Abstract
One of the major advantages of the baculovirus-insect cell system is that it is a eukaryotic system that can provide posttranslational modifications, such as protein N-glycosylation. However, this is a vastly oversimplified view, which reflects a poor understanding of insect glycobiology. In general, insect protein glycosylation pathways are far simpler than the corresponding pathways of higher eukaryotes. Paradoxically, it is increasingly clear that various insects encode and can express more elaborate protein glycosylation functions in restricted fashion. Thus, the information gathered in a wide variety of studies on insect protein N-glycosylation during the past 25 years has provided what now appears to be a reasonably detailed, comprehensive, and accurate understanding of the protein N-glycosylation capabilities of the baculovirus-insect cell system. In this chapter, we discuss the models of insect protein N-glycosylation that have emerged from these studies and how this impacts the use of baculovirus-insect cell systems for recombinant glycoprotein production. We also discuss the use of these models as baselines for metabolic engineering efforts leading to the development of new baculovirus-insect cell systems with humanized protein N-glycosylation pathways, which can be used to produce more authentic recombinant N-glycoproteins for drug development and other biomedical applications.
INTRODUCTION
Since the 1980s, the baculovirus-insect cell system has been widely utilized to produce a large variety of recombinant proteins (reviewed in references [1–4]). The baculovirus-insect cell expression system is a binary system, consisting of a recombinant baculovirus as the vector and lepidopteran insect cells or larvae as the hosts. Normally, heterologous gene expression is driven by a strong transcriptional promoter derived from a very late baculovirus gene, such as polyhedrin or p10, in conjunction with a powerful virus-encoded transcription complex (reviewed in references [5, 6]). In addition to being able to provide extremely high levels of heterologous gene expression, baculoviral vectors can accommodate large DNA inserts, which allows for the production of multi-subunit protein complexes or even virus-like particles (reviewed in references [4, 7]). Finally, the baculovirus-insect cell system is considered to be relatively safe for humans and animals. In fact, the relative biological safety of baculovirus vectors has attracted the interest of gene therapists and these viruses are now under active investigation as tools for gene delivery to mammalian cells and organisms (reviewed in references [4, 8]).
Once a baculovirus vector has entered the insect cell, host gene expression is globally inactivated and the cellular protein synthesis and processing machinery is re-directed to produce virus-encoded proteins, including the virus-encoded heterologous gene product [9–11]. Because they are eukaryotes, insect cells also can process newly synthesized proteins in many different ways. Thus, recombinant proteins typically can be folded, chemically modified, trafficked, and assembled into highly authentic, soluble end products (reviewed in references [1–4]). One widely recognized and highly attractive protein-processing capability of the baculovirus-insect cell system is protein N-glycosylation. However, it is clear that the protein glycosylation capabilities of baculovirus-infected insect cells are not equivalent to those of higher eukaryotes (reviewed in references [3, 12–21]). While native mammalian glycoproteins often have complex type N-glycans with terminal sialic acids, insect cell-derived recombinant glycoproteins usually have much simpler side chains, known as paucimannosidic N-glycans, at sites normally occupied by complex, terminally sialylated structures. In addition, insect-derived N-glycans may contain core α1,3-linked fucose residues, which are known to be allergenic (reviewed in references [20–23]). These structural differences between the major N-glycans synthesized by insect and mammalian cells are serious problems that have hindered the use of the baculovirus-insect cell expression system for the production of recombinant glycoproteins for at least some pharmaceutical applications.
Several approaches have been undertaken to begin to address this problem. One has involved developing culture conditions that might support the production of recombinant glycoproteins with more authentic, mammalian-like N-glycans by insect cell lines already being used as hosts for baculovirus vectors [24–30]. Another has involved identifying alternative, natural baculovirus-host combinations that might be able to produce recombinant glycoproteins with more authentic N-glycans [31–34]. Yet another approach has been to genetically engineer either baculovirus vectors or their hosts to encode and express mammalian genes involved in the latter steps of N-glycan processing. This approach, which will be the focus of the last part of this review, has yielded new baculovirus-insect cell systems that can produce more authentic, or “humanized” recombinant glycoproteins (reviewed in references [4, 13, 15–18]).
N-GLYCAN TRIMMING REACTIONS IN INSECT AND MAMMALIAN SYSTEMS
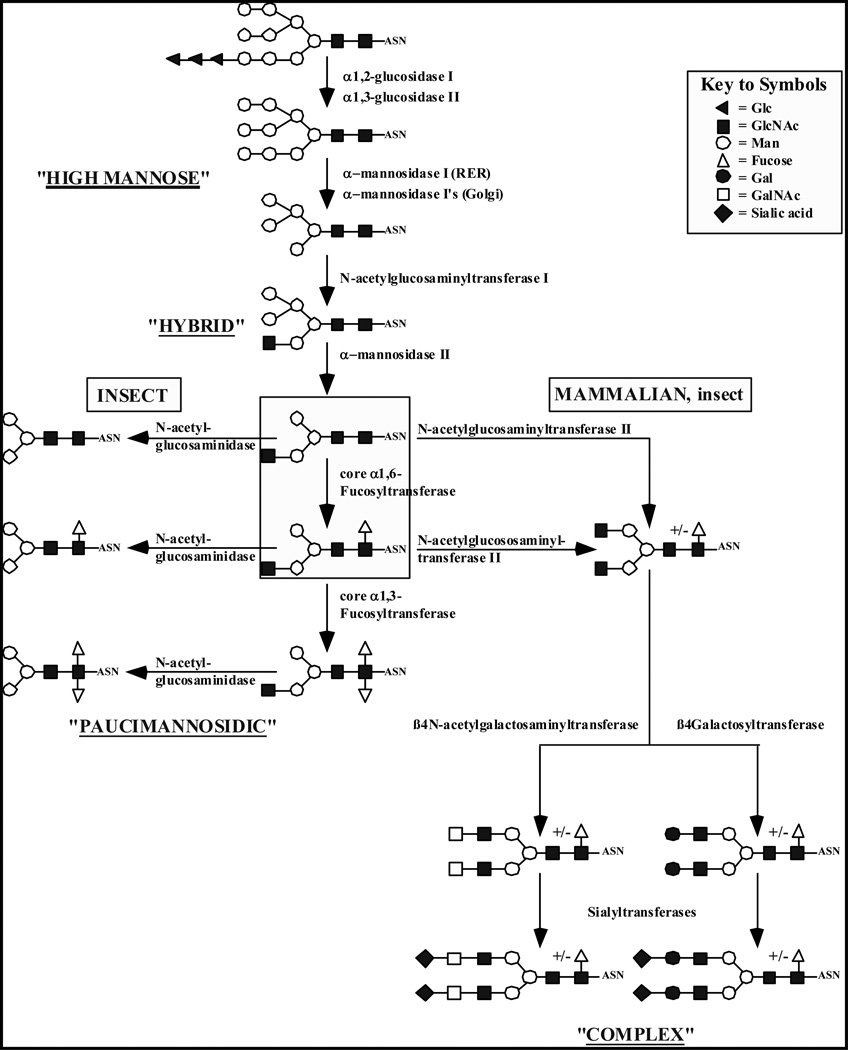
Protein N-glycosylation occurs in the endoplasmic reticulum (ER) and Golgi apparatus and generally involves the initial transfer of a pre-assembled oligosaccharide precursor to a nascent polypeptide followed by a series of N-glycan processing steps. The processing steps can be further divided into the trimming reactions, which are mediated by processing glycosidases, and the branching and elongation reactions, which are mediated by glycosyltransferases. The trimming reactions convert the oligosaccharide precursor to a smaller core structure that can be branched and elongated in various ways to produce the final oligosaccharide side-chains found on mature N-glycoproteins (reviewed in reference [35]). The oligosaccharide trimming reactions appear to be similar or identical in insect and mammalian cell systems. In contrast, there are usually significant differences in the extent of N-glycan branching and elongation obtained in these two systems (Fig. 1).
Fig. (1). Protein N-glycosylation pathways in insect and mammalian cells.
Monosaccharides are designated by their standard symbols, as defined in the key. The boxed N-glycans are the last common intermediates in the insect and mammalian pathways. The major insect cell products are paucimannosidic N-glycans with or without core fucose residues, as shown on the far left-hand side of the Figure. The major mammalian cell products are complex, terminally sialylated N-glycans, such as the biantennary N-glycans shown on the bottom right-hand side of the Figure. The Figure also accommodates the potential ability of insect cells to produce more extensively processed N-glycans like those produced by mammalian cells.
The mammalian protein N-glycosylation pathway begins when oligosaccharyltransferase, a multi-subunit enzyme complex in the ER, transfers a preassembled oligosaccharide (Glc3Man9GlcNAc2) from a lipid carrier to the asparagine residue within a specific recognition site (Asn-X-Thr/Ser) in a newly synthesized polypeptide (reviewed in references [35–38]). Immediately afterwards, the outermost α1,2-linked glucose is removed from the N-linked precursor by an ER membrane-bound enzyme, α-glucosidase I [39]. Another ER membrane-bound enzyme, α-glucosidase II, removes the next glucose residue, which is α1,3-linked, to produce the monoglucosylated core N-glycan, GlcMan9GlcNAc2 [40, 41]. This latter enzyme subsequently cleaves the last remaining α1,3-linked glucose residue from GlcMan9GlcNAc2 to produce the classic high mannose type N-glycan, Man9GlcNAc2. This structure can be either an end product, which appears as such on the final glycoprotein, or an intermediate, which is subjected to further processing. In addition to this highly conserved glucose trimming mechanism, the terminal glucose residues also can be indirectly removed from N-glycan precursors by a Golgi-associated endo-α-D-mannosidase, which is restricted to the phylum Chordata. This enzyme can cleave Glcα1,3Man, Glc2α1,3Man, or Glc3α1,3Man from mono-, di-, and triglucosylated N-glycans, respectively, to produce Man8GlcNAc2 [42–44].
Mannose trimming of deglucosylated N-glycans is typically accomplished through the concerted action of two different types of α-mannosidases, the class I and the class II α-mannosidases, which are localized in the ER and Golgi apparatus (reviewed in references [45–47]). In addition, the endo-α-mannosidase located in the Golgi apparatus and pre-Golgi intermediate compartments can cleave the glucose-substituted mannose residue from N-glycans if a glycoprotein with Glc(1–3)Man9GlcNAc2 somehow escapes the ER, as described above. The general combined effect of the class I α-mannosidases in the ER and Golgi apparatus is to remove all four α1,2-linked mannose residues from Man9GlcNAc2 and produce the key intermediate, Man5GlcNAc2. More specifically, this involves the action of two ER enzymes, known as α1,2-mannosidases I and II, and three Golgi enzymes, known as α1,2-mannosidases IA, IB, and IC (reviewed in references [45–47]). ER α-mannosidase I is a slow acting enzyme that specifically removes just one terminal mannose residue from the middle branch of the N-glycan intermediate to produce Man8GlcNAc2 isomer B. Similarly, ER α-mannosidase II removes just one terminal mannose residue from Man9GlcNAc2, but it removes the mannose residue from the α1,3-branch to produce Man8GlcNAc2 isomer C [48]. The three remaining α1,2-linked mannose residues are subsequently removed from Man8GlcNAc2 by the Golgi class I α1,2-mannosidases, each of which has its own specific substrate preferences.
The end product of the class I α-mannosidase trimming reactions, Man5GlcNAc2, is a key intermediate in the protein N-glycosylation pathway. It is the combination of subsequent trimming and elongation reactions mediated by various Golgi enzymes that ultimately converts the preceding high mannose-type N-glycans, through this intermediate, to hybrid or complex end products (reviewed in references [35, 49]). Hybrid N-glycans are defined as N-linked oligosaccharides in which the terminal α1,3-linked mannose residue is substituted by N-acetylglucosamine, but the α1,6-linked mannose residue is not. Complex N-glycans are defined as N-linked oligosaccharides in which the terminal α1,3-and α1,6-linked mannose residues are both substituted by N-acetylglucosamine residues.
It is generally accepted that the insect N-glycosylation pathway begins like the mammalian pathway, with en bloc transfer of the oligosaccharide precursor from Glc3Man9GlcNAc2-PP-Dol to an appropriate asparagine residue in a newly synthesized polypeptide [20]. This conclusion is strongly supported by studies documenting the presence of the lipid-linked N-glycan in insect cell systems [50–52]. Several lines of biochemical evidence suggest that removal of the terminal glucose residues from Glc3Man9GlcNAc2 is also accomplished in the same way in insect and mammalian cells. While genes encoding α-glucosidase I and II have not yet been cloned from insect systems, the presence of both enzyme activities has been documented in lepidopteran insect cell lines through the use of N-glycan processing inhibitors [53–56] and by direct structural analyses of insect cell-derived N-glycans [31, 57–61].
The presence of α1,2-mannosidase activities involved in mannose trimming also has been documented in lepidopteran insect cell lines [55, 62–65] and cDNAs encoding several insect α1,2-mannosidases have been cloned and functionally characterized. cDNA cloning, expression, and functional characterization clearly demonstrated that the Spodoptera frugiperda cell line, Sf9 [66], which is widely used as a host for baculovirus vectors, encodes and expresses a Golgi class I α1,2-mannosidase [67, 68]. This enzyme processes N-glycans like the mammalian α1,2-mannosidases, converting Man9GlcNAc2 to Man5GlcNAc2 through the major intermediate Man7GcNAc2 isomer C [69]. Class II α1,2-mannosidase activity also was detected in extracts of Sf21, Mb0503, and Bm-N cells [62] and was found to be associated with a Golgi-like fraction of Sf21 cells [70]. This insect class II mannosidase removes the terminal α1,3- and α1,6-linked mannose residues from GlcNAc Man5GlcNAc2 and, like its mammalian counterpart, requires the prior addition of a terminal N-acetylglucosamine residue to the α1,3 branch of Man5GlcNAc2 by GlcNAc-TI. This latter enzyme has been detected in lepidopteran insect cell lines, albeit at much lower levels than in mammalian cells [71, 72], and a Drosophila mela-nogaster GlcNAc-TI homologue has been isolated and shown to encode an enzyme with the expected biochemical activity [73]. A class II Golgi α-mannosidase homologue also was isolated from Drosophila melanogaster and shown to encode an enzyme with the expected biochemical and cell biological properties [74–76]. A gene encoding another class II α-mannosidase, designated SfManIII, was isolated from Sf9 cells and the gene product was expressed and characterized [77–79]. The results showed that this gene encodes α Golgi enzyme that hydrolyzes Man5GlcNAc2 directly to Man3Glc NAc2, without the prior addition of a terminal N-acetylglucosamine residue by GlcNAc-TI. Thus, this insect enzyme has the same substrate specificity as the α-mannosidase III activity detected in Golgi Man II knockout mice [80–82]. Finally, as expected from its apparent restriction to Chordata, no endo-α-mannosidase activity has been detected in insect cell lines [83].
N-GLYCAN FUCOSYLATION IN INSECT AND MAMMALIAN SYSTEMS
The processing reactions described above produce a trimmed N-glycan intermediate, GlcNAcMan3GlcNAc2, which is common to both mammalian and insect systems (boxed in Fig. 1). The mammalian and insect pathways both include a core α1,6-fucosyl-transferase, which can add an α1,6-linked fucose residue to the asparagine-linked N-acetylglucosamine residue of GlcNAcMan3 GlcNAc2 on some, but not all N-glycoproteins. When it occurs, this reaction gives rise to another common intermediate (also boxed in Fig. 1). However, the major insect N-glycan processing pathway clearly diverges from the mammalian pathway following the class II mannosidase trimming and core α1,6-fucosylation steps, as shown in Fig. (1).
One important difference between the insect and mammalian pathways is that at least some insect cells have a fucosyltransferase that may modify the common intermediate, GlcNAcMan3GlcNAc (α1,6Fuc)GlcNAc, to produce a core α1,3-fucosylated N-glycan [22, 84, 85]. This core α1,3-fucosyltransferase strictly requires prior catalysis by the core α1,6-fucosyltransferase [86, 87] and the resulting N-glycan product is known to be allergenic in mammals [88–92]. Another important difference between the insect and mammalian N-glycan processing pathways is that at least some insect cells have a membrane-bound, processing β-N-acetylgluco-saminidase (GlcNAcase) activity. This activity, which is also found in some nematodes [93] and plants [94], may specifically remove the terminal N-acetylglucosamine residue from GlcNAcMan3 GlcNAc(±α3/6Fuc)GlcNAc [95], thereby generating the major insect processed N-glycan products, Man3GlcNAc(±α3/6Fuc) GlcNAc, which are generally classified as paucimannosidic structures. Because removal of the terminal N-acetylglucosamine residue by this enzyme produces N-glycans that are not good substrates for GlcNAc-TII [96], these paucimannosidic N-glycans are unlikely to be further elongated.
The first cDNA experimentally shown to encode a processing GlcNAcase in any organism was recently described in Drosophila melanogaster [97]. This cDNA is derived from the fused lobes (fdl) gene and the gene product had the expected substrate specificity and other biochemical properties of a GlcNAcase involved in N-glycan processing. Interestingly, the FDL protein had an unexpected subcellular distribution, as it was not strongly localized to the Golgi apparatus and was found in other secretory pathway compartments, in the plasma membrane, and in late endosomes. A similarly unexpected localization pattern was observed with two green fluorescent protein-tagged GlcNAcases isolated from Sf9 cells, but biochemical evidence suggested that these enzymes are involved in N-glycan degradation, rather than processing [98]. Hence, a lepidopteran insect homologue of the Drosophila melanogaster fdl gene has not yet been cloned.
N-GLYCAN BRANCHING AND ELONGATION IN INSECT AND MAMMALIAN SYSTEMS
In mammalian cells the conversion of Man5GlcNAc2 to hybrid or complex N-glycan structures (Fig. 1) is initiated by Golgi N-acetylglucosaminyltransferase I (GlcNAc-TI), which transfers an N-acetylglucosamine residue in β1,2-linkage to the mannose residue on the α1,3-branch to produce GlcNAcMan5GlcNAc2 [49]. While this intermediate can be converted to a hybrid structure in various ways, a single enzyme that can commit Man5GlcNAc2 to the hybrid fate is N-acetylglucosaminyltransferase-III (GlcNAc-TIII). This enzyme adds a single N-acetylglucosamine residue to the central β1,2-linked mannose and blocks the subsequent action of Golgi α-mannosidase II [49]. Normally, Golgi α-mannosidase II performs the final mannose trimming steps, removing the terminal α1,3- and α1,6-linked mannose residues from GlcNAcMan5GlcNAc2 to produce GlcNAcMan3GlcNAc2 (reviewed in references [45, 46]). Golgi α-mannosidase II has a strict requirement for prior addition of the terminal N-acetylglucosamine residue to the α1,3-branch of Man5GlcNAc2 by GlcNAc-TI. Another interesting class II α-mannosidase activity, which has become known as α-mannosidase III, cleaves Man5GlcNAc2 directly, without the prior addition of terminal N-acetylglucosamine, to produce GlcNAcMan3GlcNAc2, thereby bypassing the requirement for Golgi α-mannosidase II [80–82].
The prior addition of a terminal N-acetylglucosamine residue to the α1,3 branch of Man5GlcNAc2 is also a prerequisite for core fucosylation by the α1,6 fucosyltransferase discussed above, which transfers an α1,6-linked fucose residue to the asparagine-linked N-acetylglucosamine residue [99]. Thus, the fucosylated core structure GlcNAcMan3GlcNAc(Fucα1,6)GlcNAc is ultimately produced via the concerted action of N-acetylglucosaminyltransferase I, Golgi α-mannosidase II, and the core α1,6 fucosyltransferase.
Another Golgi enzyme, N-acetylglucosaminyltransferase II (GlcNAc-TII), produces the first complex structure in the N-glycan processing pathway by adding a β1,2-linked N-acetylglucosamine residue to the terminal α1,6-linked mannose residue of GlcNAc Man3GlcNAc(±Fucα1,6)GlcNAc [49]. Like Golgi α-mannosidase II and core α1,6-fucosyltransferase, GlcNAc-TII also requires the prior action of GlcNAc-TI [96]. Both GlcNAc-TI and -TII are widespread in various tissue and cell types and these enzymes initiate elongation of two branches, which ultimately gives rise to common “biantennary” N-glycans. Additional branching can be initiated by one or more N-acetylglucosaminyltransferases, including GlcNAc-TIV, -TV, and/or –TVI. These enzymes ultimately yield tri-, tetra-, or penta-antennary N-glycans, but they have more highly restricted tissue distributions and/or are associated with malignant tissues [49]. The more highly branched structures produced by GlcNAc-TIV, -TV, and/or –TVI are not shown in Fig. (1) and will not be considered further.
N-glycan intermediates with terminal N-acetylglucosamine residues can be further elongated by one or more Golgi β1,4-galactosyltransferases (β4GalTs). There are at least seven human β4GalT genes encoding enzymes with various acceptor substrate specificities [100–104]. β4GalT-I to -VI can participate in glycoprotein and/or glycolipid biosynthesis [101, 105, 106], whereas β4GalT-VII is involved in proteoglycan biosynthesis [103].
Finally, many terminally galactosylated N-glycans are capped by the addition of α2,3-, α2,6, and/or α2,8-linked sialic acids in mammalian cells. The human genome encodes more than 20 different sialytransferases and 15 have been cloned and characterized [107]. All are type II-transmembrane glycoproteins of the Golgi apparatus with three consensus domains, or sialymotifs, designated L, S, and VS [108–111]. Different sialyltransferases catalyze different sialic acid linkages (α2,6, α2,3, or α2,8) and have different acceptor substrate specificities. Sialylation plays an important role in many different biological processes, such as nervous system development, turnover of circulating glycoproteins and erythrocytes, pathogen-host interactions, and immune system functions and there are correlations between sialylation and malignant transformation and cancer progression [112–117]. Sialylation is especially relevant in the context of manufacturing recombinant glycoproteins for pharmaceutical applications because it prevents carbohydrate-mediated clearance of glycoproteins from the mammalian circulatory system, which obviously impacts their pharmacokinetics and, therefore, their therapeutic efficacy [118–121].
It is important to note that mammalian cells can add several other types of monosaccharides, particularly fucose residues, to the peripheral regions of the N-glycans described above. Together with the ability to produce up to five antennae, the ability to decorate those antennae in various ways provides for a huge amount of structural and functional diversity among mammalian N-glycoprotein glycans. However, for the purposes of this review, we will focus on the relative ability of mammalian and insect cells to produce the common bi-antennary, terminally α2,3- and/or α2,6-sialylated N-glycan shown on the bottom right-hand side of Fig. (1).
Historically, it has been widely thought that insects have no terminal glycosyltransferase activities or sialic acid metabolism [122–126]. Accordingly, insect N-glycan processing pathways were considered to be limited to the production of paucimannosidic structures. The full story is not quite this simple, however, as there are now extensive biochemical, histochemical, structural, and molecular genetic evidence to suggest that at least some insects have the potential to further elongate and even terminally sialylate glycoprotein glycans. In particular, bioinformatic analyses of the Drosophila melanogaster genome have revealed that this insect has genes corresponding to many of the mammalian genes required for N-glycan branching, elongation, and sialylation. In addition, the putative biochemical functions of some of these gene products have been experimentally confirmed [73, 127–132]. Expression of the genes that could be involved in N-glycan elongation and sialylation appears to be restricted to specific tissues and/or development stages in insect systems [128, 131]. Thus, our newly-recognized genetic potential for more extensive N-glycan processing might begin to explain differences in the results obtained by various investigators who have assessed recombinant glycoprotein glycan elongation and sialylation in various insect cell lines, which are often derived from different tissues and developmental stages.
The Drosophila melanogaster genome includes homologues of GlcNAc-TI, -TII, -TIII, -TIV, -TV, and -TVI, which are the N-acetylglucosaminyltransferases involved in N-glycan branching, as described above. Among these, the fly GlcNAc-TI and -TII homologues have been cloned, characterized, and shown to have the expected biochemical activities [73, 128]. Further analysis of the developmental expression profile of the fly GlcNAc-TII gene showed that it was restricted mainly to the embryonic nervous system and to regions of the brain and imaginal discs of the eye and antennae in third instar animals [128]. Low levels of both GlcNAc-TI and TII activities have been detected in several established lepidopteran insect cell lines, as indicated above [71, 72]. However, to our knowledge, no higher order branching activities have been detected and no N-acetylglucosaminyltransferase genes have been cloned from any lepidopteran insect system to date.
The Drosophila melanogaster genome also includes three β4GalT homologues and each has been cloned and characterized [127, 130, 133]. The putative fly β4GalT gene most closely resembling human βGalT-VII (CGI170; Genbank Accession No. AE003750) was cloned, expressed, and its predicted function in the biosynthesis of the proteoglycan core linkage region was confirmed and shown to be essential for viability [133, 134]. The two other putative fly β4GalT family members (CG8536, Genbank Accession No. AAF58268 and CG14517, Genbank Accession No. AAF56843) most closely resemble human β4GalT-I to -VI, which are functionally implicated in glycoprotein and glycolipid glycan processing, as detailed above. Interestingly, expression and biochemical characterization of these latter Drosophila homologues showed that they prefer to transfer N-acetylgalactosamine, rather than galactose, from the appropriate nucleotide sugar donors to an artificial acceptor substrate [130]. This activity is consistent with the idea that these latter two fly βGalTs preferentially produce N-glycans containing peripheral GalNAcβ1,4-GlcNAc-R (LacDiNAc) rather than the Gal-β1,4-GlcNAc-R (LacNAc) disaccharide sequence commonly found at or near the non-reducing termini of mammalian N-glycans. Additional studies showed that the latter two fly β4GalT homologues are ubiquitously expressed at low levels throughout embryonic and larval development and that double mutant flies had only relatively minor neuromuscular phenotypes and no severe developmental defects [130]. Cloning and characterization of β4GalT genes from C. elegans [135] and Trichoplusia ni [129] revealed that these invertebrate gene products also preferentially transferred N-acetylgalactosamine>galactose>N-acetylgluco-samine from nucleotide sugar donor substrates to various acceptors in vitro. The Trichoplusia ni β4GalT gene product utilized artificial monosaccharides, an N-glycoprotein, and a glycolipid as in vitro acceptor substrates and was shown to function in N-glycan processing in vivo [129]. Thus, this gene product can account for the β4GalNAcT activity and low levels of β4GalT activity previously observed in a cell line derived from Trichoplusia ni [136, 137]. This gene product also can account for the presence of terminal galactose or N-acetylgalactosamine residues observed on some glycoprotein glycans produced by cell lines derived from Trichoplusia ni, particularly the BTI-Tn-5Bl-4 (High Five®; [138]) line [58, 59, 139–141].
The ability of insects to sialylate glycoprotein N-glycans has been a controversial issue in insect glycobiology and among users of the baculovirus-insect cell expression system (reviewed in references [3, 12–21]). Most evidence suggests that there is no sialic acid or sialyltransferase activity in insect cell lines and that the N-glycans of baculovirus-expressed recombinant glycoproteins are not terminally sialylated. However, other reports have provided evidence of sialic acids in insect tissues [142–145] and on baculovirus-expressed recombinant N-glycoproteins, including human plasminogen [146–148] and secreted alkaline phosphatase [25, 26, 29, 33]. In Drosophila melanogaster, sialylation was mainly restricted to the nervous system and was developmentally regulated [142]. Subsequent homology searches revealed the existence of Drosophila melanogaster genes potentially involved in sialic acid biosynthesis and utilization, including putative Neu5Ac phosphate synthase, CMP-Neu5Ac synthase, CMP-Neu5Ac/CMP antiporter, and sialyl-transferase genes [127, 131, 132]. The functions of the putative fly Neu5Ac phosphate synthase [132] and sialyltransferase [131] genes have been experimentally confirmed. The biochemical activity of the fly Neu5Ac phosphate synthase gene product indicated that it could participate in a sialic acid biosynthetic pathway in this insect. The fly sialyltransferase gene product had α2,6 sialyltransferase (ST6Gal) activity with a preference for LacDiNAc as the acceptor substrate, which is consistent with the preferred biochemical activity of the invertebrate β4GalTs, as discussed above [129, 130, 135]. Interestingly, expression of the Drosophila sialyltransferase was restricted to the embryonic central nervous system and significant expression began only by about 13–14 h of embryonic development [131]. In contrast to the results obtained with the Neu5Ac phosphate synthase and sialyltransferase, functional analysis of the putative fly CMP-Neu5Ac/CMP antiporter gene product revealed that it lacks this function and functions, instead, as a UDP-galactose/UMP and UDP-N-acetylgalactosamine/UMP antiporter [149, 150].
In summary, a significant body of accumulated evidence supports the idea that at least some insects, insect tissues, and/or insect cells have the potential to produce complex, terminally sialylated N-glycans. This is an important advance in our basic understanding of insect glycobiology, which must be appreciated. However, in the context of this volume of Current Drug Targets, it should be emphasized that the vast majority of recombinant N-glycoproteins produced in standard baculovirus-insect cell expression systems, in which Sf21, Sf9, and BTI-Tn-5Bl-4 (High Five®) cells were used as the hosts, acquired simple, non-sialylated, paucimannose side-chains at sites normally occupied by complex, sialylated N-glycans in the native mammalian products. Thus, users of the baculovirus-insect cell system should realistically anticipate being unable to produce recombinant versions of higher eukaryotic glycoproteins with fully authentic N-glycans. These users should expect that the complex, terminally sialylated N-glycans on the native products will be replaced mainly by paucimannose structures on the recombinant products. Like their native counterparts, these glycoprotein glycans are unlikely to be homogenous. Thus, the glycan profile will probably also include various proportions of partially trimmed, high mannose type structures and partially elongated hybrid or complex type structures. However, the latter will probably be relatively minor subpopulations and, even among these structures, it is highly unlikely that any will be terminally sialylated.
A CURRENT VIEW OF THE INSECT PROTEIN N-GLYCOSYLATION PATHWAY
The widespread utilization of baculovirus-insect cell system for recombinant glycoprotein production has provided a great deal of information on the protein N-glycosylation pathways of lepidopteran insect cells. One good source of information has come from investigators who have used this system to produce recombinant glycoproteins and characterized the structures of the N-glycans on their products. The advent of the baculovirus-insect cell expression system also has driven basic biochemical and molecular genetic research on insect protein N-glycosylation pathways. Finally, the completion of insect genome projects, particularly the Drosophila melanogaster genome project [151], has had a huge impact on our understanding of insect protein N-glycosylation pathways. Generally, current data suggest that the initial transfer and trimming reactions, which comprise the first half of the protein N-glycosylation pathway, are similar or identical in mammalian and insect cell systems. In contrast, it appears that the elongation reactions, which comprise the second half of the mammalian protein N-glycosylation pathway, are highly limited in insect systems (reviewed in references [3, 12–21]). Thus, the major N-glycan products of insect systems, including the N-glycans on recombinant glycoproteins produced by insect systems, have either high mannose- or paucimannose-type structures (Fig. 1). The former are identical to the high mannose N-glycans produced by mammalian cells, while the latter are trimmed and sometimes core fucosylated, but not otherwise elongated products of GlcNAcMan3GlcNAc2, a common intermediate in the insect and mammalian N-glycan processing pathways. In essence, then, paucimannosidic N-glycans are the major structural counterparts of the hybrid and complex N-glycans produced by higher eukaryotes.
A comparative model of the mammalian and insect protein N-glycosylation pathways, which accommodates all currently available data, is shown in Fig. (1). As discussed above, the first half of the pathway, involving N-glycan transfer and trimming, appears to be similar or identical in these two types of organisms. Hence, both pathways can produce identical high mannose N-glycans, which are the final end products of the processing pathway for some oligosaccharide side chains attached to some sites on some N-glycoproteins [35]. Alternatively, the high mannose N-glycans can be intermediates for further processing reactions in both cell types and these reactions can yield either one of two common intermediates, GlcNAcMan3GlcNAc[±Fuc]GlcNAc. These intermediates are then subjected to different processing reactions in the two cell types, which comprise the second halves of the insect and mammalian protein N-glycosylation pathways. In mammalian cells, the common, trimmed N-glycan intermediate is further elongated to produce a wide variety of different N-glycans, including the common bi-antennary structures shown on the bottom right-hand side of Fig. (1). In contrast, in insect cells, the major processed N-glycans are paucimannosidic structures represented by the common intermediates (in insect cells lacking a processing GlcNAcase activity), by core α1,3-fucosylated variants of the second intermediate, or by structures in which the terminal N-acetylglucosamine residue has been removed by a processing GlcNAcase, as shown on the far left-hand side of Fig. (1). In the context of this volume, it is important to emphasize that the N-glycan processing pathway of at least some insect cells can give rise to core α1,3-fucosylated structures, as these are allergenic in humans. It also is important to note that the major processed insect N-glycans lack terminal sialic acid residues, which are needed to prevent rapid clearance of glycoproteins from the mammalian circulatory system. Finally, it is important to note that the model shown in Fig. (1) is designed to accommodate the emerging fact that at least some insects and insect cells have the potential to produce more extensively elongated N-glycans, such as those found in mammalian cells. However, it should be emphasized that this branch of the N-glycosylation pathway is probably relatively minor and potentially restricted to only certain types of insects, insect tissues, and/or insect developmental stages.
The fate of any given N-glycoprotein glycan in this branched pathway model of the insect processing pathway could depend on a wide variety of different factors. One obvious factor is the nature of the glycoprotein product itself, as some products may be relatively better substrates for the processing GlcNAcase or GlcNAc-TII activities, which determine their subsequent processing fates. In addition, differential expression of the genes encoding these and downstream functions, which distinguish the two branches of the insect N-glycan processing pathway, would clearly be expected to influence the fate of a given N-glycoprotein glycan being processed in insect cells. Expression of these latter genes in insects or insect cell lines could be influenced by species, tissue type, developmental stage, and/or various environmental conditions. Thus, this branched pathway model can potentially explain many or all of the differences in the results obtained by different investigators investigating the N-glycan processing capabilities of the baculovirus-insect cell system, as these studies have utilized different recombinant glycoproteins produced by different insect cell lines derived from different species, tissues, developmental stages, and subjected to different environmental conditions.
ENGINEERING INSECT N-GLYCAN PROCESSING PATHWAYS
In the final analysis, even with a new appreciation for the extent of the N-glycan processing potential in insect systems, users of the baculovirus-insect cell system still suffer from its inability to produce recombinant glycoproteins with authentic, mammalian-like N-glycans. One approach that has been used to address this problem is to use genetic engineering methods to extend the N-glycosylation pathways of the lepidopteran insect cell lines commonly used as hosts for baculovirus expression vectors. This can be accomplished by introducing mammalian genes encoding the missing or subopti-mal processing functions into these insect cell lines. Technically, this can be done either by incorporating the mammalian genes into the viral vector genome or by incorporating them into the host cell genome. In either case, the relative timing of gene expression is important. The genes encoding the new processing functions should be expressed before the gene encoding the recombinant glycoprotein of interest is expressed. In this way, the endogenous N-glycan processing pathway in insect cells can be programmed to provide the machinery needed to produce more authentic mammalian N-glycoproteins before the genes encoding these recombinant products are expressed.
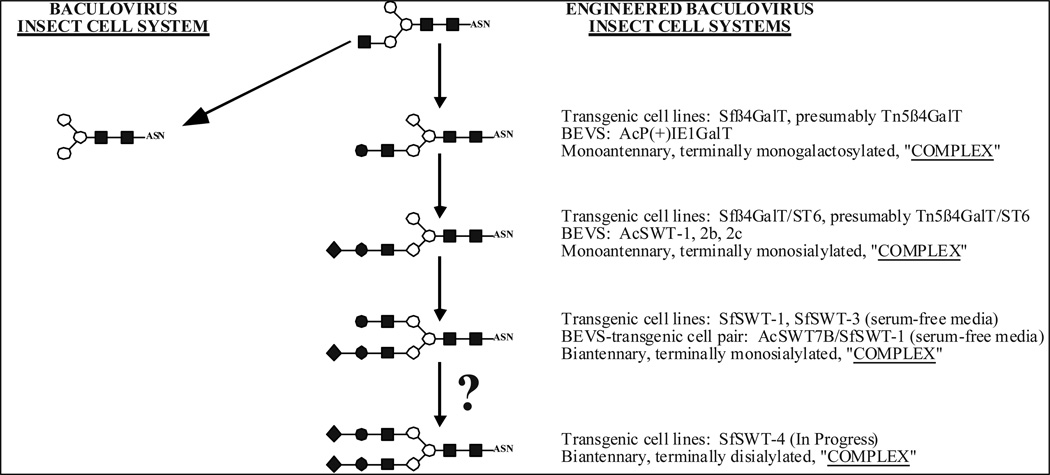
There are several published examples of baculovirus vectors that have been modified to encode mammalian N-glycan processing genes under the transcriptional control of one or more immediate early (iel) baculovirus promoters, together with a glycoprotein of interest under the control of a later baculovirus promoter [152–154]. Further, studies have shown that this type of baculovirus vector can be used to infect conventional lepidopteran insect cell lines and obtain recombinant glycoprotein products with more authentic, mammalian-like N-glycans (Fig. 2).
Fig. (2). N-glycan processing potential of modified baculovirus-insect cell systems.
Monosaccharides are designated by their standard symbols, as defined in Fig. (1). The major processed N-glycans produced by standard baculovirus-insect cell systems are the paucimannosidic structures shown on the left. Genetic engineering of baculovirus vectors and/or insect cell lines has yielded systems capable of producing the more extensively elongated N-glycan structures shown on the right.
There also are several published examples of transgenic insect cell lines that have been engineered to encode mammalian N-glycan processing genes under the transcriptional control of one or more baculovirus iel promoters. Most of these studies have involved the use of Sf9 as the parental cell line, but there also is an example of protein N-glycosylation pathway engineering by this approach in BTI-Tn-5Bl-4 (High Five®) cells. Transformation of these cell lines using a transfection and co-selection approach [155, 156] has yielded transgenic subclones that encode and express mammalian β1,4GalT (Sf β 4GalT [157]), β1,4GalT and a2,6-sialyltransferase (SF β 4Gal/ST6 [158] and Tn β 4Gal/ST6 [159]), and GlcNAc-TI, GlcNAc-TII, β 1,4-GalT, a2,6-sialyltransferase, and a2,3-sialyltrans-ferase (SfSWT-1 [160]) genes. These new cell lines have growth properties that are similar to those of the parental lines, can support baculovirus infection and foreign gene expression, and can produce recombinant glycoproteins with increasingly elongated N-glycans (Fig. 2). The most extensively elongated products are the complex, biantennary, terminally monosialylated N-glycans produced by SfSWT-1 cells [160].
Based on information available at the time, the ability of SfSWT-1 cells to sialylate recombinant glycoproteins was quite surprising. The parental cell line, Sf9, contains no detectable CMP-sialic acid, which is the donor substrate required for sialyltrans-ferase activity [161, 162]. It was subsequently discovered that recombinant glycoprotein sialylation by SfSWT-1 requires cultivating these cells in the presence of an exogenous source of sialic acid, such as fetal bovine serum or purified bovine fetuin, suggesting that they have a sialic acid salvaging pathway [163]. This requirement was subsequently overcome by transforming SfSWT-1 cells with two additional mammalian genes, which encoded a sialic acid synthase (SAS) and CMP-sialic acid synthetase (CMP-SAS). The resulting cell line, SfSWT-3, was able to produce its own CMP-sialic acid, as well as complex, biantennary, terminally monosialylated N-glycans, when cultivated in a serum-free growth medium supplemented with the sialic acid precursor, N-acetylmannosamine [164]. It is important to emphasize that these N-glycans, which are the most sophisticated structures produced by any transgenic insect cell line described to date, have no terminal sialic acid caps on the α1,6 branch (Fig. 2). The results of recent experiments designed to investigate possible reasons for this have shown that the mouse ST3GalIV gene used to produce both SfSWT-1 and SfSWT-3 cells induces no detectable sialyltransferase activity in Sf9 cells (D.L. Jarvis, R.L. Harrison, and X. Shi, unpublished data). Thus, although this gene is expressed at the RNA level in both of these cell lines, it does not encode an active gene product, which probably accounts for the absence of di-sialylated N-glycans on the recombinant glycoproteins produced using these cells so far. Efforts to create new transgenic lines in which Sf9 cells have been transformed with a mammalian gene encoding an alternative sialyltransferase are in progress in our lab (Fig. 2). Another noteworthy recent finding was that SfSWT-1 cells failed to sialylate a recombinant form of equine chorionogonadotropin during infection with a baculovirus vector [165]. This finding raised the possibility that the ability of transgenic insect cell lines engineered to sialylate recombinant glycoproteins might be product-dependent.
CONCLUSIONS AND PERSPECTIVES
During the past 25 years, the baculovirus-insect cell system has been widely utilized to produce recombinant glycoproteins. This activity has directly and indirectly advanced our understanding of protein N-glycosylation pathways in insect systems. In general, the baculovirus-insect cell system is limited by its inability to produce complex, terminally sialylated N-glycans, such as those produced by mammalian cells. At sites normally occupied by these native side-chains, recombinant forms of mammalian glycoproteins produced in the baculovirus-insect cell systems usually have paucimannosidic N-glycans, instead. This seems to be inconsistent with accumulating evidence that insect systems have the potential to produce more extensively elongated, even complex, terminally sialylated N-glycans. However, it is likely that the general lack of glycoprotein sialylation in the baculovirus-insect cell systems widely used for recombinant glycoprotein production reflects the highly specialized nature of this machinery in insect systems. In fact, available data indicate that expression of insect cognates of the mammalian genes involved in the terminal stages of N-glycan processing is restricted to certain types of insects, insect tissues, and/or insect developmental stages and/or is controlled by unknown environmental conditions. Another problem is that the recombinant glycoprotein products of some insect cell lines, particularly those derived from Trichoplusia ni, may have core α1,3-linked fucose residues that are allergenic in humans. The general inability to produce complex, sialylated N-glycans and the potential to produce allergenic carbohydrate epitopes have hindered the use of the baculovirus-insect cell system to produce recombinant glycoproteins for at least some pharmaceutical applications, particularly therapeutic applications. This has led to efforts to develop alternative virus-host combinations, environmental conditions, and transgenic virus-host systems with the ability to produce more authentic mammalian glycoprotein products. The latter approach has been the focus of work in our laboratory for the past decade and these efforts have yielded various baculovirus-insect cell systems that can routinely produce complex, terminally sialylated N-glycans. This work needs to be extended to develop systems capable of routinely producing di-sialylated products. In addition, it needs to be extended to include subtractive engineering to remove undesirable activities, such as the processing GlcNAcase and core α1,3-fucosyltransferase activities, from the system. Finally, all of these improved systems need to be used to express a much wider variety of recombinant glycoproteins to examine the breadth of their capabilities. In the end, it will be exciting to use these improved systems to produce recombinant glycoproteins for pharmaceutical applications, such as in vivo therapeutic applications, that are currently precluded by the state of the art in baculovirus-insect cell expression technology.
ACKNOWLEDGEMENTS
We sincerely thank the NIH (GM49734), NSF (BES 9814157 and BES 9818001), and the NIST-ATP program (70NANB3H 3042) for supporting research in our lab.
ABBREVIATIONS
- β4GalT
β1,4 Galatosyltransferase
- CMP-SAS
CMP sialic acid synthetase
- ER
Endoplasmic reticulum
- fdl
Fused lobes
- Fuc
Fucose
- Fuc-T8
Core α1,6-fucosyltransferase
- Gal
Galactose
- GalNAc
N-Acetylgalactosamine
- Glc
Glucose
- GlcNAc
N-Acetylglucosamine
- GlcNAcase
N-Acetylglucosaminidase
- GlcNAc-T
N-Acetylglucosaminyltransferase
- iel
Immediate early 1
- Man
Mannose
- Neu5Ac
Neuraminic acid
- SAS
Sialic acid synthase
- ST6GalI
α2,6-Sialyltransferase I
REFERENCES
- 1.Luckow VL, Summers MD. Bio/Technology. 1988;6:47–55. [Google Scholar]
- 2.O'Reilly DR, Miller LK, Luckow VA. Baculovirus expression vectors. New York: W.H. Freeman and Company; 1992. [Google Scholar]
- 3.Jarvis DL. The Baculoviruses. Plenum Press; 1997. pp. 389–431. [Google Scholar]
- 4.Kost TA, Condreay JP, Jarvis DL. Nat. Biotechnol. 2005;23:567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hefferon KL. J. Mol. Microbiol. Biotechnol. 2004;7:89–101. doi: 10.1159/000078652. [DOI] [PubMed] [Google Scholar]
- 6.Lu A, Miller LK. The Baculoviruses. Plenum Press; 1997. pp. 193–216. [Google Scholar]
- 7.Berger I, Fitzgerald DJ, Richmond TJ. Nat. Biotechnol. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 8.Kost TA, Condreay JP. Tr. Biotechnol. 2002;20:173–180. doi: 10.1016/s0167-7799(01)01911-4. [DOI] [PubMed] [Google Scholar]
- 9.Ooi BG, Miller LK. Virology. 1988;166:515–523. doi: 10.1016/0042-6822(88)90522-3. [DOI] [PubMed] [Google Scholar]
- 10.Nobiron I, O'Reilly DR, Olszewski JA. J. Gen. Virol. 2003;84:3029–3039. doi: 10.1099/vir.0.19270-0. [DOI] [PubMed] [Google Scholar]
- 11.Carstens EB, Tija ST, Doerfler W. Virology. 1979;99:386–396. doi: 10.1016/0042-6822(79)90017-5. [DOI] [PubMed] [Google Scholar]
- 12.Harrison RL, Jarvis DL. In Insect Viruses: Biotechnological Applications. 2006 pp. in press. [Google Scholar]
- 13.Jarvis DL. Virology. 2003;310:1–7. doi: 10.1016/s0042-6822(03)00120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchal I, Jarvis DL, Cacan R, Verbert A. Biol. Chem. 2001;382:151–159. doi: 10.1515/BC.2001.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvis DL, Kawar ZS, Hollister JR. Curr. Opin. Biotechnol. 1998;9:528–533. doi: 10.1016/s0958-1669(98)80041-4. [DOI] [PubMed] [Google Scholar]
- 16.Betenbaugh MJ, Tomiya N, Narang S, Hsu JT, Lee YC. Curr. Opin. Struct. Biol. 2004;14:601–606. doi: 10.1016/j.sbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Tomiya N, Narang S, Lee YC, Betenbaugh MJ. Glycoconj. J. 2004;21:343–360. doi: 10.1023/B:GLYC.0000046275.28315.87. [DOI] [PubMed] [Google Scholar]
- 18.Tomiya N, Betenbaugh MJ, Lee YC. Accts. Chem. Res. 2003;36:613–620. doi: 10.1021/ar020202v. [DOI] [PubMed] [Google Scholar]
- 19.Altmann F, Staudacher E, Wilson LB, Marz L. Glycoconj. J. 1999;16:109–123. doi: 10.1023/a:1026488408951. [DOI] [PubMed] [Google Scholar]
- 20.Marz L, Altmann F, Staudacher E, Kubelka V. Glycoproteins. Elsevier; 1995. pp. 543–563. [Google Scholar]
- 21.Wilson IBH. Curr. Opin. Struct. Biol. 2002;12:569–577. doi: 10.1016/s0959-440x(02)00367-6. [DOI] [PubMed] [Google Scholar]
- 22.Staudacher E, Altmann F, Wilson LB, Marz L. Biochim. Biophys. Acta. 1999;1473:216–236. doi: 10.1016/s0304-4165(99)00181-6. [DOI] [PubMed] [Google Scholar]
- 23.Altmann F. Glycoconj. J. 1997;14:643–646. doi: 10.1023/a:1018548812675. [DOI] [PubMed] [Google Scholar]
- 24.Joosten CE, Park TH, Shuler ML. Biotechnol. Bioengr. 2003;83:695–705. doi: 10.1002/bit.10696. [DOI] [PubMed] [Google Scholar]
- 25.Joosten CE, Shuler ML. Biotechnol. Progr. 2003;19:739–749. doi: 10.1021/bp0201049. [DOI] [PubMed] [Google Scholar]
- 26.Joosten CE, Shuler ML. Biotechnol. Progr. 2003;19:193–201. doi: 10.1021/bp025695h. [DOI] [PubMed] [Google Scholar]
- 27.Donaldson M, Wood HA, Kulakosky PC, Shuler ML. Biotechnol. Bioengr. 1999;63:255–262. doi: 10.1002/(sici)1097-0290(19990505)63:3<255::aid-bit1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 28.Donaldson M, Wood HA, Kulakosky PC, Shuler ML. Biotechnol. Progr. 1999;15:168–173. doi: 10.1021/bp9900211. [DOI] [PubMed] [Google Scholar]
- 29.Joshi L, Shuler ML, Wood HA. Biotechnol. Progr. 2001;17:822–827. doi: 10.1021/bp010071h. [DOI] [PubMed] [Google Scholar]
- 30.Estrada-Mondaca S, Delgado-Bustos LA, Ramirez OT. Biotechnol. Appl. Biochem. 2005;42:25–34. doi: 10.1042/BA20040158. [DOI] [PubMed] [Google Scholar]
- 31.Choi O, Tomiya N, Kim JH, Slavicek JM, Betenbaugh MJ, Lee YC. Glycobiology. 2003;13:539–548. doi: 10.1093/glycob/cwg071. [DOI] [PubMed] [Google Scholar]
- 32.Kulakosky PC, Hughes PR, Wood HA. Glycobiology. 1998;8:741–745. doi: 10.1093/glycob/8.7.741. [DOI] [PubMed] [Google Scholar]
- 33.Palomares L, Joosten CE, Hughes PR, Granados RR, Shuler ML. Biotechnol. Progr. 2003;19:185–192. doi: 10.1021/bp025598o. [DOI] [PubMed] [Google Scholar]
- 34.Joshi L, Davis TR, Mattu TS, Rudd PM, Dwek RA, Shuler ML, Wood HA. Biotechnol. Progr. 2000;16:650–656. doi: 10.1021/bp000057p. [DOI] [PubMed] [Google Scholar]
- 35.Kornfeld R, Kornfeld S. Ann. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 36.Schenk B, Fernandez F, Waechter CJ. Glycobiology. 2001;11:61R–70R. doi: 10.1093/glycob/11.5.61r. [DOI] [PubMed] [Google Scholar]
- 37.Yan Q, Lennarz WJ. Biochem. Biophys. Res. Comm. 1999;266:684–689. doi: 10.1006/bbrc.1999.1886. [DOI] [PubMed] [Google Scholar]
- 38.Burda P, Aebi M. Biochim. Biophys. Acta. 1999;1426:239–257. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- 39.Kalz-Fuller B, Bieberich E, Bause E. Eur. J. Biochem. 1995;231:344–351. doi: 10.1111/j.1432-1033.1995.tb20706.x. [DOI] [PubMed] [Google Scholar]
- 40.Arendt CW, Ostergaard HL. J. Biol. Chem. 1997;272:13117–13125. doi: 10.1074/jbc.272.20.13117. [DOI] [PubMed] [Google Scholar]
- 41.Trombetta ES, Simons JF, Helenius A. J. Biol. Chem. 1996;271:27509–27516. doi: 10.1074/jbc.271.44.27509. [DOI] [PubMed] [Google Scholar]
- 42.Lubas WA, Spiro RG. J. Biol. Chem. 1987;262:3775–3781. [PubMed] [Google Scholar]
- 43.Lubas WA, Spiro RG. J. Biol. Chem. 1988;263:3990–3998. [PubMed] [Google Scholar]
- 44.Roth J, Ziak M, Zuber C. Biochimie. 2003;85:287–294. doi: 10.1016/s0300-9084(03)00049-x. [DOI] [PubMed] [Google Scholar]
- 45.Trimble RB, Moremen KW, Herscovics A. Guidebook to the Secretory Pathway. Sambrook and Tooze Scientific Publishers; 1992. pp. 185–189. [Google Scholar]
- 46.Moremen KW, Trimble RG, Herscovics A. Glycobiology. 1994;4:113–125. doi: 10.1093/glycob/4.2.113. [DOI] [PubMed] [Google Scholar]
- 47.Herscovics A. Biochimie. 2001;83:757–762. doi: 10.1016/s0300-9084(01)01319-0. [DOI] [PubMed] [Google Scholar]
- 48.Weng S, Spiro RG. J. Biol. Chem. 1993;268:25656–25663. [PubMed] [Google Scholar]
- 49.Schachter H. Glycoproteins. Elsevier; 1995. pp. 153–199. [Google Scholar]
- 50.Sagami H, Lennarz WJ. J. Biol. Chem. 1987;262:15610–15617. [PubMed] [Google Scholar]
- 51.Quesada Allue LA, Belocopitow E. Eur. J. Biochem. 1978;88:529–541. doi: 10.1111/j.1432-1033.1978.tb12479.x. [DOI] [PubMed] [Google Scholar]
- 52.Parker GF, Williams PJ, Butters TD, Roberts DB. FEBS Lett. 1991;290:58–60. doi: 10.1016/0014-5793(91)81225-w. [DOI] [PubMed] [Google Scholar]
- 53.Marchal I, Mir AM, Kmiecik D, Verbert A, Cacan R. Glycobiology. 1999;9:645–654. doi: 10.1093/glycob/9.7.645. [DOI] [PubMed] [Google Scholar]
- 54.Jarvis DL, Garcia A., Jr. Virology. 1994;205:300–313. doi: 10.1006/viro.1994.1646. [DOI] [PubMed] [Google Scholar]
- 55.Jarvis DL, Summers MD. Mol. Cell. Biol. 1989;9:214–223. doi: 10.1128/mcb.9.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nairn HY, Koblet H. Arch. Virol. 1988;102:73–89. doi: 10.1007/BF01315564. [DOI] [PubMed] [Google Scholar]
- 57.Lopez M, Coddeville B, Langridge J, Plancke Y, Sautiere P, Chaabihi H, Chirat F, Harduin-Lepers A, Cerutti M, Verbert A, Delannoy P. Glycobiology. 1997;7:635–651. doi: 10.1093/glycob/7.5.635. [DOI] [PubMed] [Google Scholar]
- 58.Hsu TA, Takahashi N, Tsukamoto Y, Kato K, Shimada I, Masuda K, Whiteley EM, Fan JQ, Lee YC, Betenbaugh MJ. J. Biol. Chem. 1997;272:9062–9070. doi: 10.1074/jbc.272.14.9062. [DOI] [PubMed] [Google Scholar]
- 59.Ailor E, Takahashi N, Tsukamoto Y, Masuda K, Rahman BA, Jarvis DL, Lee YC, Betenbaugh MJ. Glycobiology. 2000;10:837–847. doi: 10.1093/glycob/10.8.837. [DOI] [PubMed] [Google Scholar]
- 60.Kubelka V, Altmann F, Kornfeld G, Marz L. Arch. Biochem. Biophys. 1994;308:148–157. doi: 10.1006/abbi.1994.1021. [DOI] [PubMed] [Google Scholar]
- 61.Nagao E, Takahashi N, Chino H. Insect Biochem. 1987;17:531–538. [Google Scholar]
- 62.Altmann F, Marz L. Glycoconj. J. 1995;12:150–155. doi: 10.1007/BF00731359. [DOI] [PubMed] [Google Scholar]
- 63.Jarvis DL, Garcia A., Jr. Virology. 1994;205:300–313. doi: 10.1006/viro.1994.1646. [DOI] [PubMed] [Google Scholar]
- 64.Ren J, Castellino FJ, Bretthauer RK. Biochem. J. 1997;15:951–956. doi: 10.1042/bj3240951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davidson DJ, Bretthauer RK, Castellino FJ. Biochemistry. 1991;30:9811–9815. doi: 10.1021/bi00105a001. [DOI] [PubMed] [Google Scholar]
- 66.Summers MD, Smith GE. Tx. Ag. Expt. Stn. Bull. No. 1987:1555. [Google Scholar]
- 67.Kawar Z, Herscovics A, Jarvis DL. Glycobiology. 1997;7:433–443. doi: 10.1093/glycob/7.3.433. [DOI] [PubMed] [Google Scholar]
- 68.Kawar Z, Jarvis DL. Insect Biochem. Mol. Biol. 2001;31:289–297. doi: 10.1016/s0965-1748(00)00121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawar Z, Romero PA, Herscovics A, Jarvis DL. Glycobiology. 2000;10:347–355. doi: 10.1093/glycob/10.4.347. [DOI] [PubMed] [Google Scholar]
- 70.Ren J, Castellino FJ, Bretthauer RK. Biochem. J. 1997;324(Pt 3):951–956. doi: 10.1042/bj3240951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Velardo MA, Bretthauer RK, Boutaud A, Reinhold B, Rein-hold VN, Castellino FJ. J. Biol. Chem. 1993;268:17902–17907. [PubMed] [Google Scholar]
- 72.Altmann F, Kornfeld G, Dalik T, Staudacher E, Glossl J. Glycobiology. 1993;3:619–625. doi: 10.1093/glycob/3.6.619. [DOI] [PubMed] [Google Scholar]
- 73.Sarkar M, Schachter H. Biol. Chem. 2001;382:209–217. doi: 10.1515/BC.2001.028. [DOI] [PubMed] [Google Scholar]
- 74.Rabouille C, Kuntz DA, Lockyer A, Watson R, Signorelli T, Rose DR, van den Heuvel M, Roberts DB. J. Cell Sci. 1999;112(Pt 19):3319–3330. doi: 10.1242/jcs.112.19.3319. [DOI] [PubMed] [Google Scholar]
- 75.Numao S, Kuntz DA, Withers SG, Rose DR. J. Biol. Chem. 2003;278:48074–48083. doi: 10.1074/jbc.M309249200. [DOI] [PubMed] [Google Scholar]
- 76.Foster JM, Yudkin B, Lockyer AE, Roberts DB. Gene. 1995;154:183–186. doi: 10.1016/0378-1119(94)00867-r. [DOI] [PubMed] [Google Scholar]
- 77.Jarvis DL, Bohlmeyer DA, Liao YF, Lomax KK, Merkle RK, Weinkauf C, Moremen KW. Glycobiology. 1997;7:113–127. doi: 10.1093/glycob/7.1.113. [DOI] [PubMed] [Google Scholar]
- 78.Kawar Z, Karaveg K, Moremen KW, Jarvis DL. J. Biol. Chem. 2001;216:16335–16340. doi: 10.1074/jbc.M100119200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Francis BR, Paquin L, Weinkauf C, Jarvis DL. Glycobiology. 2002;12:369–377. doi: 10.1093/glycob/12.6.369. [DOI] [PubMed] [Google Scholar]
- 80.Bonay P, Hughes RC. Eur. J. Biochem. 1991;197:229–238. doi: 10.1111/j.1432-1033.1991.tb15903.x. [DOI] [PubMed] [Google Scholar]
- 81.Monis E, Bonay P, Hughes R. Eur. J. Biochem. 1987;168:287–294. doi: 10.1111/j.1432-1033.1987.tb13419.x. [DOI] [PubMed] [Google Scholar]
- 82.Chui D, Oh-Eda M, Liao YF, et al. Cell. 1997;90:157–167. doi: 10.1016/s0092-8674(00)80322-0. [DOI] [PubMed] [Google Scholar]
- 83.Dairaku K, Spiro RG. Glycobiology. 1997;7:579–586. doi: 10.1093/glycob/7.4.579. [DOI] [PubMed] [Google Scholar]
- 84.Staudacher E, Altmann F, Marz L, Hard K, Kamerling JP, Vliegenthart JF. Glycoconj. J. 1992;9:82–85. doi: 10.1007/BF00731703. [DOI] [PubMed] [Google Scholar]
- 85.Fabini G, Freilinger A, Altmann F, Wilson IB. J. Biol. Chem. 2001;276:28058–28067. doi: 10.1074/jbc.M100573200. [DOI] [PubMed] [Google Scholar]
- 86.Staudacher E, Marz L. Glycoconj. J. 1998;15:355–360. doi: 10.1023/a:1006969701231. [DOI] [PubMed] [Google Scholar]
- 87.Paschinger K, Staudacher E, Stemmer U, Fabini G, Wilson IB. Glycobiology. 2005;15:463–474. doi: 10.1093/glycob/cwi028. [DOI] [PubMed] [Google Scholar]
- 88.Prenner C, Mach L, Glossl J, Marz L. Biochem. J. 1992;284(Pt 2):377–380. doi: 10.1042/bj2840377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Faye L, Gomord V, Fitchette-Laine AC, Chrispeels MJ. Analyt. Biochem. 1993;209:104–108. doi: 10.1006/abio.1993.1088. [DOI] [PubMed] [Google Scholar]
- 90.Kurosaka A, Yano A, Itoh N, Kuroda Y, Nakagawa T, Kawasaki T. J. Biol. Chem. 1991;266:4168–4172. [PubMed] [Google Scholar]
- 91.Wilson IB, Harthill JE, Mullin NP, Ashford DA, Altmann F. Glycobiology. 1998;8:651–661. doi: 10.1093/glycob/8.7.651. [DOI] [PubMed] [Google Scholar]
- 92.Bencurova M, Hemmer W, Focke-Tejkl M, Wilson IB, Altmann F. Glycobiology. 2004;14:457–466. doi: 10.1093/glycob/cwh058. [DOI] [PubMed] [Google Scholar]
- 93.Zhang W, Cao P, Chen S, Spence AM, Zhu S, Staudacher E, Schachter H. Biochem. J. 2003;372:53–64. doi: 10.1042/BJ20021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vitale A, Chrispeels MJ. J. Cell Biol. 1984;99:133–140. doi: 10.1083/jcb.99.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Altmann F, Schwihla H, Staudacher E, Glossl J, Marz L. J. Biol. Chem. 1995;270:17344–17349. doi: 10.1074/jbc.270.29.17344. [DOI] [PubMed] [Google Scholar]
- 96.Bendiak B, Schachter H. J. Biol. Chem. 1987;262:5784–5790. [PubMed] [Google Scholar]
- 97.Leonard R, Rendic D, Rabouille C, Wilson IB, Preat T, Altmann F. J. Biol. Chem. 2005 doi: 10.1074/jbc.M511023200. in press. [DOI] [PubMed] [Google Scholar]
- 98.Aumiller JJ, Hollister J, Jarvis DL. Prot. Expr. Purif. 2006 doi: 10.1016/j.pep.2005.11.026. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilson JR, Williams D, Schachter H. Biochem. Biophys. Res. Commun. 1976;72:909–916. doi: 10.1016/s0006-291x(76)80218-5. [DOI] [PubMed] [Google Scholar]
- 100.Lo NW, Shaper JH, Pevsner J, Shaper NL. Glycobiology. 1998;8:517–526. doi: 10.1093/glycob/8.5.517. [DOI] [PubMed] [Google Scholar]
- 101.Almeida R, Amado M, David L, et al. J. Biol. Chem. 1997;272:31979–31991. doi: 10.1074/jbc.272.51.31979. [DOI] [PubMed] [Google Scholar]
- 102.Furukawa K, Sato T. Biochim. Biophys. Acta. 1999;1473:54–66. doi: 10.1016/s0304-4165(99)00169-5. [DOI] [PubMed] [Google Scholar]
- 103.Almeida R, Levery SB, Mandel U, Kresse H, Schwientek T, Bennett EP, Clausen H. J. Biol. Chem. 1999;274:26165–26171. doi: 10.1074/jbc.274.37.26165. [DOI] [PubMed] [Google Scholar]
- 104.Amado M, Almeida R, Schwientek T, Clausen H. Biochim. Biophys. Acta. 1999;1473:35–53. doi: 10.1016/s0304-4165(99)00168-3. [DOI] [PubMed] [Google Scholar]
- 105.Schwientek T, Almeida R, Levery SB, Holmes EH, Bennett E, Clausen H. J. Biol. Chem. 1998;273:29331–29340. doi: 10.1074/jbc.273.45.29331. [DOI] [PubMed] [Google Scholar]
- 106.Sato T, Furukawa K, Bakker H, Van den Eijnden DH, Van Die I. Proc. Natl. Acad. Sci. U.S.A. 1998;95:472–477. doi: 10.1073/pnas.95.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. Biochimie. 2001;83:727–737. doi: 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- 108.Geremia RA, Harduin-Lepers A, Delannoy P. Glycobiology. 1997;7:v–vii. doi: 10.1093/glycob/7.2.161. [DOI] [PubMed] [Google Scholar]
- 109.Paulson JC, Colley KJ. J. Biol. Chem. 1989;264:17615–17618. [PubMed] [Google Scholar]
- 110.Datta AK, Paulson JC. J. Biol. Chem. 1995;270:1497–1500. doi: 10.1074/jbc.270.4.1497. [DOI] [PubMed] [Google Scholar]
- 111.Datta AK, Paulson JC. Ind. J. Biochem. Biophys. 1997;34:57–165. [PubMed] [Google Scholar]
- 112.Varki A. FASEB J. 1997;11:248–255. doi: 10.1096/fasebj.11.4.9068613. [DOI] [PubMed] [Google Scholar]
- 113.Kim YJ, Varki A. Glycoconj. J. 1997;14:569–576. doi: 10.1023/a:1018580324971. [DOI] [PubMed] [Google Scholar]
- 114.Dall'Olio F. Glycoconj. J. 2000;17:669–676. doi: 10.1023/a:1011077000164. [DOI] [PubMed] [Google Scholar]
- 115.Takahashi M, Tsuda T, Ikeda Y, Honke K, Taniguchi N. Glycoconj. J. 2004;20:207–212. doi: 10.1023/B:GLYC.0000024252.63695.5c. [DOI] [PubMed] [Google Scholar]
- 116.Schauer R. Glycoconj. J. 2000;17:485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 118.Grossmann M, Wong R, Teh NG, Tropea JE, East-Palmer J, Weintraub BD, Szkudlinski MW. Endocrinology. 1997;138:92–100. doi: 10.1210/endo.138.1.4897. [DOI] [PubMed] [Google Scholar]
- 119.Raju TS, Lerner L, O'Connor JV. Biotechnol. Appl. Biochem. 1996;24(Pt 3):191–194. [PubMed] [Google Scholar]
- 120.Sareneva T, Cantell K, Pyhala L, Pirhonen J, Julkunen I. J. Interferon Res. 1993;13:267–269. doi: 10.1089/jir.1993.13.267. [DOI] [PubMed] [Google Scholar]
- 121.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Warren L. Comp. Biochem. Physiol. 1963;10:153–171. doi: 10.1016/0010-406x(63)90238-x. [DOI] [PubMed] [Google Scholar]
- 123.Butters TD, Hughes RC, Vischer P. Biochim. Biophys. Acta. 1981;640:672–686. doi: 10.1016/0005-2736(81)90097-3. [DOI] [PubMed] [Google Scholar]
- 124.Hsieh P, Robbins PW. J. Biol. Chem. 1984;259:2375–2382. [PubMed] [Google Scholar]
- 125.Butters TD, Hughes RC. Biochim. Biophys. Acta. 1981;640:655–671. doi: 10.1016/0005-2736(81)90096-1. [DOI] [PubMed] [Google Scholar]
- 126.Stollar V, Stollar BD, Koo R, Harrap KA, Schlesinger RW. Virology. 1976;69:104–115. doi: 10.1016/0042-6822(76)90198-7. [DOI] [PubMed] [Google Scholar]
- 127.Altmann F, Fabini G, Ahorn H, Wilson IB. Biochimie. 2001;83:703–712. doi: 10.1016/s0300-9084(01)01297-4. [DOI] [PubMed] [Google Scholar]
- 128.Tsitilou SG, Grammenoudi S. Biochem. Biophys. Res. Comm. 2003;312:1372–1376. doi: 10.1016/j.bbrc.2003.11.059. [DOI] [PubMed] [Google Scholar]
- 129.Vadaie N, Jarvis DL. J. Biol. Chem. 2004;279:33501–33518. doi: 10.1074/jbc.M404925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Haines N, Irvine KD. Glycobiology. 2005;15:335–346. doi: 10.1093/glycob/cwi017. [DOI] [PubMed] [Google Scholar]
- 131.Koles K, Irvine KD, Panin VM. J. Biol. Chem. 2004;279:4346–4357. doi: 10.1074/jbc.M309912200. [DOI] [PubMed] [Google Scholar]
- 132.Kim K, Lawrence SM, Park J, Pitts L, Vann WF, Beten-baugh MJ, Palter KB. Glycobiology. 2002;12:73–83. doi: 10.1093/glycob/12.2.73. [DOI] [PubMed] [Google Scholar]
- 133.Vadaie N, Hulinsky RS, Jarvis DL. Glycobiology. 2002;12:589–597. doi: 10.1093/glycob/cwf074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Takemae H, Ueda R, Okubo R, Nakato H, Izumi S, Saigo K, Nishihara S. J. Biol. Chem. 2003;278:15571–15578. doi: 10.1074/jbc.M301123200. [DOI] [PubMed] [Google Scholar]
- 135.Kawar ZS, Van Die I, Cummings RD. J. Biol. Chem. 2002;277:34924–34932. doi: 10.1074/jbc.M206112200. [DOI] [PubMed] [Google Scholar]
- 136.van Die I, van Tetering A, Bakker H, van den Eijnden DH, Joziasse DH. Glycobiology. 1996;6:157–164. doi: 10.1093/glycob/6.2.157. [DOI] [PubMed] [Google Scholar]
- 137.Abdul-Rahman B, Ailor E, Jarvis D, Betenbaugh M, Lee YC. Carbohydr. Res. 2002;337:2181–2186. doi: 10.1016/s0008-6215(02)00260-4. [DOI] [PubMed] [Google Scholar]
- 138.Wickham TJ, Davis T, Granados RR, Shuler ML, Wood HA. Biotechnol. Progr. 1992;8:391–396. doi: 10.1021/bp00017a003. [DOI] [PubMed] [Google Scholar]
- 139.Park YI, Wood HA, Lee YC. Glycoconj. J. 1999;16:629–638. doi: 10.1023/a:1007029017400. [DOI] [PubMed] [Google Scholar]
- 140.Joshi L, Davis TR, Mattu TS, Rudd PM, Dwek RA, Shuler ML, Wood HA. Biotechnol. Progr. 2000;16:650–656. doi: 10.1021/bp000057p. [DOI] [PubMed] [Google Scholar]
- 141.Rudd PM, Downing AK, Cadene M, Harvey DJ, Wormald MR, Weir I, Dwek RA, Rifkin DB, Gleizes PE. Biochemistry. 2000;39:1596–1603. doi: 10.1021/bi9918285. [DOI] [PubMed] [Google Scholar]
- 142.Roth J, Kempf A, Reuter G, Schauer R, Gehring WJ. Science. 1992;256:673–675. doi: 10.1126/science.1585182. [DOI] [PubMed] [Google Scholar]
- 143.Malykh YN, Krisch B, Gerardy-Schahn R, Lapina EB, Shaw L, Schauer R. Glycoconj. J. 1999;16:731–739. doi: 10.1023/a:1007115627708. [DOI] [PubMed] [Google Scholar]
- 144.Karacali S, Kirmizigul S, Deveci R, Deveci O, Onat T, Gurcu B. Tissue and Cell. 1997;29:315–321. doi: 10.1016/s0040-8166(97)80007-9. [DOI] [PubMed] [Google Scholar]
- 145.Karacali S, Kirmizigul S, Deveci R. Invert. Repr. Dev. 1999;35:225–229. [Google Scholar]
- 146.Davidson DJ, Fraser MJ, Castellino FJ. Biochemistry. 1990;29:5584–5590. doi: 10.1021/bi00475a024. [DOI] [PubMed] [Google Scholar]
- 147.Davidson DJ, Castellino FJ. Biochemistry. 1991;30:6689–6696. doi: 10.1021/bi00241a008. [DOI] [PubMed] [Google Scholar]
- 148.Davidson DJ, Castellino FJ. Biochemistry. 1991;30:6165–6174. doi: 10.1021/bi00239a013. [DOI] [PubMed] [Google Scholar]
- 149.Aumiller JJ, Jarvis DL. Prot. Expr. Purif. 2002;26:438–448. doi: 10.1016/s1046-5928(02)00550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Segawa H, Kawakita M, Ishida N. Eur. J. Biochem. 2002;269:128–138. doi: 10.1046/j.0014-2956.2001.02632.x. [DOI] [PubMed] [Google Scholar]
- 151.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wormian JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YA, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangel-ista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibeg-wam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, Mcintosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 152.Jarvis DL, Finn EE. Nat. Biotechnol. 1996;14:1288–1292. doi: 10.1038/nbt1096-1288. [DOI] [PubMed] [Google Scholar]
- 153.Jarvis DL, Howe D, Aumiller JJ. J. Virol. 2001;75:6223–6227. doi: 10.1128/JVI.75.13.6223-6227.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Seo NS, Hollister JR, Jarvis DL. Prot. Expr. Purif. 2001;22:234–241. doi: 10.1006/prep.2001.1432. [DOI] [PubMed] [Google Scholar]
- 155.Harrison RL, Jarvis DL. Baculovirus Expression Protocols. Humana Press; 2006. pp. in press. [Google Scholar]
- 156.Jarvis DL, Fleming JA, Kovacs GR, Summers MD, Guarino LA. Bio/Technology. 1990;8:950–955. doi: 10.1038/nbt1090-950. [DOI] [PubMed] [Google Scholar]
- 157.Hollister JR, Shaper JH, Jarvis DL. Glycobiology. 1998;8:473–480. doi: 10.1093/glycob/8.5.473. [DOI] [PubMed] [Google Scholar]
- 158.Hollister JR, Jarvis DL. Glycobiology. 2001;11:1–9. doi: 10.1093/glycob/11.1.1. [DOI] [PubMed] [Google Scholar]
- 159.Breitbach K, Jarvis DL. Biotechnol. Bioengr. 2001;74:230–239. doi: 10.1002/bit.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hollister JR, Grabenhorst E, Nimtz M, Conradt HO, Jarvis DL. Biochemistry. 2002;41:15093–15104. doi: 10.1021/bi026455d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hooker AD, Green NH, Baines AJ, Bull AT, Jenkins N, Strange PG, James DC. Biotechnol. Bioengr. 1999;63:559–572. doi: 10.1002/(sici)1097-0290(19990605)63:5<559::aid-bit6>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 162.Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, Lee YC. Analyt. Biochem. 2001;293:129–137. doi: 10.1006/abio.2001.5091. [DOI] [PubMed] [Google Scholar]
- 163.Hollister JR, Conradt HO, Jarvis DL. Glycobiology. 2003;13:487–495. doi: 10.1093/glycob/cwg053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Aumiller JJ, Hollister JR, Jarvis DL. Glycobiology. 2003;13:497–507. doi: 10.1093/glycob/cwg051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Legardinier S, Klett D, Poirier JC, Combarnous Y, Cahoreau C. Glycobiology. 2005;15:776–790. doi: 10.1093/glycob/cwi060. [DOI] [PubMed] [Google Scholar]