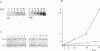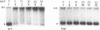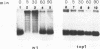Abstract
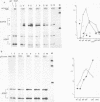
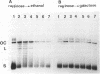
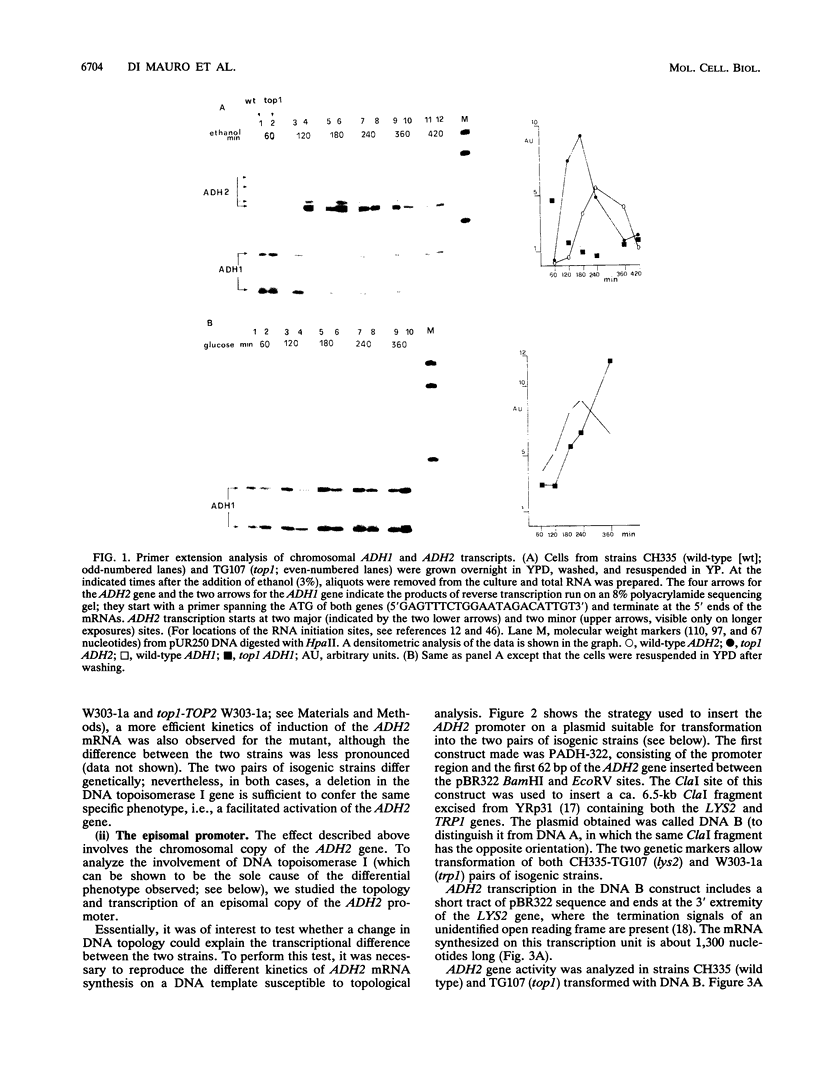
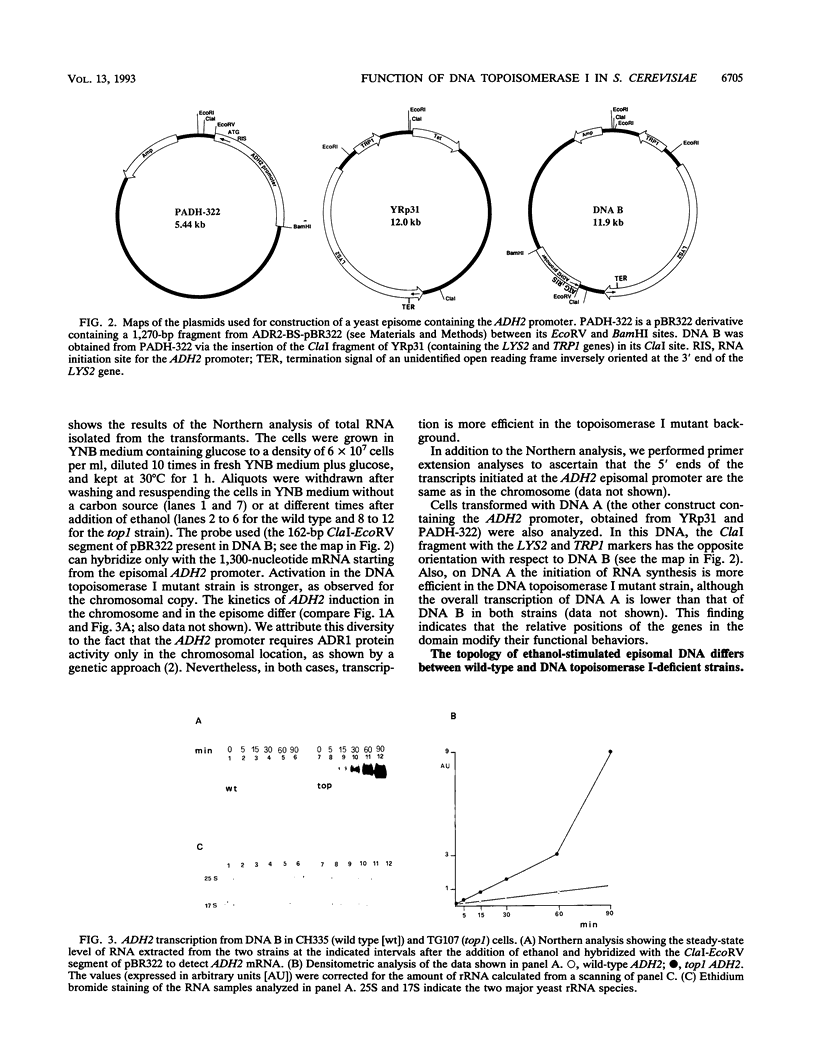
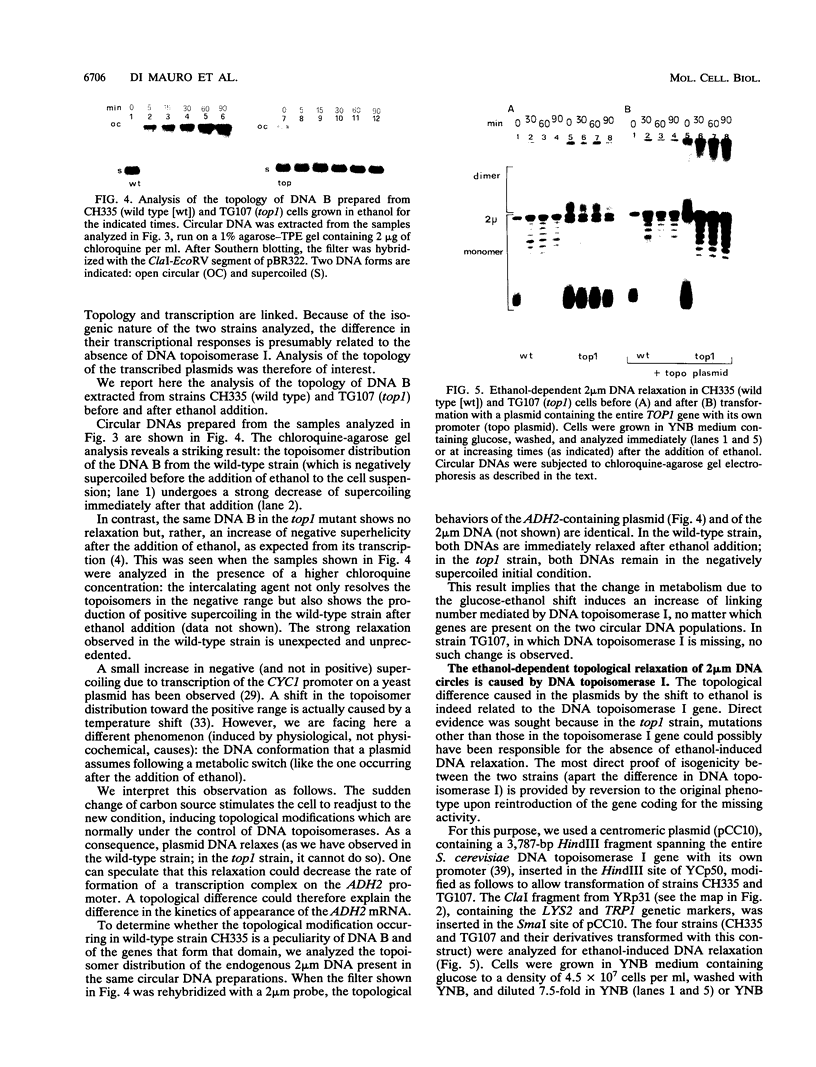
Inactivation of the nonessential TOP1 gene, which codes for Saccharomyces cerevisiae DNA topoisomerase I, affects the rate of transcription starting at the ADH2 promoter. For both the chromosomal gene and the plasmid-borne promoter, mRNA accumulation is kinetically favored in the mutant relative to a wild-type isogenic strain. The addition of ethanol causes in wild-type yeast strains a substantial increase in linking number both on the ADH2-containing plasmid and on the resident 2 microns DNA. Evidence has been obtained that such an in vivo increase in linking number depends on (i) the activity of DNA topoisomerase I and of no other enzyme and (ii) ethanol addition, not on the release from glucose repression. A direct cause-effect relationship between the change in supercoiling and alteration of transcription cannot be defined. However, the hypothesis that a metabolism-induced modification of DNA topology in a eukaryotic cell plays a role in regulating gene expression is discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balke V. L., Gralla J. D. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J Bacteriol. 1987 Oct;169(10):4499–4506. doi: 10.1128/jb.169.10.4499-4506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D. R., Young E. T. Characterization of a regulatory region upstream of the ADR2 locus of S. cerevisiae. Nature. 1982 Dec 23;300(5894):724–728. doi: 10.1038/300724a0. [DOI] [PubMed] [Google Scholar]
- Brill S. J., DiNardo S., Voelkel-Meiman K., Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. 1987 Mar 26-Apr 1Nature. 326(6111):414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Brill S. J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988 Jul 29;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Camilloni G., Caserta M., Amadei A., Di Mauro E. The conformation of constitutive DNA interaction sites for eukaryotic DNA topoisomerase I on intrinsically curved DNAs. Biochim Biophys Acta. 1991 Dec 2;1129(1):73–82. doi: 10.1016/0167-4781(91)90214-7. [DOI] [PubMed] [Google Scholar]
- Camilloni G., Di Martino E., Caserta M., di Mauro E. Eukaryotic DNA topoisomerase I reaction is topology dependent. Nucleic Acids Res. 1988 Jul 25;16(14B):7071–7085. doi: 10.1093/nar/16.14.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilloni G., Di Martino E., Di Mauro E., Caserta M. Regulation of the function of eukaryotic DNA topoisomerase I: topological conditions for inactivity. Proc Natl Acad Sci U S A. 1989 May;86(9):3080–3084. doi: 10.1073/pnas.86.9.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevali F., Caserta M., Di Mauro E. Topological modifications and template activation are induced in chimaeric plasmids by inserted sequences. J Mol Biol. 1983 Mar 25;165(1):59–77. doi: 10.1016/s0022-2836(83)80242-3. [DOI] [PubMed] [Google Scholar]
- Cherry J. R., Johnson T. R., Dollard C., Shuster J. R., Denis C. L. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell. 1989 Feb 10;56(3):409–419. doi: 10.1016/0092-8674(89)90244-4. [DOI] [PubMed] [Google Scholar]
- Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991 Dec;5(12A):2315–2326. doi: 10.1101/gad.5.12a.2315. [DOI] [PubMed] [Google Scholar]
- Christman M. F., Dietrich F. S., Fink G. R. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988 Nov 4;55(3):413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- Della Seta F., Camilloni G., Venditti S., Di Mauro E. The intrinsic topological information of the wild-type and of up-promoter mutations of the Saccharomyces cerevisiae alcohol dehydrogenase II regulatory region. J Biol Chem. 1988 Nov 5;263(31):15888–15896. [PubMed] [Google Scholar]
- Denis C. L., Audino D. C. The CCR1 (SNF1) and SCH9 protein kinases act independently of cAMP-dependent protein kinase and the transcriptional activator ADR1 in controlling yeast ADH2 expression. Mol Gen Genet. 1991 Oct;229(3):395–399. doi: 10.1007/BF00267461. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Ferguson J., Young E. T. mRNA levels for the fermentative alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. J Biol Chem. 1983 Jan 25;258(2):1165–1171. [PubMed] [Google Scholar]
- Denis C. L. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984 Dec;108(4):833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C. J., Barr G. C., Ni Bhriain N., Higgins C. F. DNA supercoiling and the anaerobic and growth phase regulation of tonB gene expression. J Bacteriol. 1988 Jun;170(6):2816–2826. doi: 10.1128/jb.170.6.2816-2826.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibel H., Philippsen P. Identification of the cloned S. cerevisiae LYS2 gene by an integrative transformation approach. Mol Gen Genet. 1983;191(1):66–73. doi: 10.1007/BF00330891. [DOI] [PubMed] [Google Scholar]
- Fleig U. N., Pridmore R. D., Philippsen P. Construction of LYS2 cartridges for use in genetic manipulations of Saccharomyces cerevisiae. Gene. 1986;46(2-3):237–245. doi: 10.1016/0378-1119(86)90408-7. [DOI] [PubMed] [Google Scholar]
- Freeman L. A., Garrard W. T. DNA supercoiling in chromatin structure and gene expression. Crit Rev Eukaryot Gene Expr. 1992;2(2):165–209. [PubMed] [Google Scholar]
- Gartenberg M. R., Wang J. C. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11461–11465. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G. N., Wang J. C. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988 Dec 2;55(5):849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Dorman C. J., Stirling D. A., Waddell L., Booth I. R., May G., Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988 Feb 26;52(4):569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnitz L., Morrison M., Young E. T. Identification and characterization of three genes that affect expression of ADH2 in Saccharomyces cerevisiae. Genetics. 1992 Oct;132(2):351–359. doi: 10.1093/genetics/132.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R. A., Wang J. C. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell. 1989 Jun 16;57(6):975–985. doi: 10.1016/0092-8674(89)90336-x. [DOI] [PubMed] [Google Scholar]
- Kim R. A., Wang J. C. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J Mol Biol. 1989 Jul 20;208(2):257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne B. I., Guarente L. Transcription by RNA polymerase II induces changes of DNA topology in yeast. Genes Dev. 1988 Jun;2(6):766–772. doi: 10.1101/gad.2.6.766. [DOI] [PubMed] [Google Scholar]
- Osheroff N., Shelton E. R., Brutlag D. L. DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA. J Biol Chem. 1983 Aug 10;258(15):9536–9543. [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989 Feb 24;56(4):521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Saavedra R. A., Huberman J. A. Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell. 1986 Apr 11;45(1):65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A., Trumpower B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990 May 25;18(10):3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. C., Brill S. J., Ju Q., Sternglanz R., Reeder R. H. Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 1992 Jul;6(7):1332–1341. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- Shure M., Pulleyblank D. E., Vinograd J. The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res. 1977;4(5):1183–1205. doi: 10.1093/nar/4.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster J., Yu J., Cox D., Chan R. V., Smith M., Young E. ADR1-mediated regulation of ADH2 requires an inverted repeat sequence. Mol Cell Biol. 1986 Jun;6(6):1894–1902. doi: 10.1128/mcb.6.6.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A. K., Young E. T. The identification and characterization of ADR6, a gene required for sporulation and for expression of the alcohol dehydrogenase II isozyme from Saccharomyces cerevisiae. Genetics. 1987 Aug;116(4):523–530. doi: 10.1093/genetics/116.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash C., Bankier A. T., Barrell B. G., Sternglanz R. Cloning, characterization, and sequence of the yeast DNA topoisomerase I gene. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4374–4378. doi: 10.1073/pnas.82.13.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townes T. M., Lingrel J. B., Chen H. Y., Brinster R. L., Palmiter R. D. Erythroid-specific expression of human beta-globin genes in transgenic mice. EMBO J. 1985 Jul;4(7):1715–1723. doi: 10.1002/j.1460-2075.1985.tb03841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. The reprogramming of transcriptional competence. Cell. 1992 May 15;69(4):573–575. doi: 10.1016/0092-8674(92)90218-2. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989 Jul 28;58(2):409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases: why so many? J Biol Chem. 1991 Apr 15;266(11):6659–6662. [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Young T., Williamson V., Taguchi A., Smith M., Sledziewski A., Russell D., Osterman J., Denis C., Cox D., Beier D. The alcohol dehydrogenase genes of the yeast, Saccharomyces cerevisiae: isolation, structure, and regulation. Basic Life Sci. 1982;19:335–361. doi: 10.1007/978-1-4684-4142-0_26. [DOI] [PubMed] [Google Scholar]
- Zechiedrich E. L., Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990 Dec;9(13):4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]