Abstract
The cell biological phenomenon of autophagy has attracted increasing attention in recent years, partly as a consequence of the discovery of key components of its cellular machinery. Autophagy plays a crucial role in a myriad of cellular functions. Autophagy has its own regulatory mechanisms, but this process is not isolated. Autophagy is coordinated with other cellular activities to maintain cell homeostasis. Autophagy is critical for a range of human physiological processes. The multifunctional roles of autophagy are explained by its ability to interact with several key components of various cell pathways. In this review, we focus on the coordination between autophagy and other physiological processes, including the ubiquitin-proteasome system (UPS), energy homeostasis, aging, programmed cell death, the immune responses, microbial invasion and inflammation. The insights gained from investigating autophagic networks should increase our understanding of their roles in human diseases and their potential as targets for therapeutic intervention.
Keywords: autophagy, apoptosis, UPS, aging, immunity, energy homeostasis, cell growth
Introduction
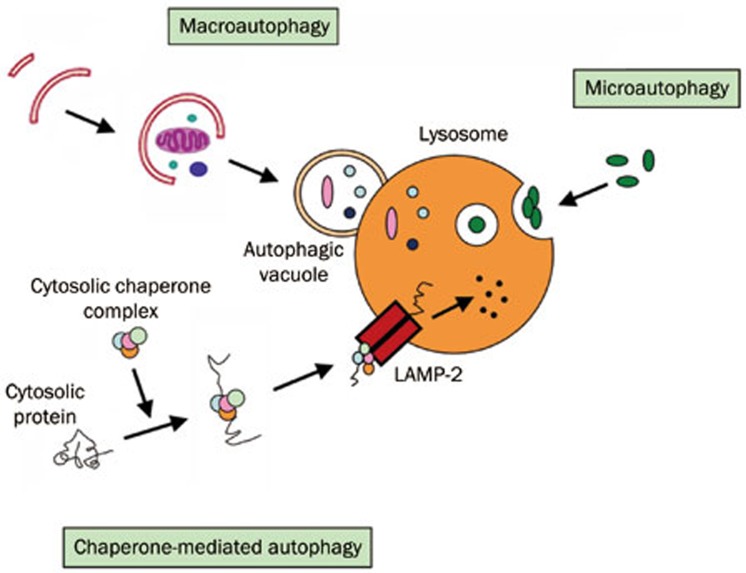
The word autophagy is derived from the Greek roots “auto” (self) and “phagy” (eating) and broadly refers to the cellular catabolic processes in which cytoplasmic materials are transported to lysosomes for degradation. Christian de Duve, who was awarded the Nobel Prize for his work on lysosomes, first used the term autophagy in 19631. There are several different types of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (Figure 1). There are also other routes for transporting cargo to the lysosomal lumen for degradation. Macroautophagy is the main route for the sequestration of cytoplasm into the lytic compartment. Macroautophagy involves the sequestration of cytoplasm by double-layered membranes to form vesicles called autophagosomes. Autophagosomes ultimately fuse with lysosomes, in which the contents are degraded. The formation of autophagosomes is initiated by class III phosphoinositide 3-kinase and autophagy-related gene Atg6 (also known as Beclin-1). In addition, two further systems are involved; these pathways are composed of the ubiquitin-like protein Atg8 (known as LC3 in mammalian cells) and the Atg4 protease on the one hand and the Atg12-Atg5-Atg16 complex on the other. In contrast, microautophagy occurs when lysosomes directly engulf cytoplasm by invagination, protrusion, or separation of the lysosomal limiting membrane. In CMA, only those proteins that have a consensus peptide sequence are recognized by the binding of an hsc70-containing chaperone/co-chaperone complex. This CMA substrate/chaperone complex then moves to the lysosomes, where the CMA receptor lysosome-associated membrane protein type-2A (LAMP-2A) recognizes it; here, the protein is unfolded and translocated across the lysosome membrane assisted by the lysosomal hsc70 on the other side2.
Figure 1.
Three main forms of autophagy.
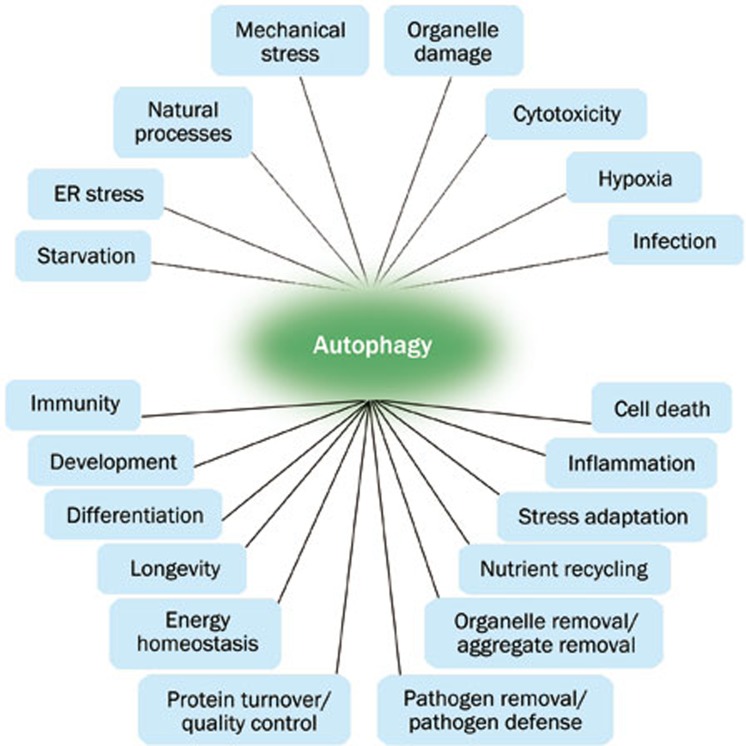
Autophagy in mammalian systems occurs under basal conditions and can be stimulated by stresses, including starvation, oxidative stress, or by treatment with the pharmacological agent rapamycin. In addition to its roles in maintaining normal cellular homeostasis by liberating nutrients from macromolecules and by assisting the clearance of misfolded proteins and damaged organelles, autophagy plays a role in the destruction of some bacteria within the cells. It has also been proposed that autophagy results in the total destruction of cells. Autophagic cell death represents one of several types of programmed cell death. Autophagy is vital in a range of physiological and pathological situations (Figure 2), including cell growth, aging, cell death, degrading disease-causing aggregate-prone proteins and clearing pathogenic bacteria3. Dysfunction in the autophagic pathway has been implicated in an increasing number of human diseases, from infectious diseases to cancer and neurodegeneration.
Figure 2.
Autophagy is vital in a range of physiological and pathological situations.
To achieve normal cell function, autophagic activity and other physiological processes regulate, compensate for and antagonize each other. In this review, we aim to discuss recent findings concerning the interactions between autophagy and other cellular processes. Although these interactions have yet to be fully defined, studies have begun to describe a growing number of cellular activities that are influenced by autophagy. The emerging evidence indicates that autophagy is a fundamental cellular process that affects other cellular activities and thus has a wider range of roles in normal physiological function and various diseases than previously thought4,5,6.
Autophagy and the ubiquitin-proteasome system (UPS)
Each cell synthesizes millions of protein molecules per minute and at the same time degrades similar numbers of proteins into peptides for further recycling into amino acids. Because large numbers of newly synthesized proteins contain errors due to mutations or misfolding, breakdown needs to be performed rapidly to prevent the accumulation of aggregation-prone protein fragments. Cells deploy two mechanisms for intracellular protein degradation: the ubiquitin-proteasome system and the autophagy-lysosome system.
The UPS and autophagy have long been viewed as independent and parallel degradation systems with no point of intersection. It was previously thought that the proteasomal and autophagic pathways target different pools of proteins, with short-lived proteins being degraded by the proteasome and large protein complexes and organelles being degraded and recycled via autophagy. This view was first challenged by the observation that mono-ubiquitination operates as a key signal in endocytosis, an important process for numerous cell functions, including lysosomal biogenesis7. Subsequently, several lines of evidence have suggested that the UPS and autophagy are functionally interrelated catabolic processes8.
Autophagy and the UPS share certain substrates and regulatory molecules and show coordinated and, in some contexts, compensatory functions. Some of the ubiquitin-recognizing molecules, or shuttle factors, are shared by both proteolytic systems. For example, p62 and ubiquilin can deliver ubiquitinated substrates to the proteasome or macroautophagy. It is now clear that a number of proteins can be degraded by both autophagy and the proteasome9,10. The neuronal protein α-synuclein, for example, can be degraded by the UPS, macroautophagy and chaperone-mediated autophagy. Furthermore, mutations that interfere with the proteasomal degradation of a protein may increase the dependency of such proteins on autophagy for their degradation, as this clearance route will then become the default. This observation is especially true for mutated proteins with an increased aggregation tendency, as oligomeric and higher-order structures will become inaccessible to the narrow proteasome barrel.
Numerous studies have reported that proteasome inhibition leads to the upregulation of autophagy. This cross-talk may be mediated by c-jun N-terminal kinase 1 (JNK1) activation following proteasome inhibition, which would be predicted to induce autophagy via Bcl-2 phosphorylation, thereby reducing the ability of Bcl-2 to bind and inhibit the function of Beclin 111,12. In vivo data further support the notion that the impairment of the proteasome leads to neurodegeneration in the fly eye. This phenotype is enhanced when essential autophagic genes are knocked down and is rescued by autophagy induction with rapamycin13.
While proteasome inhibition may induce autophagy, the converse does not occur. Indeed, the inhibition of autophagy increases the levels of proteasome substrates. This result is largely due to p62 accumulation after autophagy inhibition14. Excess p62 inhibits the clearance of ubiquitinated proteins destined for proteasomal degradation by delaying their delivery to the proteasome's proteases. Moreover, autophagy inhibition, which was previously believed to only affect long-lived proteins, also compromises the ubiquitin-proteasome system. This inhibition leads to increased levels of short-lived regulatory proteins such as p53 and the accumulation of aggregation-prone proteins, with the predicted deleterious consequences. Recently, growing amounts of data have drawn attention to p62 and its possible role in connecting autophagy with the UPS. p62 is cleared by both the UPS and autophagy and is commonly detected in ubiquitin-containing protein aggregates associated with various neurodegenerative diseases. In addition to p62, other regulators have emerged as important players in mediating the crosstalk between autophagy and UPS, including histone deacetylase 6 (HDAC6) and the FYVE-domain containing protein Alfy15. These proteins have all been found to regulate or be essential for aggresome formation.
Autophagy and energy homeostasis
Autophagy is a cellular quality control mechanism that evolved to recycle cellular waste and maintain energy homeostasis under starvation conditions. For the autophagy-lysosome pathway, the resulting breakdown products are inputs for cellular metabolism to generate energy and to build new proteins and membranes. The link between enhanced autophagy and nutrient deprivation has been well established. For example, chronic myocardial ischemia, a condition of insufficient oxygen and nutrition, activates autophagy to degrade and recycle damaged cellular structures, thereby ameliorating cardiomyocyte injury16. Autophagy provides an internal source of nutrients for energy generation and survival. A powerful promoter of metabolic homeostasis at both the cellular and whole-animal level, autophagy prevents degenerative diseases. However, autophagy does have a downside, as cancer cells exploit it to survive in nutrient-poor tumors. Autophagy is required for normal development, especially for metabolic tissues such as adipose tissue and pancreatic β-cells. Stimulating autophagy during periods of starvation is an evolutionarily conserved response to stress in eukaryotes. Under starvation conditions, the degradation of proteins and lipids allows the cell to adapt its metabolism and meet its energy needs. The physiological importance of basal autophagy in maintaining tissue homeostasis has been demonstrated in conditional brain and liver autophagy-related gene (Atg) knockout mouse models17,18. When the supply of nutrients is limited, stimulating autophagy contributes to the lysosomal recycling of nutrients to maintain protein synthesis and glucose synthesis from amino acids and to form substrates for oxidation and ATP production in the mitochondria and the inhibition of the default apoptotic pathway. In vivo studies showed that at birth, the sudden interruption of the supply of nutrients via the placenta triggers autophagy in newborn mouse tissues to maintain energy homeostasis and survival19. Moreover, starvation-induced autophagy is cytoprotective by blocking the induction of apoptosis upstream of mitochondrial events.
Some metabolic changes (ATP levels, amino acids, and insulin) may regulate autophagy. AMP-activated protein kinase (AMPK) is a crucial cellular energy sensor. Once activated by falling energy status, it promotes ATP production by increasing the activity or expression of proteins involved in catabolism while conserving ATP by switching off biosynthetic pathways. AMPK also regulates metabolic energy balance at the whole-body level20. The AMPK pathway appears to be involved in autophagy induced by nutrient deprivation, growth factor withdrawal, and hypoxia. The activation of AMPK leads to the suppression of mammalian target of rapamycin (mTOR), thereby activating autophagy. Snf1, a yeast homolog of AMPK, may be a regulator of autophagy under conditions of glucose deprivation. By contrast, autophagy is not induced by glucose depletion in fission yeast. In mammalian cells, a dominant negative mutation of AMPK and an inhibitor of AMPK both suppress autophagy, demonstrating that AMPK is important for autophagy induction, as in the budding yeast.
The inhibitory effect of amino acids on autophagy has also been well established. The signaling of amino acids in autophagic proteolysis starts with their “receptor(s)”, most likely other than transporters, at the plasma membrane, and has a novel route that is clearly distinct from the mTOR signaling pathway employed by insulin. Moreover, autophagy is also regulated by Rag proteins. Amino acids activate mTOR through Ras-related GTPase proteins21. Under nutrient-rich conditions, ER-associated Bcl-2 and Beclin 1 interact to sequester Beclin 1 from the autophagy complex22,23. This association is broken by starvation and is triggered by Bcl-2 phosphorylation by JNK1. Recruitment of the Beclin 1-PI3-kinase complex to the autophagic membrane is also dependent on the ULK1 complex. Thus, there are multiple ways that amino acids influence autophagic activity. In metazoans, cells depend on extracellular growth factors for energy homeostasis. When deinhibited by default in cells deprived of growth factors, glycogen synthase kinase-3 (GSK3) activates the acetyltransferase TIP60 by phosphorylating TIP60-Ser86. TIP60 then directly acetylates and stimulates the protein kinase ULK1, which is required for autophagy. Cells use signaling from GSK3 to TIP60 and ULK1 to regulate autophagy when deprived of serum but not glucose. These findings uncover an activating pathway that integrates protein phosphorylation and acetylation to connect growth factor deprivation to autophagy24.
Insulin is the principal hormone responsible for the control of glucose metabolism. It is synthesized in the β-cells of the islets of Langerhans as the precursor, proinsulin, which is processed to form C-peptide and insulin. It has been well established that insulin suppresses autophagy in the fed state. Insulin levels rise following a meal, causing the activation of plasma membrane insulin receptors. These receptors in turn activate downstream effectors and result in the eventual activation of mTOR kinase, one of the primary negative regulators of autophagy. Rapamycin, an inhibitor of mTOR, can activate autophagy even under nutrient-rich conditions. Autophagy is activated in the streptozotocin-induced diabetes model25. Insulin is important for autophagy regulation, but autophagy itself is also important for the homeostasis of pancreatic β cells and insulin secretion26.
The important markers activated by caloric restriction are sirtuins, a class of nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylases observed in organisms ranging from yeasts to mammals that regulate cellular metabolism through the deacetylation of several transcription factors, such as FoxO proteins, p53, and the p65 component of the NF-κB complex. The PI(3)K-PKB-FoxO signaling network provides a major intracellular hub for the regulation of cell proliferation, survival and stress resistance. Inhibition of FoxO3-mediated autophagy increases the level of apoptosis, suggesting that the induction of autophagy by FoxO3-mediated glutamine synthetase expression is important for cellular survival. These findings reveal a growth-factor-responsive network that can directly modulate autophagy through the regulation of glutamine metabolism27. NAD+ acts not only as a cofactor for cellular respiration but also as a substrate for NAD+-dependent enzymes, such as Sirtuin-1 (SIRT-1). SIRT-1 antagonizes cellular senescence in human diploid fibroblasts and protects mouse cardiac muscle against oxidative stress. If the main function of autophagic degradation seems to be cellular housekeeping, then SIRT-1 and autophagy are most likely strictly dependent on each other. Indeed, it was demonstrated that SIRT-1 deacetylates specific Atg proteins in a NAD+-dependent manner, whereas the absence of this protein in cultured cells and in embryonic and neonatal tissues increases their acetylation.
Nicotinamide phosphoribosyltransferase (Nampt, also known as visfatin), the rate-limiting enzyme in mammalian NAD+ biosynthesis, regulates NAD+ and ATP content, thereby playing an essential role in mediating cell survival by inhibiting apoptosis and stimulating autophagic flux in cardiac myocytes. Preventing downregulation of Nampt inhibits myocardial injury in response to myocardial ischemia and reperfusion. Therefore, Nampt is an essential gatekeeper of energy status and survival in cardiac myocytes28. Recent reports also indicate that autophagy serves as a stress response and may participate in the pathophysiology of cerebral ischemia. Nampt protects against ischemic stroke by inhibiting neuronal apoptosis and necrosis. Nampt promotes neuronal survival by inducing autophagy via the regulation of the TSC2-mTOR-S6K1 signaling pathway in a SIRT1-dependent manner during cerebral ischemia29. Furthermore, in multiple myeloma (MM) cells, cytotoxicity of the Nampt-specific chemical inhibitor FK866 triggers autophagy, but not apoptosis. A transcriptional-dependent (TFEB) and independent (PI3K/mTORC1) activation of autophagy mediates the cytotoxicity of FK866 on MM. Therefore, this biosynthetic pathway may be an attractive target for cancer treatment30.
Both the activation and inhibition of autophagy hold promise for improved treatment of metabolic disorders. It has been shown that autophagy in hypothalamic agouti-related peptide (AgRP) neurons regulates food intake and energy balance31. The regulation of hypothalamic autophagy could become an effective intervention in conditions such as obesity and metabolic syndrome. In diabetes, increasing autophagy may help expand β-cell mass, provide resistance against stress and enhance hepatic insulin sensitivity. In nonalcoholic fatty liver disease, increasing autophagy would not only help to remove liver fat and inflammation but also help to protect against tumorigenesis. In pancreatitis, decreasing autophagy may help to prevent or treat pancreatitis. In cardiovascular complications, increasing autophagy may safeguard plaque cells against cellular distress, particularly oxidative injury. In hypercholesterolemia and hypertriglyceridemia, increasing autophagy may help to degrade hepatic triglycerides and regulate reverse cholesterol transport (RCT) from macrophages. In obesity, a regulated decrease in autophagy could be beneficial in limiting white adipose tissue mass and lipid content.
Autophagy and cell growth
Through careful analysis of the current autophagy gene mutant mouse models, it is proposed that in mammals, autophagy may be involved in specific cytosolic rearrangements needed for proliferation, death, and differentiation during embryogenesis and postnatal development. Thus, autophagy is a process of cytosolic “renovation”, crucial in cell-fate decisions. As a response to nutrient deprivation and other cell stresses, autophagy is often induced in the context of reduced or arrested cell growth. A plethora of signaling molecules and pathways have been shown to have opposing effects on cell growth and autophagy, and the results of recent functional genomic screens support the idea that these processes might represent mutually exclusive cell fates. Understanding the ways in which autophagy and cell growth relate to one another is becoming increasingly important as new roles for autophagy in tumorigenesis and other growth-related phenomena are uncovered.
Autophagy and cell growth can inhibit one another through a variety of direct and indirect mechanisms and can be independently regulated by common signaling pathways. The central role of the mTOR pathway in regulating both autophagy and cell growth exemplifies one such mechanism. In addition, mTOR-independent signaling and other more direct connections between autophagy and cell growth also exist32. Given its central role in integrating environmental signals such as the presence of sufficient nutrients, energy, and growth factors, the PI3K/TOR signaling pathway is often viewed as the principal mediator of cellular growth control. More recently, autophagy has emerged as a crucial player in the negative regulation of cellular growth33. Although autophagy is regulated as a downstream target of TORC signaling, autophagy can also regulate cell growth in response to distinct stimuli such as amino acid depletion, energy deprivation, and ER stress, independent of PI3K/TOR signaling. Thus, TORC signaling and autophagy actually represent parallel but contrasting pathways that function together in a coordinated manner to maintain homeostasis and growth control. The loss of such control by either dysfunction of TOR signaling or autophagy likely underlies the pathogenesis of most human cancers. However, the precise mechanisms by which autophagy acts in cell growth control and tumor suppression require further investigation.
Autophagy and aging
One of the most exciting aspects of autophagy is its connection with aging and the possibility of manipulating autophagy to extend life span. Autophagy declines with age and is thought to associate with aging because it increases with the accumulation of damaged organelles and protein aggregates. The genetic inhibition of autophagy induces degenerative changes in mammalian tissues that resemble those associated with aging. In addition, normal and pathological aging are often associated with a reduced autophagic potential. Pharmacological or genetic manipulations that increase life span in model organisms often stimulate autophagy, and its inhibition compromises the longevity-promoting effects of caloric restriction, sirtuin 1 activation, the inhibition of insulin/insulin growth factor signaling, and the administration of rapamycin, resveratrol, or spermidine34.
Aging involves a deterioration of tissues and organs with the passage of time due to the progressive accumulation of malfunctioning cell components caused by oxidative damage and an age-dependent decline of turnover rate and housekeeping ability. p53 has long been viewed as a molecule that controls an evolutionary trade-off between longevity and cancer resistance: either you boost p53 to get more senescent cells, less cancer, and a shorter life, or you reduce p53 to get more cancer and a longer life. Some studies have shown that the deletion or inhibition of p53 induced autophagy in human, mouse and nematode cells. Thus, increased or reduced autophagy might be important for differences in longevity, aligning the issue of cancer with one side or another35.
Moreover, the NF-κB signaling system and the autophagic degradation pathway are crucial cellular survival mechanisms, both being well conserved during evolution. Emerging studies have indicated that the IKK/NF-κB signaling axis regulates autophagy36. Several studies have indicated that NF-κB signaling is enhanced both during aging and cellular senescence, inducing a proinflammatory phenotype. The aging process is also associated with a decline in autophagic degradation. It seems that the activity of the Beclin 1 initiation complex could be impaired with aging, as the expression of Beclin 1 decreases with age, as does the activity of type III PI3K. On the other hand, the expression of the inhibitory Bcl-2/xL proteins increases with aging. Thus, NF-κB signaling could be a potent repressor of autophagy with aging.
Autophagy declines during adulthood and is almost negligible at older age. During caloric restriction (CR), animals spend a great part of their time in a state of fasting and activated autophagy37. Thus CR prevents the age-dependent decline of autophagic proteolysis and improves the sensitivity of liver cells to stimulation of lysosomal degradation. It has been suggested primarily from work on C elegans but also some studies of human cells that SIRT-1 may exert its effects on lifespan via an effect on autophagy. mTOR is also a primary regulator of autophagy, with higher levels of mTORC1 inhibiting autophagy. SIRT-1, the closest mammalian homolog of the Sir2 yeast longevity protein, stimulates autophagy of organelles such as mitochondria in the aged mouse kidney through FoxO3 deacetylation. SIRT-1 stimulated by caloric restriction, resveratrol and cofactor NAD+ has positive effects on autophagy and on longevity38. Moreover, SIRT-1 positively regulates FoxO transcription factors and, in particular, FoxO3, which in turn is able to activate autophagy and promote longevity39. The sestrins, a family of highly inducible conserved stress-responsive proteins, have been suggested to adjust stress and metabolic responses in different organisms from Drosophila to mammals. Remarkably, sestrins induce autophagy by the inhibition of TORC1 and can also reduce ROS production by increasing the efficiency of mitochondrial respiration. Moreover, sestrins retard the appearance of age-associated cardiac pathologies40.
Autophagy and cell death
Autophagy is a cell survival mechanism that involves the degradation and recycling of cytoplasmic components. In addition, autophagy mediates cell death under specific circumstances. Autophagic cell death (type II cell death) and apoptosis (type I cell death) have taken center stage as the principal mechanisms of programmed cell death in mammalian tissues.
Although autophagy and apoptosis are markedly different processes, autophagy and apoptosis can act as partners to induce cell death in a coordinated or cooperative fashion. Several signaling pathways that are induced by common cellular stressors regulate both autophagy and apoptosis. For example, Bcl-2 phosphorylation may not only be a mechanism for regulating apoptosis and a mechanism for regulating autophagy, but may also be a mechanism for regulating the switch between the two pathways. Bcl-2 plays important roles in mitochondria dysfunction-induced apoptotic death of striatal neurons by modulating both autophagic and apoptotic processes. It is suggested that Bcl-2 also plays an essential role in limiting autophagy activation and preventing the initiation of programmed cell death41. The pro-survival role of autophagy depends on Bcl-2 under nutrition stress conditions (our unpublished data). JNK is able to trigger autophagy by targeting Bcl-2/Bcl-xL proteins and abrogating their binding to Beclin 1. Recently, Beclin 1 has been shown to be among the substrates of death-associated protein kinase (DAPK), a proapoptotic serine/threonine kinase, and its phosphorylation reduces its binding to the Bcl-2 family members, thus suggesting a possible mechanism by which DAPK may also induce autophagy42.
In addition, ceramide not only prominently induces apoptosis but also triggers autophagic cell death in malignant glioma cells via the activation of BNIP3, a proapoptotic member of the Bcl-2 family43. Sphingosine 1-phosphate-induced autophagy protects cells from death with apoptotic features during nutrient deprivation. An increase in endogenous ceramide promotes a robust accumulation of Beclin 1 and an autophagic response associated with cell death. An increase of sphingosine 1-phosphate level after starvation induces a mild accumulation of Beclin 1 and promotes cell survival by inhibiting the induction of apoptosis. Increases in the cytosolic free Ca2+ concentration not only activate proapoptotic signals but also potently induce autophagy by activating calmodulin-dependent kinase.
p53 is a tumor suppressor that regulates both autophagy and apoptosis. p53 induction contributes to excitotoxic neuronal death in rat striatum through apoptotic and autophagic mechanisms44,45,46. A complex role in the regulation of autophagy is played by p53, and p53 regulates autophagy both in a positive and in a negative fashion, depending on its subcellular localization. Studies show that p53 mediates kainic acid-induced autophagy activation and mitochondrial dysfunction in striatal neurons47. Additionally, p53 can initiate apoptosis by inducing the expression of p53-upregulated modulator of apoptosis (PUMA), which leads to the release of cytochrome c from mitochondria and apoptotic cell death48. The p53 target, damage-regulated autophagy modulator 1 (DRAM1), may regulate autophagy by participating in the fusion of autophagosomes and lysosomes. DRAM1 is required for p53-induced apoptosis, suggesting that DRAM1-dependent autophagy operates upstream of apoptosis. DRAM1 plays important roles in autophagy activation induced by mitochondria dysfunction. DRAM1 affects autophagy through augmentation of lysosomal acidification, fusion of lysosomes with autophagosomes and clearance of autophagosomes (our unpublished data). In addition, p53 affects autophagy by modulating signaling through the mTOR nutrient-sensing kinase, which controls autophagy at the initiation stage.
Autophagy proteins can play a role in cellular events that occur during apoptosis. For example, Atg5 may be a point of crosstalk between autophagic and apoptotic pathways. Basal autophagy is important for survival during neonatal nutrient deprivation and the prevention of neurodegeneration19,49. The inhibition of autophagy by Atg5 knockdown triggers apoptosis. Atg5 promotes autophagy, which is cytoprotective. However, Atg5 also has proapoptotic effects. Atg5 overexpression sensitizes tumor cells to various apoptotic stimuli. Exposure to proapoptotic stimuli activates calpain, which cleaves Atg5. The truncated Atg5 translocates from the cytosol to the mitochondria, where it associates with the antiapoptotic molecule Bcl-xL and triggers apoptosis. Another study demonstrated that the depletion of calpain impairs the induction of autophagy in response to rapamycin treatment and amino acid deprivation, thereby increasing apoptosis50. These results suggest that Atg5 is a molecular switch factor between autophagy and apoptosis. In addition, Atg5 and Beclin 1 (Atg6) can be cleaved by calpain or caspases that are activated during apoptosis51.
These findings provide important insights into the molecular cross-talk between autophagy and apoptosis. Apoptosis can begin with autophagy, which can end with apoptosis. It has been suggested that mitochondria may be central regulatory organelles that integrate both apoptosis and autophagy52. The induction of mitochondrial membrane permeabilization at a low level, below the threshold required for induction of apoptosis, results in the sequestration of damaged mitochondria in autophagic vacuoles. When mitochondrial membrane permeabilization is sufficiently high to sustain the active execution of cell death, apoptosis is induced53. Therefore, it is likely that the induction of apoptotic or autophagic cell death may depend on the level of mitochondrial membrane permeabilization. Thus, mitochondria may function as a switch between apoptosis and autophagy.
Autophagy and immunity
Research on autophagy has begun to focus on its role in the immune responses and inflammation. Related diseases include tuberculosis, infections with human immunodeficiency virus and other viruses, Salmonella, Listeria, Shigella, Toxoplasma, and inflammatory disorders such as Crohn's disease and multiple sclerosis. Moreover, similarly to cancer, neurodegenerative diseases and aging, defects in autophagy may underlie the pathogenesis of many infectious diseases and inflammatory syndromes.
Autophagy acts as an immune effector that mediates pathogen clearance. The roles of autophagy bridge both the innate and adaptive immune systems and include functions in thymic selection, antigen presentation, promotion of lymphocyte homeostasis and survival, and regulation of cytokine production. The explosive growth of the field has occurred many years after the early observational report of autophagosome-like structures detected in polymorphonuclear leukocytes infected with rickettsiae and the pioneering mechanistic work on the role of Beclin 1 in antiviral defense. This increased focus was brought on by the discovery of molecular links between autophagy and immune defense. The antiviral effects of autophagy have been extended to human immuno-deficiency virus (HIV) and roles for autophagy described in both cell-autonomous and innate and adaptive immunity against HIV54. The latest addition to the antimicrobial function of autophagy has been further strengthened by the findings that the HIV proteins Nef and Env counter autophagy: Env inhibits autophagy induction in dendritic cells (DCs), thus inhibiting DC maturation and processing and the presentation of HIV group-specific antigen (Gag), whereas Nef specifically blocks the autophagic maturation step in infected macrophages, thus protecting the HIV virus from degradation55. Moreover, T cells recognize proteolytic fragments of antigens that are presented to them on major histocompatibility complex (MHC) molecules. MHC class І molecules primarily present products of proteasomal proteolysis to CD8+ T cells, while MHC class II molecules mainly display degradation products of lysosomes for the stimulation of CD4+ T cells. Macroautophagy delivers intracellular proteins for lysosomal degradation and contributes in this fashion to the pool of MHC class II displayed peptides. Both self- and pathogen-derived MHC class II ligands are generated by this pathway. However, additional recent evidence also points to the regulation of extracellular antigen processing by macroautophagy56,57.
The connections between autophagy and conventional immunological systems include Toll-like receptors, Nod-like receptors, RIG-I-like receptors, damage-associated molecular patterns such as HMGB1, other known innate and adaptive immunity receptors and cytokines, sequestosome (p62)-like receptors that act as autophagy adapters, immunity-related GTPase IRGM, innate and adaptive functions of macrophages and dendritic cells, and differential effects on the development and homeostasis of T-lymphocyte and B-lymphocyte subsets. It is important to distinguish three principal types of contributions that autophagy imparts on the function of the immune system: specialized autophagic immune processes that are performed by the autophagic machinery at the cellular level; generic autophagic roles in cellular homeostasis; and the non-autophagic role of Atg factors.
To survive inside the host cell, bacterial pathogens have evolved a variety of mechanisms to avoid or interfere with innate immune defenses. The autophagy of foreign microorganisms, ie, xenophagy, has emerged as a powerful method of eliminating intracellular bacteria. However, there is a complex interplay between host autophagy mechanisms and pathogens. Some microorganisms have evolved strategies to evade autophagic recognition to survive and establish a persistent infection. There are two main mechanisms by which bacterial pathogens can escape from or interact with autophagic pathways. One strategy is that bacteria escape into the cytosol after breaking the host jail (L monocytogenes, S flexneri). Escape from the internalization vacuole is a bacterially driven process, and the survival of pathogens in the cytosol relies on their ability to avoid recognition and degradation by autophagy. Another strategy is bacterial survival and replication inside the pathogen-containing compartments. Several pathogens (Salmonella Typhimurium, M tuberculosis, Legionella pneumophila, Brucella abortus) can survive inside the vacuole and avoid destruction upon fusion or by preventing fusion with the lysosome. For these pathogens, escape to the cytosol is considered to be accidental. Remarkably, these bacteria can be recognized by autophagy both in the phagosome and in the cytosol, and their survival may be critically linked to their ability to co-opt the autophagic machinery.
Autophagy cross-talks with intracellular signaling molecules and effectors. Through these interactions, autophagy performs not only direct microbial degradation but also other protective mechanisms, such as lysozyme secretion, the ubiquitin-mediated pathway, and antigen presentation. In particular, the autophagic or cargo receptors have received much attention, as they play an important role in transporting ubiquitinated microbial cargos to the autophagy machinery. The autophagy pathway involves selective recognition and degradation of autophagic cargo. In selective autophagy, the modification of target proteins or intracellular bacteria with ubiquitin is necessary for their autophagic clearance. Autophagy receptors, including p62/SQSTM1, NBR1 and NDP52, can simultaneously bind intracellular ubiquitinated cargos and autophagy modifiers such as microtubule-associated protein LC358,59,60. Thus, autophagic receptors can mediate the docking of ubiquitin-marked protein aggregates to the autophagosomes to induce selective autophagic degradation of ubiquitinated proteins, organelles, and intracellular bacteria61.
It is reported that Rab8b and its downstream interacting partner, innate immunity regulator TBK-1, are required for autophagic elimination of mycobacteria in macrophages. TBK-1 is necessary for autophagic maturation. TBK-1 coordinates the assembly and function of the autophagic machinery and phosphorylates the autophagic adaptor p62 on Ser-403, a residue essential for its role in autophagic clearance. A key proinflammatory cytokine, IL-1β, induces autophagy, leading to autophagic killing of mycobacteria in macrophages; this IL-1β activity is dependent on TBK-1. Thus, TBK-1 is a key regulator of immunological autophagy and is responsible for the maturation of autophagosomes into lytic bactericidal organelles62. Rab GTPases are frequent targets of vacuole-living bacterial pathogens for appropriate trafficking of the vacuole. Bacterial effectors including VirA from non-vacuolar Shigella flexneri and EspG from extracellular Enteropathogenic Escherichia coli (EPEC) harbor TBC-like dual-finger motifs and exhibit potent RabGAP activity. Specific inactivation of Rab1 by VirA/EspG disrupts ER-to-Golgi trafficking. S flexneri intracellular persistence requires VirA TBC-like GAP activity that mediates bacterial escape from autophagy-mediated host defense63. The mitochondrial protein MAVS (also known as IPS-1, VISA, and CARDIF) interacts with RIG-I-like receptors (RLRs) to induce type I interferon (IFN-I). NLRX1 is a mitochondrial nucleotide-binding, leucine-rich repeats (NLR)-containing protein that attenuates MAVS-RLR signaling. NLRX1 suppresses vesicular stomatitis virus (VSV)-mediated type 1 IFN production but enhances autophagy during viral infection. NLRX1-mediated autophagy and IFN-I inhibition enhance VSV replication. The NLRX1-interacting partner, mitochondrial Tu translation elongation factor (TUFM), interacts with Atg5-Atg12 and Atg16L1 and has similar functions as NLRX1 in inhibiting RLR-induced IFN-I but promoting autophagy64.
Moreover, ROS maintain a balance between antibacterial autophagy and inflammation. Intracellular ROS, produced by the incomplete reduction of oxygen, can oxidize and damage various macromolecules such as proteins, lipids, and DNA, eventually causing cell death. Accumulating evidence points to a key role for ROS as signaling messengers, including the initiation of autophagy and various intracellular processes. that the coordinated interplay between autophagy activation and ROS-dependent signaling is necessary for the homeostatic regulation of innate defense and inflammation.
Future perspectives
Further studies are needed to fill in the remaining gaps in our knowledge of the relevance of autophagy to many human physiological processes and to some important diseases. It is likely that more diseases with autophagy associations will be discovered in the future. This research area is exciting, evolving and is enriched by the possibility that basic cell biology is closely linked to the understanding of disease pathogenesis and possible therapeutic strategies. However, many aspects of the crosstalk between autophagy and various physiological processes are not understood. For the proteolytic systems, autophagy and UPS, the challenge is now to decipher the code that determines targeting through one system or the other. Current findings have boosted the interest in developing pharmacological interventions that up-regulate the activity of the proteolytic systems. In light of the promising results observed on genetic manipulation of the proteolytic systems in animal models of different conformational disorders, it is predicted that compounds that can up-regulate the activity of the proteolytic systems could become an efficient future treatment for these devastating disorders. However, for a variety of cell types and bacterial pathogens, a detailed comparative survey of the composition of the various bacterial autophagosomes will be crucial for the elucidation of the molecular features unique to the various autophagic pathways. Testing the role of cytokines such as tumor necrosis factor alpha and IFNg and other physiological stimuli for their ability to induce autophagy and autophagy receptor activity will also be critical. Additionally, a major focus will be validating the molecular and cellular events analyzed in vitro during bacterial infection by in vivo experiments using the relevant animal models.
The multiple links of autophagy to cell growth, aging, cell death, immunity and inflammation await further exploration. Only by understanding the fundamental cell biology of these links will we be able to maximize our understanding of their roles in disease and the potential of autophagy modulation for the alleviation of human suffering. Novel pharmacological interventions with the ability to adjust autophagic activity in mammals should be developed. Preferably, drug candidates that directly interfere with a specific component of the biochemical pathway of autophagy are needed rather than those acting upstream of the pathway. We speculate that, similar to the way in which the initial genetic screenings in yeast transformed autophagy research, current proteomic and genomic screenings have the potential to transform research on the relationship between autophagy and physiological processes. Such a transformation would include facilitating a much deeper understanding of the molecular mechanisms of the existing known functions of autophagy through the use of the tools of modern systems biology to better understand the interactions between autophagy and physiological processes as well as signaling regulatory networks on a broad scale.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No 81000547; 30930035), the National Basic Research Program of China (No 2011CB510003), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
References
- De Duve C. The lysosome. Sci Am. 1963;208:64–72. doi: 10.1038/scientificamerican0563-64. [DOI] [PubMed] [Google Scholar]
- Penaloza C, Orlanski S, Ye Y, Entezari-Zaher T, Javdan M, Zakeri Z. Cell death in mammalian development. Curr Pharm Des. 2008;14:184–96. doi: 10.2174/138161208783378789. [DOI] [PubMed] [Google Scholar]
- Rami A. Autophagy in neurodegeneration: firefighter and/or incendiarist. Neuropathol Appl Neurobiol. 2009;35:449–61. doi: 10.1111/j.1365-2990.2009.01034.x. [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–30. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey AC, Follenzi A, Kiffin R, Zhang C, Cuervo AM. Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy. 2008;4:442–56. doi: 10.4161/auto.5654. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G, Vijayalingam S, Gibson SB. BNIP3 subfamily BH3-only proteins: mitochondrial stress sensors in normal and pathological functions. Oncogene. 2008;27:S114–27. doi: 10.1038/onc.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Pickart CM. The ubiquitin-proteasome pathway in Parkinson's disease and other neurodegenerative diseases. Trends Cell Biol. 2004;14:703–11. doi: 10.1016/j.tcb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. 2010;12:863–75. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- De Domenico I, Vaughn MB, Li L, Bagley D, Musci G, Ward DM, et al. Ferroportin-mediated mobilization of ferritin iron precedes ferritin degradation by the proteasome. EMBO J. 2006;25:5396–404. doi: 10.1038/sj.emboj.7601409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidane TZ, Sauble E, Linder MC. Release of iron from ferritin requires lysosomal activity. Am J Physiol Cell Physiol. 2006;291:C445–55. doi: 10.1152/ajpcell.00505.2005. [DOI] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem. 2009;284:2719–28. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. A novel link between autophagy and the ubiquitin-proteasome system. Autophagy. 2009;5:862–3. doi: 10.4161/auto.8840. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, et al. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–51. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- Li ZL, Lerman LO. Impaired myocardial autophagy linked to energy metabolism disorders. Autophagy. 2012;8:992–4. doi: 10.4161/auto.20285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7–deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–62. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–45. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–39. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–81. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- Wu WH, Zhang MP, Zhang F, Liu F, Hu ZX, Hu QD, et al. The role of programmed cell death in streptozotocin-induced early diabetic nephropathy. J Endocrinol Invest. 2011;34:e296–301. doi: 10.3275/7741. [DOI] [PubMed] [Google Scholar]
- Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–24. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- van der Vos KE, Eliasson P, Proikas-Cezanne T, Vervoort SJ, van Boxtel R, Putker M, et al. Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat Cell Biol. 2012;14:829–37. doi: 10.1038/ncb2536. [DOI] [PubMed] [Google Scholar]
- Hsu CP, Oka S, Shao D, Hariharan N, Sadoshima J. Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ Res. 2009;105:481–91. doi: 10.1161/CIRCRESAHA.109.203703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Guan YF, Du H, Zhai QW, Su DF, Miao CY. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy. 2012;8:77–87. doi: 10.4161/auto.8.1.18274. [DOI] [PubMed] [Google Scholar]
- Cea M, Cagnetta A, Fulciniti M, Tai YT, Hideshima T, Chauhan D, et al. Targeting NAD+ salvage pathway induces autophagy in multiple myeloma cells via mTORC1 and extracellular signal-regulated kinase (ERK1/2) inhibition. Blood. 2012;120:3519–29. doi: 10.1182/blood-2012-03-416776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, et al. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14:173–83. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–68. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YT, Tan HL, Huang Q, Ong CN, Shen HM. Activation of the PI3K-Akt-mTOR signaling pathway promotes necrotic cell death via suppression of autophagy. Autophagy. 2009;5:824–34. doi: 10.4161/auto.9099. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. SIRT1: regulation of longevity via autophagy. Cell Signal. 2009;21:1356–60. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4:870–3. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- Salminen A, Hyttinen JM, Kauppinen A, Kaarniranta K. Context-dependent regulation of autophagy by IKK-NF-kappaB signaling: impact on the aging process. Int J Cell Biol. 2012;2012:849541. doi: 10.1155/2012/849541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–55. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell MJ, Casimiro MC, Cordon-Cardo C, He X, Yeow WS, Wang C, et al. Disruption of a Sirt1-dependent autophagy checkpoint in the prostate results in prostatic intraepithelial neoplasia lesion formation. Cancer Res. 2011;71:964–75. doi: 10.1158/0008-5472.CAN-10-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Chan CH, Chen K, Guan X, Lin HK, Tong Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene. 2012;31:1546–57. doi: 10.1038/onc.2011.347. [DOI] [PubMed] [Google Scholar]
- Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–8. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Wang Y, Wu JC, Lin F, Han R, Han F, et al. Down-regulation of Bcl-2 enhances autophagy activation and cell death induced by mitochondrial dysfunction in rat striatum. J Neurosci Res. 2009;87:3600–10. doi: 10.1002/jnr.22152. [DOI] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Eisenstein M, Kimchi A. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy. 2009;5:720–2. doi: 10.4161/auto.5.5.8625. [DOI] [PubMed] [Google Scholar]
- Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–93. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dong XX, Cao Y, Liang ZQ, Han R, Wu JC, et al. p53 induction contributes to excitotoxic neuronal death in rat striatum through apoptotic and autophagic mechanisms. Eur J Neurosci. 2009;30:2258–70. doi: 10.1111/j.1460-9568.2009.07025.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gu ZL, Cao Y, Liang ZQ, Han R, Bennett MC, et al. Lysosomal enzyme cathepsin B is involved in kainic acid-induced excitotoxicity in rat striatum. Brain Res. 2006;1071:245–9. doi: 10.1016/j.brainres.2005.10.074. [DOI] [PubMed] [Google Scholar]
- Wang Y, Han R, Liang ZQ, Wu JC, Zhang XD, Gu ZL, et al. An autophagic mechanism is involved in apoptotic death of rat striatal neurons induced by the non-N-methyl-D-aspartate receptor agonist kainic acid. Autophagy. 2008;4:214–26. doi: 10.4161/auto.5369. [DOI] [PubMed] [Google Scholar]
- Dong XX, Wang YR, Qin S, Liang ZQ, Liu BH, Qin ZH, et al. p53 Mediates autophagy activation and mitochondria dysfunction in kainic acid-induced excitotoxicity in primary striatal neurons. Neuroscience. 2012;207:52–64. doi: 10.1016/j.neuroscience.2012.01.018. [DOI] [PubMed] [Google Scholar]
- Zhang XD, Wang Y, Zhang X, Han R, Wu JC, Liang ZQ, et al. p53 mediates mitochondria dysfunction-triggered autophagy activation and cell death in rat striatum. Autophagy. 2009;5:339–50. doi: 10.4161/auto.5.3.8174. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Demarchi F, Bertoli C, Copetti T, Tanida I, Brancolini C, Eskelinen EL, et al. Calpain is required for macroautophagy in mammalian cells. J Cell Biol. 2006;175:595–605. doi: 10.1083/jcb.200601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 2010;29:1717–9. doi: 10.1038/onc.2009.519. [DOI] [PubMed] [Google Scholar]
- Wang Y, Qin ZH. Molecular and cellular mechanisms of excitotoxic neuronal death. Apoptosis. 2010;15:1382–402. doi: 10.1007/s10495-010-0481-0. [DOI] [PubMed] [Google Scholar]
- Dong XX, Wang Y, Qin ZH. Molecular mechanisms of excitotoxicity and their relevance to pathogenesis of neurodegenerative diseases. Acta Pharmacol Sin. 2009;30:379–87. doi: 10.1038/aps.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–69. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–68. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CW, Fokken C, Turville SG, Lunemann A, Schmidt J, Munz C, et al. TNF-alpha induces macroautophagy and regulates MHC class II expression in human skeletal muscle cells. J Biol Chem. 2011;286:3970–80. doi: 10.1074/jbc.M110.159392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tey SK, Khanna R. Autophagy mediates transporter associated with antigen processing-independent presentation of viral epitopes through MHC class I pathway. Blood. 2012;120:994–1004. doi: 10.1182/blood-2012-01-402404. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Johansen T, Dikic I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 2009;5:732–3. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin–coated bacteria. Nat Immunol. 2009;10:1215–21. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–41. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–34. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N, Zhu Y, Lu Q, Hu L, Zheng Y, Shao F. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012;150:1029–41. doi: 10.1016/j.cell.2012.06.050. [DOI] [PubMed] [Google Scholar]
- Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933–46. doi: 10.1016/j.immuni.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]