Abstract
To perform their specific functional role, B and T lymphocytes, cells of the adaptive immune system of jawed vertebrates, need to express one (and, preferably, only one) form of antigen receptor, i.e. the immunoglobulin or T-cell receptor (TCR), respectively. This end goal depends initially on a series of DNA cis-rearrangement events between randomly chosen units from separate clusters of V, D (at some immunoglobulin and TCR loci) and J gene segments, a biomolecular process collectively referred to as V(D)J recombination. V(D)J recombination takes place in immature T and B cells and relies on the so-called RAG nuclease, a site-specific DNA cleavage apparatus that corresponds to the lymphoid-specific moiety of the VDJ recombinase. At the genome level, this recombinase's mission presents substantial biochemical challenges. These relate to the huge distance between (some of) the gene segments that it eventually rearranges and the need to achieve cell-lineage-restricted and developmentally ordered routines with at times, mono-allelic versus bi-allelic discrimination. The entire process must be completed without any recombination errors, instigators of chromosome instability, translocation and, potentially, tumorigenesis. As expected, such a precisely choreographed and yet potentially risky process demands sophisticated controls; epigenetics demonstrates what is possible when calling upon its many facets. In this vignette, we will recall the evidence that almost from the start appeared to link the two topics, V(D)J recombination and epigenetics, before reviewing the latest advances in our knowledge of this joint venture.
Keywords: allelic exclusion, chromatin, epigenetics, immunoglobulin, T-cell receptor, V(D)J recombination
Introduction; a short trip back in (adaptive) immunological time
Antigen receptor gene rearrangement and epigenetics are two indisputably complementary phenomena. Discovered in the mid-1970s1 and later known as V(D)J recombination,2,3 the former is the somatic process that reshuffles and joins together – generally in a random and imprecise manner – two or three pieces of DNA (V and J segments, or V, D and J segments) from immunoglobulin genes (in developing B lymphocytes) or T-cell receptor (TCR) genes (in developing T lymphocytes), to achieve vast repertoires of custom-made immunoglobulin and TCRs and thereby afford adaptive immunity. Over the years, systematic studies to untangle the many mysteries of V(D)J recombination and its sophisticated controls (see below) have led to numerous exciting new findings, including in the early 1990s the discovery of the Recombination Activating Genes 1 and 2 (RAG1 and RAG2), two adjacent lymphoid-specific genes that code for the core components of the long-sought VDJ recombinase (reviewed in ref. 4). Extensively covered in this Focus Issue, epigenetics conversely keeps out of all things involving the revision of germline DNA, at least so it would first seem. In short, this idiom refers to the functionally relevant modifications of, and heritable traits conveyed by the genome – in particular by the chromatin and nucleosome building block – which orchestrate regulated changes in gene expression and in cellular phenotype, yet do not involve the alteration of the underlying primary DNA sequence (e.g. ref. 5). Though coined relatively long ago,6 this field truly exploded during the past decade with a maintained exponential growth thanks to technical breakthroughs allowing depiction and interrogation at the genome scale of the so-called epigenetic code, i.e. the dynamic chromosomal marks and structural features that carry the epigenetic information. Indeed, it has now become clear that a bulk of molecular procedures (and associated enzymatic factors thereof) have an epigenetic impact that is cell-type specific and developmentally regulated. These include DNA methylation and demethylation,7,8 the active (energy-consuming) disruption or displacement of positioned nucleosomes, the exchange of nucleosomal histones and a variety of biochemical modifications of histone N-terminal tails,9–13 as well as diverse forms of non-coding transcription14,15 and three-dimensional (3D) organization of the genomic material through chromatin folding and long-range interaction via DNA looping.16,17 Over the years, many of these procedures were shown to also have an impact on V(D)J recombination.
It did not take long before V(D)J recombination and epigenetics met. A few years after the discovery of immunoglobulin gene recombination in B lymphocytes,1,18 and the subsequent description of sequential rearrangements of B-cell antigen receptor genes [i.e. immunoglobulin heavy (H) chain gene rearrangement preceding immunoglobulin light (L) chain gene rearrangement; and, within the IgH locus, DH-to-JH rearrangement preceding VH-to-DJH rearrangement19,20 (and see below)] two types of seminal observations were reported by Alt's group. First, unrearranged endogenous VH gene segments are expressed in a developmentally controlled and tissue-specific manner, with unrearranged VH expression being limited to the very early stages of B-lymphocyte differentiation and most prominent in cells undergoing VH-to-DJH rearrangement.21 Second, exogenous immunoglobulin and TCR gene segments rearrange equally well when introduced as plasmid substrates into an immature, pre-B-cell line in which endogenous immunoglobulin genes, but not TCR genes, rearrange with substrate recombination enhanced by transcription of a flanking selectable gene.22,23 Based on these observations, these authors formulated the so-called accessibility hypothesis whereby the accessibility of a locus to a unique VDJ recombinase determines the choice of the antigen receptor gene that will recombine, with chromatin structure anticipated as a key determinant of accessibility.24 Since then, evidence has accumulated in support of the accessibility hypothesis, to a ‘no doubt about it’ general agreement. Modern epigenetics has enlightened molecular features that could confidently distinguish between accessible and inaccessible immunoglobulin/TCR loci. What is more, epigenetics appears to play an even broader role in V(D)J recombination than originally anticipated, impinging not only on the direct recruitment of the RAG nuclease to discrete rearranging loci but also on its catalytic activity; and probably, through some of the molecular procedures mentioned above, also on the 3D organization of the actively recombining chromosome with important implications for the generation of antigen receptor immune repertoires.
V(D)J recombination, basic features
A succinct description of the molecular and mechanistic aspects of V(D)J recombination is provided below. Detailed reviews on these various aspects can be found elsewhere.25–27
Overall, seven distinct gene loci are normally subjected to V(D)J recombination, including three immunoglobulin genes (IgH, IgLκ and IgLλ), and four TCR genes (TCRα, TCRβ, TCRδ and TCRγ) two of which (TCRα and TCRδ) are intermingled on the same chromosome. The assembly of IgH, TCRβ and TCRδ genes is achieved from separate sets of V, D and J gene segments, whereas that of IgLκ, Iglλ, TCRα and TCRγ genes uses V and J gene segments only. Unrearranged segments are flanked on their rearranging end(s) by conserved recombination signal sequences (RSSs), which represent direct targets for the RAG nuclease. Within a given locus, V(D)J recombination is initiated following RAG binding (assisted by high mobility group proteins) to and synapsis of pairs of compatible gene segments, i.e. segments that are flanked by dissymmetric RSSs in which evolutionarily conserved 7-base-pair (bp) (heptamer) and 9-bp (nonamer) sequences are separated by either 12 or 23 bp of less-conserved sequences. [Note: Subsequent biochemical studies on RAG-mediated RSS cleavage in vitro in fact provided evidence of a two-step orchestrated ‘capture’ model of RSS synapsis involving RAG binding to a first RSS before capturing a fitting partner.28,29 Moreover, based on the detection of initial cleavage mostly occurring at 12RSSs in vivo, a ‘12RSS binding first’ model was proposed.30 However, further investigations have questioned such a universal scenario because – depending on the locus considered – preferential RAG binding to either 12RSS-associated or 23RSS-associated clusters is equally observed.31].
Of note, though theoretically 12/23 compatible, the direct joining of some gene segments such as for instance the Vβ (23RSS) and Jβ (12RSS) segments, is in fact prohibited. Termed beyond (B)12/23, this additional constraint is enforced during the V(D)J recombination reaction itself, with implications on the ordered assembly of TCRβ genes in T-cell development.32
Once brought closer together, RSSs of the paired segments are precisely cleaved by the RAG nuclease at their boundary with the coding sequences to make double-strand breaks. The neighbouring coding DNA is converted to a hairpin during breakage. Broken ends are then processed and joined with the help of ubiquitously expressed DNA repair factors, including members of the non-homologous end joining pathway (the DNA-dependent protein kinase and the Ku, Artemis, DNA ligase IV, Cernunnos/XLF and Xrcc4 proteins) and, possibly, histone H2AX and the Mre11/Rad50/Nbs1 complex.33,34 The resulting signal joint (SJ) and coding joint (CJ) products present different structures, with the former corresponding to the back-to-back fusion of RSS heptamers and the latter possibly displaying sequence variability at DNA junctions because of nucleotide deletion and non-templated nucleotide addition. V(D)J rearrangement maintains a correct reading frame in roughly one-third of cases – ‘out-of-frame’ types of CJ accounting for the remaining two-thirds – yielding one productive rearrangement (one that enables production of potentially functional immunoglobulin or TCR chains) for every three attempts on average. Importantly, concurrent analysis of the biochemistry of the RAG proteins provided evidence that these factors are also capable of transposing RSS-ended fragments into new DNA sites. Such a parallel activity helps to explain the mechanism of RAG action and supports proposals that, unexpectedly, V(D)J recombination has evolved from an ancient mobile DNA element.35,36 The importance of the role of VDJ recombinase was further highlighted when misrepair of DNA double-strand breaks produced at immunoglobulin and TCR loci was implicated in the pathogenesis of lymphoid malignancies in humans and in mice (reviewed in ref. 37; also see ref. 38).
Regulated controls of V(D)J recombination
Schematically, three levels of control appear to constrain the activity of the VDJ recombinase. As mentioned above, V(D)J recombination is accurately coordinated to the cell-lineage and developmental stage of differentiating lymphocytes. Hence, immunoglobulin gene complete rearrangements occur in early B lymphocytes whereas TCR gene rearrangements occur in early T lymphocytes. Within the B-cell lineage, IgH genes rearrange first, followed by IgL genes; likewise, within the T lineage, TCRβ genes rearrange first followed by TCRα genes. Moreover, IgH and TCRβ genes also achieve ordered recombination each beginning with a D-to-J rearrangement before being completed by the appendage of a V gene segment to the pre-formed DJ complex. Finally, at some antigen receptor gene loci including the IgH, IgLκ and TCRβ loci, V(D)J recombination appears also to be regulated in the context of allelic exclusion. This phenomenon – whereby antigen receptor chains are eventually encoded on only one of two opposite alleles – ensures the specificity of the immune response depending on antigenic selection of discrete clones of B or T cells, each restricted to expression of a homogeneous set of immunoglobulin receptors or TCR, respectively (the clonal selection theory39). In these cases, the prevailing, so-called regulated model of allelic exclusion proposes that antigen receptor V gene assembly proceeds one allele at a time and that protein products from a functionally relevant rearrangement (one that encodes IgH, IgLκ or TCRβ chains that can contribute to, respectively, the pre-B or B-cell receptors, or the pre-TCRs40,41) mediate allelic exclusion through feedback inhibition of further rearrangement at the corresponding locus. The accessibility hypothesis originally formulated to interpret cell-lineage and developmental stage specificities of V(D)J recombination was extended to the regulation of allelic exclusion assuming in particular that feedback inhibition down-modulates the accessibility of the remaining allele. Because allelically excluded B and T cells generally display DJ rearrangement on, respectively, the two IgH or the two TCRβ alleles and a functional VDJ rearrangement on only one of these, it is thought that down-modulation of the accessibility in the context of allelic exclusion would essentially impinge on V gene segments, at least at the IgH and TCRβ loci.42,43
Accessibility hypothesis: first principles and consolidation
Despite its elegance, the accessibility hypothesis – with a lack of an identified recombinase – for a while remained purely hypothetical, although it was endorsed by strong and evocative correlations (i.e. DNA and chromatin features that mimicked those accompanying gene expression or silencing). Indeed, V(D)J rearranging substrates or endogenous clusters of gene segments were found to generally display hypomethylation of scattered CpG dinucleotides and hypersensitivity to DNase and restriction endonucleases in contrast to the hypermethylation and hyposensitivity exhibited by their non-rearranging control counterparts. Furthermore, immunoglobulin and TCR loci undergoing V(D)J recombination commonly overlapped, in a lineage-specific and developmentally regulated way, with transcriptionally active domains comprised of unrearranged gene segments (hence the term germ-line transcription) – though it remained uncertain whether transcription induces or is merely a by-product of locus accessibility.24 More recently, germ-line transcription was updated, especially where heading in the opposite direction is concerned: within the IgH locus, B-cell developmentally regulated DH-JH antisense intergenic transcripts and VH antisense intergenic transcripts, each appeared to be sequentially produced before DH-to-JH recombination and during the transition from DJH-to-VHDJH recombination, respectively; and were likewise hypothesized to remodel the corresponding domains for V(D)J recombination.44,45
The accessibility hypothesis predicts the existence of accessibility control elements (ACE) at the distinct antigen receptor loci, that would be required to promote V(D)J recombination. Because of the widespread association of V(D)J recombination with germline transcription, it was anticipated that ACE would be connected to the control of the latter process. This prediction was first verified using mouse transgenesis, which demonstrated that transcriptional enhancer elements act to make a linked antigen receptor gene minilocus accessible to VDJ recombinase,46–49 a role that subsequent studies confirmed for most endogenous immunoglobulin and TCR locus enhancer elements through gene-targeted mutation in mouse embryonic stem cells.50 In addition to enhancers, such mouse knockout studies also identified endogenous immunoglobulin and TCR transcriptional promoters as potential ACEs. Generally speaking, enhancer mutation led to inhibition of V(D)J recombination on a whole-locus scale, whereas promoter mutation had a more limited regional impact;51–55 however, both resulted in chromatin remodelling as evidenced by changes in epigenetic marking.56,57 In line with these results, targeted gene mutation of transcription factors or chromatin-modifying factors, known to bind immunoglobulin and TCR ACEs, generally affected, though to varying degrees, V(D)J recombination at the corresponding loci.58–62
Finally, that cell type-specific chromatin structure indeed determines the targeting of VDJ recombinase was elegantly demonstrated by Schlissel's group.63 Using an in vitro system, these authors analysed RAG nuclease-mediated cleavage of RSSs flanking immunoglobulin and TCR gene segments in cell nuclei. They found that both the lineage-specificity and temporal ordering of gene rearrangement is reflected in the accessibility of RSSs within chromatin to in vitro cleavage, so definitively quietening any scepticism that remained about the accessibility hypothesis.
Accessibility hypothesis: the modern age
With chromatin officially holding centre stage, the time came to ascertain the molecular features that distinguish RAG accessible and inaccessible immunoglobulin and TCR loci. This was made possible by the opportunely and newly developed technical approach chromatin immunoprecipitation (ChIP), which uses antibodies raised against, in particular, specific activating or suppressing histone modifications followed by PCR amplification to accurately locate the given mark within the genome (ChIP-PCR). [Note: Later, interrogation of genomic location also used microarray hybridization or deep-sequencing of amplified ChIP DNA: referred to as (ChIP-chip) and (ChIP-seq), respectively.]
Available immunoglobulin/TCR enhancer or promoter mouse mutants also provided invaluable sources of lymphoid cell nuclei for these studies. Overall, none too surprisingly, the immunoglobulin and TCR accessible loci were found to be enriched in epigenetic marks commonly associated with gene activation [including histone H3/H4 acetylation (H3ac; H4ac); di-/tri-methylation of Lys 4 of histone H3 (H3K4me2/me3)]; whereas the inaccessible loci were mostly decorated with epigenetic marks associated with gene silencing [including di-/tri-methylation of Lys 9 or Lys 27 of histone H3 (H3K9me2 and H3K27me3, respectively)].64–69 Interestingly, hotspots of specific activating marks (H3K4me2/me3) or of marks differentially affected by enhancer mutations, for example, were identified, which may represent discrete domains perhaps important in, respectively, primary recruitment of the RAG nuclease,65,66 and the hierarchical establishment of locus-specific, chromosomal accessibility.65,69
At this point, epigenetic ChIP-based analyses were also combined with further genetic manipulations of the mouse genome (gene knockin) to intentionally modulate accessibility and V(D)J recombination at discrete antigen receptor loci, with the aim of challenging the molecular connection between and improving our mechanistic understanding of the two processes. Hence, remarkably, targeting the histone methyl transferase enzyme G9a (mediating H3K9 methylation) to chromosomal recombination substrates containing functional ACEs induced revisions in the local chromatin environment, over-rode the ACEs’ function, and crippled V(D)J recombination of linked chromosomal gene segments.70 Moreover, introducing a transcription terminator into the 5′ end of the mouse TCR-Jα locus to block transcriptional elongation effectively suppressed chromatin remodelling and Vα-to-Jα recombination of 3′ adjacent Jα segments.71 Altogether, these and other studies yielded results compatible with a scenario in which epigenetic chromatin modifications introduced during transcriptional elongation of antigen receptor genes might recruit chromatin remodelling complexes that displace or remodel nucleosomes positioned over RSSs (and thereby increase RSS accessibility to RAG proteins) or might even recruit the RAG nuclease itself, as further discussed below.
RAG2 contains a non-canonical plant homeodomain finger in a part of the protein that is dispensable for RAG-mediated DNA cleavage in vitro (RAG2 non-core region).72,73 In a number of proteins associated with epigenetic regulation, a similar domain specifically binds H3K4me2 or H3K4me3. Two groups, led respectively by M.A. Oettinger and S. Desiderio, have shown that this non-canonical plant homeodomain finger mediates direct binding of RAG2 to H3K4me2 and (preferentially) H3K4me3.74,75 The functional significance of the latter interaction was demonstrated by showing that, in vivo, (i) mutations in the plant homeodomain finger that abolished H3K4me3 recognition severely impaired V(D)J recombination, and (ii) DNA binding and recombination depended on the amount of H3K4me3 deposition. Therefore, recognition of the post-translational H3K4me3 epigenetic mark by RAG2 appears critical to V(D)J recombination. Strikingly, this may not be confined to a function in recombinase recruitment only. Indeed, building on these previous findings, biochemical investigations further indicated that recognition of H3K4me3 by RAG2 also stimulates the catalytic activity of the RAG nuclease;76,77 and, possibly, stabilizes the newly excised recombination ends within the RAG post-cleavage complex and their transfer to the non-homologous end joining repair machinery.78
In this context, what about in vivo binding of the RAG nuclease to RSS-containing domains of endogenous antigen receptor loci? This issue was beautifully addressed by Schatz's group who used transgenic mice expressing an active site mutant RAG1 protein that binds DNA normally (and interacts with RAG2) but lacks catalytic activity, so avoiding the formation of recombination products that could complicate the interpretation of ChIP data at immunoglobulin and TCR loci. They demonstrated that RAG protein binding occurs in a focal manner to small regions rich in activating histone modifications (H3Ac, H3K4me3), which they referred to as ‘recombination centres’.31 Notably, these comprised regions encompassing Igκ and TCRα J gene segments and IgH and TCRβ J and J-proximal D gene segments. Interestingly, while RAG1 binds mostly to RSS-containing regions, RAG2 binds broadly to H3K4me-rich sequences throughout the mouse genome. A later study using mutant TCRα and TCRβ alleles found that enhancers control RAG1 binding globally at Jα or Dβ-Jβ gene segments, and that promoters direct RAG1 binding locally, a profile that recapitulates the V(D)J recombinational function of these ACEs defined in previous knockout studies.79 Overall, ‘recombination centres’ were interpreted as specialized sites of high local RAG concentration that facilitate RSS binding and synapsis and help to regulate recombination order and fidelity.80 Indeed, recent analysis of DJH-recombined alleles provided evidence that DJH junctions are selectively epigenetically marked and bind RAG proteins, thereby probably permitting DJH-5′ intact RSSs to initiate the second step of IgH gene assembly.81
Accessibility in 3D
At several immunoglobulin and TCR loci, RSS synapsis poses a real challenge as V, D (or DJ), and J gene segments may be located far apart [> 1–2 Megabases (Mb)] on the chromosome. A variety of studies have implicated conformational changes of such loci as important determinants of long-distance V(D)J recombination events. 3D fluorescence in situ hybridization analyses have revealed large-scale compaction of immunoglobulin and TCR loci, which is developmentally regulated and therefore assumed to punctually facilitate the synapsis of distant RSSs.82–85 For example, the IgH locus looks compacted in pro-B cells undergoing VH-to-DJH rearrangement; whereas such compaction is released in the subsequent pre-B stage, forcing the physical separation of the distal VH genes from the proximal IgH domain at a stage where further IgH rearrangement is prevented.83 A detailed analysis of the topography of the IgH locus in pro-B cells predicted that the entire locus is organized into dynamic compartments containing clusters of loops with VH regions juxtaposed to the DH elements, mechanistically permitting long-range genomic interactions to occur.86
The molecular features that may be responsible for the developmental regulation of immunoglobulin/TCR locus compaction/de-compaction are on the verge of disclosure. We know that IgH compaction depends on the transcription factors Pax5, Ikaros and YY1.87–89 Moreover, putative regulatory sequences of conserved repeat elements with a potential role in these processes were recently discovered in the distal V gene cluster of the IgH locus.90 These ‘PAIR’ elements are bound by Pax-5 specifically in pro-B cells and subjected to Pax-5-dependent antisense transcription. They also recruit the transcription factors E2A and CTCF throughout B-cell development. Lately, the CTCF zinc finger protein and its partner, the protein complex cohesin, have attracted attention as potentially important players in the regulation of V(D)J recombination at several immunoglobulin and TCR loci, through the shaping of DNA looping interactions and their contribution to lineage-specific and developmentally ordered accessibility and germline non-coding transcription.91–95 Last but not least, detailed analysis (using RNA deep-sequencing) of germline non-coding transcription throughout the IgH locus has indicated that the majority of antisense transcripts localize around a limited number of PAIR elements; and provided evidence that this particular activity might affect the 3D chromosomal structure, bringing the distal part of the VH locus close to the domain comprising the rearranged DJH and adjacent enhancer Eμ.99 The overall emerging picture would be that of chromosomal ‘rosettes’ folding the dispersed VH gene segments around a core domain bound by the recombinase, so creating opportunities for VH-to-DJH rearrangement; with nascent non-coding transcription possibly facilitating locus compaction and the formation of macromolecular structures that serve as transcription and recombination factories99–101 (Fig. 1a).
Figure 1.
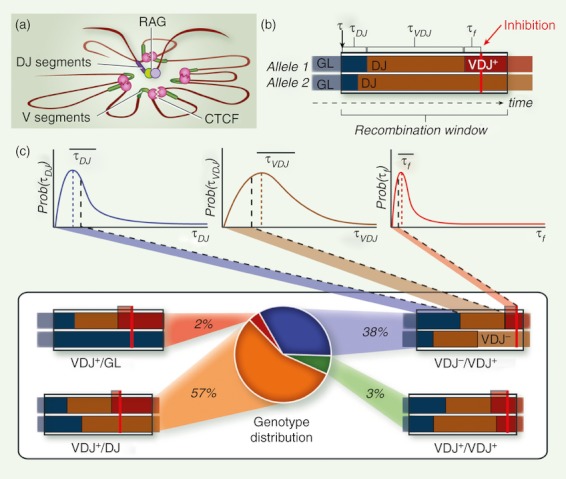
Dynamic combination of deterministic and stochastic features enforces allelic exclusion at V-D-J containing gene loci. (a) Illustration of a V-DJ recombination factory as proposed by Lucas et al. (Curr Opin Cell Biol 2011;23:318–24). The diagram illustrates rosette-like structures at an antigen receptor allele that would bring remote V gene segments in proximity to a cis-linked, RAG-loaded DJ-recombined intermediate product. At homologous alleles, stochastic proceeding of V-to-DJ recombination is assumed to mostly contribute to the V-D-J assembly time of the given locus [illustrated by the dark brown sections in (b)]. (b) Schematic depiction of stochastic modelling of DNA rearrangement at V-D-J containing antigen receptor alleles based on the Markov process formalism, as described by Farcot et al. (J Immunol 2010;185:1622–32). The recombination window defines the time period along developmental maturation of a single lymphoid cell during which all requirements for sequential rearrangements (including epigenetics) could be met (hence during which recombination of the given locus could occur). The blue, brown and red sections represent the time course of mono-allelic behaviours in a collection of individual single cells from, respectively, (i) the transition from germline (GL) to DJ-rearranged (DJ) allelic statuses, (ii) the transition from DJ-rearranged to VDJ-rearranged (VDJ) allelic statuses, and (iii) the residence of productively rearranged (VDJ+) alleles [or of non-productively rearranged (VDJ−) alleles; depicted in light brown in the upper right diagram of part (c)] till the recombination window comes to an end. Transition times τDJ and τVDJ correspond to the time lapses required in the particular case to achieve steps (i) and (ii), respectively; τf corresponds to the time lapse required to achieve feedback inhibition (red bar). (c) Statistical compilation of biallelic behaviours and their contribution to the VDJ+/DJ, VDJ−/VDJ+, VDJ+/VDJ+ and VDJ+/GL final genomic distribution. The probabilistic curves (Prob) for the τDJ, τVDJ, and τf parameters are shown on the top, with the locations of the mean values indicated ( ).
).
The control of allelic exclusion: deterministic versus stochastic
One overarching aim of 3D conformation analyses of immunoglobulin and TCR loci is a better understanding of the molecular rules that enforce allelic exclusion, a still puzzling phenomenon. It appears that allelic exclusion impinges on the assembly of V gene segments, and comprises an initiation phase to dissociate V-to-(D)J rearrangements on the two opposite alleles, followed by a maintenance phase to prohibit these events once a functional V(D)J joint has been made.42,43 In this respect, it is ostensibly appealing to consider the reversible contraction and subnuclear compartmentalization processes affecting the IgH/IgLκ and TCRβ loci in, respectively, developing pro-/pre-B and pro/pre-T cells as key regulatory mainsprings that enforce allelic exclusion (at least and most obviously during the maintenance phase).82–84 However, rather than mere locus compaction, the primary importance in enforcing allelic exclusion, at least at the IgH locus, may be that the V and D domains remain functionally separate, perhaps achieved through the insulator/DNA looping activity of CTCF–cohesin complexes.93,102 Separate studies have suggested additional mechanisms that could possibly also contribute to inducing or maintaining allelic dissociation, including (i) monoallelic epigenetic changes that may occur even before rearrangement (as reflected for example by asynchronous immunoglobulin and TCR locus replication103,104), and would eventually dispatch the two differentially packaged alleles towards either the centre of the nucleus (active allele) or heterochromatin compartments (silenced allele);105,106 (ii) pairing of homologous alleles and, following a RAG-induced DNA break on one of them, ATM-dependent repositioning of the other to pericentromeric heterochromatin;107 and (iii) discrimination between productive and non-productive alleles through differential stability of their mRNA products with a suppressive effect of the remaining stable mRNA on V(D)J recombination.108 Contrary to such deterministic procedures, however, allelic dissociation in V-to-(D)J joining was also proposed to simply rely on the stochastic accessibility of only a small fraction of alleles, as the result of either a high frequency interaction with repressive nuclear compartments including the nuclear lamina and pericentromeric heterochromatin,109 or, in a non-mutually exclusive manner, variegated transcriptional activation.110
The control of allelic exclusion: deterministic, stochastic and dynamic
Neither a purely deterministic nor a purely stochastic representation alone sufficiently accounts for allelic exclusion. Distinct from strictly mono-allelic, deterministic gene expression systems such as X chromosome inactivation or gene imprinting, allelic exclusion is a ‘faulty’ developmental process with a low but sizeable fraction of the emerging lymphoid cells being allelically included that display productive rearrangements on, e.g. the two IgH or TCRβ alleles.43 [Note: It is now clear that multiple mechanisms function in a successive manner to limit the frequency of cells with surface expression of immunoglobulin or TCR chains from productive rearrangement at both allelic copies of the corresponding loci, which depending on the locus considered may vary from 1 to 10% (ref. 43 and references therein)].
On the other hand, highly stochastic scenarios are hard to reconcile with the relatively high proportion (40–45%) of cells displaying fully rearranged, productive and non-productive, IgH or TCRβ alleles. Using TCRβ allelic exclusion as a reference system, we proposed that a dynamic combination of the two concepts, determinism and stochasticity – as modelled using the Markov chain formalism, better accounts for the distribution of TCRβ genotypes emerging from early T-cell development;111 (Fig. 1b,c). The deterministic features would simply include the basic attribute of randomness in V(D)J recombination (one-third productive / two-thirds non-productive rearrangement outcomes) and, with regards to TCRβ gene recombination, the sequential Dβ-Jβ and Vβ-DJβ rearrangements with no direct Vβ-Jβ joining (B12/23 constraint), followed by and ending with feedback inhibition. Molecular changeovers are characterized by transition rates that are expressed in terms of the mean duration of a given rearrangement step and the average time lapse to achieve inhibition, respectively. At the single cell level, however, stochasticity relies on temporal variations in the execution of these changes (in other words, noise), both between cells (extrinsic noise) and alleles (intrinsic noise), with epigenetics likely occupying the front stage. It is increasingly recognized that cell fate decisions and underlying genetic circuits are subjected to such stochastic fluctuations to generate non-genetic cellular diversity; with, in mammals, intrinsic noise in gene expression mainly depending on epigenetic-regulated chromatin changes.112,113 When integrated at the level of a whole cell-population, this model makes it possible to readily predict the distinct TCRβ cell genotypes and allelic exclusion/inclusion profiles from wild-type and mutant mice.111 In keeping with the same concepts, we anticipate that corresponding features at other gene loci could likewise be interpreted. Indeed, within the transcription and recombination factories evoked above, VH-to-DJH recombination might putatively proceed by way of dynamic and stochastic interactions involving on the one hand the VH domain-folded ‘rosettes’ and on the other hand the inner RAG-loaded loci comprising the DJH intermediate CJ products, respectively.99–101
Conclusions
The long-standing joint venture between V(D)J recombination and epigenetics tells a win–win story. Epigenetics has already contributed to ascertaining and clarifying the regulated recruitment of a common recombinase to RSS substrates that vary according to modulated changes in gene accessibility. Deeper examination of these controls has also offered a unique insight into the epigenetic multifaceted potential to promoting accurate interconnection between widely separated gene partners within an intricate chromosomal landscape. The application of increasingly sophisticated genomic approaches to investigate these issues should continue to disclose as yet unforeseen biological resources that enforce the V(D)J recombination–epigenetics partnership. Mathematical modelling based on the dynamics of epigenetic-driven changes allied to (by nature) randomness in V(D)J recombination outcomes may concur and uncover the true intricacy of regulatory controls on such as allelic exclusion. Advances could also arise from the investigation of apparently conflicting results such as those regarding the generation of the TCRα repertoire as analysed by fluorescence in situ hybridization in single cells or deep-sequencing from whole αβ T-cell populations, which either suggested a co-ordinate bidirectional trimming mechanism that relies on the proximity of Vα and Jα gene segments or the formation of intralocus loops whereby all Vα gene segments have equal opportunity to recombine.85,114 More generally, the development of elaborate tools to improve our understanding of V(D)J recombination events,115 or the structural organization of immunoglobulin/TCR chromosomal DNA,111,116,117 may shed more light on individual differences118 in immune repertoire development.
Acknowledgments
Work in the P.F. laboratory is supported by institutional grants from Inserm and CNRS, and by grants from the Commission of the European Communities, the Agence Nationale de la Recherche, the Institut National du Cancer, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale and the Fondation Princesse Grace de la Principauté de Monaco. B.F. was supported by a Marie Curie EU fellowship (PIOF-GA-2009-235741) and by CNRS in the framework of the project PEPS Physique Théorique et ses Interfaces. We apologize to those colleagues whose work we could not acknowledge owing to space restrictions.
Disclosures
The authors declare no conflict of interest.
References
- 1.Hozumi N, Tonegawa S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc Natl Acad Sci U S A. 1976;73:3628–32. doi: 10.1073/pnas.73.10.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alt FW, Oltz EM, Young F, Gorman J, Taccioli G, Chen J. VDJ recombination. Immunol Today. 1992;13:306–14. doi: 10.1016/0167-5699(92)90043-7. [DOI] [PubMed] [Google Scholar]
- 3.Schatz DG. V(D)J recombination. Immunol Rev. 2004;200:5–11. doi: 10.1111/j.0105-2896.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- 4.Schatz DG, Baltimore D. Uncovering the V(D)J recombinase. Cell. 2004;116:S103–6. doi: 10.1016/s0092-8674(04)00042-x. 102 p following S106. [DOI] [PubMed] [Google Scholar]
- 5.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–96. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 7.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 8.Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu Rev Genet. 2012;46:419–41. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- 9.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 10.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–91. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 11.Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–6. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Mohn F, Schubeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25:129–36. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Turner BM. The adjustable nucleosome: an epigenetic signaling module. Trends Genet. 2012;28:436–44. doi: 10.1016/j.tig.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–81. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dostie J, Bickmore WA. Chromosome organization in the nucleus – charting new territory across the Hi-Cs. Curr Opin Genet Dev. 2012;22:125–31. doi: 10.1016/j.gde.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Fudenberg G, Mirny LA. Higher-order chromatin structure: bridging physics and biology. Curr Opin Genet Dev. 2012;22:115–24. doi: 10.1016/j.gde.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brack C, Tonegawa S. Variable and constant parts of the immunoglobulin light chain gene of a mouse myeloma cell are 1250 nontranslated bases apart. Proc Natl Acad Sci U S A. 1977;74:5652–6. doi: 10.1073/pnas.74.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981;27:381–90. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- 20.Alt FW, Yancopoulos GD, Blackwell TK, et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–19. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yancopoulos GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–81. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 22.Yancopoulos GD, Blackwell TK, Suh H, Hood L, Alt FW. Introduced T cell receptor variable region gene segments recombine in pre-B cells: evidence that B and T cells use a common recombinase. Cell. 1986;44:251–9. doi: 10.1016/0092-8674(86)90759-2. [DOI] [PubMed] [Google Scholar]
- 23.Blackwell TK, Moore MW, Yancopoulos GD, Suh H, Lutzker S, Selsing E, Alt FW. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986;324:585–9. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- 24.Alt FW, Blackwell TK, DePinho RA, Reth MG, Yancopoulos GD. Regulation of genome rearrangement events during lymphocyte differentiation. Immunol Rev. 1986;89:5–30. doi: 10.1111/j.1600-065x.1986.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 25.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–32. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 26.Schatz DG, Spanopoulou E. Biochemistry of V(D)J recombination. Curr Top Microbiol Immunol. 2005;290:49–85. doi: 10.1007/3-540-26363-2_4. [DOI] [PubMed] [Google Scholar]
- 27.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 28.Jones JM, Gellert M. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J. 2002;21:4162–71. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mundy CL, Patenge N, Matthews AG, Oettinger MA. Assembly of the RAG1/RAG2 synaptic complex. Mol Cell Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curry JD, Geier JK, Schlissel MS. Single-strand recombination signal sequence nicks in vivo: evidence for a capture model of synapsis. Nat Immunol. 2005;6:1272–9. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- 31.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–31. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sleckman BP. Lymphocyte antigen receptor gene assembly: multiple layers of regulation. Immunol Res. 2005;32:253–8. doi: 10.1385/IR:32:1-3:253. [DOI] [PubMed] [Google Scholar]
- 33.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 35.Fugmann SD. The origins of the Rag genes – from transposition to V(D)J recombination. Semin Immunol. 2010;22:10–6. doi: 10.1016/j.smim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hencken CG, Li X, Craig NL. Functional characterization of an active Rag-like transposase. Nat Struct Mol Biol. 2012;19:834–6. doi: 10.1038/nsmb.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dadi S, Le Noir S, Asnafi V, Beldjord K, Macintyre EA. Normal and pathological VDJ recombination: contribution of the understanding of human lymphoid malignancies. Adv Exp Med Biol. 2009;650:180–94. doi: 10.1007/978-1-4419-0296-2_15. [DOI] [PubMed] [Google Scholar]
- 38.Deriano L, Chaumeil J, Coussens M, et al. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471:119–23. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burnet FM. The Clonal Selection Theory of Acquired Immunity. Cambridge: The University Press; 1959. [Google Scholar]
- 40.Kisielow P, von Boehmer H. Development and selection of T cells: facts and puzzles. Adv Immunol. 1995;58:87–208. doi: 10.1016/s0065-2776(08)60620-3. [DOI] [PubMed] [Google Scholar]
- 41.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–8. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 42.Mostoslavsky R, Alt FW, Rajewsky K. The lingering enigma of the allelic exclusion mechanism. Cell. 2004;118:539–44. doi: 10.1016/j.cell.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 43.Brady BL, Steinel NC, Bassing CH. Antigen receptor allelic exclusion: an update and reappraisal. J Immunol. 2010;185:3801–8. doi: 10.4049/jimmunol.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan G, Chakalova L, Fraser PJ, Corcoran AE. Antisense intergenic transcription in V(D)J recombination. Nat Immunol. 2004;5:630–7. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 45.Bolland DJ, Wood AL, Afshar R, Featherstone K, Oltz EM, Corcoran AE. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Mol Cell Biol. 2007;27:5523–33. doi: 10.1128/MCB.02407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrier P, Krippl B, Blackwell TK, et al. Separate elements control DJ and VDJ rearrangement in a transgenic recombination substrate. EMBO J. 1990;9:117–25. doi: 10.1002/j.1460-2075.1990.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capone M, Watrin F, Fernex C, Horvat B, Krippl B, Scollay R, Ferrier P. TCRα and TCRβ gene enhancers confer tissue- and stage-specificity on V(D)J recombination events. EMBO J. 1993;12:4335–46. doi: 10.1002/j.1460-2075.1993.tb06118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lauzurica P, Krangel MS. Temporal and lineage-specific control of T cell receptor α/δ gene rearrangement by T cell receptor α and δ enhancers. J Exp Med. 1994;179:1913–21. doi: 10.1084/jem.179.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lauzurica P, Krangel MS. Enhancer-dependent and -independent steps in the rearrangement of a human T cell receptor δ transgene. J Exp Med. 1994;179:43–55. doi: 10.1084/jem.179.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 51.Bories JC, Demengeot J, Davidson L, Alt FW. Gene-targeted deletion and replacement mutations of the T-cell receptor β-chain enhancer: the role of enhancer elements in controlling V(D)J recombination accessibility. Proc Natl Acad Sci U S A. 1996;93:7871–6. doi: 10.1073/pnas.93.15.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouvier G, Watrin F, Naspetti M, Verthuy C, Naquet P, Ferrier P. Deletion of the mouse T-cell receptor β gene enhancer blocks αβ T-cell development. Proc Natl Acad Sci U S A. 1996;93:7877–81. doi: 10.1073/pnas.93.15.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villey I, Caillol D, Selz F, Ferrier P, de Villartay JP. Defect in rearrangement of the most 5′ TCR-J α following targeted deletion of T early α (TEA): implications for TCR α locus accessibility. Immunity. 1996;5:331–42. doi: 10.1016/s1074-7613(00)80259-9. [DOI] [PubMed] [Google Scholar]
- 54.Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW. Function of the TCR α enhancer in αβ and γδ T cells. Immunity. 1997;7:505–15. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- 55.Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the Dβ1 gene segment at the TCR β locus by a germline promoter. Immunity. 1999;10:313–22. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- 56.Mathieu N, Hempel WM, Spicuglia S, Verthuy C, Ferrier P. Chromatin remodeling by the T cell receptor (TCR)-β gene enhancer during early T cell development: implications for the control of TCR-β locus recombination. J Exp Med. 2000;192:625–36. doi: 10.1084/jem.192.5.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitehurst CE, Schlissel MS, Chen J. Deletion of germline promoter PD β1 from the TCR β locus causes hypermethylation that impairs D β1 recombination by multiple mechanisms. Immunity. 2000;13:703–14. doi: 10.1016/s1074-7613(00)00069-8. [DOI] [PubMed] [Google Scholar]
- 58.Lazorchak AS, Schlissel MS, Zhuang Y. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin κ locus in pre-B cells. Mol Cell Biol. 2006;26:810–21. doi: 10.1128/MCB.26.3.810-821.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. Regulation of T cell receptor β gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity. 2007;27:871–84. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 60.Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, Singh H. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–45. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 61.D'Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010;11:240–9. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou XF, Yu J, Chang M, Zhang M, Zhou D, Cammas F, Sun SC. TRIM28 mediates chromatin modifications at the TCRα enhancer and regulates the development of T and natural killer T cells. Proc Natl Acad Sci U S A. 2012;109:20083–88. doi: 10.1073/pnas.1214704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanhope-Baker P, Hudson KM, Shaffer AL, Constantinescu A, Schlissel MS. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–97. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- 64.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–8. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 65.Spicuglia S, Kumar S, Yeh J-H, Vachez E, Chasson L, Gorbatch S, Cautres J, Ferrier P. Promoter activation by enhancer-dependent and -independent loading of activator and coactivator complexes. Mol Cell. 2002;10:1479–87. doi: 10.1016/s1097-2765(02)00791-8. [DOI] [PubMed] [Google Scholar]
- 66.Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A. 2003;100:11577–82. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Espinoza CR, Feeney AJ. The extent of histone acetylation correlates with the differential rearrangement frequency of individual VH genes in pro-B cells. J Immunol. 2005;175:6668–75. doi: 10.4049/jimmunol.175.10.6668. [DOI] [PubMed] [Google Scholar]
- 68.Espinoza CR, Feeney AJ. Chromatin accessibility and epigenetic modifications differ between frequently and infrequently rearranging V(H) genes. Mol Immunol. 2007;44:2675–85. doi: 10.1016/j.molimm.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chakraborty T, Perlot T, Subrahmanyam R, et al. A 220-nucleotide deletion of the intronic enhancer reveals an epigenetic hierarchy in immunoglobulin heavy chain locus activation. J Exp Med. 2009;206:1019–27. doi: 10.1084/jem.20081621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Osipovich O, Milley R, Meade A, Tachibana M, Shinkai Y, Krangel MS, Oltz EM. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nat Immunol. 2004;5:309–16. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- 71.Abarrategui I, Krangel MS. Regulation of T cell receptor-α gene recombination by transcription. Nat Immunol. 2006;7:1109–15. doi: 10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
- 72.Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–91. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elkin SK, Ivanov D, Ewalt M, et al. A PHD finger motif in the C terminus of RAG2 modulates recombination activity. J Biol Chem. 2005;280:28701–10. doi: 10.1074/jbc.M504731200. [DOI] [PubMed] [Google Scholar]
- 74.Matthews AG, Kuo AJ, Ramon-Maiques S, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–10. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–71. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol Cell. 2009;34:535–44. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grundy GJ, Yang W, Gellert M. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc Natl Acad Sci U S A. 2010;107:22487–92. doi: 10.1073/pnas.1014958107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang G, Dhar K, Swanson PC, Levitus M, Chang Y. Real-time monitoring of RAG-catalyzed DNA cleavage unveils dynamic changes in coding end association with the coding end complex. Nucleic Acids Res. 2012;40:6082–96. doi: 10.1093/nar/gks255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ji Y, Little AJ, Banerjee JK, Hao B, Oltz EM, Krangel MS, Schatz DG. Promoters, enhancers, and transcription target RAG1 binding during V(D)J recombination. J Exp Med. 2010;207:2809–16. doi: 10.1084/jem.20101136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–63. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 81.Subrahmanyam R, Du H, Ivanova I, et al. Localized epigenetic changes induced by DH recombination restricts recombinase to DJH junctions. Nat Immunol. 2012;13:1205–12. doi: 10.1038/ni.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–62. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 83.Roldán E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok JA. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Skok JA, Gisler R, Novatchkova M, Farmer D, de LW, Busslinger M. Reversible contraction by looping of the Tcrα and Tcrβ loci in rearranging thymocytes. Nat Immunol. 2007;8:378–87. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 85.Shih HY, Krangel MS. Distinct contracted conformations of the Tcra/Tcrd locus during Tcra and Tcrd recombination. J Exp Med. 2010;207:1835–41. doi: 10.1084/jem.20100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jhunjhunwala S, van Zelm MC, Peak MM, et al. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–79. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–22. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu H, Schmidt-Supprian M, Shi Y, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–89. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reynaud D, Demarco A, Reddy L, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–36. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ebert A, McManus S, Tagoh H, et al. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–87. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 91.Degner SC, Verma-Gaur J, Wong TP, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci U S A. 2011;108:9566–71. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seitan VC, Hao B, Tachibana-Konwalski K, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–71. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guo C, Yoon HS, Franklin A, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–30. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ribeiro de Almeida C, Stadhouders R, de Bruijn MJ, et al. The DNA-binding protein CTCF limits proximal Vκ recombination and restricts κ enhancer interactions to the immunoglobulin κ light chain locus. Immunity. 2011;35:501–13. doi: 10.1016/j.immuni.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 95.Shih HY, Verma-Gaur J, Torkamani A, Feeney AJ, Galjart N, Krangel MS. Tcra gene recombination is supported by a Tcra enhancer- and CTCF-dependent chromatin hub. Proc Natl Acad Sci U S A. 2012;109:E3493–502. doi: 10.1073/pnas.1214131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seitan VC, Krangel MS, Merkenschlager M. Cohesin, CTCF and lymphocyte antigen receptor locus rearrangement. Trends Immunol. 2012;33:153–9. doi: 10.1016/j.it.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chaumeil J, Skok JA. The role of CTCF in regulating V(D)J recombination. Curr Opin Immunol. 2012;24:153–9. doi: 10.1016/j.coi.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ribeiro de Almeida C, Stadhouders R, Thongjuea S, Soler E, Hendriks RW. DNA-binding factor CTCF and long-range gene interactions in V(D)J recombination and oncogene activation. Blood. 2012;119:6209–18. doi: 10.1182/blood-2012-03-402586. [DOI] [PubMed] [Google Scholar]
- 99.Verma-Gaur J, Torkamani A, Schaffer L, Head SR, Schork NJ, Feeney AJ. Noncoding transcription within the Igh distal VH region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci U S A. 2012;109:17004–9. doi: 10.1073/pnas.1208398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jhunjhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138:435–48. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lucas JS, Bossen C, Murre C. Transcription and recombination factories: common features? Curr Opin Cell Biol. 2011;23:318–24. doi: 10.1016/j.ceb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bates JG, Cado D, Nolla H, Schlissel MS. Chromosomal position of a VH gene segment determines its activation and inactivation as a substrate for V(D)J recombination. J Exp Med. 2007;204:3247–56. doi: 10.1084/jem.20071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mostoslavsky R, Singh N, Tenzen T, et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–5. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- 104.Farago M, Rosenbluh C, Tevlin M, et al. Clonal allelic predetermination of immunoglobulin-κ rearrangement. Nature. 2012;490:561–5. doi: 10.1038/nature11496. [DOI] [PubMed] [Google Scholar]
- 105.Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, Bergman Y. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 106.Cedar H, Bergman Y. Choreography of Ig allelic exclusion. Curr Opin Immunol. 2008;20:308–17. doi: 10.1016/j.coi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 107.Hewitt SL, Yin B, Ji Y, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–64. doi: 10.1038/ni.1735. Published erratum appeared in 2009 Nat. Immunol. 2010:1034 and 2010 Nat. Immunol. 2011:2355-2356002E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lutz J, Heideman MR, Roth E, et al. Pro-B cells sense productive immunoglobulin heavy chain rearrangement irrespective of polypeptide production. Proc Natl Acad Sci U S A. 2011;108:10644–9. doi: 10.1073/pnas.1019224108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schlimgen RJ, Reddy KL, Singh H, Krangel MS. Initiation of allelic exclusion by stochastic interaction of Tcrβ alleles with repressive nuclear compartments. Nat Immunol. 2008;9:802–9. doi: 10.1038/ni.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liang H-E, Hsu L-Y, Cado D, Schlissel MS. Variegated transcriptional activation of the immunoglobulin κ locus in pre-b cells contributes to the allelic exclusion of light-chain expression. Cell. 2004;118:19–29. doi: 10.1016/j.cell.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 111.Farcot E, Bonnet M, Jaeger S, Spicuglia S, Fernandez B, Ferrier P. TCRβ allelic exclusion in dynamical models of V(D)J recombination based on allele independence. J Immunol. 2010;185:1622–32. doi: 10.4049/jimmunol.0904182. [DOI] [PubMed] [Google Scholar]
- 112.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–73. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–25. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Genolet R, Stevenson BJ, Farinelli L, Osteras M, Luescher IF. Highly diverse TCRα chain repertoire of pre-immune CD8+ T cells reveals new insights in gene recombination. EMBO J. 2012;31:1666–78. doi: 10.1038/emboj.2012.48. Addendum, EMBO J31, 4247-4248 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Murugan A, Mora T, Walczak AM, Callan CG., Jr Statistical inference of the generation probability of T-cell receptors from sequence repertoires. Proc Natl Acad Sci U S A. 2012;109:16161–6. doi: 10.1073/pnas.1212755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ndifon W, Gal H, Shifrut E, et al. Chromatin conformation governs T-cell receptor Jβ gene segment usage. Proc Natl Acad Sci U S A. 2012;109:15865–70. doi: 10.1073/pnas.1203916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thuderoz F, Simonet MA, Hansen O, et al. Numerical modelling of the V-J combinations of the T cell receptor TRA/TRD locus. PLoS Comput Biol. 2010;6:e1000682. doi: 10.1371/journal.pcbi.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Murray JM, Messier T, Rivers J, O'Neil JPl, Walker VE, Vacek PM, Finette BA. V(D)J Recombinase-Mediated TCR β locus gene usage and coding joint processing in peripheral T cells during perinatal and pediatric development. J Immunol. 2012;189:2356–64. doi: 10.4049/jimmunol.1200382. [DOI] [PubMed] [Google Scholar]


