Abstract
Aim:
To investigate whether sequestosome 1/p62 (p62), a key cargo adaptor protein involved in both the ubiquitin-proteasome system and the autophagy-lysosome system, could directly regulate autophagy in vitro.
Methods:
HEK 293 cells or HeLa cells were transfected with p62-expressing plasmids or siRNA targeting p62. The cells or the cell lysates were subsequently subjected to immunofluorescence assay, immunoprecipitation assay, or immunoblot analysis. In vitro pulldown assay was used to study the interaction of p62 with Bcl-2.
Results:
Overexpression of p62 significantly increased the basal level of autophagy in both HEK 293 cells and HeLa cells, whereas knockdown of p62 significantly decreased the basal level of autophagy. In vitro pulldown assay showed that p62 directly interacted with Bcl-2. It was observed in HeLa cells that p62 co-localized with Bcl-2. Furthermore, knockdown of p62 in HEK 293 cells significantly increased the amount of Beclin 1 that co-immunoprecipitated with Bcl-2.
Conclusion:
p62 induces autophagy by disrupting the association between Bcl-2 and Beclin 1.
Keywords: autophagy, sequestosome 1/p62 (p62), LC3, Bcl-2, Beclin 1, HEK 293 cell, HeLa cell, in vitro pulldown assay
Introduction
The ubiquitin-proteasome system and autophagy-lysosome system are the two major degradation systems for cell homeostasis1. Macroautophagy (hereafter referred to as autophagy) is primarily involved in the degradation of substrate proteins or organelles such as mitochondria2. The first step of autophagy is the formation of a double-membrane structure called the autophagosome (including initiation and vesicle nucleation), the second step is autophagosome expansion and closure, and the final step is autophagosome fusion with the lysosome. Ultimately, substrates are degraded in the lysosome3,4,5. Previous reports have suggested that autophagy is implicated in tumorigenesis, aging and many aging-associated diseases6,7. Suppression of autophagy in neuronal cells can cause neurodegeneration in mice8,9. Additionally, drugs that stimulate autophagy, such as rapamycin ester temsirolimus, can improve the motor performance of mice with spinocerebellar ataxia type 310.
Sequestosome 1/p62 (p62), a scaffold protein that possesses a PB1 (Phox and Bem1p-1) domain and a UBA (ubiquitin-associated) domain as well as TRAF6 and light chain 3 (LC3, Atg8 in yeast) binding sequences, is a key cargo adaptor protein involved in both the ubiquitin-proteasome system and the autophagy-lysosome system11,12,13,14. p62 interacts with ubiquitinated substrates and LC3, a well-known autophagy effector and marker protein, to mediate autophagic degradation of substrates13,14. p62 itself is an autophagy substrate that is involved in many diseases. It has been shown to have a protective role in cellular models of Huntington's disease and amyotrophic lateral sclerosis (ALS)15,16. Furthermore, p62 mutations are associated with ALS and Paget's disease of the bone17,18,19,20. Importantly, p62 plays multiple roles in tumorigenesis. In autophagy-deficient tumor cells, p62 accumulates, resulting in an alteration of the NF-κB pathway to promote tumorigenesis21. Moreover, multiple tumors developed in mice with a mosaic deletion of Atg5 or a liver-specific Atg7 deficiency22, further suggesting an association between p62 and tumors. Accumulation of p62 also activates Nrf2 to transactivate anti-oxidative gene expression to promote cell survival23.
Although p62 is a cargo adaptor protein involved in autophagy, and autophagy is closely associated with many diseases, it is still unclear whether p62 can directly regulate autophagy. In our study, we investigated the role of p62 in the regulation of autophagy. Our results show that p62 can enhance autophagy activity by disrupting the interactions between Bcl-2 and Beclin 1.
Materials and methods
Plasmid constructs
The construction of full-length plasmids with EGFP-Bcl-2 and Flag-Beclin 1 has been described previously24,25. Full-length human p62 was cloned using PCR products amplified from a human fetal brain cDNA library with the primers 5′- CCCAAGCTTACCATGGCGTCGCTCACCGTG-3′ and 5′- ACGCGTCGACTCACAACGGCGGGGGATGCTTTGA-3′ and subsequently inserted into the pEGFPN3 (Clontech) and p3xFLAG-myc-CMV™-24 vectors (Sigma) at Hind III and Sal I sites. pET-21a-p62 was created by subcloning p62 cDNA excised from the p62-EGFP plasmid into the pET-21a vector at EcoR I/Sal I sites. GST-Bcl-2 was created by subcloning Bcl-2 cDNA excised from the EGFP-Bcl-2 plasmid into the pGEX-5x-1 vector at EcoR I/Sal I sites.
Cell culture, plasmids and si-RNA transfection
Human embryonic kidney 293 (HEK293) cells and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) containing 10% fetal bovine serum (FBS) (Gibco). For DNA and siRNA transfection, cells were transfected with plasmids or siRNAs using TurboFect (Fermentas) or RNAiMAX (Invitrogen), respectively. Human siRNA sequences were si-p62 sense #1: CATGTCCTACGTGAAGGATGATT, sense #2: GCATTGAAGTTGATATCGATTT; and si-Bcl-2 sense: GCATGCGGCCTCTGTTTGATT. The negative control (si-nc) was a non-targeting oligonucleotide. siRNA oligonucleotides were purchased from GenePharma (Shanghai, China).
Immunoblots and antibodies
Cell extracts were prepared in 1×RIPA lysis buffer (25 mmol/L Tris-HCl, pH 7.6, 150 mmol/L NaCl, 1% NP-40 and 1% sodium deoxycholate) with the PI (protease inhibitor cocktail) (Roche). The lysates were separated by 10%–15% SDS-PAGE and transferred onto a PVDF membrane (Millipore). Immunoblot analyses were performed using the following antibodies: mouse monoclonal antibodies against Flag, Flag-HRP (Sigma), GFP (Santa Cruz) or p62 (Santa Cruz); rabbit polyclonal antibodies against Bcl-2 (Epitomics), Beclin 1 (Abcam) or GFP26. The secondary antibodies used were sheep anti-mouse IgG-HRP and anti-rabbit IgG-HRP antibodies (Amersham Pharmacia Biotech). Proteins were visualized using an ECL detection kit (Amersham Pharmacia Biotech).
Immunoprecipitation
1×RIPA buffer was used to prepare cell lysates. RIPA-insoluble debris was removed after centrifugation at 12 000 r/min for 30 min at 4 °C. The supernatants were subjected to immunoprecipitation with rabbit polyclonal anti-GFP antibodies coupled to protein G Sepharose (Roche) overnight at 4 °C. The protein G Sepharose was washed with 1×RIPA buffer six times and then eluted with SDS sample buffer for immunoblot analysis.
In vitro pulldown assay
An aliquot containing approximately 20 μg of GST or GST-Bcl-2 expressed by E coli was incubated with 30 μL of glutathione agarose beads (Pharmacia) for 30 min at 4 °C. After being washed two times with ice-cold 1×PBS, the beads were incubated with approximately 40 μg of p62 expressed by E coli strain BL21 for 3 h at 4 °C. Finally, the beads were washed five times with the ice-cold 1×PBS. Bound proteins were eluted with SDS loading buffer for immunoblot analysis.
Fluorescent microscopy
HeLa cells were washed with 1×PBS and fixed with 4% paraformaldehyde for 10 min at room temperature and then treated with 0.25% Triton X-100 for 5 min. After blocking with 5% fetal bovine serum for 30 min, cells were incubated with anti-p62 (Enzo Life) and anti-Flag antibody (Sigma), then with Alexa Fluor 594 (red) and Alexa Fluor 350 (blue) conjugated secondary antibodies (Invitrogen) for 2 h. The cells were visualized using an IX71 inverted system microscope (Olympus).
Statistical analysis
Western blot densitometry analyses of immunoblots from three independent experiments were performed with Photoshop 7.0 software (Adobe). Final data analyses were performed with Origin 6.0 (Originlab).
Results
p62 can enhance basal autophagy
The conversion of LC3 type I (LC3-I) to LC3 type II (LC3-II) reflects autophagosome formation and autophagy activation27,28. To determine whether p62 is involved in autophagy regulation, we examined the levels of LC3-I/II after p62 was knocked down or overexpressed in HeLa cells. Surprisingly, LC3-II levels decreased when we knocked down p62 using si-p62 siRNA (Figure 1A) but increased when we overexpressed p62 in HeLa cells (Figure 1B). To further confirm the effects of p62 on LC3 conversion and to ascertain whether there was cell-type specificity, we knocked down and overexpressed p62 in HEK293 cells and also observed that p62 affected LC3 conversion (Figure 1C, 1D, and 1E). Taken together, these data suggest that p62 itself can activate autophagy in cell lines (Figure 1).
Figure 1.
p62 activates autophagy. (A) Knockdown of p62 inhibited autophagy in HeLa cells. HeLa cells were transfected with si-nc (negative control siRNA) or si-p62 (combination of #1 and #2 si-p62) for 48 h, and then cell lysates were subjected to immunoblot analysis using LC3, tubulin and p62 antibodies. (B) Autophagy was activated after overexpression of Flag-p62 in HeLa cells. Flag or Flag-p62 was transfected into HeLa cells for 48 h. The cell lysates were then subjected to immunoblot analysis using LC3, tubulin, and Flag antibodies. (C) Knockdown of p62 decreased autophagy in HEK293 cells. HEK293 cells were transfected with si-nc (negative control siRNA) or si-p62 (combination of #1 and #2 si-p62) for 48 h, and then cell lysates were subjected to immunoblot analysis using LC3, tubulin and p62 antibodies. (D and E) Autophagy was activated when Flag-p62 and p62-EGFP were overexpressed in HEK293 cells. Flag, Flag-p62, EGFP, or p62-EGFP was transfected into HEK293 cells for 48 h. The cell lysates were then subjected to immunoblot analysis using LC3, tubulin, Flag and GFP antibodies. Mean±SEM. n=3. bp<0.05.
p62 activates autophagy in a Bcl-2 dependent manner
We have shown that p62 can activate autophagy (Figure 1). As Beclin 1/class III phosphatidylinositol 3-kinase (PI3K) complex is a key component for autophagosome nucleation5,29, we examined whether p62 could affect the function of this component. When p62 was knocked down or overexpressed in HeLa cells, the protein levels of Beclin 1 remained unchanged (Figure 2A and 2B), indicating that p62 does not activate autophagy by influencing Beclin 1 levels. It was previously reported that Beclin 1 and Bcl-2 function together to regulate autophagy by interacting with each other30,31. We next investigated whether p62 regulates autophagy through Bcl-2. We overexpressed p62 in Bcl-2 knockdown cells. The effect of p62 on autophagy induction was abrogated by Bcl-2 knockdown (Figure 2C). These results suggest that p62 activates autophagy in a Bcl-2-dependent manner.
Figure 2.
p62 activates autophagy in a Bcl-2 dependent manner. (A) HeLa cells were transfected with si-nc or si-p62 for 48 h, and then cell lysates were subjected to immunoblot analysis using Beclin 1, LC3, tubulin, and p62 antibodies. (B) Flag or Flag-p62 was transfected into HeLa cells for 48 h. The cell lysates were then subjected to immunoblot analysis using Beclin 1, LC3, tubulin, and Flag antibodies. (C) HeLa cells were transfected with si-nc or si-Bcl-2 for 24 h and then were transfected with Flag or Flag-p62 for another 48 h. Finally, the cell lysates were subjected to immunoblot analysis using LC3, tubulin, Bcl-2, and Flag antibodies. Mean±SEM. n=3. bp<0.05.
p62 interacts with and co-localizes with Bcl-2
To further explore how p62 activates autophagy through Bcl-2, we performed various biochemical and immunofluorescent assays. The in vitro pulldown assay showed that p62 directly interacted with Bcl-2 (Figure 3A). To confirm this interaction, we transfected Flag-p62 into HEK293 cells and performed co-immunoprecipitation experiments. p62 was co-immunoprecipitated when Bcl-2 was immunoprecipitated using anti-Bcl-2 antibodies (Figure 3B). Moreover, p62 co-localized with Bcl-2 in HeLa cells (Figure 3C), suggesting that p62 interacts with Bcl-2.
Figure 3.
Direct association between p62 and Bcl-2. (A) p62 directly interacted with Bcl-2. GST or GST-tagged Bcl-2 bound to glutathione agarose beads was incubated with pET-21a-p62. After incubation, the beads were washed, and bound proteins were subjected to immunoblot analysis using GST or p62 antibodies. (B) Flag-p62 interacted with endogenous Bcl-2 in HEK293 cells. FLAG-tagged p62 was transfected into HEK293 cells. Cells were collected 48 h after transfection and subjected to immunoprecipitation analysis. The inputs and immunoprecipitates were then subjected to immunoblot analysis using antibodies against Bcl-2 and FLAG-HRP. (C) Flag-Bcl-2 co-localized with endogenous p62 in HeLa cells. HeLa cells were transfected with Flag-Bcl-2 for 48 h. Cells were then fixed and subjected to an immunofluorescent assay.
p62 regulates the interaction between Bcl-2 and Beclin 1
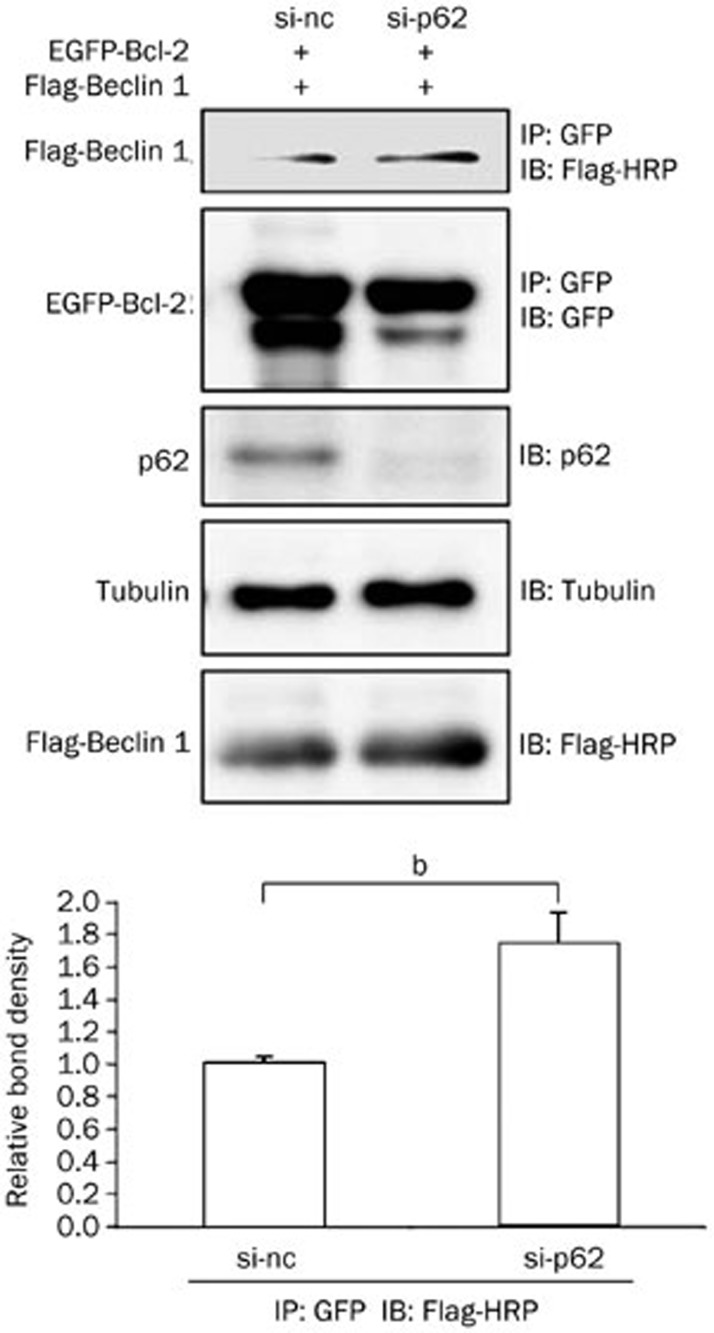
Because p62 can interact with Bcl-2, and Bcl-2 is a well-known inhibitor of autophagy that acts through interaction with Beclin 1, we next examined whether p62 affects the association between Bcl-2 and Beclin 1. In p62 knockdown cells, a greater amount of Beclin 1 co-immunoprecipitated with Bcl-2 (Figure 4), suggesting that p62 can block the association between Bcl-2 and Beclin 1.
Figure 4.
p62 regulates the interaction between Bcl-2 and Beclin 1. HEK293 cells were first transfected with si-nc or si-p62 for 24 h and then transfected with Flag-Beclin 1 together with EGFP or EGFP-Bcl-2 for another 48 h. The cells were then collected and subjected to immunoprecipitation analysis. The inputs and immunoprecipitants were subjected to immunoblot analysis using antibodies against GFP, FLAG-HRP, or tubulin. Mean±SEM. n=3. bp<0.05.
Discussion
p62 is a cargo receptor that is involved in autophagy and other cellular pathways. It is generally believed that p62 is an adaptor protein for the autophagic degradation of aggregate-prone or misfolded proteins and organelles, and that p62 itself is a selective autophagy substrate targeted by the autophagy-lysosome degradation system through its interaction with LC313. In this study, we demonstrated that p62 can directly induce autophagy in cultured cells (Figure 1).
Beclin 1/class III PI3K complex plays an important role in autophagosome nucleation29. From our observations, p62 does not induce autophagy by directly affecting Beclin 1/class III PI3K pathway, because the protein levels of Beclin 1 did not change when we knocked down or overexpressed p62 (Figure 2A and 2B). However, p62 binds to Bcl-2 and affects the protein interaction between Bcl-2 and Beclin 1 (Figures 3 and 4), suggesting that p62 regulates autophagy through its direct association with Bcl-2. Given that the anti-apoptotic protein Bcl-2 can bind to Beclin 1 to disturb the Beclin 1-class III PI3K interaction and Beclin 1-associated class III PI3K activity30,31, Bcl-2 and Beclin 1 cooperate together to regulate the vesicle nucleation step of the autophagy process. We found that autophagy was activated when Bcl-2 was knocked down (Figure 2C), which is consistent with other studies30,31. We further observed that p62 induces autophagy activation, which can be blocked by Bcl-2 knockdown (Figure 2C), suggesting that p62 induces autophagy in a Bcl-2-dependent manner.
Because a better understanding of the physiological function of p62 may require the identification of p62 binding partners, in the current study we identified a novel p62 binding protein involved in Beclin 1/class III PI3K machinery. The question that remains is how p62 affects the interaction between Bcl-2 and Beclin 1. We speculate that there are three possibilities: (1) p62 interacts with Bcl-2 to influence Beclin 1/Bcl-2 complex formation; (2) because the association between Bcl-2 and Beclin 1 may require a specific conformation of Bcl-2, p62 may cause a conformational change in Bcl-2 after they interact and subsequently facilitate the dissociation of Bcl-2 from Beclin 1, which may promote Beclin 1-class III PI3K interaction and/or Beclin 1-associated class III PI3K activity; and (3) posttranslational modification such as ubiquitination and phosphorylation of Bcl-2 is very important for Bcl-2 function24,28,32. For example, ERK1-mediated phosphorylation of Bcl-2 promotes autophagy by dissociating Bcl-2 and Beclin 127,33. Because p62 can regulate the activity of various kinases such as PKC and ERK34,35,36, it is possible that p62 may affect the phosphorylation of Bcl-2 through its interaction with Bcl-2 and then modulate Bcl-2- and Beclin 1-mediated autophagy. The precise molecular mechanism underlying the modification of the interaction between Bcl-2 and Beclin 1 driven by p62 needs to be further explored in the future.
In summary, we find that p62 directly binds to Bcl-2 to disturb the Bcl-2 and Beclin 1 interaction, thereby stimulating autophagy through the Bcl-2- and Beclin 1-mediated pathway. Our findings define a novel mechanism in which mammalian cells can use the key autophagic cargo adaptor protein p62 to regulate autophagy activity. Given that p62 is known to be involved in crosstalk between cellular protein quality control systems such as the proteasomal and lysosomal degradation systems37,38, as well as diverse aspects of cellular pathways involved in cell death, apoptosis, tumorigenesis, metabolism and inflammation21,39, this autophagic mechanism may help us to extend our knowledge of p62 function.
Author contribution
Liang ZHOU, Zheng YING, and Guang-hui WANG designed the research; Liang ZHOU, Hong-feng WANG, Hai-gang REN, Dong CHEN, Feng GAO, Qing-song HU, Chen FU, and Ran-jie XU performed the research; Liang ZHOU, Zheng YING, and Guang-hui WANG analyzed the data; Liang ZHOU, Zheng YING, and Guang-hui WANG wrote the paper.
Acknowledgments
This work was supported in part by the National High-Tech Research and Development Program of China 973 Projects (2011CB504102), and the National Natural Sciences Foundation of China (91132723 and 31200803).
References
- Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2:a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy in protein and organelle turnover. Cold Spring Harb Symp Quant Biol. 2011;76:397–402. doi: 10.1101/sqb.2011.76.011023. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–9. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–65. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- Burman C, Ktistakis N. Autophagosome formation in mammalian cells. Semin Immunopathol. 2010;32:397–413. doi: 10.1007/s00281-010-0222-z. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–40. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- Yen WL, Klionsky DJ. How to live long and prosper: autophagy, mitochondria, and aging. Physiology (Bethesda) 2008;23:248–62. doi: 10.1152/physiol.00013.2008. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Menzies FM, Huebener J, Renna M, Bonin M, Riess O, Rubinsztein DC. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133:93–104. doi: 10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diazmeco M, Wooten M. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Tung YT, Hsu WM, Lee H, Huang WP, Liao YF. The evolutionarily conserved interaction between LC3 and p62 selectively mediates autophagy-dependent degradation of mutant huntingtin. Cell Mol Neurobiol. 2010;30:795–806. doi: 10.1007/s10571-010-9507-y. [DOI] [PubMed] [Google Scholar]
- Gal J, Strom AL, Kwinter DM, Kilty R, Zhang J, Shi P, et al. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111:1062–73. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey JR, Ralston SH, Hocking LJ, Sheppard PW, Ciani B, Searle MS, et al. Loss of ubiquitin-binding associated with paget's disease of bone p62 (SQSTM1) mutations. J Bone Miner Res. 2005;20:619–24. doi: 10.1359/JBMR.041205. [DOI] [PubMed] [Google Scholar]
- Rea SL, Walsh JP, Ward L, Yip K, Ward BK, Kent GN, et al. A novel mutation (K378X) in the sequestosome 1 gene associated with increased NF-kappaB signaling and Paget's disease of bone with a severe phenotype. J Bone Miner Res. 2006;21:1136–45. doi: 10.1359/jbmr.060405. [DOI] [PubMed] [Google Scholar]
- Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–6. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- Hocking LJ, Lucas GJ, Daroszewska A, Mangion J, Olavesen M, Cundy T, et al. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget's disease. Hum Mol Genet. 2002;11:2735–9. doi: 10.1093/hmg/11.22.2735. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–84. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Gao F, Li B, Wang H, Xu Y, Zhu C, et al. Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J Biol Chem. 2010;285:38214–23. doi: 10.1074/jbc.M110.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Hu Q, Wang H, Man N, Ren H, Wen L, et al. Omi/HtrA2 is a positive regulator of autophagy that facilitates the degradation of mutant proteins involved in neurodegenerative diseases. Cell Death Differ. 2010;17:1773–84. doi: 10.1038/cdd.2010.55. [DOI] [PubMed] [Google Scholar]
- Ying Z, Wang H, Fan H, Wang G. The endoplasmic reticulum (ER)-associated degradation system regulates aggregation and degradation of mutant neuroserpin. J Biol Chem. 2011;286:20835–44. doi: 10.1074/jbc.M110.200808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–5. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad N, Iyer AK, Manosroi A, Wang L, Rojanasakul Y. Superoxide-mediated proteasomal degradation of Bcl-2 determines cell susceptibility to Cr(VI)-induced apoptosis. Carcinogenesis. 2008;29:1538–45. doi: 10.1093/carcin/bgn137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–92. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GY, Nigro P, Fujiwara K, Abe JI, Berk BC. p62 Binding to protein kinase C zeta regulates tumor necrosis factor alpha-induced apoptotic pathway in endothelial cells. Arterioscler Thromb Vasc Biol. 2012;32:2974–80. doi: 10.1161/ATVBAHA.112.300054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Pfluger PT, Kim JY, Nogueiras R, Duran A, Pages G, et al. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep. 2010;11:226–32. doi: 10.1038/embor.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19:1576–86. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–8. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–27. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. Feedback on fat: p62-mTORC1-autophagy connections. Cell. 2011;147:724–7. doi: 10.1016/j.cell.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]