interactions between Shewanella and α-FeOOH. Science. 2001;292:1360–1363. doi: 10.1126/science.1059567. [DOI] [PubMed] [Google Scholar]
- 23.Yongsunthon R., Lower S.K. Force measurements between a bacterium and another surface In Situ. Adv. Appl. Microbiol. 2005;58C:97–124. doi: 10.1016/S0065-2164(05)58003-1. [DOI] [PubMed] [Google Scholar]
- 24.Meister A., Gabi M., Zambelli T. FluidFM: combining atomic force microscopy and nanofluidics in a universal liquid delivery system for single cell applications and beyond. Nano Lett. 2009;9:2501–2507. doi: 10.1021/nl901384x. [DOI] [PubMed] [Google Scholar]
- 25.Dorig P., Stiefel P., Zambelli T. Force-controlled spatial manipulation of viable mammalian cells and micro-organisms by means of FluidFM technology. Appl. Phys. Lett. 2010;97:023701. [Google Scholar]
- 26.Ducker W.A., Senden T.J., Pashley R.M. Direct measurement of colloidal forces using an atomic force microscope. Nature. 1991;353:239–241. [Google Scholar]
- 27.Lee H., Lee B.P., Messersmith P.B. A reversible wet/dry adhesive inspired by mussels and geckos. Nature. 2007;448:338–341. doi: 10.1038/nature05968. [DOI] [PubMed] [Google Scholar]
- 28.Lee H., Scherer N.F., Messersmith P.B. Single-molecule mechanics of mussel adhesion. Proc. Natl. Acad. Sci. USA. 2006;103:12999–13003. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brubaker C.E., Messersmith P.B. The present and future of biologically inspired adhesive interfaces and materials. Langmuir. 2012;28:2200–2205. doi: 10.1021/la300044v. [DOI] [PubMed] [Google Scholar]
- 30.Lee H., Dellatore S.M., Messersmith P.B. Mussel-inspired surface chemistry for multifunctional coatings. Science. 2007;318:426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang S., Elimelech M. Bioinspired single bacterial cell force spectroscopy. Langmuir. 2009;25:9656–9659. doi: 10.1021/la902247w. [DOI] [PubMed] [Google Scholar]
- 32.Doyle R.J. Contribution of the hydrophobic effect to microbial infection. Microbes Infect. 2000;2:391–400. doi: 10.1016/s1286-4579(00)00328-2. [DOI] [PubMed] [Google Scholar]
- 33.Andre G., Kulakauskas S., Dufrêne Y.F. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat. Commun. 2010;1:27. doi: 10.1038/ncomms1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaussart A., Alsteens D., Dufrêne Y.F. Single-molecule imaging and functional analysis of Als adhesins and mannans during Candida albicans morphogenesis. ACS Nano. 2012;6:10950–10964. doi: 10.1021/nn304505s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andre G., Deghorain M., Dufrene Y.F. Fluorescence and atomic force microscopy imaging of wall teichoic acids in Lactobacillus plantarum. ACS Chem. Biol. 2011;6:366–376. doi: 10.1021/cb1003509. [DOI] [PubMed] [Google Scholar]
- 36.Alsteens D., Dupres V., Dufrêne Y.F. Structure, cell wall elasticity and polysaccharide properties of living yeast cells, as probed by AFM. Nanotechnology. 2008;19:384005. doi: 10.1088/0957-4484/19/38/384005. [DOI] [PubMed] [Google Scholar]
- 37.Francius G., Lebeer S., Dufrêne Y.F. Detection, localization, and conformational analysis of single polysaccharide molecules on live bacteria. ACS Nano. 2008;2:1921–1929. doi: 10.1021/nn800341b. [DOI] [PubMed] [Google Scholar]
- 38.Dettmann W., Grandbois M., Gaub H.E. Differences in zero-force and force-driven kinetics of ligand dissociation from beta-galactoside-specific proteins (plant and animal lectins, immunoglobulin G) monitored by plasmon resonance and dynamic single molecule force microscopy. Arch. Biochem. Biophys. 2000;383:157–170. doi: 10.1006/abbi.2000.1993. [DOI] [PubMed] [Google Scholar]


