To the Editor: The pathogenesis of Crohn's disease (CD) is not known: the currently accepted hypothesis is that genetic susceptibility and environmental factors cooperate to trigger the chronic inflammation typical of the disease. Recent evidence implicating the gut microbiome in CD development has generated interest in its characterization (1). Previous studies have shown that bacterial diversity is reduced in CD patients, that bacterial populations in CD patients differ in relation to phenotype, and that the microbial composition differs significantly between inflamed and non-inflamed mucosal areas (2).
Nutritional therapy is effective in pediatric CD; it induces remission of mucosal inflammatory reactions and exerts a beneficial effect on the child's nutritional status and growth (3). It has been suggested that nutritional therapy may modify the fecal microflora in CD children (4). In an attempt to address this hypothesis, we characterized the entire gut mucosal microbiome in a child with CD at diagnosis and after nutritional therapy, and in a sex- and age-matched subject not affected by CD.
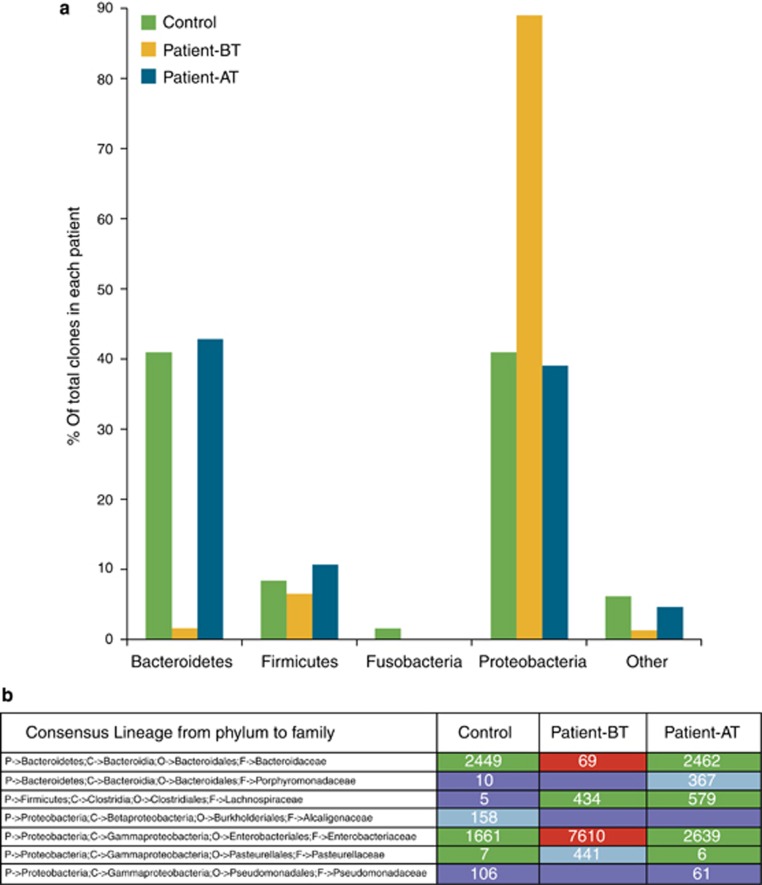
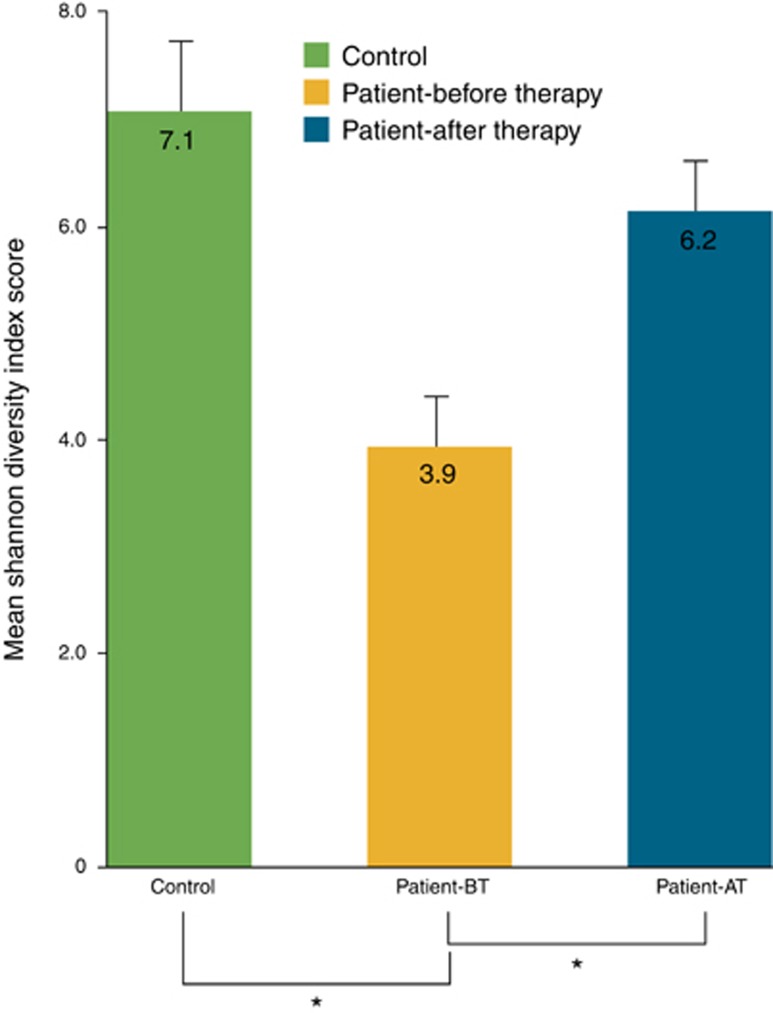
This 14-year-old boy was diagnosed with active CD (Pediatric Crohn's Disease Activity Index, PCDAI=50). After colonoscopy, he underwent nutritional therapy consisting of a daily powder constituted by proteins, antioxidants, and anti-inflammatory fats (Alicalm formula, Nutricia Advanced Medical Nutrition) for 8 weeks. After this time, a clinical re-evaluation revealed disease remission (PCDAI=0). Endoscopic ileum mucosal samples at diagnosis and after therapy were collected for DNA extraction. An ileum tissue sample was obtained from a 15-year-old boy with a gut polyp and without family history of CD. We used a 16S rRNA next-generation sequencing strategy to investigate the gut microbiome composition in these three ileum samples. A specific amplicon was designed to cover the V4–V6 variable regions of the bacterial 16S rRNA genes and amplified using hybrid-tagged primers. Ultra-deep amplicon sequencing was performed on the 454 Genome Sequencer FLX instrument (Roche Life Science, Branford, CT). Data were analyzed using the QIIME community analysis pipeline (5). High-quality filtered sequences were used to identify the operational taxonomic units (OTUs): 705, 1,328, and 2,171 OTUs were obtained in the patient before and after therapy, and in the control subject, respectively. On the basis of the taxonomic assignment of the OTUs, we characterized the ileum microbiome of our samples at five phylogenetic levels (from phylum to genus). As shown in Figure 1, Proteobacteria were more abundant, and Bacteroidetes less abundant, in our CD patient before therapy than in the control. Interestingly, the composition of the ileum microbiome in the patient after therapy was virtually the same as in the control. Moreover, our data support the reduced bacterial diversity of the CD microbiome. In fact, the Shannon Diversity Index Score was significantly lower (P<0.05) in our CD patient before therapy than after therapy and also when compared with the control (Figure 2).
Figure 1.
Composition of the ileum microbiome characterized in the control subject and in the Crohn's disease (CD) patient before therapy (patient-BT) and after therapy (patient-AT) by next-generation sequencing of the 16S rRNAs. (a) Phylum-level classification shows the reduction of Bacteroidetes and the significant prevalence of Proteobacteria in the patient-BT vs. the control and the patient-AT. (b) The heatmap table highlights in brownish red the most significant alterations in the bacterial composition of gut microbiome detected in the CD patient-BT; the numerical figures indicate the number of bacterial families sequenced in each patient.
Figure 2.
The mean Shannon Diversity Index Score. Values were significantly lower before therapy (patient-before therapy (patient-BT)) compared with values obtained after treatment (patient after therapy (patient-AT)) and with values obtained in the control subject (*P<0.05).
Although our findings were obtained in one case–control study, and therefore may be considered preliminary, they strongly suggest that nutritional therapy can improve the inflammatory status of CD by restoring the composition of the mucosal microbiome. This case of CD gut microbiome dysbiosis that responded to nutritional therapy can be considered proof-of-concept to evaluate a similar approach in other pediatric clinical laboratories and may serve to prompt multicenter studies and possible clinical trials.
Acknowledgments
We thank Jean Ann Gilder (Scientific Communication srl., Naples) for editing the text, and Vittorio Lucignano, CEINGE–Biotecnologie Avanzate, for technical assistance related to graphics.
Guarantor of the article: Francesco Salvatore, MD, PhD.
Specific author contributions: V.D., A.S., L.S., and F.S. designed the project; A.S., E.M., and M.M. enrolled the study subjects, performed the endoscopy, treated and monitored the patient during follow-up; V.D. and V.P. performed the next-generation sequencing experiment; V.D. and G.C. performed the bioinformatic analysis; V.D., L.S., and F.S. analyzed the data and wrote the manuscript. All the authors revised and approved the final draft.
Financial support: This work was supported by grant L.5/95 (to F.S.) from Regione Campania; grant PS 35-126/Ind and grant PON01_02589 (MICROMAP) 2012 from the Ministry of University and Research (both to F.S.); grant RF-2010-2318372 from the Ministry of Health (to F.S.).
Potential competing interests: None.
References
- Neuman MG, Nanau RM. Inflammatory bowel disease: role of diet, microbiota, life style. Transl Res. 2012;160:29–44. doi: 10.1016/j.trsl.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Walker AW, Sanderson JD, Churcher C, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day AS, Whitten KE, Sidler M, et al. Systematic review: nutritional therapy in paediatric Crohn's disease. Aliment Pharmacol Ther. 2008;27:293–307. doi: 10.1111/j.1365-2036.2007.03578.x. [DOI] [PubMed] [Google Scholar]
- Lionetti P, Callegari M, Cavicchi M, et al. Enteral nutrition-induced remission is associated with profound modification of the intestinal microflora in Crohn's disease. J Pediatr Gastroenterol Nutr. 2004;39:S106. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]