Abstract
Docetaxel-related neutropenia was associated with polymorphisms in the drug transporters ABCC2 and SLCO1B3 in Japanese cancer patients. We hypothesized that this association is because of reduced docetaxel clearance, associated with polymorphisms in those genes. We studied 64 US cancer patients who received a single cycle of 75 mg/m2 of docetaxel monotherapy. We found that the ABCC2 polymorphism at rs-12762549 trended to show a relationship with reduced docetaxel clearance (P = 0.048), but not with neutropenia. There was no significant association of the SLCO1B3 polymorphisms with docetaxel clearance or neutropenia. We conclude that the relationship between docetaxel-associated neutropenia and polymorphisms in drug transporters identified in Japanese patients was not confirmed in this cohort of US cancer patients.
Keywords: ABCC2 (pharmacogenetics), docetaxel, neutropenia, pharmacokinetics, SLCO1B3
Docetaxel, a semisynthetic taxane, is a cytotoxic agent used widely for the treatment of breast, non-small-cell lung and prostate cancer [1,2]. Previous pharmacokinetic studies have evaluated both the erythromycin breath test and genetic variation in drug-metabolizing enzymes and transporters as determinants of the variability in docetaxel clearance [3], but the major determinants of this variability are incompletely defined. The differential expression and function of polymorphic drug-metabolizing enzymes and/or transporters at the sites of drug elimination could play a major role in this variability. CYP3A4 and CYP3A5 are the primary enzymes involved in hepatic oxidation of docetaxel to its major metabolite, C-13-hydroxydocetaxel [3,4]. Hepatocellular uptake of taxanes is regulated, at least in part, by the solute carrier OATP1B3 (SLCO1B3; OATP8) [5], whereas the ATP-binding cassette (ABC) transporters P-glycoprotein (ABCB1) and MRP2 (ABCC2; cMOAT) [6] are involved in the secretion of taxanes from the liver into the bile [7]. Neutropenia is the dose-limiting toxicity of docetaxel [2,3]. To identify the genetic factors determining the risk of docetaxel-induced neutropenia, Kiyotani et al. [8] carried out a retrospective case–control study of 140 Japanese cancer patients who received docetaxel monotherapy, with 84 cases of grade 3/4 neutropenia and 56 controls (no neutropenia). The researchers identified a strong association of grade 3/4 neutropenia with the SNPs rs-12762549 in ABCC2 and rs-11045585 in SCLO1B3. The presence of polymorphisms in both genes yielded a seven-fold (95% confidence interval 2.95–16.59) increase in the odds ratio for developing docetaxel-associated neutropenia. As docetaxel-associated neutropenia has been related to drug exposure (AUC) [3], we hypothesized that rs-12762549 in ABCC2 and rs-11045585 in SLCO1B3 are associated with reduced docetaxel clearance and neutropenia.
We studied this hypothesis retrospectively in a subgroup of patients who originally participated in CALGB 9871 (Alliance) [9] and who consented to have DNA collected for studies of genes involved in docetaxel disposition and pharmacodynamics. The protocol was approved by local Institutional Review Boards. Details of the study design, patient eligibility criteria, docetaxel regimen, neutropenia monitoring, docetaxel concentration measurement, and population pharmacokinetic modeling using NONMEM have been described previously [9,10]. All patients in this retrospective study received a single intravenous dose of 75 mg/m2 docetaxel. The original study found no difference in docetaxel clearance with ethnicity; thus, this patient population was considered a single subgroup.
Blood samples (4.5 ml) for genotyping were collected in citrated tubes and DNA was isolated using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, Minnesota, USA) according to the manufacturer's instructions. Genotyping of ABCC2 and SLCO1B3 polymorphisms was carried out at the University of California San Francisco. The genotyping of de-identified DNA samples was approved by the University of California San Francisco Institutional Review Board. TaqMan SNP genotyping was carried out for rs-12762549 (ABCC2) and rs-11045585 (SLCO1B3). TaqMan SNP genotyping 40 × primer/probe assays were purchased from Applied Biosystems (Foster City, California, USA; ABI assay ID numbers C_11214917_10 for rs-12762549 and C_31106434_10 for rs-11045585). Three additional polymorphisms selected for strong linkage disequilibrium to the target SNPs in Asians were also genotyped. Primers and probes were purchased from Applied Biosystems [ABI assay ID numbers C_31980850 for rs-11190298 (ABCC2) and C_25766123 for rs-16923270 (SLCO1B3)] and were custom designed by Applied Biosystems for SLCO1B3 rs-4149155
Forward primer: 5′-TTGTAGGAAGAACAGAGTATATAGGCATA-3′.
Reverse primer: 5′-CAGATGTATTTGATCTACTCTTCTCTCCCTAT-3′.
VIC-labeled reporter 1: 5′-CAGAGGGAAGAAAGAGT-3′.
FAM-labeled reporter 2: 5′-ACAGAGGGAATAAAGAGT-3′.
The 5 μl reactions contained 5 ng of genomic DNA, 1 × TaqMan Universal Master Mix and 1 × primer/probe mix (900 nmol/l final concentration of primers and 200 μmol/l final concentration of FAM-labeled and VIC-labeled probes). PCR and post-reads were performed on a 7900 Real-Time PCR system (Applied Biosystems, Foster City, California. USA). Amplification conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Negative controls and random sample duplicates were included on each plate for quality control purposes. All SNPs were tested for deviation from Hardy–Weinberg equilibrium using the χ2-test. Haplotypes were statistically inferred using the program PHASE.
The primary objective of this retrospective study was to investigate the association of the ABCC2 polymorphism (rs-12762549) and the SLCO1B3 polymorphism (rs-11045585) with docetaxel clearance. The specific hypotheses, on the basis of the results of Kiyotani et al. [8], were that carriers of rs-12762549 or rs-11045585 have reduced docetaxel clearance and an increased risk of neutropenia, compared with their reference genotypes. Each hypothesis was tested using the Wilcoxon rank-sum test at the one-sided marginal level of 0.025. A secondary objective was to investigate the association between drug transporter genotypes and neutropenia. To this end, the Wilcoxon test for absolute neutrophil count nadir and Fisher's test for neutropenic event (toxicity grade) were used. All secondary and exploratory analyses were carried out using a two-sided marginal level of 0.05 and not adjusted for multiple testing.
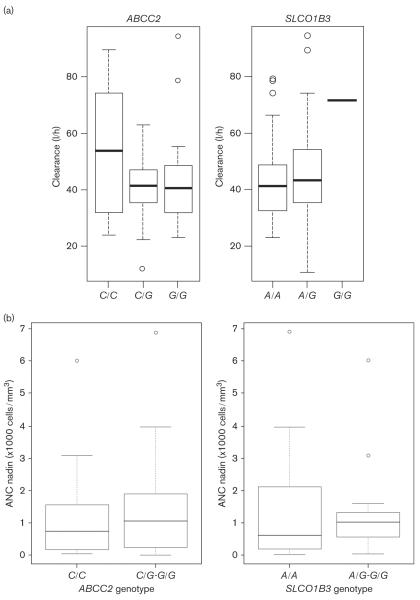
Ninety-nine patients were originally enrolled in CALGB 9871; of these, 64 patients had DNA samples available for pharmacogenetic analysis with concomitant docetaxel pharmacokinetic and neutropenia data. The demographics of this patient subgroup are shown in Table 1. Patients with rs-12762549 showed some evidence of reduced docetaxel clearance compared with those patients with the reference genotype (left panel in Fig. 1a, P = 0.048), although this did not achieve the predefined level of statistical significance (P < 0.025). There was no association between rs-11045585 and docetaxel clearance (right panel Fig. 1a, P = 0.799). There was no association between the absolute neutrophil count nadir and either the ABCC2 genotype (P = 0.86) or the SLCO1B3 genotype (P = 0.92), Fig. 1b. Additional analyses indicated no relationship between these drug transporter genotypes and the occurrence of grade 3/4 neutropenia. Three additional polymorphisms (rs-4149155, rs-16923270, rs-11190298) were also tested for their association with docetaxel clearance and neutropenia, on the basis of the observation that the two tag SNPs identified in the Kiyotani et al.'s [8] study had different linkage disequilibrium patterns in Caucasians and African-Americans compared with the Asian population. However, in our study population, no association was found between any of these SNPs and docetaxel clearance or neutropenia (data not shown). Post-hoc power analyses for the SLCO1B3 polymorphism to define an odds ratio of 6 for ≥ grade 3 neutropenia yielded a power of 0.84 (one sided α = 0.025); for the ABCC2 polymorphism, the power was 0.33 (one sided α = 0.025) to define an odds ratio of 3 for ≥ grade 3 neutropenia.
Table 1.
Patient demographics for CALGB 60805 (Alliance) (a subgroup of patients from CALGB 9871: Alliance)
| African-American | Caucasian | Total | |
|---|---|---|---|
| Number of patients enrolled | 20 | 44 | 64 |
| Median age (years) | 59.5 (range 42–70) | 62.5 (range 42–79) | 62 (range 42–79) |
| Sex | |||
| Male | 13 (65%) | 33 (75%) | 46 (72%) |
| Female | 7 (35%) | 11 (25%) | 18 (28%) |
| Performance status | |||
| 0 | 2 (10%) | 7 (16%) | 9 (14%) |
| 1 | 15 (75%) | 24 (55%) | 39 (61%) |
| 2 | 3 (15%) | 12 (27%) | 15 (23%) |
| Unknown | 0 (0%) | 1 (2%) | 1 (2%) |
| Tumor types | |||
| Lung | 12 (60%) | 30 (68%) | 42 (66%) |
| Breast | 0 (0%) | 1 (2%) | 1 (2%) |
| GI and pancreas | 3 (15%) | 3 (7%) | 6 (9%) |
| Head and neck | 1 (5%) | 1 (2%) | 2 (3%) |
| Prostate | 1 (5%) | 1 (2%) | 2 (3%) |
| Other | 3 (15%) | 8 (18%) | 11 (17%) |
| Docetaxel dose −75 mg/m2 | 20 (100%) | 44 (100%) | 64 (100%) |
| Median baseline WBC×103/μl (range) | 8.9 (2.7–18.7) | 6.7 (2.5–12.8) | 7.0 (2.5–18.7) |
Fig. 1.
(a) Box and whisker plots of docetaxel clearance in patients who received a single intravenous dose of 75 mg/m2 of docetaxel and ABCC2 (left panel) and SLCO1B3 (right panel) genotype. The horizontal lines represent the median values of docetaxel clearance for each genotype. For ABCC2 ?? vs CG/GG rs-12762549, the Wilcoxon–Mann–Whitney rank-sum test (P = 0.048). For the SLCO1B3 AA vs AG/GG rs-11045585, the Wilcoxon–Mann–Whitney rank-sum test (P = 0.799). (b) Box and whisker plots of the nadir absolute neutrophil count (ANC) and ABCC2 rs-12762549 (left panel) and SLCO1B3 rs-11045585 (right panel) genotype. The Wilcoxon–Mann–Whitney rank-sum test (P = 0.861 and 0.922) for the ABCC2 CC vs. CG/GG and SLCO1B3 AA vs. AG/GG genotypes, respectively.
This study investigated whether the polymorphisms in ABCC2 (rs-12762549) and in SLCO1B3 (rs-11045585) [11,12] that were previously associated with docetaxel-induced neutropenia in Japanese cancer patients [8] were similarly associated with docetaxel clearance and hematopoietic toxicity in US Caucasian and African-American patients enrolled in CALGB 9871. We observed decreased docetaxel clearance in association with the ABCC2 polymorphism, but this did not result in greater neutropenia. Similarly, in CALGB 9871 [9], we did not establish a pharmacokinetic–pharmacodynamic relationship for docetaxel-associated neutropenia. Given the findings of Kiyotani et al. [8], this is weak evidence in support of a relationship between the ABCC2 rs-12762549 polymorphism and docetaxel clearance. Furthermore, our study did not identify an association of SLCO1B3 polymorphisms with docetaxel clearance or hematologic toxicity. To further elucidate whether our findings may have been confounded by the derivation of the docetaxel clearance using population pharmacokinetics models, we added the transporter genotypes to the Bruno docetaxel NONMEM model [10], but this did not yield additional docetaxel pharmacokinetic–drug transporter genotype associations (data not shown). Confounding results of the association of drug transporter polymorphisms with docetaxel pharmacokinetics highlight the importance of replication of pharmacogenetic results. Baker et al. [13] studied 92 White US and European cancer patients receiving docetaxel therapy (at different doses, either as monotherapy or in combination therapy) and found no relationship between docetaxel clearance and ABCC2, SLCO1B3, or ABCB1 genotypes. The relationships between transporter genotype and neutropenia were not explored in this study because of the variation in the docetaxel dose used and the use of combination chemotherapy. In a population of Asian nasopharyngeal carcinoma patients treated with weekly docetaxel monotherapy (n = 54), an influence of functional polymorphisms in SLCO1B3 and ABCB1 with interindividual variability in docetaxel clearance was reported [14]. Although there are conflicting reports on the association of the ABCB1 genotype with docetaxel clearance [13–17] and toxicity [18], the prevailing evidence does not support a significant role for ABCB1 (P-glycoprotein) polymorphisms in determining docetaxel clearance.
There are a number of possible explanations why the previous Japanese data [8] were not replicated in this present study. Selection bias cannot be ruled out in any retrospective study with small cohorts; furthermore, the doses of docetaxel administered to Japanese patients were not defined. Differences in the linkage disequilibrium patterns for ABCC2 and SLCO1B3 were considered in the current study and tested by the inclusion of three additional SNPs. There was no evidence that this was the reason for nonreplication of the previous findings in our US population. Interestingly, the minor allele frequencies in the study patients for rs-11045585 and rs-1276259 were 0.148 and 0.460, respectively, similar to the minor allele frequencies in HapMap CEU Environmental differences between study populations; variations in treatment paradigms and concomitant medications may all affect the phenotypes studied and therefore any genetic associations related to drug exposures. It is not possible to identify which one (or combination) of these factors may have influenced the current findings. It is possible that ongoing genome-wide association studies of docetaxel will provide additional insights into the variable toxicity of this agent.
Acknowledgements
The research for CALGB 60805 and 9871 (Alliance) was supported, in part, by grants from the National Cancer Institute (CA31946) to the Alliance for Clinical Trials in Oncology (Monica M. Bertagnolli, MD, Chair) and to the Alliance Statistical Center (Daniel J. Sargent, PhD, CA33601). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Support for this study also came from CA31956, CA 44691, CA23108, GM61393, EB001975, and the Pharmacogenetics Research Network to Dr Deanna L. Kroetz (GM61390) and Dr Mark J. Ratain (GM61393).
The following institutions participated in this study: Duke University Medical Center, Durham, NC, Jeffrey Crawford, MD, supported by CA47577; Memorial Sloan-Kettering Cancer Center, New York, NY, Clifford A. Hudis, MD, supported by CA77651; Roswell Park Cancer Institute, Buffalo, NY, Ellis Levine, MD, supported by CA59518; Southeast Cancer Control Consortium Inc. CCOP, Goldsboro, NC, James N. Atkins, MD, supported by CA45808; State University of New York Upstate Medical University, Syracuse, NY, Stephen L. Graziano, MD, supported by CA21060; The Ohio State University Medical Center, Columbus, OH, Clara D. Bloomfield, MD, supported by CA77658; University of California at San Diego, San Diego, CA, Barbara A. Parker, MD, supported by CA11789; University of Texas Southwestern Medical Center, Dallas, TX, supported by CA37347; University of California at San Francisco, San Francisco, CA, Charles J. Ryan, MD, supported by CA60138; University of Chicago, Chicago, IL, Hedy L. Kindler, MD, supported by CA41287; University of Maryland Greenebaum Cancer Center, Baltimore, MD, Martin Edelman, MD, supported by CA31983; Wake Forest University School of Medicine, Winston-Salem, NC, David D. Hurd, MD, supported by CA03927.
Footnotes
Conflicts of interest M.J.R. declares that he has acted as a consultant to Sanofi-Aventis (not relating to docetaxel). For the remaining authors there are no conflicts of interest.
References
- 1.Fulton B, Spencer CM. Docetaxel. A review of its pharmacodynamic and pharmacokinetic properties and therapeutic efficacy in the management of metastatic breast cancer. Drugs. 1996;51:1075–1092. doi: 10.2165/00003495-199651060-00011. [DOI] [PubMed] [Google Scholar]
- 2.Eisenhauer EA, Vermorken JB. The taxoids. Comparative clinical pharmacology and therapeutic potential. Drugs. 1998;55:5–30. doi: 10.2165/00003495-199855010-00002. [DOI] [PubMed] [Google Scholar]
- 3.Baker SD, Sparreboom A, Verweij J. Clinical pharmacokinetics of docetaxel: recent developments. Clin Pharmacokinet. 2006;45:235–252. doi: 10.2165/00003088-200645030-00002. [DOI] [PubMed] [Google Scholar]
- 4.Eichelbaum M, Burk O. CYP3A genetics in drug metabolism. Nat Med. 2001;7:285–287. doi: 10.1038/85417. [DOI] [PubMed] [Google Scholar]
- 5.Smith NF, Acharya MR, Desai N, Figg WD, Sparreboom A. Identification of OATP1B3 as a high-affinity hepatocellular transporter of paclitaxel. Cancer Biol Ther. 2005;4:815–818. doi: 10.4161/cbt.4.8.1867. [DOI] [PubMed] [Google Scholar]
- 6.Bardelmeijer HA, Ouwehand M, Buckle T, Huisman MT, Schellens JH, Beijnen JH, van Tellingen O. Low systemic exposure of oral docetaxel in mice resulting from extensive first-pass metabolism is boosted by ritonavir. Cancer Res. 2002;62:6158–6164. [PubMed] [Google Scholar]
- 7.Huisman MT, Chhatta AA, van Tellingen O, Beijnen JH, Schinkel AH. MRP2 (ABCC2) transports taxanes and confers paclitaxel resistance and both processes are stimulated by probenecid. Int J Cancer. 2005;116:824–829. doi: 10.1002/ijc.21013. [DOI] [PubMed] [Google Scholar]
- 8.Kiyotani K, Mushiroda T, Kubo M, Zembutsu H, Sugiyama Y, Nakamura Y. Association of genetic polymorphisms in SLCO1B3 and ABCC2 with docetaxel-induced leucopenia. Cancer Sci. 2008;99:967–972. doi: 10.1111/j.1349-7006.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis LD, Miller AA, Rosner GL, Dowell JE, Valdivieso M, Relling MV, et al. Cancer and Leukemia Group B. A comparison of the pharmacokinetics and pharmacodynamics of docetaxel between African-American and Caucasian cancer patients: CALGB 9871. Clin Cancer Res. 2007;13:3302–3311. doi: 10.1158/1078-0432.CCR-06-2345. [DOI] [PubMed] [Google Scholar]
- 10.Bruno R, Vivler N, Vergniol JC, De Phillips SL, Montay G, Sheiner LB. A population pharmacokinetic model for docetaxel (Taxotere): model building and validation. J Pharmacokinet Biopharm. 1996;24:153–172. doi: 10.1007/BF02353487. [DOI] [PubMed] [Google Scholar]
- 11.Hirouchi M, Suzuki H, Itoda M, Ozawa S, Sawada J, Ieiri I, et al. Characterization of the cellular localization, expression level, and function of SNP variants of MRP2/ABCC2. Pharm Res. 2004;21:742–748. doi: 10.1023/b:pham.0000026422.06207.33. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz UI, Meyer zu Schwabedissen HE, Tirona RG, Suzuki A, Leake BF, Mokrab Y, et al. Identification of novel functional organic anion-transporting polypeptide 1B3 polymorphisms and assessment of substrate specificity. Pharmacogenet Genomics. 2011;21:103–114. doi: 10.1097/FPC.0b013e328342f5b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker SD, Verweij J, Cusatis GA, van Schaik RH, Marsh S, Orwick SJ, et al. Pharmacogenetic pathway analysis of docetaxel elimination. Clin Pharmacol Ther. 2009;85:155–163. doi: 10.1038/clpt.2008.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chew SC, Singh O, Chen X, Ramasamy RD, Kulkarni T, Lee EJ, et al. The effects of CYP3A4, CYP3A5, ABCB1, ABCC2, ABCG2 and SLCO1B3 single nucleotide polymorphisms on the pharmacokinetics and pharmacodynamics of docetaxel in nasopharyngeal carcinoma patients. Cancer Chemother Pharmacol. 2011;67:1471–1478. doi: 10.1007/s00280-011-1625-9. [DOI] [PubMed] [Google Scholar]
- 15.Bosch TM, Huitema AD, Doodeman VD, Jansen R, Witteveen E, Smit WM, et al. Pharmacogenetic screening of CYP3A and ABCB1 in relation to population pharmacokinetics of docetaxel. Clin Cancer Res. 2006;12:5786–5793. doi: 10.1158/1078-0432.CCR-05-2649. [DOI] [PubMed] [Google Scholar]
- 16.Zamboni WC, Combest AJ, Deloia JA, Edwards RP, Bridges AS, Zamboni BA, et al. Pharmacologic and phenotypic study of docetaxel in patients with ovarian or primary peritoneal cancer. Cancer Chemother Pharmacol. 2011;68:1255–1262. doi: 10.1007/s00280-011-1609-9. [DOI] [PubMed] [Google Scholar]
- 17.de Graan AJ, Lancaster CS, Obaidat A, Hagenbuch B, Elens L, Friberg LE, et al. Influence of polymorphic OATP1B-type carriers on the disposition of docetaxel. Clin Cancer Res. 2012;18:4433–4440. doi: 10.1158/1078-0432.CCR-12-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran A, Jullien V, Alexandre J, Rey E, Rabillon F, Girre V, et al. Pharmacokinetics and toxicity of docetaxel: role of CYP3A, MDR1, and GST polymorphisms. Clin Pharmacol Ther. 2006;79:570–580. doi: 10.1016/j.clpt.2006.02.003. [DOI] [PubMed] [Google Scholar]