Abstract
Autophagy is a cell self-digestion process via lysosomes that clears “cellular waste”, including aberrantly modified proteins or protein aggregates and damaged organelles. Therefore, autophagy is considered a protein and organelle quality control mechanism that maintains normal cellular homeostasis. Dysfunctional autophagy has been observed in ageing tissues and several ageing-associated diseases. Lifespan of model organisms such as yeast, worms, flies, and mice can be extended through promoting autophagy, either by genetic manipulations such as over-expression of Sirtuin 1, or by administrations of rapamycin, resveratrol or spermidine. The evidence supports that autophagy may play an important role in delaying ageing or extending lifespan. In this review, we summarize the current knowledge about autophagy and its regulation, outline recent developments ie the genetic and pharmacological manipulations of autophagy that affects the lifespan, and discuss the role of autophagy in the ageing-related diseases.
Keywords: autophagy, ageing, ageing-associated diseases, cancer, neurodegenerative diseases, Sirtuin 1, p53, rapamycin, resveratrol, spermidine
Introduction
Ageing is a complex process in which continuous accumulations of damages to molecules, organelles, cells, tissues and organs lead to a progressive decline in function and rise in vulnerability to diseases and eventually to organism death. Autophagy is a catabolic process involving the degradation of cellular components through the lysosomal machinery. It is a tightly regulated process that plays a role in cell growth, development, and homeostasis that maintain a balance between the synthesis, degradation, and subsequent recycling of cellular products.
The hypothesis that autophagy is involved in delaying ageing and extending lifespan is partially supported by three lines of evidence: First, the abundance of autophagy related proteins and autophagic activity decline over ageing. One of the common characteristics of senescent cells is the accumulation of abnormal proteins in the cytosol. Although several different intracellular proteolytic systems contribute to total rates of protein degradation, lysosomes are the proteolytic system likely most affected by ageing. A decrease in macroautophagy, chaperone-mediated autophagy, and some forms of endocytosis (receptor-mediated endocytosis) occur in most tissues of old organisms1. Second, a microarray-based genetic screen for genes that function in the regulation of chronological lifespan in yeast revealed that a number of mutants defective for autophagy are short-lived2. Last, autophagy is required for the lifespan-extending effect of genetic and pharmacological manipulations in several organisms. Thus, autophagy may play an important role in delaying ageing and extending lifespan.
Autophagy and its regulation
Autophagy is a highly conserved cellular mechanism for degradation and recycle of long-lived proteins and damaged organelles. Depending on the route of cargo delivery, autophagy is classified into three types: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA). Macroautophagy is the best characterized type and is the focus of this review (hereafter referred to autophagy). Our knowledge of autophagy regulation first came from the genetic study of the yeast. Up to now 35 AuTophagy-related Genes (ATG) have been identified in the yeast, and 16 ATG genes have orthologues in human3. These ATG gene products function at different stages of autophagy process and can be classified into 5 major functional groups: (i) the ULK1 kinase complex, (ii) the Atg9 cycling complex, (iii) the Vps34/class III PI3-kinase (PI3K) complexes, (iv) the lipid-binding Atg18 homolog, and (v) the ubiquitin-like proteins Atg12 and Atg8/LC3 and their conjugation systems4. The process of autophagy is composed of multiple sequential steps: induction, elongation/autophagsome formation and maturation/lysosomal degradation. Atg1/ULK1 kinase complex (Atg1, Atg13, FIP200, and Atg101) assembly at the isolated membrane called phagophore is the early event of autophagy induction5. Subsequently, the Beclin-1/Class III PI3K complex [including Beclin 1, PI3K Vacuolar protein sorting 34 (Vps34), p150, Atg14, UV irradiation resistance-associated tumor suppressor gene (UVRAG)] will be recruited to the isolated membrane to initialize the nucleation and elongation of autophagosome6. Autophagosomes are then fused with lysosomes to form autophagolysosome for degradation. Several proteins including Rab7, TECtonin β-Propeller Repeat containing 1 (TECPR1) and LAMP2 are involved in this step7,8,9.
Although it normally eliminates unselectively bulky proteins or organelles, autophagy is sometimes involved in degradation of specific proteins (such as p62/SQSTM1) or organelles, such as aggrephagy (for protein aggregates), mitophagy (for mitochondria), pexophagy (for peroxisomes), reticulophagy (for ER), xenophagy (for pathogens)10. Autophagy can be induced in response to diverse stresses and the induction process is highly regulated. Its regulation network is complex. One of the key regulators of autophagy in mammalian cells is mTOR (mammalian target of rapamycin) kinase, which suppresses autophagy in conditions of nutrient and growth factor repletion. mTOR is activated by signal transducers including class I phoshatidylinositol-3-kinases (PI3Ks) and Akt in response to insulin, insulin-like growth factor (IGF) and other growth signals, and inhibited by AMP-activated protein kinase (AMPK) in response to reduced ATP levels11.
Autophagy occurs constitutively at basal levels in many cell types to ensure the homoestatic turnover of long-lived proteins and organelles. Moreover, autophagy is upregulated: (i) when cells need to mobilize intracellular nutrients, as occurring during glucose and /or amino acid deprivation; (ii) when cells need to clear potentially toxic cytoplasmic materials including damaged organelles, aggregates of misfolded proteins, or invading microbes11; and (iii) when cells are under various stress conditions such as oxidative stress12, ER stress13, and proteasome inhibition14,15. Technically, autophagy can be upregulated by genetic or pharmacolgical manipulations such as transgenic overexpression of sirtuin 1, knockdown of p53, administration of rapamycin, resveratrol and spermidine.
Genetic and pharmacologic manipulations that affect longevity and autophagy
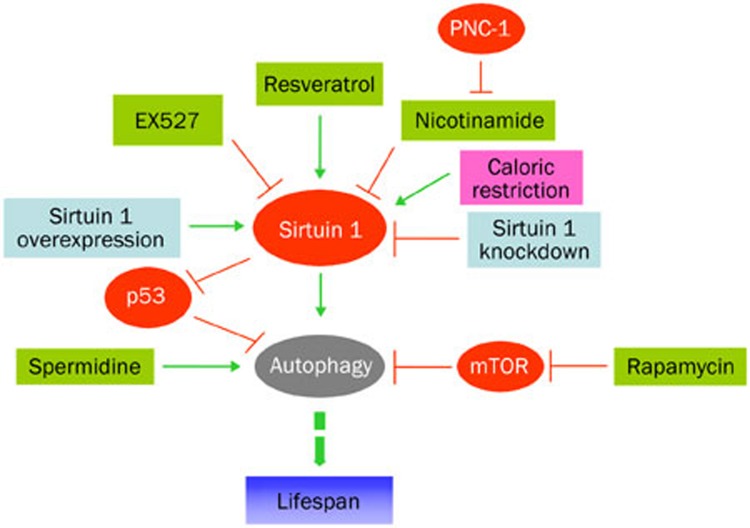
Longevity and autophagy in model organisms can be affected via direct manipulation of gene expression such as knockdown or knockout of Sirtuin 1, p53, or via indirect regulation of gene expression such as administration of rapamycin, resveratrol, or spermidine. Figure 1 shows the hypothetical modes of genetic and pharmacologic manipulations in the regulation of longevity and autophagy.
Figure 1.
Hypothetical modes of genetic and pharmacologic manipulations in the regulation of longevity and autophagy. Sirtuin 1 is a nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase that can be activated by caloric restriction, by depletion of its negative regulators (such as nicotinamide that can be depleted by overexpression of pnc-1, which encodes a pyrazinamidase/nicotinamidase (C elegan), by pharmacological activators, in particular resveratrol. Its activity can be inhibited by pharmacological inhibitors such as EX527. Autophagy can be induced by upregulation of sirtuin 1, by downregulation of p53, or by administration of spermidine or rapamycin. These measures eventually lead to organismal longevity.
Sirtuin 1
Sirtuin 1, a phylogenetically conserved NAD+-dependent deacetylase16, is an important in vivo autophagy regulator. The transient increase in Sirtuin 1 expression is sufficient to stimulate basal rates of autophagy. Sirtuin1 can form a molecular complex with several essential components of the autophagy machinery, including autophagy related proteins (such as, Atg5, Atg7, and Atg8). It also can directly deacetylate these components in vitro. Sirtuin 1 knockout mice partially resemble the phenotypes of Atg5−/− mice, including the accumulation of damaged organelles, disruption of energy homeostasis, and early perinatal mortality. These results suggest that the Sirtuin 1 deacetylase is an important in vivo regulator of autophagy and provide a link between sirtuin 1 function and the overall cellular response to limited nutrients17.
Sirtuin 1 has also been demonstrated to delay ageing and extend lifespan through inducing autophagy. Activation of the Sirtuin 1 by three different approaches (overexpression, pharmacological activation with resveratrol, and depletion of its negative regulator nicotinamide) extends lifespan through the induction of autophagy. Conversely, deactivation of Sirtuin 1 (by knockdown, knockout or pharmacological inhibition) prevents the induction of autophagy and the improvement of organismal or cellular survival by resveratrol (an indirect activator of Sirtuin 1), nutrient starvation (in human cells) or caloric restriction (in C elegans)18. Thus, Sirtuin 1 is also a major regulator of longevity in part through autophagy.
Apart from the deacetylation of autophagy-related proteins (Atg5, Atg7, and Atg8), sirtuin 1 also deacetylates lysine residues in histone 1, histone 3 and histone 4, indicating a role for sirtuin 1 in the regulation of transcription and genomic stability via chromatin modifications15. In addition, sirtuin 1 controls many key pathways that are associated with its beneficial effects on metabolism and delaying ageing. They include downregulation of p53 activity, suppression of nuclear factor-κB (NF-κB)-mediated inflammatory pathways, modulation of forkhead box protein O (FOXO) transcription factors, suppression of adipogenesis pathways mediated by peroxisome proliferator-activated receptor-α (PPARα), activation of PPARα co-activator 1α (PGC1α), and promotion of insulin secretion through the suppression of mitochondrial uncoupling protein 2 in pancreatic β-cells19. Thus, sirtuin 1 may delay ageing and extend lifespan through other molecular mechanisms besides autophagy. Sirtuin 1 functions at a regulatory crossroad between nutrient sensing, energy metabolism and genome stability19.
Caloric restriction may be a physiological inducer of autophagy3 . Caloric restriction is also the only intervention that is known to retard ageing in most organisms and delay the onset of disease and functional decline in mammals19. Dietary restriction has been shown to extend life span in yeast, worms, flies, and mammals. Autophagy is required for dietary restriction-mediated life span extension in C elegans since the lifespan extending effects were compromised by knockdown of autophagy genes. Although there is a debate as to whether caloric restriction will be as effective in humans as it is in short-lived research models, data from non-human primates previously suggested that caloric restriction can improve the quality of life, reduce the risk of disease and delay mortality19. Interestingly, a most recent report revisits the same question and provides conflicting evidence that caloric restriction in non-human primates failed to offer any benefit in slowing ageing process20. Deletion of sirtuin 1 in lower organisms appears to interfere with the beneficial effects of caloric restriction in some experimental settings. However, it is noteworthy that sirtuin 1-independent lifespan extension in response to caloric restriction has also been demonstrated in yeast and worms19. Thus, sirtuin 1 is required for the induction of autophagy, while caloric restriction-mediated lifespan extension remains debatable especially with emerging evidence and the role of sirtuin 1 in caloric restriction mediated lifespan extension needs further clarification.
p53
P53 is a well-characterized tumor suppressor. The down-regulation of p53 is associated with autophagy induction and longevity. Simultaneous promotion of lifespan extension and autophagy is also observed in C elegans following knockdown of the p53 orthologue, CEP-1, while the beneficial effects are abolished by depleting beclin 1/ATG6. In addition, lysine 382 of p53 is one known target of Sirtuin 1. Deacetylation of this residue by Sirtuin 1 decreases the activity and half-life of p53, accompanied by increase in cell su rvival under a variety of DNA-damaging conditions21.
Rapamycin
Rapamycin is the best-characterized pharmacological inducer of autophagy by inhibiting TORC1. It prolongs lifespan in various organisms including mice. In worms and yeast, rapamycin extends lifespan only when autophagy is induced. Rapamycin cannot extend the chronological lifespan of yeast mutants that lack the essential autophagy genes ATG1 or ATG722. In C elegans, the beneficial effects of rapamycin on longevity are lost when the essential autophagy genes bec-1 (the worm ortholog of mammalian gene Atg6/beclin 1) or vps34 are knocked down23. These results indicate that rapamycin induce lifespan extension by inducing autophagy in worms and yeast. In mice, it remains to be determined whether rapamycin prolongs the lifespan of mice by inducing autophagy, although rapamycin fed late in life extends lifespan in genetically heterogeneous (out-bred) mice. Rapamycin may extend lifespan by postponing death from cancer, by retarding mechanisms of ageing, or both24. Rapamycin-induced autophagy is independent of sirtuin 1 in human cells and in C elegans, suggesting that rapamycin and sirtuin 1 promote autophagy through distinct, non-overlapping mechanisms25.
Resveratrol
Resveratrol is a polyphenol mainly found in red grape skin. It has been shown to extend the lifespan in diverse organisms such as yeast, worms and flies21,26. An additional study showed that resveratrol extends lifespan in a short-lived species of fish19. It also improves health and survival of mice on a high-calorie diet27. However, it is noteworthy that sirtuin 1 overexpression or resveratrol fails to extend the lifespan of mice on a normal diet. Resveratrol significantly increased the level of sirtuin 1 in diverse cells including human cells, but failed to extend lifespan under the condition of the lack of sirtuin 1. These results indicate that the lifespan-extending effect of resveratrol is dependent on sirtuin 119,21.
As discussed above, sirtuin 1 is an important regulator of autophagy and longevity. Resveratrol also induces autophagy in human cancer cells and C elegans, and the effect was fully prevented with genetically impaired sirtuin 1 or treatment of pharmacological inhibitor (EX527)28. Resveratrol only prolonged the lifespan of autophagy-proficient C elegans, whereas these beneficial effects on longevity were abolished by the knockdown of the essential autophagic modulator beclin 1. Although it may extend lifespan through pathways such as suppressing rDNA recombination21, resveratrol has been shown to promote longevity (or at least partly) through sirtuin-1-dependent induction of autophagy28.
It was shown that resveratrol has many effects that are consistent with sirtuin1 activation. For example, it improves insulin sensitivity, inhibits tumour growth, suppresses inflammation, promotes cardiovascular health and protects against neurodegenerative diseases19.
Spermidine
The decreases in the levels of the main polyamines (putrescine and spermidine) were observed in different mammalian organs with ageing29. The spermidine has also been shown to delay ageing and promote longevity in yeast, flies and worms. A diet enriched in physiologically relevant polyamines (eg putrescine, spermidine and spermine) also increases lifespan and health span in mice30.
Spermidine treatment also induces autophagy in model systems including yeasts, flies, nematodes and mammalian cells. Polyamine depletion decreases yeast lifespan and increases necrosis. Spermidine-mediated lifespan extension was abolished in yeast, flies and worms if autophagy is blocked by knockout or knockdown of the essential autophagy genes, ATG7, BECN1. These results indicate that spermidine promotes ageing-delaying or lifespan extension through autophagy-dependent manner31.
Deletion of sirtuin 1 or any other sirtuin did not abrogate the ability of spermidine to extend chronological lifespan, indicating that spermidine promotes autophagy and then induces lifespan extension by other pathways rather than sirtuin 1. Spermidine inhibits histone acetylase, while resveratrol activates the histone deacetylase sirtuin 1 to confer cytoprotein/longevity. These results indicate the essential role of protein hypoacetylation in autophagy control and in the regulation of longevity.
Autophagy and ageing-related diseases
Autophagy plays vital roles in multiple biological functions from development to cell survival. Dysfunction of autophagy has been linked to a variety of ageing-related diseases including cancer and neurodegenerative diseases32.
Autophagy and cancer
Emerging evidence reveals the connection between autophagy and cancer. One of the most important lines of evidence came from the study of the autophagy gene BECN1. Monallelic deletion of BECN1 was associated with high frequency of tumors in multiple tissues including breast, ovarian and prostate33. Heterozygous deletion of BECN1 mice showed increased susceptibility to multiple tumors34,35. However, there is also evidence that autophagy is a mechanism by which solid tumor cells survive from hypoxic and metabolic stresses36,37. Despite recent efforts, it is still inconclusive about what exact role autophagy plays in the different stages of carcinogenesis.
Early studies suggested that autophagy suppresses the initiation of tumorigenesis. In addition to BECN1 gene that is monoallelic deleted in multiple cancers, genetic deletions or mutations in other autophagy associated genes including UVRAG, ATG2B, ATG5, ATG9B, and ATG12, were also observed in multiple types of cancers37,38. These reports highlight the genetic links between autophagy and tumorigenesis. It is not clear how autophagy defects may contribute to the initiation of tumor; at least two hypothetic mechanisms may be involved. First, autophagy maintains cellular homeostasis by constantly digesting misfolded proteins and damaged organelles (eg mitochondria), while disturbing the normal function of autophagy leads to accumulation of harmful metabolic end products, oxidized molecular and damaged organelles. As consequence of disrupted autophagy, the cells suffer damages in chromosomal DNA, causing genomic instability. Second, inhibition of autophagy may deregulate cell proliferation process or stimulate necrotic cell death, which cause release of cellular contents to the surrounding environments and initiate inflammation responses that promote tumor developments39.
In addition, autophagy can also serve as a survival mechanism for tumors. Tumors are basically under increased hypoxia stress and nutrition deprivation due to rapid proliferation rate and limited blood supply40. Under extremely harsh metabolic stress, tumors rely on autophagy to provide nutrient and keep survival41 . For certain cancer types with apoptosis defects, autophagy is an important survival mechanism to keep cell viability through long-term metabolic stresses such as glucose deprivation and hypoxia36,42.
Targeting autophagy as therapeutical strategy against cancer attracts wide interests. It is not only because mounting genetic evidence links autophagy to cancer, but also multiple anti-cancer drugs indeed activate autophagy response (For details please refer to previous review43). Autophagy inhibitors such as 3-MA and hydroxychloroquine sensitize tumors to the anti-cancer drugs, while autophagy inducers like rapamycin inhibits tumor proliferation44 and sensitizes tumors to the radiation45. Current knowledge supports the view that modulation of autophagy can be a therapeutic strategy against cancers. Future work will be needed to determine the time points and direction (induce or inhibit) of autophagy modulation for different types of tumors and at different stages.
Autophagy and neurodegenerative diseases
Neurodegenerative diseases are pathological conditions in nervous system characterized by progressive neuron loss, normally accompanied by accumulation of abnormal protein aggregates in the affected regions. The initial observation linking autophagy to neurodegenerative diseases is the presence of abnormal autophagosomes in the affected neurons of neurodegenerative diseases46. However, it is unclear whether the autophagosome formation is a consequence of impaired autophagic clearance or enhanced autophagy induction. Furthermore, it remains to be determined under those conditions whether neuronal autophagy represents protective or deleterious mechanisms. Two seminal studies using genetic mouse models lacking essential autophagy genes in the brain unequivocally demonstrate neuroprotective function of basal autophagy47,48.
Protein aggregation, a common feature of several major neurodegenerative diseases such as Alzheimer's, Parkinson's and Huntington's disease, is the consequence of accelerated protein deposition or/and reduced turnover. Numerous disease-related protein species tend to form toxic oligomers that evoke a wide range of cell stresses including calcium flux, ER stress and oxidative stress that eventually lead to neuronal loss. Our study using the transgenic mouse model with essential autophagy gene (Atg7) deletion in TH positive neurons suggests that impairment of autophagy may be associated with accumulation of endogenous α-syn and LRRK2 proteins in the dopaminergic neurons, implying that dysfunction of autophagy may be one of the pathogenic mechanisms of PD49. The emerging evidence that autophagy is neuroprotective through the clearance of unwanted protein complexes (including disease protein oligomers) makes autophagy a promising drug target, and indeed inspired a new round of drug discovery for the autophagy enhancers as therapeutic agents against neurodegenerative diseases such as Parkinson's disease and Huntington's disease. A number of chemical autophagy inducers have been identified to promote the clearance of pathogenic protein aggregates relevant to neurodegenerative diseases (summarized in Table 1).
Table 1. Autophagy modulator in neurodegenerative disease models.
| Autophagy inducer | Mechanism of action | Neurodegenerative disease models |
|---|---|---|
| Rapamycin | mTOR inhibition | Reduction of polyglutamine and polyalanine aggregation45; Ameliorates polyglutamine aggregates and toxicity in animal models46; Alpha-synuclein degradation47; Ameliorates cognitive deficits and reduces amyloid-beta AD mouse model of Alzheimer's disease48. |
| Small molecular enhancers (SMERs) | mTOR-independent | Mutant Huntingtin (Htt) and mutant A53T α-synuclein clearance49. |
| Lithium | Inositol Monophosphatase Inhibition | Mutant Htt aggregation/toxicity amelioration49. |
| N10-substituted phenoxazine | mTOR-independent | Decreased the accumulation of diffuse and aggregated Htt50. Mutant Htt and α-synuclein reduction51,52; Ameliorates dopaminergic and tau pathology53; |
| Trehalose | mTOR-independent | Enhanced cellular degradation of prions54. |
| Latrepirdine | mTOR-dependent | Improved learning behavior and reduction of Aβ42 and α-synuclein55,56. |
| Corynoxine B | mTOR-independent, Beclin 1-indepent | WT and mutant α-synuclein reduction57,58. |
| 17-AAG | Unknown | Wild type and mutant α-synuclein reduction59. |
Neurons are highly specialized cell type which relies on vigorous autophagy to remove misfolded proteins and damaged organelles for keeping viability. The regulation of autophagy in the neurons is different from other cell types, given the observations that: i) lipidation form of LC3 and autophagosomes are scarce in-neurons at basal level65; ii) nutrition starvation, the most robust autophagy inducer for most cell types, poorly induce autophagy in-brain66. Understanding the regulatory mechanism of neuronal autophagy will enable the design and discovery of the neuron-specific autophagy enhancers which regulate the autophagic activity within safe range. However, autophagy is a complex process including initiation, elongation and maturation steps, and thus simply evoking the formation of autophagosome may not be sufficient for treating the diseases with defects in cargo turnover such as AD and frontotemporal dementia67,68. It is thus desirable to restore the lysosome function while inducing autophagy biosynthesis. Thus combination of autophagy inducers and lysosome biogenesis enhancers is a better strategy for fighting the neurodegerative diseases.
References
- Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–13. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, Boeke JD, et al. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 2010;6:e1000921. doi: 10.1371/journal.pgen.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–9. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex — at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–62. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Saftig P, Beertsen W, Eskelinen EL. LAMP-2: a control step for phagosome and autophagosome maturation. Autophagy. 2008;4:510–2. doi: 10.4161/auto.5724. [DOI] [PubMed] [Google Scholar]
- Chen D, Fan W, Lu Y, Ding X, Chen S, Zhong Q. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell. 2012;45:629–41. doi: 10.1016/j.molcel.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–96. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Galluzzi L, Kepp O, Criollo A, Maiuri MC, Tavernarakis N, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY) 2009;1:961–70. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152–62. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Dunner K, Jr, McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451–62. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge PF, Zhang JZ, Wang XF, Meng FK, Li WC, Luan YX, et al. Inhibition of autophagy induced by proteasome inhibition increases cell death in human SHG-44 glioma cells. Acta Pharmacol Sin. 2009;30:1046–52. doi: 10.1038/aps.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–9. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–9. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R. Are sirtuins viable targets for improving healthspan and lifespan. Nat Rev Drug Discov. 2012;11:443–61. doi: 10.1038/nrd3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–21. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, et al. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8:353–69. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity. Nat Cell Biol. 2010;12:842–6. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–9. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalabrino G, Ferioli ME. Polyamines in mammalian ageing: an oncological problem, too? A review. Mech Ageing Dev. 1984;26:149–64. doi: 10.1016/0047-6374(84)90090-3. [DOI] [PubMed] [Google Scholar]
- Soda K, Dobashi Y, Kano Y, Tsujinaka S, Konishi F. Polyamine-rich food decreases age-associated pathology and mortality in aged mice. Exp Gerontol. 2009;44:727–32. doi: 10.1016/j.exger.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Kim SS, et al. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol. 2009;217:702–6. doi: 10.1002/path.2509. [DOI] [PubMed] [Google Scholar]
- Vakkila J, Lotze MT. Inflammation and necrosis promote tumour growth. Nat Rev Immunol. 2004;4:641–8. doi: 10.1038/nri1415. [DOI] [PubMed] [Google Scholar]
- Harris AL. Hypoxia — a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–29. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–7. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, Ryan KM. Autophagy and cancer — issues we need to digest. J Cell Sci. 2012;125:2349–58. doi: 10.1242/jcs.093708. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, et al. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–46. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- Cao C, Subhawong T, Albert JM, Kim KW, Geng L, Sekhar KR, et al. Inhibition of mammalian target of rapamycin or apoptotic pathway induces autophagy and radiosensitizes PTEN null prostate cancer cells. Cancer Res. 2006;66:10040–7. doi: 10.1158/0008-5472.CAN-06-0802. [DOI] [PubMed] [Google Scholar]
- Lee JA. Autophagy in neurodegeneration: two sides of the same coin. BMB Rep. 2009;42:324–30. doi: 10.5483/bmbrep.2009.42.6.324. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, et al. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of alpha–synuclein and LRRK2 in the brain. J Neurosci. 2012;32:7585–93. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate–prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–17. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–13. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- Spilman P, Podlutskaya N, Hart MJ, Debnath J, Gorostiza O, Bredesen D, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer's disease. PLoS One. 2010;5:e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3:331–8. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci U S A. 2010;107:16982–7. doi: 10.1073/pnas.1004498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–52. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro JA, Rodriguez L, Casarejos MJ, Solano RM, Gomez A, Perucho J, et al. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol Dis. 2010;39:423–38. doi: 10.1016/j.nbd.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Aguib Y, Heiseke A, Gilch S, Riemer C, Baier M, Schatzl HM, et al. Autophagy induction by trehalose counteracts cellular prion infection. Autophagy. 2009;5:361–9. doi: 10.4161/auto.5.3.7662. [DOI] [PubMed] [Google Scholar]
- Steele JW, Lachenmayer ML, Ju S, Stock A, Liken J, Kim SH, et al. Latrepirdine improves cognition and arrests progression of neuropathology in an Alzheimer's mouse model Mol Psychiatry 2012. doi: 10.1038/mp.2012.106 [DOI] [PMC free article] [PubMed]
- Steele JW, Ju S, Lachenmayer ML, Liken J, Stock A, Kim SH, et al. Latrepirdine stimulates autophagy and reduces accumulation of alpha-synuclein in cells and in mouse brain Mol Psychiatry 2012. doi: 10.1038/mp.2012.115 [DOI] [PMC free article] [PubMed]
- Lu JH, Tan JQ, Durairajan SS, Liu LF, Zhang ZH, Ma L, et al. Erratum to: Lu JH, Tan JQ, Durairajan SSK, Liu LF, Zhang ZH, Ma L, et al. Isorhynchophylline, a natural alkaloid, promotes the degradation of alpha-synuclein in neuronal cells via inducing autophagy. Autophagy. 2012;8:864–6. doi: 10.4161/auto.8.1.18313. [DOI] [PubMed] [Google Scholar]
- Lu JH, Tan JQ, Durairajan SS, Liu LF, Zhang ZH, Ma L, et al. Isorhynchophylline, a natural alkaloid, promotes the degradation of alpha-synuclein in neuronal cells via inducing autophagy. Autophagy. 2012;8:98–108. doi: 10.4161/auto.8.1.18313. [DOI] [PubMed] [Google Scholar]
- Riedel M, Goldbaum O, Schwarz L, Schmitt S, Richter-Landsberg C. 17-AAG induces cytoplasmic alpha-synuclein aggregate clearance by induction of autophagy. PLoS One. 2010;5:e8753. doi: 10.1371/journal.pone.0008753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Tsvetkov AS, Finkbeiner S. Protein turnover and inclusion body formation. Autophagy. 2009;5:1037–8. doi: 10.4161/auto.5.7.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Gao FB. Inhibition of autophagy induction delays neuronal cell loss caused by dysfunctional ESCRT-III in frontotemporal dementia. J Neurosci. 2009;29:8506–11. doi: 10.1523/JNEUROSCI.0924-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28:6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]