The composition of the subgingival bacterial microbiota is a critical determinant in the health status of periodontal tissues. Gram-negative anaerobes such as Porphyromonas gingivalis are well-established periodontal pathogens, and high numbers of these bacteria are found in the subgingival sulcus of patients with chronic periodontitis (43). However, a burgeoning pool of evidence indicates that these organisms have a far more complex relationship with the host than merely as pathogens. Gram-negative anaerobes are frequently present in the oral cavity of periodontally healthy individuals (23, 44, 92, 121, 127, 128, 170), and indeed health is the most common status of the human gingiva despite years of exposure to a large microbial burden. Periodontal organisms thus appear to have co-evolved with their host to maintain an ecologically balanced association whereby minimal harm is inflicted on, or by, either party. Disease will only ensue when this interaction becomes unbalanced, an event that has been termed an ecological catastrophe (83). Organisms such as P. gingivalis may thus be more accurately characterized as accidental, or host-adapted, pathogens.
In the subgingival compartment, epithelial cells represent a major host interface for colonizing organisms; hence, the interaction between gingival epithelial cells and periodontal bacteria will contribute to the success or failure of colonization, and to the maintenance of health or disease in the host. Undeniably, in the case of P. gingivalis, an intricate and multithreaded relationship exists between the organism and gingival epithelial cells, which, under optimal conditions, results in stable cohabitation, with both bacteria and host cells responding and adapting to the presence of their partner to maintain a state of health. In the event that this relationship becomes perturbed, for example because of an increase in bacterial burden or an inappropriate immune response, the periodontal disease process can be initiated (13).
The ability to adapt in response to the host environment is reflected in the genetic diversity found within many species of periodontal bacteria. Bacteria are masters of adaptation, and at the genetic level are able to rapidly modify and share DNA. For example, there are significant levels of genetic variation among P. gingivalis strains, and many studies have linked this genetic variability to virulence potential (7, 14, 18, 38, 67, 85, 108). Genetic variability among strains is common in bacteria with long-term carrier states, possibly arising from the co-evolutionary dynamic of host–pathogen interactions (62, 123). Genetic variation can produce lineages of bacteria with `good or evil' personalities, in that some are more virulent and associated with disease, whereas other strains of the same species behave in a more commensal manner. In this review, we will discuss the current understanding of pathogenic and commensal aspects of bacterial interactions with periodontal tissues, with a specific focus on P. gingivalis intracellular invasion and molecular modulation of host cells.
Interactions of periodontal bacteria with epithelial cells observed in vivo
Tissue destruction, mediated either by the host or by bacteria, is a hallmark of periodontal disease, and with the consequent loss of barrier function it is not surprising that periodontal bacteria are frequently detected within gingival tissues (1, 20, 41, 101, 116, 131–133, 152–154, 157). While tissue invasion (intercellular invasion) is an almost inevitable corollary of the disease process, a number of oral bacteria have been observed to locate inside host cells (intracellular invasion) both in the presence and in the absence of disease. Fluorescence in situ hybridization, combined with confocal microscopy, has established that buccal epithelial cells from healthy individuals contain a polymicrobial intracellular microbiota that includes the periodontal bacteria P. gingivalis, Tannerella forsythia, Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, Prevotella intermedia, Eikenella corrodens and Treponema denticola (128, 129). Importantly, these colonized epithelial cells are not necrotic or apoptotic, but remain viable (127). In tissue samples from patients with periodontitis, electron microscopy has demonstrated the presence of periodontal bacterial within epithelial cells (165); and immunohistochemistry revealed that these intracellular bacteria include P. gingivalis (122). Similar microscopic techniques have shown P. gingivalis, T. forsythia, A. actinomycetemcomitans and T. denticola within gingival and buccal epithelial cells from both healthy individuals and patients with periodontitis (22).
Residence within host cells provides bacteria with a nutrient-rich, generally reducing environment that is partially protected from the host immune system. Accessing this secure niche may be critical in the early stages of sulcus colonization by periodontal bacteria, as low numbers of bacteria are particularly susceptible to clearance by immune mechanisms. While not immediately contributing to disease, invasive bacteria may use the intracellular locale to safely persist and replicate. In disease states, intracellular bacteria are less likely to be physically removed by scaling and root planing (61) and are more resistant to antibiotics (36). Furthermore, this intracellular population could constitute a reservoir of bacteria for the repopulation of treated subgingival sites. The ability to invade and persist in host cells is evidently an important factor in the overall disease process, and P. gingivalis strains isolated from disease sites possess greater invasion capabilities in vitro than strains from healthy sites (58). Collectively, these observations indicate that an intracellular location is an integral component of the lifestyle of many periodontal bacteria, whether in a healthy or diseased host, and probably contributes to the chronic nature of periodontal disease.
Models to study intracellular invasion
Epithelial tissues are structurally diverse and range from simple, single-layered gut or glandular epithelia to the complex, stratified epithelia that form the body surface, including the oral cavity. While all epithelia have features and functions in common, they also exhibit many tissue-specific properties. The gingival epithelium is a stratified and squamous tissue, and is composed of oral, sulcular and junctional epithelium. The periodontally relevant sulcular and junctional epithelia are neither keratinized nor terminally differentiated, unlike other oral epithelial cells (118). As the differentiation status and tissue of origin can affect bacteria–epithelium interactions (66, 81, 91, 117), the most relevant models for periodontal bacteria will involve cells derived from, or with characteristics of, junctional or sulcular epithelium. A number of in vitro tissue culture models have been developed to study periodontal bacteria–epithelium interactions, including primary gingival epithelial cells (73, 74), transformed epithelial cells (46) and multilayers (4, 110, 140). Primary gingival epithelial cells, obtained from gingival explants, express keratin and differentiation markers characteristic of the junctional epithelium, and are naturally senescent (105, 106). Transformed lines derived from gingival epithelial cells are also poorly differentiated and respond similarly to oral bacteria (46, 104), without potential confounding influences of patient to patient variability (64). The KB and HEp-2 cell lines were originally thought to be derived from human oral epidermal carcinomas, and have been used as model systems for studies of periodontal disease. However, these cell lines are now known to have arisen from HeLa cell contamination, and as such are less relevant for studies of the oral cavity.
Utilization of orally derived model systems has demonstrated that several species of periodontal bacteria can internalize within epithelial cells through a bacterially directed process. Invasive organisms include P. gingivalis (Fig. 1), A. actinomycetemcomitans, T. forsythia, F. nucleatum and P. intermedia (Table 1) (6, 33, 45, 54, 55, 72–77, 86, 87, 130, 136). In addition, consistent with the multispecies etiology of periodontal disease, bacteria can co-operate with one another to facilitate invasion. For example, P. gingivalis (or its outer membrane vesicles) enhance the invasion of T. forsythia into epithelial cells (55). F. nucleatum can transport noninvasive Streptococcus cristatus into epithelial cells through the formation of co-adhered dual-species consortia (35). F. nucleatum can also enhance the invasion of P. gingivalis, although in this case the synergistic effect results from the interaction of F. nucleatum with the host cells (135). These in vitro observations emphasize the ecological nature of periodontal disease, in which multiple species act in concert. Periodontal bacteria can also enhance the invasion of Pseudomonas aeruginosa into epithelial cells, which may provide a mechanistic basis for the epidemiological association between periodontal disease and respiratory tract infections (109).
Fig. 1.
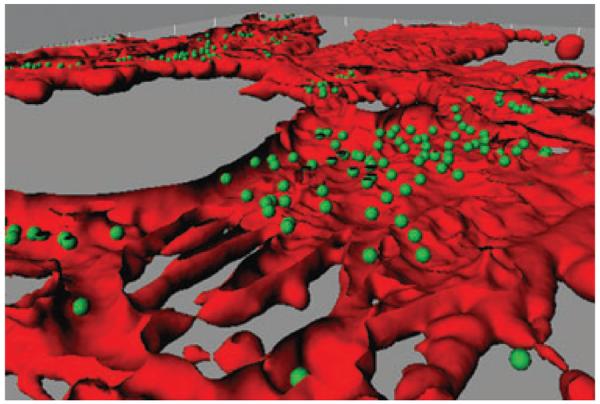
Invasion of human epithelial cells with Porphyromonas gingivalis. Confocal image of gingival epithelial cells [stained with TRITC-phalloidin (red)] infected with P. gingivalis [stained with FITC (green)]. The image was analyzed using Imaris version 5.0.1 software. A Z-stack of the x–y sections was converted to composite images using the iso surface and spot detection functions of the surpass option. The section view in the x and y axes was created using the clipping function. The image was generated by Masae Kuboniwa, Osaka University, Japan.
Table 1.
Invasive periodontal bacteria, invasion effectors and host cytoskeletal requirements for internalization
| Species | Invasins | Cytoskeletal requirements |
|---|---|---|
| Porphyromonas gingivalis | Major fimbriae (FimA) | Actin polymerization |
| Gingipains | Microtubule activity | |
| Phosphoserine phosphatase (SerB) | ||
| Aggregatibacter actinomycetemcomitans | Fimbriae | Actin polymerization |
| Membrane vesicles | ||
| Extracellular amorphous material | ||
| Api surface proteins/autotransporters | ||
| Fusobacterium nucleatum | FadA surface protein | Actin polymerization |
| Lam adhesin | Microtubule activity | |
| Prevotella intermedia | Type C fimbriae | Actin polymerization |
| Tannerella forsythia | BspA leucine-rich surface protein | |
| S-layer |
The initial interaction with epithelial cells
Attachment or close physical association between bacteria and epithelial cells can be a prelude to internalization. Engagement of membrane receptors by bacterial surface ligands allows recalibration of the cellular machinery to mediate pathogen entry into these nonphagocytic host cells. Many invasive bacterial species manipulate host cell receptors to activate their uptake, and oral pathogens are no exception to this paradigm (37). The most intensively studied of the invasive oral bacteria is P. gingivalis, and this subgingival resident will be the focus of the remainder of this review.
The mechanisms of P. gingivalis adhesion to, and invasion of, epithelial cells are multifaceted and involve a number of effector molecules (Fig. 2). In terms of initial binding, the predominant adhesins are the major fimbriae (168, 174), which are composed of the FimA structural subunit protein along with minor proteins FimC, D and E (100). The FimA subunit directly engages αvβ3 and α5β1 integrins on the epithelial surface (93, 174), and this interaction initiates an integrin-associated signaling cascade that triggers bacterial internalization (174). The focal adhesion adaptor and signaling proteins paxillin and focal adhesion kinase (FAK) are recruited to sites of P. gingivalis attachment (176), and the resulting information flow converges on the cytoskeletal architecture. Both actin microfilament and microtubule structures are remodeled to accommodate the entry of P. gingivalis (73, 174, 176). Integrin-mediated internalization may take place in lipid raft entry platforms that signal through the Rho GTPase, Rac1 (156, 161).
Fig. 2.
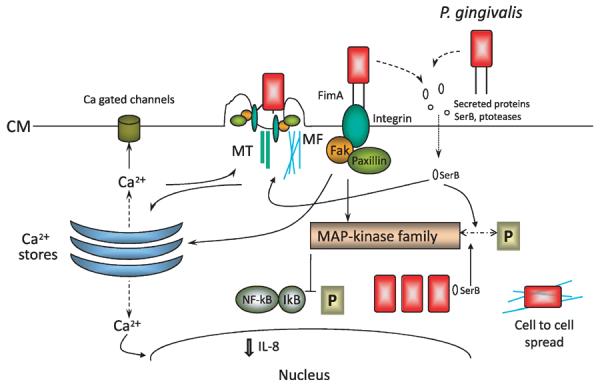
Model of interactions between Porphyromonas gingivalis and gingival epithelial cells that are associated with internalization. Proximity to gingival epithelial cells induces P. gingivalis to secrete proteins such as the SerB serine phosphatase. SerB enters gingival epithelial cells where it dephosphorylates target proteins, including mitogen-activated protein kinase family members, which in turn prevent NF-κB activation. SerB activity culminates in a reduction of interleukin-8 production and in the remodeling of microfilament and microtubule cytoskeletal architecture. Adhesion of P. gingivalis is mediated by the long (FimA) fimbriae that engage integrins and induce the formation of focal adhesin complexes and integrin-dependent signaling. Calcium ions (Ca2+) are released from intracellular stores, a signaling event that also funnels through the cytoskeletal structure, and the cytoskeletal re-arrangements allow P. gingivalis to enter the host cell. P. gingivalis cells rapidly locate in the perinuclear area where they replicate and utilize microfilaments to spread to adjacent gingival epithelial cells. CM, cytoplasmic membrane; IL-8, interleukin-8; IκB, inhibitor of κB; MAP, mitogen-activated protein kinase; MF, micro-filament; MT, microtubule; NF-κB, nuclear factor-κB; P, phosphate.
The strength of the initial adherence between P. gingivalis and the host epithelial cell can vary. The major (long) fimbriae are the primary bacterial adhesins, and there are at least six alleles of the fimA gene (fimA I, Ib, II, III, IV and V) distributed among strains worldwide (3, 95). In studies of P. gingivalis isolates from healthy and diseased individuals, major fimbriae composed of type Ib, II or IV are more commonly associated with periodontal disease, whereas type I, III, or V fimbriae are more often found in P. gingivalis strains colonizing healthy patients (3, 89, 90). The fimA II allele has been shown to result in stronger adherence to epithelial cell receptor α5β1 integrin, compared with other fimA types (94, 96).
While FimA–integrin interactions constitute the predominant means of P. gingivalis adherence and entry into gingival epithelial cells, FimA binding to intercellular adhesion molecule (ICAM) 1 can also initiate invasion into HeLa cells (156), and this mechanism could also play a role in gingival cells. Furthermore, invasion occurs in the absence of FimA, albeit less efficiently. Therefore, other invasins are operational, and these include the gingipain proteases (103, 119). The gingipains are a family of three arginine/lysine-specific proteases (RgpA, RgpB and Kgp) that are found in the outer membrane of the bacteria and are also secreted into the surrounding environment. The gingipain proteases have both enzymatic and structural functions that are integral to the successful adherence of P. gingivalis (19, 145). Gingipain protease activity has been shown to improve P. gingivalis binding to gingival cells by modifying matrix proteins and revealing epithelial surface cryptitopes (68, 69, 158), and protease-deficient mutants show diminished invasion efficiency (112). Structurally, the RgpA and Kgp gingipains possess hemagglutinin/adhesin domains that are involved in the attachment of P. gingivalis to epithelial cells (17, 114). The adhesin domain of RgpA associates with fibronectin and with the α5β1 integrin receptor for fibronectin on gingival fibroblasts (142), which can lead to internalization and nuclear targeting of this gingipain protease (141). RgpA can interact with clatherin, and the purified protein can be internalized via a clathrin-dependent endocytosis pathway in HeLa (HEp-2) cells (12). As RgpA proteins are present on the surface of P. gingivalis, this molecule may act as an adhesin and allow internalization of P. gingivalis cells via an alternative pathway to that driven by FimA–integrin interactions.
Internalization
The interaction of P. gingivalis FimA fimbriae with epithelial cell surface integrins initiates a cellular response that recruits FAK and paxillin to the cytoplasmic membrane at the bacterial attachment site (174, 176). The resulting protein–protein interactions among integrin, FAK and paxillin produce a phosphorylation-regulated signaling scaffold that activates Rho-family GTPases, enzymes which play a central role in initiating downstream signaling cascades and regulating cytoskeletal dynamics (25, 47). The subsequent actin and microtubule remodeling, the recruitment of lipid raft components, and host-cell phosphorylation activity are all required for internalization of P. gingivalis (73, 75, 138, 161, 167, 176). The invasion process is complete in approximately 15 min and ultimately results in the perinuclear localization of the bacteria (11).
Entry of P. gingivalis into host cells results in the reprogramming of major host-cell signaling pathways. Consistent with activation by integrin-initiated Rho cascades, components of mitogen-activated protein kinase pathways are selectively targeted for regulation by internalized P. gingivalis. Extracellular signal-regulated kinase 1/2 and c-Jun N-terminal kinase activities are down-regulated and up-regulated, respectively (167). Kinase regulation occurs in a dose-dependent manner and requires metabolically active bacteria, implying that the regulation of host mitogen-activated protein kinases requires the production of bacterial effectors. Invasion by P. gingivalis also induces a transient increase in epithelial cell cytosolic calcium concentrations (10, 57). Calcium signaling can regulate a variety of cellular functions; and during invasion with bacterial pathogens, calcium levels can influence cytokine expression and modulate intracellular trafficking and cytoskeletal activities (159). Calcium levels impact P. gingivalis manipulation of host cells, as over-expression of the calcium-binding protein, calprotectin, in gingival epithelial cells inhibits invasion (99).
Although P. gingivalis does not possess the type III secretion machinery that injects bacterial invasion effectors directly into the host cell cytoplasm (56, 98, 134), it does secrete a distinct set of proteins upon encountering the epithelial cell environment (112). Among these is a HAD family serine phosphatase (SerB), that is active on host cell phosphoproteins and influences P. gingivalis entry and survival (48, 160). Microarray analysis found that SerB impacts the transcriptional profile of gingival epithelial cells, with pathways involving the actin cytoskeleton among those significantly overpopulated with differentially regulated genes (48). Moreover, a SerB mutant of P. gingivalis is defective in actin remodeling and in internalization, and interaction between gingival epithelial cells and purified SerB protein results in actin re-arrangements, an increase in the F/G actin ratio, and disruption of microtubule dynamics. Thus, SerB can interact with signaling pathways that regulate gene expression, cytoskeletal dynamics and ultimately affect P. gingivalis internalization and survival. One could presume that the net effect of signaling pathway manipulation is alteration of the host cellular physiology, restructuring the cytoskeleton to direct bacterial uptake and perinuclear localization, and ultimately crafting a protected intracellular niche for these fastidious organisms.
Adaptation of P. gingivalis to the intracellular environment
It hardly bears mentioning that the mammalian intracellular environment is quite distinct from both bacterial culture medium and the subgingival sulcus. Intracellular bacteria will experience distinct nutritional and physical conditions and, as is facile for bacteria, will adapt to these conditions through gene and protein regulation. Global molecular approaches to examine bacterial transcriptional and proteomic changes have provided insights into this complex process.
Transcriptional profiling by microarrays has been adopted to examine global gene regulation during the process of P. gingivalis invasion. Hosogi & Duncan (50) found that during attachment to HeLa (HEp-2) cells, genes encoding oxidative stress-response components and heat shock proteins were up-regulated, indicating that P. gingivalis bacteria on the surface of host cells experience oxidative stress and produce heat shock proteins to maintain protein function and viability. A study of gene expression of P. gingivalis within endothelial cells (125), demonstrated that internalized bacteria regulate pathways relating to energy metabolism, protein synthesis and transport through the outer membrane. Differential display reverse transcription-polymerase chain reaction confirmed that inside epithelial cells P. gingivalis regulates the expression of membrane transporters, and that loss of the corresponding gene products impairs bacterial invasive ability (113). Although functional roles for these transporters have yet to be defined, it is likely that they will affect the import/export of cations and nutrients.
Protein expression has been compared between internal and external P. gingivalis using whole-cell quantitative proteomic analyses (169). Interestingly, several classical virulence factors, including FimA, RgpA/B and Kgp, show decreased expression in internalized P. gingivalis. While FimA is required for optimal adherence and to initiate invasion pathways, production of this protein is evidently superfluous once the bacteria reach the intracellular milieu. Tight control of gingipain production is also a prudent maneuver for bacteria attempting to establish an intracellular niche, as gingipains are potent proteases and their over-expression could result in excessive damage to the interior of the host cell. Internal P. gingivalis also down-regulate a number of hemin-acquisition systems, and thus iron may not be limiting in the intracellular environment.
Establishment of a stable relationship between the host cell and internalized bacteria is stressful for both host and pathogen. For P. gingivalis, the transition from extracellular to intracellular environments evidently requires a drastic overhaul of the proteins and enzymes required for survival. As is common for bacteria adapting to changing conditions, multiple systems exist to mediate the disposal of toxic products, degrade inactive proteins and to assist in the expression and folding of new proteins. Intracellular P. gingivalis up-regulates the production of stress-associated proteins such as peroxidases, components of the Clp family and heat shock proteins such as HtrA. Deletion of the clpB or clpP genes has a negative impact on bacterial survival in gingival epithelial cells (16, 177, 181), and mutation of htrA results in an increased sensitivity to hydrogen peroxide and a decreased survival in animal infection models (126, 178).
In terms of bacterial metabolism, in internalized bacteria there is an increased abundance of proteins comprising the energy pathway leading from asparagine/aspartate amino acids to ATP. The pathway producing propionate shows an increased abundance of component proteins, while a tendency towards decreased abundance of proteins is observed for the pathway leading to butyrate production. As propionate is a less potent inducer of apoptosis than butyrate (84), this metabolic shift could also minimize damage to the host cells. The translational machinery, including ribosomal proteins and transfer RNA synthetases, shows a significant increase in expression, as do proteins responsible for transcription. In total, approximately 50% of the expressed proteome is differentially regulated by intracellular P. gingivalis. Overall, these results suggest that the intracellular environment, while initially stressful, is energy rich for P. gingivalis and consequently it is advantageous for the organism to undergo major adaptations that permit entry into, and use of the metabolic substrates available within, the host cell.
Intracellular localization
Intracellular bacteria must avoid host cell defenses located within the cytoplasm in order to establish long-term residence in the host cell. A primary defense organelle is the acid-containing lysosome, which is the `garbage disposal' system of the eukaryotic cell. Membrane-bound vacuoles containing cytoplasmic debris fuse with lysosomes, which degrade the material contained within the vacuole by exposure to lysosomal acids and enzymes. Once inside the cell, bacteria must act swiftly to prevent exposure to lysosomes. Two cytoplasmic membrane-trafficking systems converge on lysosomes: the autophagic pathway and the endosomal pathway. Intracellular pathogens use varying approaches to manipulate these membrane trafficking systems. Some pathogens remain within a membrane-bound vacuole and express effectors to block fusion with lysosomes. Other organisms escape from the vacuole and subsist freely in the cytoplasm. P. gingivalis is capable of invading multiple cell types (30), and accumulating evidence indicates that it uses different strategies to evade lysosomes in epithelial cells and endothelial cells.
Gingival epithelial cells are the main host tissue in contact with P. gingivalis, thus eons of host–pathogen contact and adaptation have resulted in a finely tuned relationship. P. gingivalis traversing the epithelial cell outer membrane must initially be encompassed within a compartment derived from the host membrane (34, 74, 139, 140). Once inside the cell, however, P. gingivalis escapes and survives unbound in the cytoplasm, where it remains viable for extended periods of time and even replicates (73, 74, 79, 111). Ultimately, these nonmotile bacteria localize to the perinuclear region of the host cell, an area densely packed with endoplasmic reticulum. One could hypothesize that the endoplasmic reticulum contents act as an excellent nutrient source for these proteolytic bacteria, and they may target this location to feed off proteins produced by the endoplasmic reticulum during host cell translation.
Although the oral cavity is the natural home of P. gingivalis, these bacteria can disseminate intra-vascularly during the transient bacteremias that result from mastication or oral hygiene procedures. Under these circumstances, P. gingivalis will be in contact with the endothelial cells that line the vessels of the circulatory system. P. gingivalis can adhere to and invade endothelial cells, although at a lower frequency than gingival epithelial cells (29, 120). Variation between the frequencies of invasion into epithelial and endothelial cells may be related to differences in surface receptors and signal transduction pathways. Microscopic evidence illustrates differences in the intracellular localization route of P. gingivalis between epithelial and endothelial cells. In endothelial cells, P. gingivalis ultimately traffics to autophagosomes, distinctive double-membraned vacuoles that are part of the autophagic pathway (31, 32). Once in these membrane compartments, the bacteria block fusion with lysosomes and probably use protein debris trafficked through the autophagic pathway as their nutrient source (9, 124). P. gingivalis persists in these autophagosome vacuoles, and inflammation and cell damage can result from the accumulation of high loads of these intracellular bacteria. It has been speculated that the invasion of coronary artery endothelial cells by oral bacteria may be a contributing factor to the link between periodontal disease and cardiovascular disease (28).
Phenotype of colonized cells
The stress associated with maintaining an intracellular bacterial burden results in significant phenotypic changes in the infected host cells. There is a spectrum of physiological and morphological outcomes that may result from bacterial invasion, with the ultimate destiny of the host cell being dependent on the characteristics of the invading P. gingivalis strain, the total bacterial burden and the host cell type. Perhaps one of the most critical bacterial characteristics is the production of gingipain virulence factors. Gingipain proteases are secreted to make protein nutrients available for the asaccharolytic P. gingivalis. High levels of these bacterial enzymes can damage host cells and connective tissue, and can induce apoptosis (145). Hence, strains of P. gingivalis that produce high levels of gingipains will be more cytotoxic (60, 102, 143, 144, 149, 166), whereas less proteolytic strains, and strains that can control protease production appropriately, are able to establish a more commensal relationship with the host by regulating apoptotic signaling pathways to prevent cell death (82, 97, 163, 172, 175).
Exposure of host cells to high-protease-secreting P. gingivalis, or to high numbers of bacteria, results in cell rounding and loss of attachment as a result of gingipain cleavage of cadherins and integrins (143). Gingipains can also penetrate the host cell (102), where gingipain protease activity is sufficient to activate pro-apoptotic molecules such as caspase-3, caspase-8, caspase-9, Bid and Bax (149). Additional damage to periodontal tissues can result from the activation of matrix metalloproteases by P. gingivalis gingipains (26, 27, 42, 115) and the destruction of paxillin and other focal adhesion components (49, 63, 96). Conversely, the anti-apoptotic phenotype induced by low-protease-secreting P. gingivalis, or by challenge with lower numbers of bacteria, is associated with activation of the phosphatidylinositol 3-kinase/Akt and Janus kinase/STAT pathways, up-regulation of anti-apoptosis genes Bcl-2 and survivin, and inhibition of cytochrome c release and of caspase-3 activity (Fig. 3) (82, 172). More recently, P. gingivalis has been shown to interfere with ATP-induced apoptotic pathways via the secretion of an ATP-hydrolyzing enzyme that is a homolog of nucleoside diphosphate kinase (175). ATP scavenging by P. gingivalis inhibits apoptosis by preventing ATP ligation of P2X7 purinergic receptors (Fig. 3).
Fig. 3.
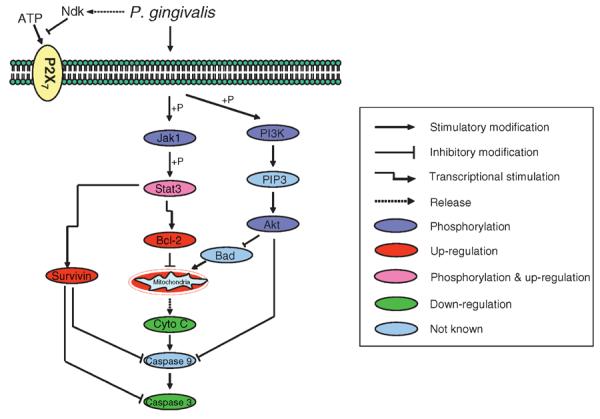
Summary of major apoptotic pathways modulated by Porphyromonas gingivalis in gingival epithelial cells to suppress apoptotic cell death. Jak1; P; PI3K; PIP3; Stat3.
Bacterial interference with cellular physiology can also impact the host cell cycle, by either activating or inhibiting cell cycle progression. In gingival epithelial cells, invasion with P. gingivalis results in increased proliferation, which is associated with accelerated progression through the S-phase (Fig. 4) (70). Up-regulating the rate of cell division may be a mechanism to maintain a reservoir of bacterially infected cells, in response to the high rate of cell turnover in the junctional epithelium (13). Host cell division may also allow bacterial cells to replicate without creating an overwhelming intracellular bacterial burden. In disease states, loss of cell cycle control could impact wound healing in the periodontal pocket, thus facilitating bacterial penetration of the periodontal tissues.
Fig. 4.
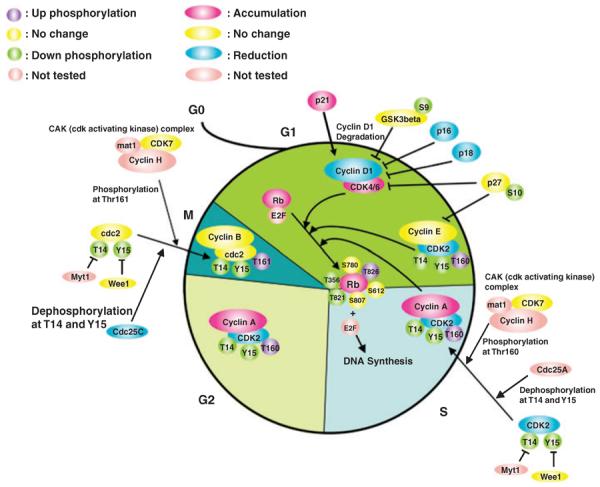
Porphyromonas gingivalis impacts cell cycle control in gingival epithelial cells. Schematic representation of cell cycle pathways modulated by P. gingivalis infection. The pointed arrow indicates molecular interactions resulting in activation; the flat arrow indicates molecular interactions resulting in inhibition. Reprinted with permission from Microbes and Infection 2008, 10:122-128, Copyright Elsevier (70).
Invasion of periodontal tissues
As discussed, subversion of host physiology is a complex and stressful process for both the host cell and the invading pathogen. Once a stable relationship is achieved, it is understandable that P. gingivalis would attempt to prolong the life span of its host cell by blocking apoptosis and stimulating proliferation. The host cell does not, however, necessarily achieve a state of immortality; ultimately, intracellular bacteria must have a strategy to access new environments. P. gingivalis has been detected in the periodontal connective tissue, implying that cell-to-cell spread of bacteria is a common event.
In a recent study, Yilmaz et al. (173) presented a new in vitro model system to study bacterial cell-to-cell transmission. In this system, P. gingivalis bacteria (labeled with red fluorescence) are allowed to invade cultured gingival epithelial cells (labeled with blue fluorescence). After 24 h of co-culture, the infected blue gingival cells are mixed with uninfected green gingival cells. Using fluorescence microscopy, the investigators were able to clearly demonstrate transmission of bacteria from infected to noninfected host cells. Transmission from cell to cell is mediated by a membranous projection with a structural scaffold composed of actin filaments. Initiation of the spreading mechanism occurs at the highest frequency after 24 h of invasion, indicating that cell-to-cell transmission is a late-stage strategy of invasive P. gingivalis (171). This intracellular transmission does not appear to affect host cell viability and may be a prominent mechanism for the spread of invasive bacteria in the `stealth' mode. By moving deeper into the epithelial layers, P. gingivalis can ensure access to viable, nonshedding epithelial cells.
In a three-dimensional cellular model for bacterial dissemination, Andrian et al. (4) demonstrated that P. gingivalis can spread through the upper layers of gingival epithelial cells and can also penetrate the basement membrane into connective tissues. P. gingivalis gingipain proteases are capable of degrading matrix and tight junction components, destroying the physical barriers formed by extracellular connective tissue and cellular adhesion (2, 4, 146, 147). A P. gingivalis gingipain mutant was also able to invade the upper gingival layers, but was unable to access the connective tissue layer (4). The ability to disseminate beyond the initial site of infection is a characteristic of pathogenic bacteria in general, and we can anticipate that P. gingivalis strains able to penetrate the basement membrane and approach the alveolar bone may be more likely to exacerbate the bone loss associated with periodontal disease.
Impact on innate immune surveillance
An expected advantage of residing in an intracellular niche is avoidance of the host immune response. However, during adherence and invasion bacteria are exposed to innate immune-surveillance systems. Gingival epithelial cells express toll-like receptors and other surface pattern-recognition receptors along with intracellular recognition systems such as NODs (150). A robust proinflammatory cytokine and chemokine response would thus be expected following an interaction between the gingival epithelium and the periodontal bacteria. In many instances, such as with F. nucleatum, this is indeed the case (164). P. gingivalis can also induce the expression of proinflammatory immune mediators from gingival epithelial cells (5, 71, 137); however, the inflammatory phenotype of P. gingivalis is much more subtle and nuanced. P. gingivalis often suppresses or evades various components of innate immunity, a feature that has led to its characterization as a stealth-like pathogen (24, 40). For example, gingival epithelial cells do not express CD14 (a co-receptor for toll-like receptor 2) on the surface and thus respond poorly to P. gingivalis FimA (39), which may limit inflammatory responses to fimbriated, invasive P. gingivalis. A more pro-active role for P. gingivalis in dampening innate immune responses can be seen from its ability to suppress transcription of the interleukin-8 gene in gingival epithelial cells, and thus inhibit expression of this chemokine (51–53, 59, 164). Moreover, P. gingivalis can antagonize interleukin-8 secretion following stimulation of epithelial cells with other common plaque constituents, a phenomenon known as localized chemokine paralysis (24). A reduction in interleukin-8 levels, along with the down-regulation of intercellular adhesion molecule-1 (52, 80) will impair neutrophil infiltration of gingival tissues, and consequently debilitate local innate immunity and eventually disrupt the ecological balance between the host and the subgingival microbiota, contributing to the initiation of disease activity.
Mechanistically, invasive P. gingivalis inhibit the activity of the transcription factor NF-κB through the SerB-mediated disruption of signaling pathways (48). In addition, P. gingivalis proteases impair inflammatory responses through the degradation of cytokines, chemokines and their receptors (8, 15, 88, 107, 148, 155, 179, 180). However, consistent with the bipolar personality of P. gingivalis, gingipain RgpA–Kgp complexes can penetrate the gingival connective tissue and stimulate the secretion of proinflammatory mediators (102). Moreover, the activation of protease-activated receptors PAR-1 and PAR-2 by P. gingivalis proteases can both down-regulate the production of interleukin-8 (162) and up-regulate the production of interleukin-6 (78). Clearly the pro- or anti-inflammatory status of gingival tissues in the presence of P. gingivalis is highly context dependent.
Gingival epithelial cells can protect themselves against microbial challenge by the production of antimicrobial peptides, such as human beta-defensins (40), and intracellular antimicrobial compounds, such as calprotectin (99). As with some cytokine responses, there is no obvious trend for human beta-defensin regulation by P. gingivalis. In various studies, human beta-defensins 1, 2 and 3, which are produced by gingival epithelial cells, have been found to be all up-regulated, variously up-regulated or not regulated (21, 40, 151, 164). While bacterial strain differences and heterogeneity in epithelial cell receptor expression (65) may account for some of these differences, it is also possible that the bacterial load plays an important role. At a high number of P. gingivalis (possibly corresponding to advanced disease), the secreted proteases may overwhelm host cells and obscure the biological activity of other molecules, whereas at lower bacterial numbers (more equivalent to gingival health), the full range of P. gingivalis host physiology subversion may be observed.
Conclusions
Periodontitis presents with a wide spectrum of clinical severity, and this complex disease phenotype is produced by variations in host susceptibility as well as variations in the composition and virulence of the oral bacterial microbiota. Investigation of the host- pathogen interaction at the cellular level has begun to reveal the behaviors of periodontal bacteria that contribute to the disease process. For many oral bacteria, the ability to invade host cells and establish an intracellular niche is a critical survival mechanism. As exemplified by P. gingivalis, this initially innocuous relationship with a host cell can potentially shift to a more sinister one.
From the host–bacteria interactions described here, it may seem as if P. gingivalis is suffering from multiple-personality disorder. However, this pathogenic variability is consistent with the genetic diversity of P. gingivalis. Strains with more potent combinations of virulence factors are capable of causing cell damage or death, whereas other, less virulent, strains behave in a more commensal manner. Thus, the specific virulence attributes of the invading bacterial strain are key to determining the final host cell outcome. Further investigation of these virulence traits will facilitate the development of the next generation of periodontal therapies.
References
- 1.Allenspach-Petrzilka GE, Guggenheim B. Bacterial invasion of the periodontium; an important factor in the pathogenesis of periodontitis? J Clin Periodontol. 1983;10:609–617. doi: 10.1111/j.1600-051x.1983.tb01299.x. [DOI] [PubMed] [Google Scholar]
- 2.Amano A. Disruption of epithelial barrier and impairment of cellular function by Porphyromonas gingivalis. Front Biosci. 2007;12:3965–3974. doi: 10.2741/2363. [DOI] [PubMed] [Google Scholar]
- 3.Amano A, Kuboniwa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79:1664–1668. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 4.Andrian E, Grenier D, Rouabhia M. In vitro models of tissue penetration and destruction by Porphyromonas gingivalis. Infect Immun. 2004;72:4689–4698. doi: 10.1128/IAI.72.8.4689-4698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrian E, Grenier D, Rouabhia M. Porphyromonas gingivalis lipopolysaccharide induces shedding of syndecan-1 expressed by gingival epithelial cells. J Cell Physiol. 2005;204:178–183. doi: 10.1002/jcp.20287. [DOI] [PubMed] [Google Scholar]
- 6.Asakawa R, Komatsuzawa H, Kawai T, Yamada S, Goncalves RB, Izumi S, Fujiwara T, Nakano Y, Suzuki N, Uchida Y, Ouhara K, Shiba H, Taubman MA, Kurihara H, Sugai M. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol Microbiol. 2003;50:1125–1139. doi: 10.1046/j.1365-2958.2003.03748.x. [DOI] [PubMed] [Google Scholar]
- 7.Asano H, Ishihara K, Nakagawa T, Yamada S, Okuda K. Relationship between transmission of Porphyromonas gingivalis and fimA type in spouses. J Periodontol. 2003;74:1355–1360. doi: 10.1902/jop.2003.74.9.1355. [DOI] [PubMed] [Google Scholar]
- 8.Banbula A, Bugno M, Kuster A, Heinrich PC, Travis J, Potempa J. Rapid and efficient inactivation of IL-6 gingi-pains, lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Biochem Biophys Res Commun. 1999;261:598–602. doi: 10.1006/bbrc.1999.1075. [DOI] [PubMed] [Google Scholar]
- 9.Belanger M, Rodrigues PH, Dunn WA, Jr, Progulske-Fox A. Autophagy: a highway for Porphyromonas gingivalis in endothelial cells. Autophagy. 2006;2:165–170. doi: 10.4161/auto.2828. [DOI] [PubMed] [Google Scholar]
- 10.Belton CM, Goodwin PC, Fatherazi S, Schubert MM, Lamont RJ, Izutsu KT. Calcium oscillations in gingival epithelial cells infected with Porphyromonas gingivalis. Microbes Infect. 2004;6:440–447. doi: 10.1016/j.micinf.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Belton CM, Izutsu KT, Goodwin PC, Park Y, Lamont RJ. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell Microbiol. 1999;1:215–223. doi: 10.1046/j.1462-5822.1999.00022.x. [DOI] [PubMed] [Google Scholar]
- 12.Boisvert H, Duncan MJ. Clathrin-dependent entry of a gingipain adhesin peptide and Porphyromonas gingivalis into host cells. Cell Microbiol. 2008;10:2538–2552. doi: 10.1111/j.1462-5822.2008.01228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosshardt DD, Lang NP. The junctional epithelium: from health to disease. J Dent Res. 2005;84:9–20. doi: 10.1177/154405910508400102. [DOI] [PubMed] [Google Scholar]
- 14.Califano JV, Arimoto T, Kitten T. The genetic relatedness of Porphyromonas gingivalis clinical and laboratory strains assessed by analysis of insertion sequence (IS) element distribution. J Periodontal Res. 2003;38:411–416. doi: 10.1034/j.1600-0765.2003.00665.x. [DOI] [PubMed] [Google Scholar]
- 15.Calkins CC, Platt K, Potempa J, Travis J. Inactivation of tumor necrosis factor-alpha by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. J Biol Chem. 1998;273:6611–6614. doi: 10.1074/jbc.273.12.6611. [DOI] [PubMed] [Google Scholar]
- 16.Capestany CA, Tribble GD, Maeda K, Demuth DR, Lamont RJ. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J Bacteriol. 2008;190:1436–1446. doi: 10.1128/JB.01632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T, Duncan MJ. Gingipain adhesin domains mediate Porphyromonas gingivalis adherence to epithelial cells. Microb Pathog. 2004;36:205–209. doi: 10.1016/j.micpath.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Chen T, Hosogi Y, Nishikawa K, Abbey K, Fleischmann RD, Walling J, Duncan MJ. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J Bacteriol. 2004;186:5473–5479. doi: 10.1128/JB.186.16.5473-5479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen T, Nakayama K, Belliveau L, Duncan MJ. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect Immun. 2001;69:3048–3056. doi: 10.1128/IAI.69.5.3048-3056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christersson LA, Wikesjo UM, Albini B, Zambon JJ, Genco RJ. Tissue localization of Actinobacillus actinomycetemcomitans in human periodontitis. II. Correlation between immunofluorescence and culture techniques. J Periodontol. 1987;58:540–545. doi: 10.1902/jop.1987.58.8.540. [DOI] [PubMed] [Google Scholar]
- 21.Chung WO, Hansen SR, Rao D, Dale BA. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J Immunol. 2004;173:5165–5170. doi: 10.4049/jimmunol.173.8.5165. [DOI] [PubMed] [Google Scholar]
- 22.Colombo AV, da Silva CM, Haffajee A, Colombo AP. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J Periodontal Res. 2007;42:236–243. doi: 10.1111/j.1600-0765.2006.00938.x. [DOI] [PubMed] [Google Scholar]
- 23.Colombo AV, Silva CM, Haffajee A, Colombo AP. Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J Med Microbiol. 2006;55:609–615. doi: 10.1099/jmm.0.46417-0. [DOI] [PubMed] [Google Scholar]
- 24.Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121(Pt 15):2435. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeCarlo AA, Grenett HE, Harber GJ, Windsor LJ, Bodden MK, Birkedal-Hansen B, Birkedal-Hansen H. Induction of matrix metalloproteinases and a collagen-degrading phenotype in fibroblasts and epithelial cells by secreted Porphyromonas gingivalis proteinase. J Periodontal Res. 1998;33:408–420. doi: 10.1111/j.1600-0765.1998.tb02337.x. [DOI] [PubMed] [Google Scholar]
- 27.DeCarlo AA, Jr, Windsor LJ, Bodden MK, Harber GJ, Birkedal-Hansen B, Birkedal-Hansen H. Activation and novel processing of matrix metalloproteinases by a thiol-proteinase from the oral anaerobe Porphyromonas gingivalis. J Dent Res. 1997;76:1260–1270. doi: 10.1177/00220345970760060501. [DOI] [PubMed] [Google Scholar]
- 28.Deshpande RG, Khan M, Genco CA. Invasion strategies of the oral pathogen Porphyromonas gingivalis: implications for cardiovascular disease. Invasion Metastasis. 1998;18:57–69. doi: 10.1159/000024499. [DOI] [PubMed] [Google Scholar]
- 29.Deshpande RG, Khan MB, Genco CA. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect Immun. 1998;66:5337–5343. doi: 10.1128/iai.66.11.5337-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorn BR, Burks JN, Seifert KN, Progulske-Fox A. Invasion of endothelial and epithelial cells by strains of Porphyromonas gingivalis. FEMS Microbiol Lett. 2000;187:139–144. doi: 10.1111/j.1574-6968.2000.tb09150.x. [DOI] [PubMed] [Google Scholar]
- 31.Dorn BR, Dunn WA, Jr, Progulske-Fox A. Invasion of human coronary artery cells by periodontal pathogens. Infect Immun. 1999;67:5792–5798. doi: 10.1128/iai.67.11.5792-5798.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorn BR, Dunn WA, Jr, Progulske-Fox A. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect Immun. 2001;69:5698–5708. doi: 10.1128/IAI.69.9.5698-5708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorn BR, Leung KL, Progulske-Fox A. Invasion of human oral epithelial cells by Prevotella intermedia. Infect Immun. 1998;66:6054–6057. doi: 10.1128/iai.66.12.6054-6057.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duncan MJ, Nakao S, Skobe Z, Xie H. Interactions of Porphyromonas gingivalis with epithelial cells. Infect Immun. 1993;61:2260–2265. doi: 10.1128/iai.61.5.2260-2265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards AM, Grossman TJ, Rudney JD. Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infect Immun. 2006;74:654–662. doi: 10.1128/IAI.74.1.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eick S, Pfister W. Efficacy of antibiotics against period-ontopathogenic bacteria within epithelial cells: an in vitro study. J Periodontol. 2004;75:1327–1334. doi: 10.1902/jop.2004.75.10.1327. [DOI] [PubMed] [Google Scholar]
- 37.Ellen RP. Perturbation and exploitation of host cell cytoskeleton by periodontal pathogens. Microbes Infect. 1999;1:621–632. doi: 10.1016/s1286-4579(99)80062-8. [DOI] [PubMed] [Google Scholar]
- 38.Enersen M, Olsen I, van Winkelhoff AJ, Caugant DA. Multilocus sequence typing of Porphyromonas gingivalis strains from different geographic origins. J Clin Microbiol. 2006;44:35–41. doi: 10.1128/JCM.44.1.35-41.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eskan MA, Hajishengallis G, Kinane DF. Differential activation of human gingival epithelial cells and monocytes by Porphyromonas gingivalis fimbriae. Infect Immun. 2007;75:892–898. doi: 10.1128/IAI.01604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Z, Weinberg A. Role of bacteria in health and disease of periodontal tissues. Periodontol 2000. 2006;40:50–76. doi: 10.1111/j.1600-0757.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 41.Frank RM. Bacterial penetration in the apical pocket wall of advanced human periodontitis. J Periodontal Res. 1980;15:563–573. doi: 10.1111/j.1600-0765.1980.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 42.Grayson R, Douglas CW, Heath J, Rawlinson A, Evans GS. Activation of human matrix metalloproteinase 2 by gingival crevicular fluid and Porphyromonas gingivalis. J Clin Periodontol. 2003;30:542–550. doi: 10.1034/j.1600-051x.2003.00301.x. [DOI] [PubMed] [Google Scholar]
- 43.Griffen AL, Becker MR, Lyons SR, Moeschberger ML, Leys EJ. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol. 1998;36:3239–3242. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haffajee AD, Cugini MA, Tanner A, Pollack RP, Smith C, Kent RL, Jr, Socransky SS. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 45.Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, Genco RJ. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Handfield M, Mans JJ, Zheng G, Lopez MC, Mao S, Progulske-Fox A, Narasimhan G, Baker HV, Lamont RJ. Distinct transcriptional profiles characterize oral epithelium–microbiota interactions. Cell Microbiol. 2005;7:811–823. doi: 10.1111/j.1462-5822.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- 47.Harburger DS, Calderwood DA. Integrin signalling at a glance. J Cell Sci. 2009;122(Pt 2):159. doi: 10.1242/jcs.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasegawa Y, Tribble GD, Baker HV, Mans JJ, Handfield M, Lamont RJ. Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect Immun. 2008;76:2420–2427. doi: 10.1128/IAI.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hintermann E, Haake SK, Christen U, Sharabi A, Quaranta V. Discrete proteolysis of focal contact and adherens junction components in Porphyromonas gingivalis-infected oral keratinocytes: a strategy for cell adhesion and migration disabling. Infect Immun. 2002;70:5846–5856. doi: 10.1128/IAI.70.10.5846-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hosogi Y, Duncan MJ. Gene expression in Porphyromonas gingivalis after contact with human epithelial cells. Infect Immun. 2005;73:2327–2335. doi: 10.1128/IAI.73.4.2327-2335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang GT, Haake SK, Kim JW, Park NH. Differential expression of interleukin-8 and intercellular adhesion molecule-1 by human gingival epithelial cells in response to Actinobacillus actinomycetemcomitans or Porphyromonas gingivalis infection. Oral Microbiol Immunol. 1998;13:301–309. doi: 10.1111/j.1399-302x.1998.tb00711.x. [DOI] [PubMed] [Google Scholar]
- 52.Huang GT, Kim D, Lee JK, Kuramitsu HK, Haake SK. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via antagonistic mechanisms. Infect Immun. 2001;69:1364–1372. doi: 10.1128/IAI.69.3.1364-1372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang GT, Zhang HB, Dang HN, Haake SK. Differential regulation of cytokine genes in gingival epithelial cells challenged by Fusobacterium nucleatum and Porphyromonas gingivalis. Microb Pathog. 2004;37:303–312. doi: 10.1016/j.micpath.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 54.Ikegami A, Chung P, Han YW. Complementation of the fadA mutation in Fusobacterium nucleatum demonstrates that the surface-exposed adhesin promotes cellular invasion and placental colonization. Infect Immun. 2009;77:3075–3079. doi: 10.1128/IAI.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by “Tannerella forsythia”. Infect Immun. 2006;74:5023–5028. doi: 10.1128/IAI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishiguro I, Saiki K, Konishi K. PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system. FEMS Microbiol Lett. 2009;292:261–267. doi: 10.1111/j.1574-6968.2009.01489.x. [DOI] [PubMed] [Google Scholar]
- 57.Izutsu KT, Belton CM, Chan A, Fatherazi S, Kanter JP, Park Y, Lamont RJ. Involvement of calcium in interactions between gingival epithelial cells and Porphyromonas gingivalis. FEMS Microbiol Lett. 1996;144:145–150. doi: 10.1111/j.1574-6968.1996.tb08521.x. [DOI] [PubMed] [Google Scholar]
- 58.Jandik KA, Belanger M, Low SL, Dorn BR, Yang MC, Progulske-Fox A. Invasive differences among Porphyromonas gingivalis strains from healthy and diseased periodontal sites. J Periodontal Res. 2008;43:524–530. doi: 10.1111/j.1600-0765.2007.01064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ji S, Kim Y, Min BM, Han SH, Choi Y. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J Periodontal Res. 2007;42:503–510. doi: 10.1111/j.1600-0765.2007.00974.x. [DOI] [PubMed] [Google Scholar]
- 60.Johansson A, Kalfas S. Characterization of the proteinase-dependent cytotoxicity of Porphyromonas gingivalis. Eur J Oral Sci. 1998;106:863–871. doi: 10.1046/j.0909-8836.1998.eos106405.x. [DOI] [PubMed] [Google Scholar]
- 61.Johnson JD, Chen R, Lenton PA, Zhang G, Hinrichs JE, Rudney JD. Persistence of extracrevicular bacterial reservoirs after treatment of aggressive periodontitis. J Periodontol. 2008;79:2305–2312. doi: 10.1902/jop.2008.080254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jolley KA, Wilson DJ, Kriz P, McVean G, Maiden MC. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol Biol Evol. 2005;22:562–569. doi: 10.1093/molbev/msi041. [DOI] [PubMed] [Google Scholar]
- 63.Katz J, Yang QB, Zhang P, Potempa J, Travis J, Michalek SM, Balkovetz DF. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infect Immun. 2002;70:2512–2518. doi: 10.1128/IAI.70.5.2512-2518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kinane DF, Galicia JC, Gorr SU, Stathopoulou PG, Benakanakere M. P. gingivalis interactions with epithelial cells. Front Biosci. 2008;13:966–984. doi: 10.2741/2736. [DOI] [PubMed] [Google Scholar]
- 65.Kinane DF, Shiba H, Stathopoulou PG, Zhao H, Lappin DF, Singh A, Eskan MA, Beckers S, Waigel S, Alpert B, Knudsen TB. Gingival epithelial cells heterozygous for Toll-like receptor 4 polymorphisms Asp299Gly and Thr399ile are hypo-responsive to Porphyromonas gingivalis. Genes Immun. 2006;7:190–200. doi: 10.1038/sj.gene.6364282. [DOI] [PubMed] [Google Scholar]
- 66.Klumpp DJ, Forrestal SG, Karr JE, Mudge CS, Anderson BE, Schaeffer AJ. Epithelial differentiation promotes the adherence of type 1-piliated Escherichia coli to human vaginal cells. J Infect Dis. 2002;186:1631–1638. doi: 10.1086/345557. [DOI] [PubMed] [Google Scholar]
- 67.Koehler A, Karch H, Beikler T, Flemmig TF, Suerbaum S, Schmidt H. Multilocus sequence analysis of Porphyromonas gingivalis indicates frequent recombination. Microbiology. 2003;149:2407–2415. doi: 10.1099/mic.0.26267-0. [DOI] [PubMed] [Google Scholar]
- 68.Kontani M, Kimura S, Nakagawa I, Hamada S. Adherence of Porphyromonas gingivalis to matrix proteins via a fimbrial cryptic receptor exposed by its own argininespecific protease. Mol Microbiol. 1997;24:1179–1187. doi: 10.1046/j.1365-2958.1997.4321788.x. [DOI] [PubMed] [Google Scholar]
- 69.Kontani M, Ono H, Shibata H, Okamura Y, Tanaka T, Fujiwara T, Kimura S, Hamada S. Cysteine protease of Porphyromonas gingivalis 381 enhances binding of fimbriae to cultured human fibroblasts and matrix proteins. Infect Immun. 1996;64:756–762. doi: 10.1128/iai.64.3.756-762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuboniwa M, Hasegawa Y, Mao S, Shizukuishi S, Amano A, Lamont RJ, Yilmaz O. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10:122–128. doi: 10.1016/j.micinf.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kusumoto Y, Hirano H, Saitoh K, Yamada S, Takedachi M, Nozaki T, Ozawa Y, Nakahira Y, Saho T, Ogo H, Shimabukuro Y, Okada H, Murakami S. Human gingival epithelial cells produce chemotactic factors interleukin-8 and monocyte chemoattractant protein-1 after stimulation with Porphyromonas gingivalis via toll-like receptor 2. J Periodontol. 2004;75:370–379. doi: 10.1902/jop.2004.75.3.370. [DOI] [PubMed] [Google Scholar]
- 72.Lamont RJ, Bevan CA, Gil S, Persson RE, Rosan B. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol Immunol. 1993;8:272–276. doi: 10.1111/j.1399-302x.1993.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 73.Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lamont RJ, Oda D, Persson RE, Persson GR. Interaction of Porphyromonas gingivalis with gingival epithelial cells maintained in culture. Oral Microbiol Immunol. 1992;7:364–367. doi: 10.1111/j.1399-302x.1992.tb00637.x. [DOI] [PubMed] [Google Scholar]
- 75.Lamont RJ, Yilmaz O. In or out: the invasiveness of oral bacteria. Periodontol 2000. 2002;30:61–69. doi: 10.1034/j.1600-0757.2002.03006.x. [DOI] [PubMed] [Google Scholar]
- 76.Lee SW, Sabet M, Um HS, Yang J, Kim HC, Zhu W. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene. 2006;371:102–111. doi: 10.1016/j.gene.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 77.Li L, Matevski D, Aspiras M, Ellen RP, Lepine G. Two epithelial cell invasion-related loci of the oral pathogen Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2004;19:16–25. doi: 10.1046/j.0902-0055.2003.00102.x. [DOI] [PubMed] [Google Scholar]
- 78.Lourbakos A, Potempa J, Travis J, D'Andrea MR, Andrade-Gordon P, Santulli R, Mackie EJ, Pike RN. Arginine-specific protease from Porphyromonas gingivalis activates protease-activated receptors on human oral epithelial cells and induces interleukin-6 secretion. Infect Immun. 2001;69:5121–5130. doi: 10.1128/IAI.69.8.5121-5130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Madianos PN, Papapanou PN, Nannmark U, Dahlen G, Sandros J. Porphyromonas gingivalis FDC381 multiplies and persists within human oral epithelial cells in vitro. Infect Immun. 1996;64:660–664. doi: 10.1128/iai.64.2.660-664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Madianos PN, Papapanou PN, Sandros J. Porphyromonas gingivalis infection of oral epithelium inhibits neutrophil transepithelial migration. Infect Immun. 1997;65:3983–3990. doi: 10.1128/iai.65.10.3983-3990.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mans JJ, Baker HV, Oda D, Lamont RJ, Handfield M. Distinctive characteristics of transcriptional profiles from two epithelial cell lines upon interaction with Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2006;21:261–267. doi: 10.1111/j.1399-302X.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 82.Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, Stavropoulos MF, Yilmaz O, Lamont RJ. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9:1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 84.Matthews GM, Howarth GS, Butler RN. Short-chain fatty acid modulation of apoptosis in the Kato III human gastric carcinoma cell line. Cancer Biol Ther. 2007;6:1051–1057. doi: 10.4161/cbt.6.7.4318. [DOI] [PubMed] [Google Scholar]
- 85.Menard C, Mouton C. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect Immun. 1995;63:2522–2531. doi: 10.1128/iai.63.7.2522-2531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meyer DH, Fives-Taylor PM. The role of Actinobacillus actinomycetemcomitans in the pathogenesis of periodontal disease. Trends Microbiol. 1997;5:224–228. doi: 10.1016/S0966-842X(97)01055-X. [DOI] [PubMed] [Google Scholar]
- 87.Meyer DH, Lippmann JE, Fives-Taylor PM. Invasion of epithelial cells by Actinobacillus actinomycetemcomitans: a dynamic, multistep process. Infect Immun. 1996;64:2988–2997. doi: 10.1128/iai.64.8.2988-2997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mikolajczyk-Pawlinska J, Travis J, Potempa J. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 1998;440:282–286. doi: 10.1016/s0014-5793(98)01461-6. [DOI] [PubMed] [Google Scholar]
- 89.Missailidis CG, Umeda JE, Ota-Tsuzuki C, Anzai D, Mayer MP. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004;19:224–229. doi: 10.1111/j.1399-302X.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 90.Miura M, Hamachi T, Fujise O, Maeda K. The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J Periodontal Res. 2005;40:147–152. doi: 10.1111/j.1600-0765.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 91.Mooney A, Byrne C, Clyne M, Johnson-Henry K, Sherman P, Bourke B. Invasion of human epithelial cells by Campylobacter upsaliensis. Cell Microbiol. 2003;5:835–847. doi: 10.1046/j.1462-5822.2003.00325.x. [DOI] [PubMed] [Google Scholar]
- 92.Moore WE, Moore LV. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 93.Nakagawa I, Amano A, Inaba H, Kawai S, Hamada S. Inhibitory effects of Porphyromonas gingivalis fimbriae on interactions between extracellular matrix proteins and cellular integrins. Microbes Infect. 2005;7:157–163. doi: 10.1016/j.micinf.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 94.Nakagawa I, Amano A, Kuboniwa M, Nakamura T, Kawabata S, Hamada S. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect Immun. 2002;70:277–285. doi: 10.1128/IAI.70.1.277-285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakagawa I, Amano A, Ohara-Nemoto Y, Endoh N, Morisaki I, Kimura S, Kawabata S, Hamada S. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J Periodontal Res. 2002;37:425–432. doi: 10.1034/j.1600-0765.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- 96.Nakagawa I, Inaba H, Yamamura T, Kato T, Kawai S, Ooshima T, Amano A. Invasion of epithelial cells and proteolysis of cellular focal adhesion components by distinct types of Porphyromonas gingivalis fimbriae. Infect Immun. 2006;74:3773–3782. doi: 10.1128/IAI.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakhjiri SF, Park Y, Yilmaz O, Chung WO, Watanabe K, El-Sabaeny A, Park K, Lamont RJ. Inhibition of epithelial cell apoptosis by Porphyromonas gingivalis. FEMS Microbiol Lett. 2001;200:145–149. doi: 10.1111/j.1574-6968.2001.tb10706.x. [DOI] [PubMed] [Google Scholar]
- 98.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nisapakultorn K, Ross KF, Herzberg MC. Calprotectin expression in vitro by oral epithelial cells confers resistance to infection by Porphyromonas gingivalis. Infect Immun. 2001;69:4242–4247. doi: 10.1128/IAI.69.7.4242-4247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nishiyama S, Murakami Y, Nagata H, Shizukuishi S, Kawagishi I, Yoshimura F. Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology. 2007;153:1916–1925. doi: 10.1099/mic.0.2006/005561-0. [DOI] [PubMed] [Google Scholar]
- 101.Noiri Y, Ozaki K, Nakae H, Matsuo T, Ebisu S. An immunohistochemical study on the localization of Porphyromonas gingivalis, Campylobacter rectus and Actinomyces viscosus in human periodontal pockets. J Periodontal Res. 1997;32:598–607. doi: 10.1111/j.1600-0765.1997.tb00937.x. [DOI] [PubMed] [Google Scholar]
- 102.O'Brien-Simpson NM, Pathirana RD, Walker GD, Reynolds EC. Porphyromonas gingivalis RgpA-Kgp proteinase-adhesin complexes penetrate gingival tissue and induce proinflammatory cytokines or apoptosis in a concentration-dependent manner. Infect Immun. 2009;77:1246–1261. doi: 10.1128/IAI.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O'Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Pept Sci. 2003;4:409–426. doi: 10.2174/1389203033487009. [DOI] [PubMed] [Google Scholar]
- 104.Oda D, Bigler L, Lee P, Blanton R. HPV immortalization of human oral epithelial cells: a model for carcinogenesis. Exp Cell Res. 1996;226:164–169. doi: 10.1006/excr.1996.0215. [DOI] [PubMed] [Google Scholar]
- 105.Oda D, Dale BA, Bourekis G. Human oral epithelial cell culture. II. Keratin expression in fetal and adult gingival cells. In Vitro Cell Dev Biol. 1990;26:596–603. doi: 10.1007/BF02624209. [DOI] [PubMed] [Google Scholar]
- 106.Oda D, Watson E. Human oral epithelial cell culture I. Improved conditions for reproducible culture in serum-free medium. In Vitro Cell Dev Biol. 1990;26:589–595. doi: 10.1007/BF02624208. [DOI] [PubMed] [Google Scholar]
- 107.Oleksy A, Banbula A, Bugno M, Travis J, Potempa J. Proteolysis of interleukin-6 receptor (IL-6R) by Porphyromonas gingivalis cysteine proteinases (gingipains) inhibits interleukin-6-mediated cell activation. Microb Pathog. 2002;32:173–181. doi: 10.1006/mpat.2002.0491. [DOI] [PubMed] [Google Scholar]
- 108.Ozmeric N, Preus NR, Olsen I. Genetic diversity of Porphyromonas gingivalis and its possible importance to pathogenicity. Acta Odontol Scand. 2000;58:183–187. doi: 10.1080/000163500429190. [DOI] [PubMed] [Google Scholar]
- 109.Pan Y, Teng D, Burke AC, Haase EM, Scannapieco FA. Oral bacteria modulate invasion and induction of apoptosis in HEp-2 cells by Pseudomonas aeruginosa. Microb Pathog. 2009;46:73–79. doi: 10.1016/j.micpath.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 110.Papaioannou W, van Steenberghe D, Cassiman JJ, Dierickx K, Quirynen M. Adhesion of Porphyromonas gingivalis to cultured pocket epithelium: mono- and multi-layered. Clin Oral Investig. 2003;7:162–166. doi: 10.1007/s00784-003-0217-4. [DOI] [PubMed] [Google Scholar]
- 111.Papapanou PN, Sandros J, Lindberg K, Duncan MJ, Niederman R, Nannmark U. Porphyromonas gingivalis may multiply and advance within stratified human junctional epithelium in vitro. J Periodontal Res. 1994;29:374–375. doi: 10.1111/j.1600-0765.1994.tb01237.x. [DOI] [PubMed] [Google Scholar]
- 112.Park Y, Lamont RJ. Contact-dependent protein secretion in Porphyromonas gingivalis. Infect Immun. 1998;66:4777–4782. doi: 10.1128/iai.66.10.4777-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Park Y, Yilmaz O, Jung IY, Lamont RJ. Identification of Porphyromonas gingivalis genes specifically expressed in human gingival epithelial cells by using differential display reverse transcription-PCR. Infect Immun. 2004;72:3752–3758. doi: 10.1128/IAI.72.7.3752-3758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pathirana RD, O'Brien-Simpson NM, Visvanathan K, Hamilton JA, Reynolds EC. The role of the RgpA-Kgp proteinase-adhesin complexes in the adherence of Porphyromonas gingivalis to fibroblasts. Microbiology. 2008;154:2904–2911. doi: 10.1099/mic.0.2008/019943-0. [DOI] [PubMed] [Google Scholar]
- 115.Pattamapun K, Tiranathanagul S, Yongchaitrakul T, Kuwatanasuchat J, Pavasant P. Activation of MMP-2 by Porphyromonas gingivalis in human periodontal ligament cells. J Periodontal Res. 2003;38:115–121. doi: 10.1034/j.1600-0765.2003.01650.x. [DOI] [PubMed] [Google Scholar]
- 116.Pekovic DD, Fillery ED. Identification of bacteria in immunopathological mechanisms of human periodontal diseases. J Periodontal Res. 1984;19:329–351. doi: 10.1111/j.1600-0765.1984.tb01007.x. [DOI] [PubMed] [Google Scholar]
- 117.Plotkowski MC, de Bentzmann S, Pereira SH, Zahm JM, Bajolet-Laudinat O, Roger P, Puchelle E. Pseudomonas aeruginosa internalization by human epithelial respiratory cells depends on cell differentiation, polarity, and junctional complex integrity. Am J Respir Cell Mol Biol. 1999;20:880–890. doi: 10.1165/ajrcmb.20.5.3408. [DOI] [PubMed] [Google Scholar]
- 118.Pollanen MT, Salonen JI, Uitto VJ. Structure and function of the tooth-epithelial interface in health and disease. Periodontol 2000. 2003;31:12–31. doi: 10.1034/j.1600-0757.2003.03102.x. [DOI] [PubMed] [Google Scholar]
- 119.Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003;4:397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- 120.Progulske-Fox A, Kozarov E, Dorn B, Dunn W, Jr, Burks J, Wu Y. Porphyromonas gingivalis virulence factors and invasion of cells of the cardiovascular system. J Periodontal Res. 1999;34:393–399. doi: 10.1111/j.1600-0765.1999.tb02272.x. [DOI] [PubMed] [Google Scholar]
- 121.Quirynen M, Vogels R, Pauwels M, Haffajee AD, Socransky SS, Uzel NG, van Steenberghe D. Initial subgingival colonization of `pristine' pockets. J Dent Res. 2005;84:340–344. doi: 10.1177/154405910508400409. [DOI] [PubMed] [Google Scholar]
- 122.Rautemaa R, Jarvensivu A, Kari K, Wahlgren J, DeCarlo A, Richardson M, Sorsa T. Intracellular localization of Porphyromonas gingivalis thiol proteinase in periodontal tissues of chronic periodontitis patients. Oral Dis. 2004;10:298–305. doi: 10.1111/j.1601-0825.2004.01021.x. [DOI] [PubMed] [Google Scholar]
- 123.Robinson K, Loughlin MF, Potter R, Jenks PJ. Host adaptation and immune modulation are mediated by homologous recombination in Helicobacter pylori. J Infect Dis. 2005;191:579–587. doi: 10.1086/427657. [DOI] [PubMed] [Google Scholar]
- 124.Rodrigues PH, Belanger M, Dunn W, Jr, Progulske-Fox A. Porphyromonas gingivalis and the autophagic pathway: an innate immune interaction? Front Biosci. 2008;13:178–187. doi: 10.2741/2668. [DOI] [PubMed] [Google Scholar]
- 125.Rodrigues PH, Progulske-Fox A. Gene expression profile analysis of Porphyromonas gingivalis during invasion of human coronary artery endothelial cells. Infect Immun. 2005;73:6169–6173. doi: 10.1128/IAI.73.9.6169-6173.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roy F, Vanterpool E, Fletcher HM. HtrA in Porphyromonas gingivalis can regulate growth and gingipain activity under stressful environmental conditions. Microbiology. 2006;152:3391–3398. doi: 10.1099/mic.0.29147-0. [DOI] [PubMed] [Google Scholar]
- 127.Rudney JD, Chen R. The vital status of human buccal epithelial cells and the bacteria associated with them. Arch Oral Biol. 2006;51:291–298. doi: 10.1016/j.archoralbio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 128.Rudney JD, Chen R, Sedgewick GJ. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J Dent Res. 2005;84:59–63. doi: 10.1177/154405910508400110. [DOI] [PubMed] [Google Scholar]
- 129.Rudney JD, Chen R, Zhang G. Streptococci dominate the diverse flora within buccal cells. J Dent Res. 2005;84:1165–1171. doi: 10.1177/154405910508401214. [DOI] [PubMed] [Google Scholar]
- 130.Sabet M, Lee SW, Nauman RK, Sims T, Um HS. The surface (S-) layer is a virulence factor of Bacteroides forsythus. Microbiology. 2003;149:3617–3627. doi: 10.1099/mic.0.26535-0. [DOI] [PubMed] [Google Scholar]
- 131.Saglie FR, Carranza FA, Jr, Newman MG, Cheng L, Lewin KJ. Identification of tissue-invading bacteria in human periodontal disease. J Periodontal Res. 1982;17:452–455. doi: 10.1111/j.1600-0765.1982.tb02024.x. [DOI] [PubMed] [Google Scholar]
- 132.Saglie FR, Marfany A, Camargo P. Intragingival occurrence of Actinobacillus actinomycetemcomitans and Bacteroides gingivalis in active destructive periodontal lesions. J Periodontol. 1988;59:259–265. doi: 10.1902/jop.1988.59.4.259. [DOI] [PubMed] [Google Scholar]
- 133.Saglie FR, Smith CT, Newman MG, Carranza FA, Jr, Pertuiset JH, Cheng L, Auil E, Nisengard RJ. The presence of bacteria in the oral epithelium in periodontal disease. II. Immunohistochemical identification of bacteria. J Periodontol. 1986;57:492–500. doi: 10.1902/jop.1986.57.8.492. [DOI] [PubMed] [Google Scholar]
- 134.Saiki K, Konishi K. Identification of a Porphyromonas gingivalis novel protein sov required for the secretion of gingipains. Microbiol Immunol. 2007;51:483–491. doi: 10.1111/j.1348-0421.2007.tb03936.x. [DOI] [PubMed] [Google Scholar]
- 135.Saito A, Inagaki S, Kimizuka R, Okuda K, Hosaka Y, Nakagawa T, Ishihara K. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2008;54:349–355. doi: 10.1111/j.1574-695X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- 136.Sakakibara J, Nagano K, Murakami Y, Higuchi N, Nakamura H, Shimozato K, Yoshimura F. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology. 2007;153:866–876. doi: 10.1099/mic.0.29275-0. [DOI] [PubMed] [Google Scholar]
- 137.Sandros J, Karlsson C, Lappin DF, Madianos PN, Kinane DF, Papapanou PN. Cytokine responses of oral epithelial cells to Porphyromonas gingivalis infection. J Dent Res. 2000;79:1808–1814. doi: 10.1177/00220345000790101301. [DOI] [PubMed] [Google Scholar]
- 138.Sandros J, Madianos PN, Papapanou PN. Cellular events concurrent with Porphyromonas gingivalis invasion of oral epithelium in vitro. Eur J Oral Sci. 1996;104:363–371. doi: 10.1111/j.1600-0722.1996.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 139.Sandros J, Papapanou P, Dahlen G. Porphyromonas gingivalis invades oral epithelial cells in vitro. J Periodontal Res. 1993;28:219–226. doi: 10.1111/j.1600-0765.1993.tb01072.x. [DOI] [PubMed] [Google Scholar]
- 140.Sandros J, Papapanou PN, Nannmark U, Dahlén G. Porphyromonas gingivalis invades human pocket epithelium in vitro. J Periodontal Res. 1994;29:62–69. doi: 10.1111/j.1600-0765.1994.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 141.Scragg MA, Alsam A, Rangarajan M, Slaney JM, Shepherd P, Williams DM, Curtis MA. Nuclear targeting of Porphyromonas gingivalis W50 protease in epithelial cells. Infect Immun. 2002;70:5740–5750. doi: 10.1128/IAI.70.10.5740-5750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Scragg MA, Cannon SJ, Rangarajan M, Williams DM, Curtis MA. Targeted disruption of fibronectin-integrin interactions in human gingival fibroblasts by the RI protease of Porphyromonas gingivalis W50. Infect Immun. 1999;67:1837–1843. doi: 10.1128/iai.67.4.1837-1843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sheets SM, Potempa J, Travis J, Casiano CA, Fletcher HM. Gingipains from Porphyromonas gingivalis W83 induce cell adhesion molecule cleavage and apoptosis in endothelial cells. Infect Immun. 2005;73:1543–1552. doi: 10.1128/IAI.73.3.1543-1552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sheets SM, Potempa J, Travis J, Fletcher HM, Casiano CA. Gingipains from Porphyromonas gingivalis W83 synergistically disrupt endothelial cell adhesion and can induce caspase-independent apoptosis. Infect Immun. 2006;74:5667–5678. doi: 10.1128/IAI.01140-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sheets SM, Robles-Price AG, McKenzie RM, Casiano CA, Fletcher HM. Gingipain-dependent interactions with the host are important for survival of Porphyromonas gingivalis. Front Biosci. 2008;13:3215–3238. doi: 10.2741/2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Smalley JW, Birss AJ, Shuttleworth CA. The degradation of type I collagen and human plasma fibronectin by the trypsin-like enzyme and extracellular membrane vesicles of Bacteroides gingivalis W50. Arch Oral Biol. 1988;33:323–329. doi: 10.1016/0003-9969(88)90065-9. [DOI] [PubMed] [Google Scholar]
- 147.Smalley JW, Shuttleworth CA, Birss AJ. Collagenolytic activity of the extracellular vesicles of Bacteroides gingivalis W50 and an avirulent variant W50 / BE1. Arch Oral Biol. 1989;34:579–583. doi: 10.1016/0003-9969(89)90098-8. [DOI] [PubMed] [Google Scholar]
- 148.Stathopoulou PG, Benakanakere MR, Galicia JC, Kinane DF. The host cytokine response to Porphyromonas gingivalis is modified by gingipains. Oral Microbiol Immunol. 2009;24:11–17. doi: 10.1111/j.1399-302X.2008.00467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Stathopoulou PG, Galicia JC, Benakanakere MR, Garcia CA, Potempa J, Kinane DF. Porphyromonas gingivalis induce apoptosis in human gingival epithelial cells through a gingipain-dependent mechanism. BMC Microbiol. 2009;9:107. doi: 10.1186/1471-2180-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sugawara Y, Uehara A, Fujimoto Y, Kusumoto S, Fukase K, Shibata K, Sugawara S, Sasano T, Takada H. Toll-like receptors, NOD1, and NOD2 in oral epithelial cells. J Dent Res. 2006;85:524–529. doi: 10.1177/154405910608500609. [DOI] [PubMed] [Google Scholar]
- 151.Taguchi Y, Imai H. Expression of beta-defensin-2 in human gingival epithelial cells in response to challenge with Porphyromonas gingivalis in vitro. J Periodontal Res. 2006;41:334–339. doi: 10.1111/j.1600-0765.2006.00879.x. [DOI] [PubMed] [Google Scholar]
- 152.Takarada H, Cattoni M, Sugimoto A, Rose GG. Ultra-structural studies of human gingiva. I. The upper part of the pocket epithelium in chronic periodontitis. J Periodontol. 1974;45:30–42. doi: 10.1902/jop.1974.45.1.30. [DOI] [PubMed] [Google Scholar]
- 153.Takarada H, Cattoni M, Sugimoto A, Rose GG. Ultra-structural studies of human gingiva. II. The lower part of the pocket epithelium in chronic periodontitis. J Periodontol. 1974;45:155–169. doi: 10.1902/jop.1974.45.3.155. [DOI] [PubMed] [Google Scholar]
- 154.Takeuchi H, Sumitani M, Tsubakimoto K, Tsutsui M. Oral microorganisms in the gingiva of individuals with periodontal disease. J Dent Res. 1974;53:132–136. doi: 10.1177/00220345740530010601. [DOI] [PubMed] [Google Scholar]
- 155.Tam V, O'Brien-Simpson NM, Chen YY, Sanderson CJ, Kinnear B, Reynolds EC. The RgpA-Kgp proteinase-adhesin complexes of Porphyromonas gingivalis inactivate the Th2 cytokines interleukin-4 and interleukin-5. Infect Immun. 2009;77:1451–1458. doi: 10.1128/IAI.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tamai R, Asai Y, Ogawa T. Requirement for intercellular adhesion molecule 1 and caveolae in invasion of human oral epithelial cells by Porphyromonas gingivalis. Infect Immun. 2005;73:6290–6298. doi: 10.1128/IAI.73.10.6290-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Thiha K, Takeuchi Y, Umeda M, Huang Y, Ohnishi M, Ishikawa I. Identification of periodontopathic bacteria in gingival tissue of Japanese periodontitis patients. Oral Microbiol Immunol. 2007;22:201–207. doi: 10.1111/j.1399-302X.2007.00354.x. [DOI] [PubMed] [Google Scholar]
- 158.Tokuda M, Duncan M, Cho MI, Kuramitsu HK. Role of Porphyromonas gingivalis protease activity in colonization of oral surfaces. Infect Immun. 1996;64:4067–4073. doi: 10.1128/iai.64.10.4067-4073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.TranVan Nhieu G, Clair C, Grompone G, Sansonetti P. Calcium signalling during cell interactions with bacterial pathogens. Biol Cell. 2004;96:93–101. doi: 10.1016/j.biolcel.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 160.Tribble GD, Mao S, James CE, Lamont RJ. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc Natl Acad Sci USA. 2006;103:11027–11032. doi: 10.1073/pnas.0509813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tsuda K, Furuta N, Inaba H, Kawai S, Hanada K, Yoshimori T, Amano A. Functional analysis of alpha5beta1 integrin and lipid rafts in invasion of epithelial cells by Porphyromonas gingivalis using fluorescent beads coated with bacterial membrane vesicles. Cell Struct Funct. 2008;33:123–132. doi: 10.1247/csf.08012. [DOI] [PubMed] [Google Scholar]
- 162.Uehara A, Naito M, Imamura T, Potempa J, Travis J, Nakayama K, Takada H. Dual regulation of interleukin-8 production in human oral epithelial cells upon stimulation with gingipains from Porphyromonas gingivalis. J Med Microbiol. 2008;57:500–507. doi: 10.1099/jmm.0.47679-0. [DOI] [PubMed] [Google Scholar]
- 163.Urnowey S, Ansai T, Bitko V, Nakayama K, Takehara T, Barik S. Temporal activation of anti- and pro-apoptotic factors in human gingival fibroblasts infected with the periodontal pathogen, Porphyromonas gingivalis: potential role of bacterial proteases in host signalling. BMC Microbiol. 2006;6:26. doi: 10.1186/1471-2180-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 164.Vankeerberghen A, Nuytten H, Dierickx K, Quirynen M, Cassiman JJ, Cuppens H. Differential induction of human beta-defensin expression by periodontal commensals and pathogens in periodontal pocket epithelial cells. J Periodontol. 2005;76:1293–1303. doi: 10.1902/jop.2005.76.8.1293. [DOI] [PubMed] [Google Scholar]
- 165.Vitkov L, Krautgartner WD, Hannig M. Bacterial internalization in periodontitis. Oral Microbiol Immunol. 2005;20:317–321. doi: 10.1111/j.1399-302X.2005.00233.x. [DOI] [PubMed] [Google Scholar]
- 166.Wang PL, Shirasu S, Shinohara M, Daito M, Oido M, Kowashi Y, Ohura K. Induction of apoptosis in human gingival fibroblasts by a Porphyromonas gingivalis protease preparation. Arch Oral Biol. 1999;44:337–342. doi: 10.1016/s0003-9969(99)00002-3. [DOI] [PubMed] [Google Scholar]
- 167.Watanabe K, Yilmaz O, Nakhjiri SF, Belton CM, Lamont RJ. Association of mitogen-activated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect Immun. 2001;69:6731–6737. doi: 10.1128/IAI.69.11.6731-6737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Weinberg A, Belton CM, Park Y, Lamont RJ. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1997;65:313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xia Q, Wang T, Taub F, Park Y, Capestany CA, Lamont RJ, Hackett M. Quantitative proteomics of intracellular Porphyromonas gingivalis. Proteomics. 2007;7:4323–4337. doi: 10.1002/pmic.200700543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol. 2000;27:648–657. doi: 10.1034/j.1600-051x.2000.027009648.x. [DOI] [PubMed] [Google Scholar]
- 171.Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology. 2008;154:2897–2903. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yilmaz O, Jungas T, Verbeke P, Ojcius DM. Activation of the phosphatidylinositol 3-kinase / Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2004;72:3743–3751. doi: 10.1128/IAI.72.7.3743-3751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yilmaz O, Verbeke P, Lamont RJ, Ojcius DM. Intercellular spreading of Porphyromonas gingivalis infection in primary gingival epithelial cells. Infect Immun. 2006;74:703–710. doi: 10.1128/IAI.74.1.703-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yilmaz O, Watanabe K, Lamont RJ. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 2002;4:305–314. doi: 10.1046/j.1462-5822.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 175.Yilmaz O, Yao L, Maeda K, Rose TM, Lewis EL, Duman M, Lamont RJ, Ojcius DM. ATP scavenging by the intracellular pathogen Porphyromonas gingivalis inhibits P2X7-mediated host-cell apoptosis. Cell Microbiol. 2008;10:863–875. doi: 10.1111/j.1462-5822.2007.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yilmaz O, Young PA, Lamont RJ, Kenny GE. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology. 2003;149:2417–2426. doi: 10.1099/mic.0.26483-0. [DOI] [PubMed] [Google Scholar]
- 177.Yuan L, Rodrigues PH, Belanger M, Dunn W, Jr, Progulske-Fox A. The Porphyromonas gingivalis clpB gene is involved in cellular invasion in vitro and virulence in vivo. FEMS Immunol Med Microbiol. 2007;51:388–398. doi: 10.1111/j.1574-695X.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 178.Yuan L, Rodrigues PH, Belanger M, Dunn WA, Jr, Progulske-Fox A. Porphyromonas gingivalis htrA is involved in cellular invasion and in vivo survival. Microbiology. 2008;154:1161–1169. doi: 10.1099/mic.0.2007/015131-0. [DOI] [PubMed] [Google Scholar]
- 179.Yun PL, Decarlo AA, Collyer C, Hunter N. Hydrolysis of interleukin-12 by Porphyromonas gingivalis major cysteine proteinases may affect local gamma interferon accumulation and the Th1 or Th2 T-cell phenotype in periodontitis. Infect Immun. 2001;69:5650–5660. doi: 10.1128/IAI.69.9.5650-5660.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang J, Dong H, Kashket S, Duncan MJ. IL-8 degradation by Porphyromonas gingivalis proteases. Microb Pathog. 1999;26:275–280. doi: 10.1006/mpat.1998.0277. [DOI] [PubMed] [Google Scholar]
- 181.Zhang Y, Wang T, Chen W, Yilmaz O, Park Y, Jung IY, Hackett M, Lamont RJ. Differential protein expression by Porphyromonas gingivalis in response to secreted epithelial cell components. Proteomics. 2005;5:198–211. doi: 10.1002/pmic.200400922. [DOI] [PMC free article] [PubMed] [Google Scholar]


