Abstract
The motor head of kinesin carries out microtubule binding, ATP hydrolysis, and force generation. Despite a high level of sequence and structural conservation, subtle variations in subdomains of the motor head determine family-specific properties. In particular, both Kinesin-1 (Kin-1) and Kinesin-5 (Kin-5) walk processively to the microtubule plus-end, yet show distinct motility characteristics suitable for their functions. We studied chimeric Kin-1/Kin-5 constructs with a combination of single molecule motility assays and molecular dynamics simulations to demonstrate that Kin-5 possesses a force-generating element similar to Kin-1, i.e., the cover-neck bundle. Furthermore, the Kin-5 neck linker makes additional contacts with the core of the motor head via loop L13, which putatively compensates for the shorter cover-neck bundle of Kin-5. Our results indicate that Kin-1 is mechanically optimized for individual cargo transport, whereas Kin-5 does not necessarily maximize its mechanical performance. Its biochemical rates and enhanced force sensitivity may instead be beneficial for operation in a group of motors. Such variations in subdomains would be a strategy for achieving diversity in motility with the conserved motor head.
Introduction
Kinesins are microtubule-associated motor proteins that are responsible for a wide variety of cellular processes, such as vesicle trafficking, mitotic spindle organization and maintenance, and microtubule depolymerization (1). They have a conserved motor head (2) whose ATPase domain is very similar to those of other major classes of proteins, including myosins and G-proteins (3–5). In certain kinesin families (Kinesin-1, -3, and -5, family names from Lawrence et al. (6)), the motor head and the neck stalk are connected via the neck linker domain that is crucial for force generation and stepping motion (7). An important issue regarding kinesin domain organization is how structurally similar components are fine-tuned to give rise to motility characteristics that meet the functional needs for each kinesin family.
Here, we investigate the basis of divergent motility characteristics among kinesin families by utilizing Kinesin-1 (Kin-1) and Kinesin-5 (Kin-5), which are two closely related motors with dissimilar biophysical characteristics. Kin-1 was chosen as it has generally been used as the model motor from which much of our understanding of the kinesin mechanochemical cycle has been derived. Kin-5, on the other hand, has been widely studied due to its action in spindle formation during mitosis (8) and is a therapeutically relevant target for cancer treatment (9–14).
The motor heads of Kin-1 and -5 have ∼44% sequence identity, and are structurally very similar (based on aligning Protein Data Bank (15) structures PDB:2Y65 for Drosophila melanogaster Kin-1 (16) and PDB:1Q0B for Homo sapiens Kin-5 (17); Fig. 1, and see Text S1 in the Supporting Material). Nevertheless, these motors differ greatly in their motile characteristics, and are not functionally equivalent. For example, replacing the motor head of Kin-5 with that of Kin-1 or of the nonprocessive Kinesin-10, led to collapse of mitotic spindles into bundles of microtubules in Xenopus egg extract (18). Kin-1 is highly processive with unloaded run lengths of a few microns, unloaded velocities of 500–700 nm/s, and a stall force of 5–7 pN (19–23). In comparison, Kin-5 is weakly processive, with an unloaded run length of only ∼60 nm and approximately one-fifth as fast with an unloaded velocity of ∼100 nm/s (24). Kin-5 has been found to resist loads (before dissociation from the microtubule) ranging from 1.5 to 7 pN (24,25). Unlike the clear stalls of Kin-1, Kin-5 does not appreciably slow before abruptly detaching from the microtubule. A dimeric construct with the Kin-5 motor head and Kin-1 coiled-coil stalk was reported to have a dissociation force of 4.6 pN, with an increased processivity (1.9 μm) (26).
Figure 1.
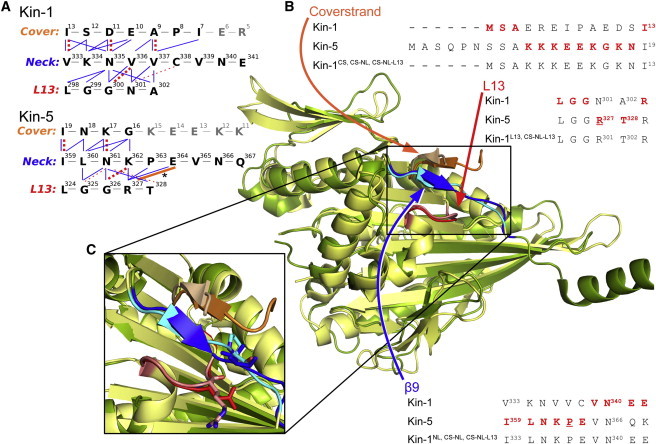
Structural alignment between Kin-1 (dark green; PDB:2Y65 (16)), and Kin-5 (light yellow; PDB:1Q0B (17)). (A) Contact map between the three domains in Kin-1 and Kin-5. Bonds are assessed from the crystal structures, and are as follows: (thick dashed) backbone hydrogen bond; (thin dashed) hydrogen bond between the backbone and a side chain; (thin solid) nonpolar interaction; and (thick solid) R327-E364 salt bridge in Kin-5 (also denoted with an asterisk). Contact maps for the other constructs tested in the neck-pulling simulations are shown in Fig. S1. (B) Sequences of the respective elements for Kin-1 and Kin-5 along with the resulting chimeric sequence. β9 comprises the first seven residues of the neck linker (up to V339 in Kin-1). (C) Magnified view of the three domains. (Lighter colors) Domains of Kin-5. The conserved V337 (neck linker) and N301 (L13) of Kin-1 (darker color) and the corresponding P363 and R327 in Kin-5 (lighter color) are shown in stick representations.
Although interaction between the Kin-5 motor head and other proteins may play a role (27,28), tuning of motility characteristics, such as unloaded velocity, processivity, and force generation, may dominate family-specific function. These features are in turn determined by variations in the relevant domains of the motor (29). For Kin-1, motility is driven by the forward bias of the neck linker, which is precipitated by the formation of a β-sheet named the cover-neck bundle (CNB) between the cover strand (β0) at the N-terminal end of the motor head, and the first half of the neck linker (β9) (21,30). This structural rearrangement occurs upon binding of ATP to the microtubule-bound motor head (31). After the forward motion driven by the CNB, the unbound head searches for the new leading position to bind on the microtubule (29,32–34). Based on sequence alignment and structural comparison, it has been suggested that the CNB mechanism for force generation applies across kinesin families including Kin-1, -3, and -5 (30,35).
Loop 13 (L13), found on the core motor domain located beneath the neck linker (Fig. 1), may also influence mechanical transition of the neck linker. We have previously observed that neck-linker binding to the motor head can be blocked in simulations where an ADP rather than ATP Kin-1 structure is used (30). This effect is due to L13 adopting a conformation in the ADP state that occludes the binding pocket of the neck linker. Additionally, alanine mutations of two conserved glycine residues of L13 reduce the microtubule gliding velocity of Kin-1 by 100-fold (7).
To quantitatively understand the roles of the cover strand, neck linker, and L13, we systematically studied the motile characteristics of chimeras containing different combinations of these domains from Kin-1 and -5. We find that these domains are interchangeable between Kin-1 and -5. Kinetic analysis using a three-state model (24) reveals quantitative differences between chimeras, consistent with differences in their amino-acid composition and interdomain contacts. Interestingly, the use of the fully paired Kin-5 CNB unit rescues processivity defects that develop when the Kin-5 neck linker is used alone. Also, we find that the Kin-5 L13 reduces force sensitivity when combined with the Kin-5 CNB, which is likely due to stronger contacts between the neck linker and L13. These results were further supported by a suite of molecular dynamics (MD) simulations including computational pulling, relaxation, and tug-of-war sampling. In MD simulations, L13 of Kin-5 indeed holds the neck linker and resists neck-linker detachment, whereas, in Kin-1, the neck linker interacts far less strongly with L13. We develop a model where Kin-5 relies on contacts between the neck linker and L13 to compensate for a shorter CNB. Together, these results show how subtle variations in these mechanical elements lead to different motility behaviors of kinesin families suited for their functions in the cell.
Materials and Methods
Construction of chimeras
All kinesin constructs are derivatives of the WC2 plasmid coding for amino acids 1–401 of D. melanogaster Kin-1 heavy chain (DmK401) (36) followed by the Escherichia coli biotin carboxyl carrier protein and a 6× histidine tag. The cover-strand mutation replaced residues 4–12 of DmK401 with residues 10–18 of H. sapiens Kin-5 (coding sequence 5′-AAGAAGAAGGAGGAGAAGGGCAAGAAC-3′). The neck-linker mutation replaced residues 333–338 of DmK401 with residues 359–364 of H. sapiens Kin-5 (coding sequence 5′-ATCCTGAACAAGCCGGAG-3′). Both the cover strand and neck-linker mutations were generated by standard round-the-horn mutagenesis. The L13 mutation was generated by replacing residues N301 and A302 of DmK401 with residues R327 and T328 of H. sapiens Kin-5. Mutagenesis was carried out by QuikChange mutagenesis (Stratagene, La Jolla, CA) with the primer pair 5′-GGAGTCGCTGGGAGGCAGAACACGCACAACCATCGTC-3′ and 5′-GACGATGGTTGTGCGTGTTCTGCCTCCCAGCGACTCC-3′. Mutated plasmids were transformed into ER2566 chemically competent cells. Mutations were confirmed by sequencing of the entire protein-reading frame. Kinesin constructs were expressed and purified as described previously in Khalil et al. (21).
Sample preparation
Samples for the unloaded and optical tweezers motility assays were created by diluting kinesin from a stock frozen at −80°C into assay buffer composed of PEM80 (80 mM PIPES, 1 mM EGTA, 4 mM MgCl2) and 0.1 mM DTT, 20 μM taxol, 1 mM ATP, 50 mM potassium acetate, 0.2 mg/mL casein, pH 6.9. We used 0.44-μm diameter streptavidin-coated beads (Spherotech, Lake Forest, IL) which were diluted and washed with phosphate-buffered saline, then sonicated. Diluted kinesin solutions (∼1–10 pM concentration) were incubated with the washed beads for 1 h at 4°C (giving ∼0–5 kinesin molecules per bead).
Flow cells of ≈15 μL were constructed by attaching a KOH-etched, poly-L-lysine-coated coverslip to a glass slide using double-sided tape. Microtubules (bovine; Cytoskeleton, Denver, CO) diluted 150-fold in PEM80, 20 μM taxol were introduced into the flow cell and incubated for 10 min. The flow cell was washed with 20 μL PEM80, then incubated with PEM80 supplemented with 2 mg/mL casein for 5 min. After another wash with PEM80 and then with assay buffer, experiments were begun by introducing the diluted kinesin solution into the flow cell.
Unloaded motility assay
The unloaded motility assay was performed by trapping a freely diffusing bead and placing it near a microtubule. The analysis of single molecules was ensured by using data from experiments where approximately half or less of observed beads were motile after being tested on three different microtubules (37). For beads that did show motility near a microtubule, the trap was shuttered upon the start of a run. A custom LABVIEW program (National Instruments, Austin, TX) was used to automate the trap-shuttering process. The bead was monitored using video-enhanced differential-interference contrast microscopy. Custom-written MATLAB (Natick, MA) scripts were used to analyze the unloaded motility data. The tracking scripts were calibrated using data taken from beads tethered to microtubules via kinesin in the presence of adenylyl-imidodiphosphate (AMP-PNP) and moving the piezo stage at a predetermined velocity and distance. Unloaded run length was calculated by fitting a single exponential to the histogram of run distances for each motor (see Fig. S3, C and D, in the Supporting Material).
Optical tweezers motility assay
The motility assay under load was carried out using a stationary optical tweezers instrument with separate trapping and detection systems. The instrument was calibrated as previously done (21,38,39), and a trap stiffness of ∼0.05 pN/nm was used for all experiments. The same bead motility criteria for the unloaded motility assay were also used for the assay under load. Single molecule behavior was further confirmed by appearance of distinct 8-nm steps, records that snapped back to baseline, a single peak in the distributions of stall forces (distributions well fit with single Gaussians; see Fig. S3 A) as well as general agreement of our Kin-1 data with that of previously published single molecule studies (21,37,40). Voltage data (corresponding to the displacement of the bead from the center of the trap) from the photosensitive device were low-pass-filtered at 1 kHz and sampled at 2 kHz.
The motility data was analyzed using custom-written MATLAB scripts as done in Khalil et al. (21). Stalling events were identified from the raw data traces with criteria of having a stall force ≥0.7 pN, stall plateau time ≥0.1 s, snapback velocity ≥500 nm/s, and a return to baseline after motor release from the microtubule. The force-velocity behavior of the motors was calculated by dividing the stalling events into 15-ms windows, over which the velocity and average force were computed. These data were then divided into 1-pN force bins to which the three-state kinetic model (Eqs. 1 and 2) was fit. The velocities at which the motors released from the microtubule (stall velocity) were calculated from slopes of lines manually fit to the ends of runs. These velocities were then normalized by dividing by the unloaded velocity for each construct.
Molecular dynamics
The PDB structures were converted into the CHARMM (41,42) format with the param22 force field (43). The GBMV II implicit solvent model (44) available in CHARMM was used for all simulations. To construct chimeras, Kin-1 and Kin-5 structures were aligned using the online CE Calc Two Chains utility (45). The residues and coordinates of the Kin-1 domain to be mutated were replaced by the corresponding ones from the aligned Kin-5 structure. Loops missing from the PDB files (e.g., residue 244–259 in PDB:2Y65) were generated using the MODLOOP server (46,47). Contacts were analyzed with the software LIGPLOT (48).
Neck-pulling simulations were carried out as described in Hwang et al. (30). Briefly, the structure was initially energy-minimized and heated from 100 K to 300 K over 10 ps. For pulling the neck, harmonic constraints were applied at the microtubule binding sites (Cα atoms of residue 148–151 (β5), 244–261 (L11), and 279–287 (L12)) with a spring constant of 10 kcal/(mol·Å2). A force of 440 pN was applied to the neck helix (distributed over the Cα atoms of residues 345–362). The pulling direction in all simulations was set to be equivalent to that of the axis used for PDB:1MKJ, for consistency. For simulations using PDB:1MKJ as the base structure, the structure was energy-minimized and heated, after which the pulling axis was calculated by using the COOR HELIX command in CHARMM on the Cα atoms of the neck-helix residues 337–347. In the case of simulations with PDB:2Y65 as the base structure, the neck helix of PDB:2Y65 was pulled along an axis that was analogous to the axis used for the PDB:1MKJ simulations. This axis was calculated by taking the average pulling axis (along the PDB:1MKJ neck helix, calculated with the COOR HELIX command, as described above) of eight simulations done with PDB:1MKJ. All pulling simulations were performed at 300 K for 400–800 ps. A 2-fs integration time was used. The CNB relaxation simulations (see Fig. S4 B) were performed at 300 K by releasing the pulling force from a structure where the CNB was fully extended away from the motor head, while retaining harmonic constraints on the microtubule binding domains of the motor head. The simulation lasted 1.6 ns for each construct. The force histograms for isolated CNBs were calculated using the method detailed in Hwang et al. (30).
Results
Structural comparison between Kin-1 and 5
We first compared D. melanogaster Kin-1 (PDB:2Y65) and Homo sapiens Kin-5 (PDB:1Q0B) structures. Additionally, the structures of H. sapiens Kin-1 (PDB:1MKJ) and D. melanogaster Kin-5 (PDB:2WBE) were analyzed (see Fig. S1) for comparison to previous studies done with PDB:1MKJ (49), and to examine a Kin-5 structure with additional visible cover-strand residues (PDB:2WBE). See Text S1 in the Supporting Material for our choice of structures and additional comparisons. All structures are in the ATP-like state with the neck linker docked to the motor head (Fig. 1). The cover strand of Kin-5 is six residues longer than that of Kin-1, and the sequence conservation (23%) is weaker than that of the motor head as a whole (44%). The Kin-5 cover strand is also overall positively charged due to its five lysine residues. Only the C-terminal five residues of the Kin-1 cover strand participate in the CNB formation, whereas the role of the N-terminal portion of the cover strand is unclear. As previously noted, the CNB is a transient structure that is formed only during the force-generating ATP-state (30), thus ATP-like structures should give the most probable CNB length. The CNB length visible in x-ray structures, however, may also vary depending on the crystallization conditions. For L13, although only two residues differ between the two motors; the sequence mismatch greatly changes the loop’s characteristics. R327 and T328 of Kin-5 makes L13 more positively charged and polar compared to N301 and A302 of Kin-1. Additionally, R327 and T328 are bulkier than N301 and A302, which may impact the neck-linker docking because L13 is positioned beneath β9 (Fig. 1).
The N-terminal half of the neck linker that forms the CNB (β9) has 38% sequence identity between Kin-1 and Kin-5, close to the value for the whole motor head. The most prominent difference is the presence of a proline, P363, in the neck linker of Kin-5. Because the β-sheet propensity of proline is very low (the second lowest among the 20 amino acids (50)), the CNB of Kin-5 is shorter than that of Kin-1. The CNB of Kin-1 is five-residues long (cover strand/β9: A9–I13/V333–V337), whereas it is three residues in Kin-5 (K17–I19/I359–N361), as defined by the formation of backbone hydrogen bonds between the cover strand and the neck linker (Fig. 1 and see Fig. S1). Considering its conformational bias (30), a longer CNB would be preferred for force generation, whereas the length would be ultimately limited by entropic penalty for forming an extended β-sheet. The presence of proline in β9 indicates that Kin-5 actually avoids formation of a long CNB. This could be due to its functional requirement to work as a group of motors, where an extensive CNB formation, although beneficial for force generation, may interfere with cooperative behavior, and is akin to the case where the α-helical neck of Kin-5 apparently downregulates motor motility (51).
We measured the forces generated by isolated CNBs of Kin-1 and Kin-5 by using the tug-of-war sampling simulation method that we used earlier for Kin-1 (a stiff harmonic potential is applied to the C-terminal valine of β9 in the CNB, and the fluctuations of the Cα atom are used to calculate the force applied by the CNB at various sampling locations (30,52)). Forces generated by the CNB of Kin-5 (using PDB:1Q0B) are indeed lower compared to forces by Kin-1 (see Fig. S2), likely due to CNB shortening by the proline of Kin-5 β9. To compensate for the shorter CNB, Kin-5 may employ additional interactions in its force-generating machinery. We found a salt bridge between E364 (β9) and R327 (L13), both of which are highly conserved in the Kin-5 family (Fig. 1), assists with stronger binding of the neck linker to the motor head. Compared to Kin-1, the forward motion of the neck linker in Kin-5 may be achieved by the CNB over a shorter interval, after which specific interaction with L13 and subsequently the latching action of the C-terminal half of the neck linker (β10) on the motor head (30) completes the neck-linker binding. Once specific bonds form between the neck linker and L13, it would be more difficult to pull back the neck linker by the applied load compared to the case where the neck linker is not yet attached to the motor head. It is thus likely that Kin-5 has less load dependence in its stepping motion, and consequently in its velocity.
The above results allow us to formulate a model for the relative contributions of Kin-5 structural elements to the motile behavior of the chimeric motors.
-
1.
The cover strand, by itself, should not greatly affect motility, as the contacts it makes with β9 to form the CNB are backbone hydrogen bonds, which can be formed by a wide variety of amino acids.
-
2.
β9 likely will have several impacts on the ability of the motor to generate force, as the proline residue limits the length of the CNB, which is critical to force production.
-
3.
The CNB unit, comprised of the matched Kin-5 cover strand and β9, should rescue motility defects caused by either cover strand or β9 swaps, as the two elements that comprise the CNB must have evolved to optimally work together.
-
4.
L13 of Kin-5 should affect motor behavior in two opposing ways. It should slow down the forward motion of the CNB by its bulky side chains, but once the neck linker is bound to the motor head, L13 may help to stabilize and reduce the force sensitivity of the Kin-5 CNB by forming specific bonds with the neck linker.
Design of chimeras
With the above considerations, we created chimeric motor constructs that swapped the cover strand, β9, and L13 from Kin-1 with the corresponding elements from Kin-5. Chimeric constructs of Kin-1 and Kin-5 have been used previously (18,51,53), but the specific roles of the CNB or L13 were not foci of those works. The chimeras were built on the extensively characterized K401 construct of the Drosophila melanogaster Kin-1 (DmK) (21,54–56), with the Kin-5 elements following the amino-acid sequence of H. sapiens Kin-5. Kin-1 based chimeras were chosen because a strong consensus on the biophysical properties of the Kin-5 motor head has not yet been reached, as they have for Kin-1 (especially DmK), thus conclusions from Kin-1 based chimeras are supported by a stronger foundation of studies. Furthermore, using a common motor head for the chimeras allows us to study only the effects of the domains related to force generation, whereas other factors such as catalytic activity, microtubule binding, and the neck stalk characteristics are expected to be similar.
Six constructs were tested, including individual component swaps and higher-order combinations of functional units such as the CNB, where, for example, both the cover strand and β9 are changed. The naming convention used for chimeras makes use of superscripts indicating the structural elements derived from Kin-5; for example, Kin-1CS denotes the DmK401 motor with the cover strand (β0) sequence from Kin-5. The five other constructs are: Kin-1NL, Kin-1L13, Kin-1CS·NL, and Kin-1CS·NL·L13 (Fig. 1). For the chimeras containing cover-strand mutations, the sequence from Kin-5 (MASQPNSSAKKKEEKGKNI) was truncated to SSAKKKEEKGKNI (amino acids 7–19; the N-terminal serine was then changed to the start codon, methionine) to match the length of the Kin-1 cover strand (13 amino acids) (Fig. 1 B). Because only three residues at the C-terminal end of the cover strand participate in the CNB formation for Kin-5, it is unlikely that the truncation at the N-terminal end would have any large effect in the chimera. The sequence for the neck linker chimera, ILNKPEVN, includes all of β9 from Kin-5. The L13 chimeras have the double-mutation N301R and A302T.
Motility characteristics of chimeras
Our single-molecule optical tweezers assays (Fig. 2 A) show that each chimera is motile with an 8-nm step size, resists force, and comes to a stall before dissociation from the microtubule, as evidenced by the plateaus before dissociation in kinesin run traces (Fig. 2 A), and the strong low-velocity peak in their velocity distributions at the time of microtubule release (see Fig. S3 B). However, the mutations in each chimera had different effects on stall forces, unloaded velocities, and run lengths (Fig. 2 B). None of the chimeras experienced motility defects as severely as the Kin-1 construct with the cover-strand deletion (21), thus the cover strand and the neck linker from Kin-5 likely form a CNB when used either together or in combination with the Kin-1 elements.
Figure 2.
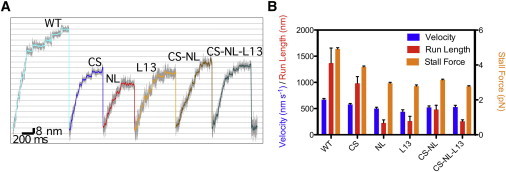
Differences between the constructs in optical trap stall force experiments. (A) Representative run traces from stall force experiments. All constructs are motile, take 8-nm steps, and slow to a stall before dissociating from the microtubule (see plateaus in motor traces before return to the baseline level). Legends denote wild-type (WT) and chimeras with mutations in corresponding domains (e.g., CS denotes Kin-1CS). The same legend convention is used in all figures below. (B) Unloaded velocity (first bar), run length (second bar), and the stall force (third bar) for each motor construct. Values for these quantities are in Table 1.
CNB formation in chimeras
The wild-type (WT) motor had a stall force of ∼5 pN, consistent with our previous measurement (21). Among the chimeras, Kin-1CS behaved the closest to the WT, with the stall force and unloaded run-length reduced, respectively, by ∼80% and 70% compared to those of WT. Because the cover strand interacts with the neck linker primarily by forming backbone hydrogen bonds (Fig. 1 and see Fig. S1) (30), changes in its amino-acid sequence from that of Kin-1 to Kin-5 should not affect its behavior or contacts with the neck linker in any major way, which was also seen in our kinetic analysis below.
Impact of Neck Linker changes
On the other hand, Kin-1NL had among the lowest unloaded velocities, run lengths, and stall forces. As mentioned above, proline in the Kin-5 β9 causes the CNB in Kin-1NL to be shorter, putatively limiting the amount of force that can be generated.
Rescue of matched CNB unit
The matched Kin-5 CNB construct (Kin-1CS·NL) had slightly higher unloaded velocity and run length about two times longer compared to Kin-1NL. Similarly, Kin-1CS·NL has a slightly higher stall force compared to Kin-1NL. Given the wide variations in reported values of stall forces in different Kin-5 constructs (1.5–7 pN) (24–26), it is unclear if the introduction of Kin-5 domains into Kin-1 causes the motors to behave more like Kin-5 than Kin-1 with respect to force generation, or if these domains disrupt Kin-1’s ability to produce force. Analysis of motor kinetics, discussed below, provides insight into this question.
Influence of L13
Inclusion of the Kin-5 L13 had a strong effect on the motor’s ability to generate force and walk processively. Like Kin-1NL, Kin-1L13 had among the lowest unloaded velocities, run lengths, and stall forces. The bulkier and charged Kin-5 L13 may impede docking of the neck linker in Kin-1L13, thus reducing force generation capability. Previously, it had been seen that force generation could be decoupled from ATP hydrolysis by mutating the conserved glycine residues of L13 to alanine (7), which presumably made L13 less flexible and protrude into the path of CNB folding (30). By interfering with the ability of the CNB to fold forward, the Kin-5 L13 may also affect the motor head gating, which is dependent on the orientation of the neck linker (57). Interestingly, the unloaded run length and stall force of the Kin-1CS·NL·L13 construct were nearly identical to that of Kin-1L13 (Fig. 2 B).
Among the motility characteristics, the run length varied the most (Fig. 2 B), which suggests that the probability of detaching from the microtubule is affected. However, because the same Kin-1 motor head was used, the microtubule binding affinity per se should be similar in the chimeras. Instead, slower CNB formation or incomplete neck linker docking may reduce the likelihood of the unbound, moving head to find and bind to the next microtubule site, which would affect the duty ratio and hence the processivity.
The unloaded velocity of the chimeras was the motility parameter that changed the least. In fact, all of the motors tested here had unloaded velocities within the expected range for Kin-1 motors. This may be because the ATP binding site of the motors remains the same, so that the ATPase activity is minimally affected by the mutations. At saturating ATP and zero force, both the rates of ATP binding and of the mechanical transition are much faster than that of biochemical transitions, thus the rate-limiting step is biochemical, and should be similar for all of the motors tested. Correspondingly, the unloaded velocity should vary little.
Analysis of the force-velocity curves
To further elucidate the effect of swapping elements of the force generation machinery, we use the three-state kinetic model of the form (24)
| (1) |
with the force-dependent rate of the mechanical component of the mechanochemical cycle
| (2) |
Here, v is the velocity of the motor under an applied force F; δ1 = 8.2 nm is the step size of kinesin; k1 and k−1 are the rates of ATP binding and unbinding, respectively; k20 is the unloaded mechanical rate; k3 is the rate of ATP hydrolysis; δ2 is the distance to the transition for the mechanical portion of the cycle; and kBT (T = 300 K) is thermal energy. The force sensitivity in this model is represented by δ2, as it scales the force-dependent decay of the mechanical rate, k20. Larger values of δ2 indicate a more force-sensitive motor.
Fig. 3 and Table 1, respectively, show the force-velocity curves for the motors tested and the parameters used for fitting the data. Although Kin-1CS behaved very similar to the WT motor, our kinetic analysis reveals that its unloaded mechanical rate (k20) is approximately one-third of the WT value. The cover strand of Kin-1 is electrostatically net negative and β9 is net positive, whereas in Kin-1CS, the cover strand and β9 are both positive, which may contribute to the slower CNB formation and manifest as a lower k20. But once the CNB forms by making backbone hydrogen bonds between the cover strand and the neck linker, its forward conformational bias would be relatively insensitive to whether the cover strand sequence is from Kin-1 or Kin-5. In fact, the force sensitivity of Kin-1CS (δ2) is nearly identical to that of the WT.
Figure 3.
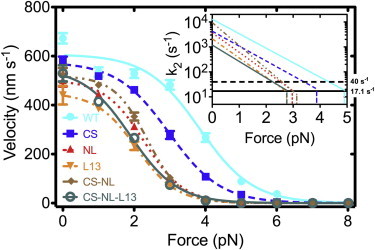
The velocity of each motor as a function of applied force. (Lines) Fits to the data by the three-state kinetic model. (Inset) Mechanical rate (k2) versus F (Eq. 2) in log-normal scale. (Thick horizontal solid line) Average mechanical rate at stall for all motors (17.1 s−1; k2stall in Table S1 shows individual values). (Thick horizontal dashed line) Rate-limiting biochemical step for Kin-1 (40 s−1). The system becomes mechanically limited above the force at which k2 crosses this line (see Text S2 in the Supporting Material). (Thin vertical lines) Stall force of each motor.
Table 1.
Results of the single molecule measurements of the motors at saturating ATP
| Constructs | Unloaded Velocitya,c |
Run lengthb,c |
Stall forcea,d |
Three-state modelb,d |
|||||
|---|---|---|---|---|---|---|---|---|---|
|
k1e |
k−1e |
k20 |
k3 |
δ2 |
vmax |
||||
| (nm/s) | (nm) | (pN) | (μM−s)−1 | (s−1) | (s−1) | (s−1) | (nm) | (nm/s) | |
| WT | 671 ± 21 | 1370 ± 287 | 4.92 ± 0.08 | 2 | 120 | 12,900 ± 6540 | 76.86 ± 1.88 | 5.50 ± 0.51 | 630 ± 15 |
| Kin-1CS | 579 ± 18 | 982 ± 129 | 3.89 ± 0.05 | 2 | 120 | 4260 ± 2060 | 72.74 ± 2.45 | 5.65 ± 0.60 | 597 ± 20 |
| Kin-1NL | 501 ± 26 | 225 ± 59 | 2.95 ± 0.05 | 2 | 120 | 3640 ± 2200 | 63.21 ± 2.19 | 7.35 ± 0.99 | 518 ± 21 |
| Kin-1L13 | 440 ± 38 | 263 ± 89 | 2.79 ± 0.06 | 2 | 120 | 2120 ± 1410 | 56.43 ± 2.97 | 7.27 ± 1.18 | 463 ± 24 |
| Kin-1CS·NL | 522 ± 28 | 482 ± 83 | 3.15 ± 0.04 | 2 | 120 | 9440 ± 6320 | 65.36 ± 2.19 | 8.69 ± 1.10 | 536 ± 18 |
| Kin-1CS·NL·L13 | 529 ± 33 | 258 ± 27 | 2.78 ± 0.03 | 2 | 120 | 1190 ± 573 | 69.42 ± 3.98 | 6.22 ± 0.85 | 569 ± 33 |
Values are mean ± standard error.
Values are fitted value ± standard deviation.
n = 35, 34, 45, 24, 38, 50.
n = 185, 364, 292, 153, 422, 585, for each of the six constructs listed top to bottom in the left column of the table.
Globally fit, values from Valentine and Gilbert (61).
Kin-1L13 and Kin-1CS·NL·L13 had the two lowest values of k20, which are six and eight times smaller than the respective non-L13 containing motor, WT and Kin-1CS·NL. Because the Kin-5 L13 has bulkier and more charged residues, it would cause slower neck linker docking, which is in agreement with a previous study showing that Kin-5 neck linker docking is ∼10 times slower than Kin-1 neck linker docking (58). More extensive contacts between L13 and the neck linker in Kin-5 may also reduce the sensitivity to load, as evidenced by the smaller value of δ2 (6.2 nm) for Kin-1CS·NL·L13 compared to 8.7 nm for Kin-1CS·NL. However, the interaction between L13 and the neck linker seems rather specific to Kin-5, because Kin-1L13 has a higher force sensitivity (δ2 = 7.3 nm) than WT. The WT had the smallest δ2 (5.5 nm), indicating that it is mechanically optimized.
MD simulation of chimeras
To gain atomistic insight into the effect of the Kin-5 components on behavior of the chimeric motors, we performed MD simulations on each of the above constructs as well as on Kin-5. Two sets of simulations were run with PDB:2Y65 (D. melanogaster Kin-1) and PDB:1Q0B (H. sapiens Kin-5), and with PDB:1MKJ (59) (H. sapiens Kin-1) and PDB:2WBE (60) (D. melanogaster Kin-5), to test for generalizability of our findings. In our computational unbinding simulations, the neck helix of the motor in the ATP-like state is pulled, and the unbinding of the CNB from the motor head is followed (30). This allowed us to determine whether the CNB makes specific contacts with the motor head, especially with L13, and the relative strengths of the contacts among the chimeras tested.
The extent of the neck-linker unbinding was quantified by the distance between the conserved asparagine in the middle of the neck linker and the glycine on the motor head that form a hydrogen bond when the neck linker is docked to the motor head, termed the asparagine latch (Fig. 4) (30). The retention of the asparagine latch contact, which is outside of the regions that are modified in the chimeras, indicates that the motor head is not destabilized or that contacts outside of the regions explicitly covered by this study are not perturbed. The results for the PDB:1MKJ/2WBE chimeras are shown in Fig. S4 A. We performed 10 neck-pulling simulations to ensure the observed behaviors are robust (see Fig. S4, C and D). In all cases, the CNB stayed intact until the neck helix had been fully pulled away from the motor head. For PDB:1MKJ/2WBE constructs, we performed additional simulations where the unbound neck linker from the neck-pulling simulations was released. The CNB moved toward the motor head whereas simulations with a deleted cover strand showed no biased motion of the neck linker (see Fig. S4 B), as we previously observed for Kin-1 (30). This result shows that the conformational bias that is key to CNB function is present in Kin-5, thus a functional CNB is part of the motor activity of Kin-5.
Figure 4.
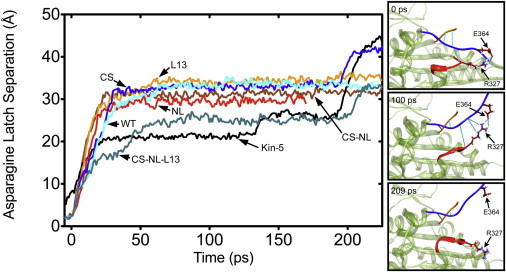
(Left) Example trajectories of neck-pulling simulations (see Fig. S4). (Vertical axis) Backbone hydrogen-oxygen distance of G83-N340 (residue numbers are for Kin-1; they are G96-N366 in Kin-5). In the case of WT Kin-1, breakage of this bond leads to full release of the neck linker, hence it is named the asparagine latch (30). Each trajectory was aligned to the moment when the asparagine latch released (backbone H–O distance became >2.4 Å for >5 ps, and remained >2.4 Å). Kin-5 does not appear to form an asparagine latch (G96 and N366 are not as close as in Kin-1), so a cutoff distance of 13 Å was used to align the Kin-5 data with the release of the asparagine latch of Kin-1 based motors. Only for Kin-1CS·NL·L13 and Kin-5 that contain the Kin-5 L13 and β9, there is a plateau before the full release of the neck linker at ∼190 ps, which is due to the R327-E364 bond (see Fig. 1A). (Right) Snapshots of the Kin-1CS·NL·L13 construct. (Straight lines) Hydrogen bonds between the cover strand and β9 (CNB), and between L13 and β9. At 100 ps, the R327–E364 bond prevents the neck linker from full unbinding.
We also found that additional interactions between L13 and β9 of Kin-5 (side panels in Fig. 4), in particular, between R327 (L13) and E364 (β9), alter the unbinding trajectory of the neck linker. In the absence of this bond, the neck linker fully extends after the hydrogen bond between G96 and N366 (the asparagine latch) breaks. In contrast, when the R327-E364 bond is present, a plateau in the extension trace is witnessed even after the asparagine latch breaks. The plateaus were most evident for constructs containing all three elements, e.g., Kin-1CS·NL·L13 and the Kin-5 structure, further supporting specificity of the bond in Kin-5 (see Fig. S4).
Concluding Discussion
Our results suggest that there is indeed some degree of modularity to the force generation machinery of kinesin, such that the CNB as a whole or in part may be transferred from Kin-5 to Kin-1 while maintaining a functioning motor. These results also indicate that Kin-1 and Kin-5 use the same basic mechanism for force generation, which is supported by a recent study employing cryo-electron microscopy and fluorescent spectroscopy that suggests the CNB of human Kin-5 forms upon ATP binding (35).
Synergistic behavior of mechanical elements
Changes in the force-generation machinery manifested themselves in the motile characteristics of the chimeric motors, and follow the biophysical considerations that we set out in the Results.
Robustness and specificity of CNB
The minimum requirement for the cover strand is that it is able to form an extended β-sheet with the neck linker through backbone hydrogen bonds. This allows for flexibility in the choice of amino acids that comprise the cover strand. The force sensitivity of Kin-1CS is similar to that of WT, indicating that once the motor is in the force-generating conformation, the cycle proceeds as it does for Kin-1. In contrast, the neck-linker swap had much greater effects on motility, as seen by the greatly reduced stall force and processivity of Kin-1NL. The reduction in stall force corresponds well with the translation of the peak in the distribution of forces generated by the isolated Kin-5 CNB to lower forces in our simulation (57% that of WT; see Fig. S2). These results support that the proline of the Kin-5 neck linker limits the length of the CNB, thus reducing the amount of force that can be generated. The chimera with the fully matched CNB unit from Kin-5 (Kin-1CS·NL) rescued some of the motility defects of Kin-1NL, namely by doubling the processivity and modestly increasing the stall force (Table 1 and Fig. 2 B). The high unloaded mechanical rate of WT and Kin-1CS·NL, k20, indicate that motors with matched CNBs have mechanical components that operate most efficiently.
Interestingly, in prior studies the Kin-1 construct with two glycine mutations in the cover strand (denoted here by Kin-12G) impaired motility more severely than Kin-1CS (21), and was much closer in properties to that of Kin-1NL reported here, suggesting that the key force producing neck linker interactions are with the cover strand and not the motor head. In Kin-12G, one of the glycine mutations was positioned such that the N-terminal backbone hydrogen bond (between residues A9 and V337; Fig. 1) may not form appropriately, because glycine has the lowest β-sheet propensity (50). P363 in β9 affects the same backbone hydrogen bond, and indeed Kin-1NL has a very similar stall force (2.95 pN) to that of Kin-12G (3.02 pN; see Table S1 in the Supporting Material).
L13 Interactions
The L13 mutations appear to affect motor function in a number of ways such as by making specific contacts with the Kin-5 neck linker, or by hampering the neck linker docking to the head. For Kin-1L13 and Kin-1CS·NL·L13, the stall force and processivity was greatly diminished. In addition to steric and electrostatic interactions, the charged and polar character of the Kin-5 L13 will generate a greater hydration force where water molecules between the neck linker and L13 have to be removed during the neck-linker docking event. The use of L13 for steric control has been demonstrated by alanine mutations (7) and in simulations using an ADP-like structure of the motor head (30). Here we further show that despite possibly slowing the binding of the neck linker to the motor head, the Kin-5 L13 makes the binding of the Kin-5 CNB to the motor head more mechanically robust through specific contacts between L13 and the matched neck linker, as seen by the reduction in force sensitivity (δ2) of Kin-1CS·NL·L13 (Table 1), and by our molecular dynamics simulations (Fig. 4).
Dependence of stall force on ATPase
Formation of a shorter CNB and impediment of neck-linker docking by L13 may seem like paradoxical optimizations, similar to the reduction of motility by Kin-5′s neck stalk (51), but they may be compensated by Kin-5 carrying out its biochemical events much more slowly than that of Kin-1. Even though the cover strand, neck linker, and L13 may be the major elements that are directly involved in force generation, biochemical events such as ATP hydrolysis also play a critical role in determining the mechanical properties of the motor as a whole. In particular, if chemical events occur faster than mechanical events, force generation may not fully develop; thus, one way to increase force would be to slow down the former. Kin-1 has a limiting biochemical rate (Pi release, 40 s−1) that is ∼3 times faster than that of Kin-5 (ATP hydrolysis, 13.5 s−1) (61). Thus, it is possible that the reduced stall forces of the chimers could be in part due to slower Kin-5 mechanical components paired with faster Kin-1 biochemical rates. We estimate that the chimeric motors tested here may have been capable of producing ∼22% more force on average (∼3.5 pN), if the biochemical rate of the motor was more like that of Kin-5 (see Text S2 in the Supporting Material for detailed calculations).
Autonomous force generation versus guided diffusion
This study also elucidates a strategy where kinesins fine-tune mechanical transitions by balancing the CNB-mediated autonomous force generation and formation of specific contacts between the moving element (neck linker) and the motor head. Kin-1 has a relatively long CNB, formation of which launches the unbound head from the lagging to the leading position on the microtubule upon ATP binding. Because the CNB effectively resists external load (30), Kin-1 has low sensitivity to force (small δ2; Table 1), a desired property for an independently walking motor. By orienting the neck linker in the forward direction, CNB formation may also play a role in motor head gating, because neck-linker orientation and tension are important for maintaining the asynchrony of ATPase cycles in the two motor heads (57).
On the other hand, Kin-5 relies less on its shorter CNB but adds specific contacts between the neck linker and L13. Due to the short-ranged nature of these interactions, motion of the neck linker would be more diffusive and demonstrate a higher sensitivity to external force, as seen in chimeras without proper contacts with L13. In another extreme, the microtubule minus-end-directed motor Ncd, a member of the Kinesin-14 family, has no neck linker or CNB, but instead has an α-helical neck that continues from the neck stalk (62). Our recent study suggests that its neck carries out a lever-arm motion without any autonomous force generation component and moves diffusively, guided by forming and breaking a series of intermediate contacts between the neck and the motor head (63). How the autonomous force generation and diffusion guided by specific contacts are partitioned in other kinesin families and motors remains to be elucidated.
Acknowledgments
This work was supported by the National Institutes of Health grant No. R01GM087677, the Singapore-MIT Alliance for Research and Technology (SMART) program, a National Defense Science and Engineering Graduate Fellowship (NSDEG) from the Department of Defense (W.R.H.), and the National Science Foundation Graduate Research Fellowship under grant No. 1122374 (W.R.H. and M.L.W.).
Contributor Information
Wonmuk Hwang, Email: hwm@tamu.edu.
Matthew J. Lang, Email: matt.lang@vanderbilt.edu.
Supporting Material
References
- 1.Hirokawa N., Noda Y., Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 2.Vale R.D., Fletterick R.J. The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 3.Kull F.J., Vale R.D., Fletterick R.J. The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J. Muscle Res. Cell Motil. 1998;19:877–886. doi: 10.1023/a:1005489907021. [DOI] [PubMed] [Google Scholar]
- 4.Vale R.D., Milligan R.A. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 5.Woehlke G., Schliwa M. Walking on two heads: the many talents of kinesin. Nat. Rev. Mol. Cell Biol. 2000;1:50–58. doi: 10.1038/35036069. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence C.J., Dawe R.K., Wordeman L. A standardized kinesin nomenclature. J. Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Case R.B., Rice S., Vale R.D. Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr. Biol. 2000;10:157–160. doi: 10.1016/s0960-9822(00)00316-x. [DOI] [PubMed] [Google Scholar]
- 8.Sawin K.E., LeGuellec K., Mitchison T.J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- 9.Huszar D., Theoclitou M.-E., Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 2009;28:197–208. doi: 10.1007/s10555-009-9185-8. [DOI] [PubMed] [Google Scholar]
- 10.Koller E., Propp S., Murphy C. Use of a chemically modified antisense oligonucleotide library to identify and validate Eg5 (kinesin-like 1) as a target for antineoplastic drug development. Cancer Res. 2006;66:2059–2066. doi: 10.1158/0008-5472.CAN-05-1531. [DOI] [PubMed] [Google Scholar]
- 11.Bhat K.M., Setaluri V. Microtubule-associated proteins as targets in cancer chemotherapy. Clin. Cancer Res. 2007;13:2849–2854. doi: 10.1158/1078-0432.CCR-06-3040. [DOI] [PubMed] [Google Scholar]
- 12.Yan Y., Sardana V., Kuo L.C. Inhibition of a mitotic motor protein: where, how, and conformational consequences. J. Mol. Biol. 2004;335:547–554. doi: 10.1016/j.jmb.2003.10.074. [DOI] [PubMed] [Google Scholar]
- 13.DeBonis S., Skoufias D., Kozielski F. In vitro screening for inhibitors of the human mitotic kinesin Eg5 with antimitotic and antitumor activities. Mol. Cancer Ther. 2004;3:1079–1090. [PubMed] [Google Scholar]
- 14.Kozielski F., Skoufias D.A., Thiede B. Proteome analysis of apoptosis signaling by S-trityl-L-cysteine, a potent reversible inhibitor of human mitotic kinesin Eg5. Proteomics. 2008;8:289–300. doi: 10.1002/pmic.200700534. [DOI] [PubMed] [Google Scholar]
- 15.Berman H.M., Westbrook J., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaan H.Y.K., Hackney D.D., Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333:883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan Y., Sardana V., Kuo L.C. Inhibition of a mitotic motor protein: where, how, and conformational consequences. J. Mol. Biol. 2004;335:547–554. doi: 10.1016/j.jmb.2003.10.074. [DOI] [PubMed] [Google Scholar]
- 18.Cahu J., Surrey T. Motile microtubule crosslinkers require distinct dynamic properties for correct functioning during spindle organization in Xenopus egg extract. J. Cell Sci. 2009;122:1295–1300. doi: 10.1242/jcs.044248. [DOI] [PubMed] [Google Scholar]
- 19.Block S.M. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys. J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coppin C.M., Pierce D.W., Vale R.D. The load dependence of kinesin’s mechanical cycle. Proc. Natl. Acad. Sci. USA. 1997;94:8539–8544. doi: 10.1073/pnas.94.16.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalil A.S., Appleyard D.C., Lang M.J. Kinesin’s cover-neck bundle folds forward to generate force. Proc. Natl. Acad. Sci. USA. 2008;105:19247–19252. doi: 10.1073/pnas.0805147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnitzer M.J., Visscher K., Block S.M. Force production by single kinesin motors. Nat. Cell Biol. 2000;2:718–723. doi: 10.1038/35036345. [DOI] [PubMed] [Google Scholar]
- 23.Svoboda K., Block S.M. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 24.Valentine M.T., Fordyce P.M., Block S.M. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 2006;8:470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korneev M.J., Lakämper S., Schmidt C.F. Load-dependent release limits the processive stepping of the tetrameric Eg5 motor. Eur. Biophys. J. 2007;36:675–681. doi: 10.1007/s00249-007-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakämper S., Thiede C., Schmidt C.F. The effect of monastrol on the processive motility of a dimeric kinesin-5 head/kinesin-1 stalk chimera. J. Mol. Biol. 2010;399:1–8. doi: 10.1016/j.jmb.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Eckerdt F., Eyers P.A., Maller J.L. Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr. Biol. 2008;18:519–525. doi: 10.1016/j.cub.2008.02.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma N., Titus J., Wadsworth P. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J. Cell Biol. 2011;195:87–98. doi: 10.1083/jcb.201106149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang W., Lang M.J. Mechanical design of translocating motor proteins. Cell Biochem. Biophys. 2009;54:11–22. doi: 10.1007/s12013-009-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang W., Lang M.J., Karplus M. Force generation in kinesin hinges on cover-neck bundle formation. Structure. 2008;16:62–71. doi: 10.1016/j.str.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Sindelar C.V., Downing K.H. An atomic-level mechanism for activation of the kinesin molecular motors. Proc. Natl. Acad. Sci. USA. 2010;107:4111–4116. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guydosh N.R., Block S.M. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461:125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori T., Vale R.D., Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z., Thirumalai D. Dissecting the kinematics of the kinesin step. Structure. 2012;20:628–640. doi: 10.1016/j.str.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 35.Goulet A., Behnke-Parks W.M., Moores C.A. The structural basis of force generation by the mitotic motor kinesin-5. J. Biol. Chem. 2012;287:44654–44666. doi: 10.1074/jbc.M112.404228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berliner E., Young E.C., Gelles J. Failure of a single-headed kinesin to track parallel to microtubule protofilaments. Nature. 1995;373:718–721. doi: 10.1038/373718a0. [DOI] [PubMed] [Google Scholar]
- 37.Block S.M., Goldstein L.S.B., Schnapp B.J. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 38.Lang M.J., Asbury C.L., Block S.M. An automated two-dimensional optical force clamp for single molecule studies. Biophys. J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svoboda K., Block S.M. Biological applications of optical forces. Annu. Rev. Biophys. Biomol. Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- 40.Thoresen T., Gelles J. Processive movement by a kinesin heterodimer with an inactivating mutation in one head. Biochemistry. 2008;47:9514–9521. doi: 10.1021/bi800747e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brooks B.R., Brooks C.L., 3rd, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brooks B.R., Bruccoleri R.E., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 43.MacKerell A.D., Jr., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 44.Lee M.S., Salsbury J., Brooks C.L., III Novel generalized Born methods. J. Chem. Phys. 2002;116:10606–10614. [Google Scholar]
- 45.Shindyalov I.N., Bourne P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 46.Fiser A., Do R.K.G., Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiser A., Sali A. MODLOOP: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 48.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 49.Hwang W., Lang M., Karplus M. Force generation in kinesin hinges on cover-neck bundle formation. Structure. 2008;16:62–71. doi: 10.1016/j.str.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Kim C.A., Berg J.M. Thermodynamic β-sheet propensities measured using a zinc-finger host peptide. Nature. 1993;362:267–270. doi: 10.1038/362267a0. [DOI] [PubMed] [Google Scholar]
- 51.Kalchishkova N., Böhm K.J. The role of kinesin neck linker and neck in velocity regulation. J. Mol. Biol. 2008;382:127–135. doi: 10.1016/j.jmb.2008.06.092. [DOI] [PubMed] [Google Scholar]
- 52.Hwang W. Calculation of conformation-dependent biomolecular forces. J. Chem. Phys. 2007;127:175104. doi: 10.1063/1.2784557. [DOI] [PubMed] [Google Scholar]
- 53.Kalchishkova N., Bohm K.J. On the relevance of the core helix α6 to kinesin activity generation. Biophys. Rev. Lett. 2009;4:63–75. [Google Scholar]
- 54.Gelles J., Berliner E., Anderson K. Structural and functional features of one- and two-headed biotinated kinesin derivatives. Biophys. J. 1995;68:276S–282S. [PMC free article] [PubMed] [Google Scholar]
- 55.Gilbert S.P., Johnson K.A. Expression, purification, and characterization of the Drosophila kinesin motor domain produced in Escherichia coli. Biochemistry. 1993;32:4677–4684. doi: 10.1021/bi00068a028. [DOI] [PubMed] [Google Scholar]
- 56.Subramanian R., Gelles J. Two distinct modes of processive kinesin movement in mixtures of ATP and AMP-PNP. J. Gen. Physiol. 2007;130:445–455. doi: 10.1085/jgp.200709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clancy B.E., Behnke-Parks W.M., Block S.M. A universal pathway for kinesin stepping. Nat. Struct. Mol. Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenfeld S.S., Xing J., King P.H. Docking and rolling, a model of how the mitotic motor Eg5 works. J. Biol. Chem. 2005;280:35684–35695. doi: 10.1074/jbc.M506561200. [DOI] [PubMed] [Google Scholar]
- 59.Sindelar C.V., Budny M.J., Cooke R. Two conformations in the human kinesin power stroke defined by x-ray crystallography and EPR spectroscopy. Nat. Struct. Biol. 2002;9:844–848. doi: 10.1038/nsb852. [DOI] [PubMed] [Google Scholar]
- 60.Bodey A.J., Kikkawa M., Moores C.A. 9-Ångström structure of a microtubule-bound mitotic motor. J. Mol. Biol. 2009;388:218–224. doi: 10.1016/j.jmb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 61.Valentine M.T., Gilbert S.P. To step or not to step? How biochemistry and mechanics influence processivity in kinesin and Eg5. Curr. Opin. Cell Biol. 2007;19:75–81. doi: 10.1016/j.ceb.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Endow S.A., Kull F.J., Liu H. Kinesins at a glance. J. Cell Sci. 2010;123:3420–3424. doi: 10.1242/jcs.064113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lakkaraju S.K., Hwang W. Hysteresis-based mechanism for the directed motility of the Ncd motor. Biophys. J. 2011;101:1105–1113. doi: 10.1016/j.bpj.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parke C.L., Wojcik E.J., Worthylake D.K. ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism. J. Biol. Chem. 2010;285:5859–5867. doi: 10.1074/jbc.M109.071233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


