SUMMARY
Circadian clocks are coupled to metabolic oscillations through nutrient-sensing pathways. Nutrient flux into the hexosamine biosynthesis pathway triggers covalent protein modification by O-linked β-D-N-acetylglucosamine (O-GlcNAc). Here we show that the hexosamine/O-GlcNAc pathway modulates peripheral clock oscillation. O-GlcNAc transferase (OGT) promotes expression of BMAL1/CLOCK target genes and affects circadian oscillation of clock genes in vitro and in vivo. Both BMAL1 and CLOCK are rhythmically O-GlcNAcylated and this protein modification stabilizes BMAL1 and CLOCK by inhibiting their ubiquitination. In vivo analysis of genetically modified mice with perturbed hepatic OGT expression shows aberrant circadian rhythms of glucose homeostasis. These results establish the counteraction between O-GlcNAcylation and ubiquitination as a key mechanism that regulates the circadian clock and suggest a crucial role for O-GlcNAc signaling in transducing nutritional signals to the core circadian timing machinery.
INTRODUCTION
Almost all mammalian cells contain a self-sustained circadian (about 24-h) clock that runs in tight synchrony with environmental cues, including light and food (Bass and Takahashi, 2010). While the master pacemaker residing in the hypothalamic suprachiasmatic nucleus (SCN) is entrained directly by light, peripheral circadian oscillators can be entrained by diurnal feeding (Schibler and Sassone-Corsi, 2002). Among various macronutrients, glucose is a particularly potent entraining cue for peripheral clocks (Stephan and Davidson, 1998). Cellular nutrient sensors such as nuclear receptors have been proposed as mechanisms for entrainment by food (Asher and Schibler, 2011; Yang, 2010; Yang et al., 2006), but the molecular basis for glucose-mediated entrainment remains a mystery.
Circadian timekeeping occurs at the cellular level by virtue of transcriptional-translational auto-regulatory feedback loops (Mohawk et al., 2012). The transcriptional activators BMAL1 and CLOCK drive expression of Period (Per1 and Per2) and Cryptochrome (Cry1 and Cry2) genes. PER and CRY proteins accumulate progressively and in turn inhibit BMAL1/CLOCK activity, thus generating the approximate 24-h cycle of clock gene expression. The pace of oscillation of this auto-feedback loop is controlled by various regulatory mechanisms, including post-translational modifications of clock proteins (Bass and Takahashi, 2010).
Cells also possess a distinct form of post-translational modification that is highly sensitive to nutrient availability. Glucose flux via the hexosamine biosynthesis pathway leads to intracellular glycosylation by addition of β-D-N-acetylglucosamine (GlcNAc) to many cytoplasmic and nuclear proteins at the hydroxyl groups of serine and threonine residues (Hanover et al., 2012; Hart et al., 2011). This widespread and dynamic glycosylation is mediated by O-linked GlcNAc transferase (OGT) and O-GlcNAcase (OGA), which catalyzes sugar addition and removal, respectively. O-GlcNAc modification is increasingly recognized as a key regulator of diverse cellular processes. O-GlcNAcylation of a number of transcription factors mediates the effects of glucose on transcription of genes involved in key metabolic processes (Hart et al., 2011). A recent study has shown that O-GlcNAcylation links the cardiomyocyte circadian clock to metabolic outputs (Durgan et al., 2011). In Drosophila, O-GlcNAcylation of the PER protein has been shown to contribute to setting the clock speed (Kim et al., 2012). The present study provides the direct evidence that glucose availability regulates cellular clock oscillation through the hexosamine/O-GlcNAc pathway. We further demonstrate that BMAL1 and CLOCK are key targets of O-GlcNAcylation that in turn prevents degradation of these proteins by inhibiting their ubiquitination. Accordingly, this work establishes a new mechanism for metabolic entrainment of the circadian clock by covalent modification of core clock components.
RESULTS
The hexosamine/O-GlcNAc pathway modulates cellular clock oscillation
In light of the important role of food-derived signals for peripheral clock entrainment, we examined whether the nutrient-sensing hexosamine pathway affects circadian oscillation. After dexamethasone synchronization, U2OS cells (U2OS-B6) stably expressing a Bmal1:luciferase reporter construct were grown in high (25 mM) glucose culture medium containing D-luciferin and monitored by the real-time bioluminescence recording system. Addition of azaserine, an inhibitor of hexosamine biosynthesis (Figure S1A), increases the period length and decreases the amplitude of Bmal1 oscillation (Figure 1A). D-glucosamine is able to fuel the cellular pool of UDP-GlcNAc, the donor substrate of O-GlcNAcylation (Figure S1A). Addition of D-glucosamine dramatically delays the phase of Bmal1 oscillation (Figure S1B). These data indicate a role for the hexosamine pathway in circadian regulation.
Figure 1. The hexosamine/O-GlcNAc pathway modulates cellular clock oscillation.
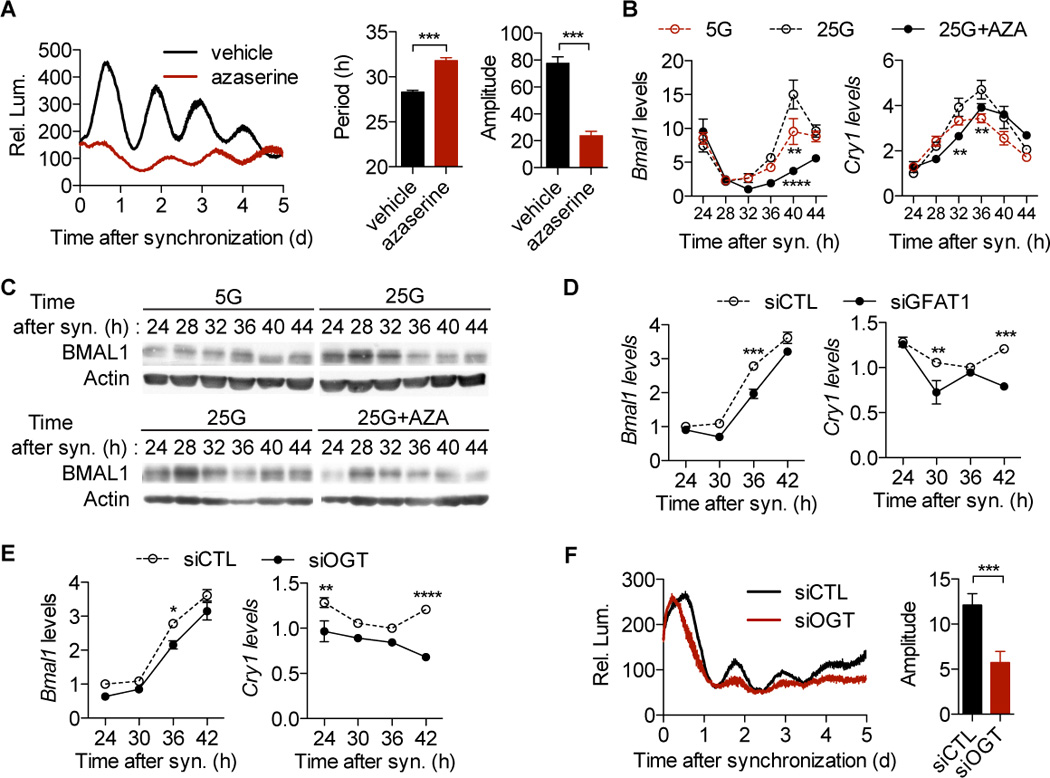
(A) Average real-time bioluminescence from synchronized U2OS-B6 cells stably expressing a Bmal1:luc construct in the presence or absence of azaserine (n = 5). The vertical bar represents relative luminescene (Rel. Lum.). Circadian parameters were calculated by JTK_CYCLE. (B) RT-qPCR analysis of synchronized U2OS-B6 cells (n = 3). 5G/25G, 5/25 mM glucose; AZA, azaserine. (C) Immunoblot analysis of synchronized U2OS-B6 cells (n = 3 per lane). (D) RT-qPCR analysis of synchronized U2OS-B6 cells transfected with GFAT1 siRNA (n = 3). (E) RT-qPCR analysis (n = 3) and (F) average real-time bioluminescence (n = 5) of OGT knockdown on clock oscillation in synchronized U2OS-B6 cells. siCTL, scrambled siRNA; siGFAT1, GFAT1 siRNA; siOGT, OGT siRNA. All data are shown as mean ± SEM. *P < 0.05, **P <0.01, ***P < 0.001, ****P <0.0001, post-hoc Bonferroni’s test or two-tailed Student's t test.
To substantiate our observations, we examined the oscillation of endogenous clock genes in synchronized U2OS-B6 cells. Compared with low (5 mM) glucose, high glucose increases the amplitude of Bmal1 and Cry1 mRNA oscillation, whereas azaserine suppresses them (Figure 1B). Immunoblot analysis shows that low glucose and azaserine also decrease BMAL1 protein levels as compared with high glucose (Figure 1C). Although the glucose concentrations do not affect the phase of Bmal1 mRNA cycling (Figure 1B), low glucose delays the phase of BMAL1 protein accumulation (Figure 1C), suggesting that glucose can regulate BMAL1 levels post-transcriptionally.
GFAT1 is the first and rate-limiting enzyme in hexosamine biosynthesis and the target of azaserine. The siRNA-mediated knockdown of GFAT1 decreases expression of Bmal1 and Cry1 (Figures 1D and S1C), which is also seen in the cells transfected with OGT siRNA (Figures 1E and S1D). Consistently, OGT knockdown reduces BMAL1 protein abundance (Figure S1E). Furthermore, OGT knockdown dramatically decreases the amplitude of the Bmal1:luciferase rhythm (Figure 1F). Taken together, these data demonstrate that the hexosamine/O-GlcNAc pathway regulates cellular clock oscillation.
OGT promotes expression of BMAL1/CLOCK target genes
To further dissect the circadian function of hexosamine signaling, we examined the rhythmicity of expression of key genes in this pathway. In mouse livers, diurnal levels of Gfat1 transcripts are ultradian with a 12-hr period, whereas Ogt transcripts oscillate in a circadian manner (Figure 2A). Oga expression exhibits a weak diurnal rhythm (Figure S2A).
Figure 2. OGT promotes expression of BMAL1/CLOCK target genes.
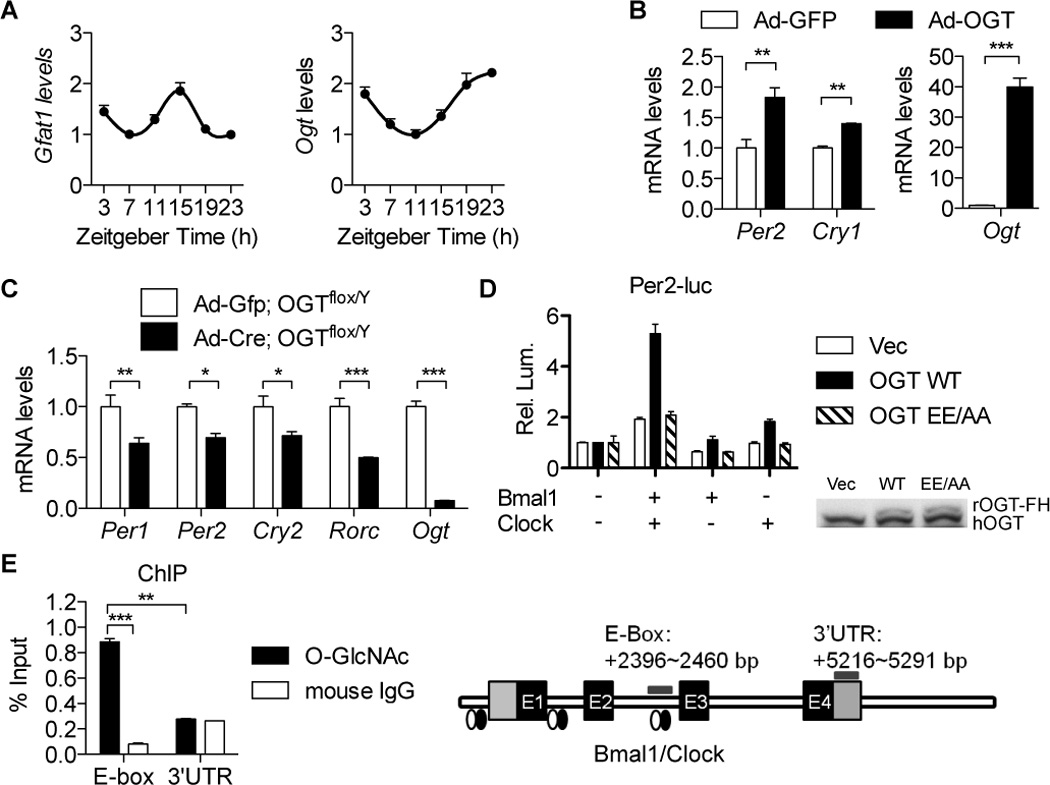
(A) Diurnal levels of Gfat1 and Ogt mRNA in mouse livers (n = 4). Data were normalized to that of u36b4. (B) RT-qPCR analysis of U2OS cells infected with the adenovirus expressing OGT (n = 3). (C) RT-qPCR analysis of OGTflox/Y mouse primary hepatocytes transduced with the adenoviral vector expressing Cre recombinase (n = 3). (D) Per2-luciferase assays of HeLa cells transiently expressing OGT, Myc-BMAL1, and Myc-CLOCK constructs (n = 3). GFP was used to equalize the total plasmid amount. Luminenscence signals were normalized to that of GFP control groups. Immunoblot analysis of cell lysates is shown to confirm overexpression of OGT. (E) Chromatin immunoprecipitation (ChIP)-qPCR analysis of mouse primary hepatocytes using an O-GlcNAc antibody (n = 3). ChIP with mouse IgG was used as the negative control. The diagram of assayed DNA regions is shown on the right. qPCR signals were normalized to those from genomic DNA inputs. All data are shown as mean ± SEM. *P <0.05, **P < 0.01, ***P < 0.001, post-hoc two-tailed Student's t test.
To investigate whether rhythmic hexosamine signaling affects expression of clock genes, we analyzed the endogenous gene expression in U2OS cells transiently expressing GFP or OGT from recombinant adenovirus vectors. The results show that OGT significantly increases expression of Per2 and Cry1 (Figure 2B). In contrast, Cre-induced homologous recombination in OGTflox/Y mouse primary hepatocytes that eliminates OGT expression decreases expression of BMAL1/CLOCK target genes, including Per1, Per2, Cry1 and Rorc genes (Figure 2C). Luciferase reporter assays using a Per2-luciferase construct reveal that OGT promotes BMAL1/CLOCK-mediated activation of Per2 transcription, whereas the catalytically dead OGTE899A/E900A (OGTEE/AA) has no effect (Figures 2D, S2B and S2C). These results indicate that OGT increases BMAL1/CLOCK transcriptional activity by an enzymatic mechanism, and thereby promotes expression of BMAL1/CLOCK target genes.
Rhythmic O-GlcNAcylation stabilizes BMAL1 and CLOCK by inhibiting their ubiquitination
Chromatin immunoprecipitation using an anti-O-GlcNAc antibody reveals that O-GlcNAcylated proteins associate with the known E-box elements (Ripperger and Schibler, 2006), the conserved motif recognized by BMAL1/CLOCK, in the promoter of the circadian gene Dbp (Figure 2E). This suggests that the BMAL1/CLOCK complex itself could be O-GlcNAcylated. To test this possibility, we immunoprecipitated epitope-tagged BMAL1 and CLOCK proteins expressed in synchronized U2OS cells and assayed the O-GlcNAc levels of BMAL1 and CLOCK over a complete circadian cycle. Immunoblot analysis reveals that both BMAL1 and CLOCK are O-GlcNAcylated rhythmically (Figure 3A). O-GlcNAcylation of endogenous CLOCK proteins is decreased by azaserine treatment (Figure S3A), supporting the importance of the hexosamine pathway in circadian regulation.
Figure 3. O-GlcNAcylation stabilizes CLOCK and BMAL1 by inhibiting ubiquitination.
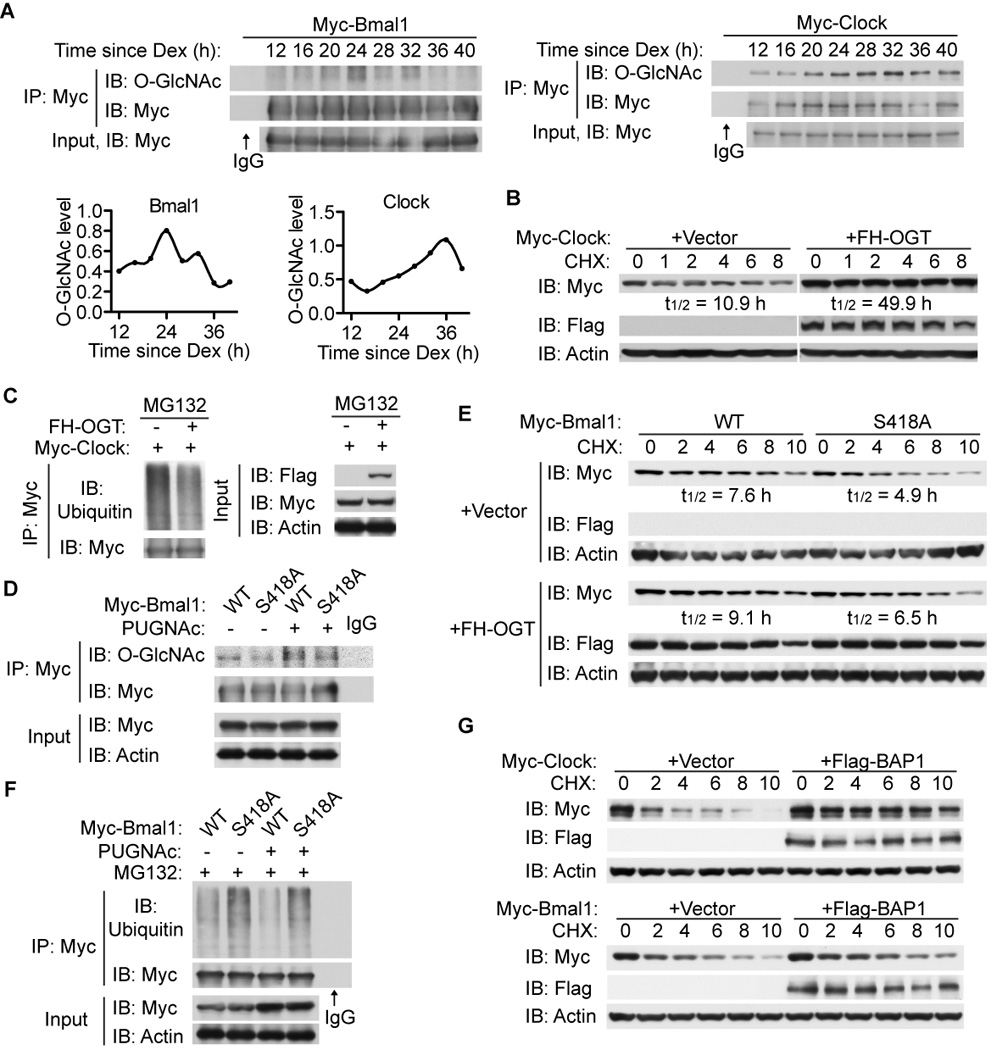
(A) Circadian O-GlcNAc levels of BMAL1/CLOCK in synchronized U2OS cells transiently expressing Myc-tagged BMAL1 or CLOCK. O-GlcNAc levels normalized to levels of BMAL1 and CLOCK proteins are shown below the BMAL1 blot. (B–C) HEK 293T cells were transfected with Myc-CLOCK in the absence or presence of Flag/HA (FH)-tagged OGT. (B) Immunoblot analysis of CLOCK upon cycloheximide (CHX) treatment. Half-lives of Clock are shown. (C) Immunoblot analysis of ubiquitination of CLOCK. Cells were pre-treated with MG132 and subjected to immunoprecipitation. (D) HEK 293T cells were transfected with Myc-tagged wildtype (WT) or S418A mutant BMAL1. O-GlcNAcylation of BMAL1 was determined when pre-treated with or without PUGNAc was determined. (E) Stability of WT and S418A BMAL1 in the absence or presence of FH-OGT was determined by CHX treatment of transfected HEK 293T cells. (F) Immunoblot analysis of ubiquitination of BMAL1 WT and S418A in HEK 293T cells. Cells were pre-treated with MG132 and subjected to immunoprecipitation. (G) Immunoblot analysis of Myc-tagged BMAL1/CLOCK upon CHX treatment in the presence or absence of Flag-BAP1.
It has been known that OGT can modulate protein stability (Dey et al., 2012; Ruan et al., 2012). Whether OGT regulates CLOCK stability was tested by treating HEK293T cells expressing Myc-tagged CLOCK in the presence or absence of exogenous OGT with cycloheximide (Figure 3B). OGT overexpression increases the estimated half-life of CLOCK proteins. In line with this, we found that OGT overexpression decreases the steady-state ubiquitination of CLOCK (Figure 3C).
As shown in Figure 3A, BMAL1 is also O-GlcNAcylated. The dbOGAP bioinformatic database predicts serine 418 (S418) in BMAL1 as a putative modification site (Wang et al., 2011) (Figure S3B). Mutation of S418 to alanine decreases but does not abolish O-GlcNAcylation of BMAL1 (Figure 3D). To investigate whether O-GlcNAc modification on BMAL1 regulates its stability, Myc-tagged wildtype (WT) BMAL1 or S418A mutant was transiently expressed in HEK293T cells in the presence or absence of exogenous OGT and treated with cycloheximide (Figure 3E). The BMAL1 S418A mutant degrades faster than the WT protein, and OGT overexpression increases the half-life of BMAL1, as it does on CLOCK (Figures 3B and 3E). To test whether O-GlcNAcylation regulates BMAL1 stability by inhibiting ubiquitination, HEK293T cells transiently expressing Myc-tagged BMAL1 WT or S418A were treated with the proteasome inhibitor MG132 in the presence or absence of the OGA inhibitor PUGNAc. Elevation of global O-GlcNAc levels by PUGNAc leads to decreased ubiquitination, and BMAL1 S418A has more attached ubiquitins than WT (Figure 3F). Per2-luciferase reporter assays show that BMAL1 S418A exhibits impaired transcriptional activity compared to WT when co-expressed with CLOCK (Figure S3C).
The nuclear deubiquitinase BRCA1-associated protein 1 (BAP1) has recently been characterized as an OGT-binding protein that removes the ubiquitin markers on other associated proteins (Dey et al., 2012; Ruan et al., 2012). It follows that OGT-targeted proteins are likely to be regulated by BAP1. Co-expression of BAP1 in HEK293T cells transiently expressing Myc-tagged BMAL1 or CLOCK reveals that BAP1 stabilizes both proteins (Figure 3G). Based on these results, we conclude that OGT stabilizes BMAL1 and CLOCK through direct O-GlcNAcylation which prevents ubiquitination and subsequent degradation.
Hepatic manipulation of OGT perturbs the diurnal rhythm of glucose homeostasis
In line with the results from U2OS cells (Figure 1C), immunoprecipitation analysis of O-GlcNAcylated proteins from mouse livers shows diurnal variations in O-GlcNAcylation of BMAL1 and CLOCK that peaks in the fed/dark phase (Figure 4A). Consistently, O-GlcNAcylation of hepatic BMAL1/CLOCK is increased by refeeding, confirming that O-GlcNAcylation of clock proteins is responsive to food availability (Figure S4A). To determine whether O-GlcNAcylation regulates circadian clocks in vivo, we generated liver-specific OGT overexpression mice by tail-vein injection of recombinant adenovirus (Figure 4B). Analysis of circadian transcripts in livers of these mice shows that OGT overexpression advances the phase of Bmal1 and Clock and increases expression levels of Per2, Cry1, Rorγ, and Dbp during the peak phase (Figures 4B and S4B, and Table S1), supporting the notion that O-GlcNAcylation increases BMAL1/CLOCK-mediated E-box-dependent transcription.
Figure 4. OGT regulates expression of clock genes and glucose homeostasis in mouse livers.
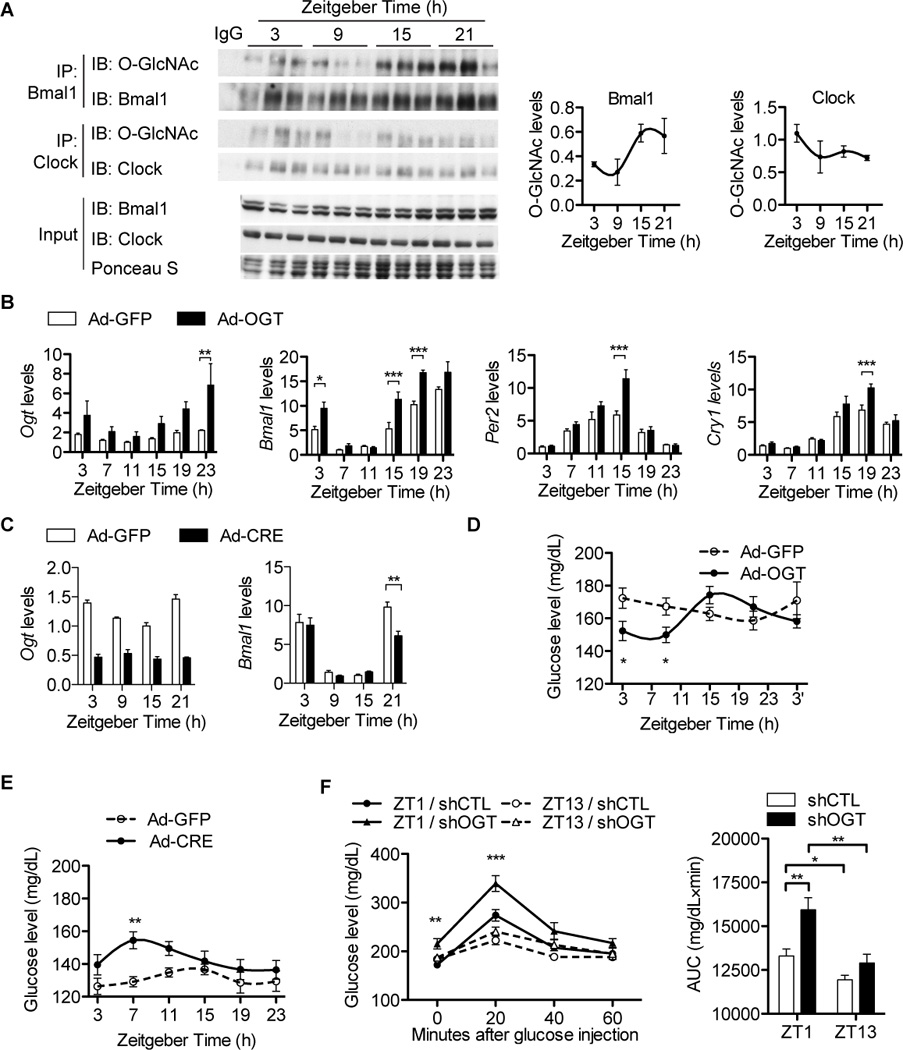
(A) Diurnal O-GlcNAc profiles of BMAL1/CLOCK in mouse livers (n = 3 per time point). Results of densitometry analysis are shown on the right. (B) Diurnal gene expression profiles of male mouse livers transduced with Ad-OGT (n = 4 per time point). Data were normalized to u36b4. (C) Diurnal gene expression profiles of Ogt-floxed female mouse livers transduced with Ad-Cre (n = 3 per time point). Data were normalized to Gapdh. (D) Diurnal plasma glucose levels in male mice overexpressing OGT in livers (n = 7). (E) Diurnal plasma glucose levels in female OGTflox/flox mice overexpressing Cre in livers (n = 12). (F) Diurnal plasma glucose responses to intraperitoneal glucose tolerance tests (GTT) at week 12 in male mice expressing scrambled-shRNA (shCTL) or OGT-shRNA (shOGT) in the liver (n = 7). Average areas under curve (AUC) of GTT curves are shown on the right. All data are shown as mean ± SEM. *P <0.05, **P < 0.01, ***P < 0.001, post-hoc Bonferroni’s multiple comparison test.
To study the effects of O-GlcNAc deficiency on clock oscillation, we generated liver-specific OGT knockout mice by tail-vein injection of the recombinant adenovirus expressing Cre recombinase into OGT floxed mice. Immunoblot analysis shows that BMAL1 and CLOCK exhibit decreased O-GlcNAc levels in mouse livers (Figure S4C). The oscillation of Bmal1 transcripts exhibits decreased amplitude, due to reduced peak levels (Figure 4C). However, Per2 and Cry1 oscillation is unchanged (Figure S4D), suggesting the existence of compensatory mechanisms. Together, these results indicate that O-GlcNAcylation regulates circadian rhythms of clock gene expression in vivo.
Nutrient-dependent peripheral clock entrainment allows metabolic tissues to optimize the timing of their metabolic processes. Accordingly, we examined whether glucose-responsive O-GlcNAc signaling in the liver affects diurnal rhythms of glucose metabolism. The results show that overexpression of OGT boosts the diurnal rhythm of blood glucose whereas control mice maintain a weak diurnal variation of circulating glucose (Figure 4D). Knockout of OGT advances the circulating glucose rhythm by 6–8 h and induces hyperglycemia in the daytime (Figure 4E). To assess the circadian metabolic effects of reduced O-GlcNAc signaling, we assayed the circadian responses of OGT deficient animals to intraperitoneal injection of a bolus of glucose. While control mice exhibit diurnal changes in glucose tolerance, depletion of OGT exacerbates the already poor glucose tolerance at ZT1 (1h after light–on), which is not seen at ZT13 (Figure 4F). Gluconeogenesis is known to be circadian. RT-qPCR analysis of liver transcripts shows that rhythmic expression of gluconeogenic genes is perturbed by OGT overexpression or depletion (Figures S4B and S4D). This indicates that O-GlcNAc signaling is important for diurnal regulation of glucose metabolism in vivo and support the conclusion that OGT acts as a nutrient-sensing mediator that resets peripheral circadian clocks.
DISCUSSION
Here we have shown that the hexosamine/O-GlcNAc pathway regulates the circadian clock in peripheral tissues. It has been known for a decade that diurnal variation in nutrient availability can override the light/dark cycle to entrain circadian rhythms in peripheral tissues (Damiola et al., 2000; Stokkan et al., 2001). How metabolic signals entrain the circadian clock remains a central question in circadian biology (Bass and Takahashi, 2010). Among macronutrients, glucose has a prominent role in metabolic entrainment (Hirota et al., 2002; Stephan and Davidson, 1998). Extracellular glucose levels modulate intracellular UDP-GlcNAc and subsequent O-GlcNAc levels through the hexosamine biosynthesis pathway (Figure S1A). OGT overexpression increases the amplitude of clock oscillation in vivo (Figure 4B) whereas OGT knockout decreases O-GlcNAcylation and protein abundance of BMAL1 and CLOCK and decreases Bmal1 oscillation (Figures 4C and S4B). Notably, depletion of OGT in liver fails to perturb oscillation of the core oscillator genes Per and Cry (Figure S4D). This conundrum may be explained by the dominant effect of cryptic oscillating systemic cues (Hughes et al., 2012; Kornmann et al., 2007). For instance, Hughes et al. found that recovery of the SCN clock in clock mutant mice is sufficient to re-establish the circadian rhythm of the liver clock. Whether O-GlcNAc signaling is integral to food entrainment in peripheral clocks is an important subject for further investigation.
We further demonstrate that O-GlcNAcylation on BMAL1 and CLOCK prevents their protein degradation by inhibiting ubiquitination. The control of BMAL1/CLOCK protein stability is emerging as a critical layer of regulation on the amplitude and phase of clock oscillation (Cardone et al., 2005; Lee et al., 2008; Sahar et al., 2010; Stratmann et al., 2012). We have demonstrated that O-GlcNAcylation stabilizes BMAL1/CLOCK and thereby increases BMAL1/CLOCK-mediated transcription of genes in the negative limb of the clock, such as Per and Cry (Figures 3 and 4). O-GlcNAcylation of PER and other components could further stabilize the negative limb (Kim et al., 2012). Together, our study helps establish a framework for understanding the crosstalk between different protein modifications on the positive limb of the circadian clock and provide a novel mechanism for food entrainment.
In physiological context, perturbation of the O-GlcNAc signaling in liver affects the diurnal rhythm of glucose homeostasis (Figures 4D–4F). OGT has been established as a suppressor of insulin signaling and a mediator of glucorticoid transrepression and gluconeogenesis (Dentin et al., 2008; Housley et al., 2009; Li et al., 2012; Ruan et al., 2012; Yang et al., 2008). Thus, changes in plasma glucose rhythm are likely due to the combined effects of OGT on the circadian clock and nutrient/hormone signaling.
In summary, the present study establishes the crosstalk between O-GlcNAcylation and ubiquitination as a key molecular mechanism underlying metabolic entrainment of the circadian clock, supporting the concept that various post-translational modifications on the clock proteins integrate environmental and physiological cues to control circadian rhythms. Diurnal rhythms of O-GlcNAc signaling have broad implications for the circadian regulation of physiological processes in peripheral tissues, and the O-GlcNAc cycling enzymes OGT and OGA are thus potential drug targets for treating disorders at the interface of nutrient metabolism and circadian rhythms.
EXPERIMENTAL PROCEDURES
Cell culture
U2OS, HeLa, and HEK 293T cells were maintained in high glucose DMEM with 10% fetal bovine serum (FBS). U2OS-B6 cells were maintained in high glucose DMEM with 2 µg/ml of Puromycin (Sigma) and 10% FBS. Primary hepatocytes were isolated by Yale Liver Center Core Facility and plated in DMEM with 10% FBS, 2 mM sodium pyruvate, 1 µM dexamethasome, 0.1 µM insulin on Collagen I coated plates. U2OS-B6 cells were transfected by lipofectamine 2000 (Invitrogen). U2OS, HeLa, and HEK 293T cells were transfected with FuGENE HD (Promega). For time course studies of BMAL1/CLOCK O-GlcNAc modifications, U2OS cells were transfected upon confluence, cultured for two days, then shocked by 100 nM dexamethosone for 90 min and switched to fresh high glucose DMEM medium with 10% FBS. For expression assays, primary hepatocytes and U2OS cells were infected with adenoviruses in serum-free DMEM medium containing 0.5% BSA. Azaserine (20 µM), D-glucosamine (5 mM), PUGNAc (10 µM, 16 h), MG132 (20 µM, 4 h), Cycloheximide (100 µg/ml) were added to the cultures as indicated.
Real-time recordings of bioluminescence
48 hours after transfection, cells were shocked for 90 min at 37°C in a final concentration of 100 nM dexamethasone. Following dexamethasone shock, the medium was replaced with high glucose Phenol Red-free DMEM (Gibco, supplemented with 10% FBS, 10 mM HEPES pH 7.3, non-essential amino acids, sodium pyruvate, and 100 µM D-Luciferin). The plate was sealed with a plastic cover and was read in a temperature-controlled TECAN infinite M200 Luminometer and iControl Software (Tecan Group, Ltd) (Vollmers et al., 2008). Luminescence for each well was integrated over 5 seconds and read at 30-minute intervals for 5 days at a temperature setting of 37 °C. Lumicycle data were statistically assessed for rhythmicity using JTK_Cycle (Hughes et al., 2010) using a period length window of 18–40 hours. Three days of data were used for analyses, spanning 24–96 h after changing to the assay culture media. JTK_Cycle was implemented in R (x64 v2.12.1) (Hughes et al., 2010; Miyazaki et al., 2011). All scripts are available on demand.
RNA extraction, cDNA synthesis, and real-time quantitative PCR
Procedures were described previously (Ruan et al., 2012). Q-PCR data were normalized to either u36b4 or Gapdh as indicated. Primer sequences were listed in Table S2.
Antibodies, immunoprecipitation and immunoblotting
Anti-Flag (F3165) and anti-β-Actin (A5441) antibodies were from Sigma. Anti-Bmal1 (A302-616A) and anti-Clock (A302-617A) were from Bethyl Laboratories. Anti-O-GlcNAc (RL2, ab2739) and anti-OGT (ab50270) were from Abcam. Anti-Ub (P4D1, sc-8017) and anti-Myc (9E10, sc-40) antibodies were from Santa Cruz Biotechnology. Anti-HA antibody (12CA5) was from Roche. Procedures for immunoprecipitation and immunoblotting assays were described previously (Ruan et al., 2012).
Chromatin immunoprecipitation
Procedures were described previously (Ruan et al., 2012). The 3′ UTR of Dbp was used as the negative control. A small aliquot of untreated sonicated chromatin was reverse cross-linked and used as the total input DNA control.
Animal studies
All procedures have been approved by the Institutional Animal Care and Use Committee of Yale University. Male C57Bl/6 mice (10-week old) were purchased from NCI/NIH. Female OGTflox/flox (5-months old) mice were generated previously (Shafi et al., 2000; Watson et al., 2010). Mice were maintained under 12h/12h light/dark cycle with free access to food and water. Recombinant adenoviruses (2 × 109 plaque forming units (pfu) for males, 5 × 108 pfu for females) was delivered by systemic tail-vein injection to mice. 3–6 days after viral infection, mice were subjected for glucose tolerance tests. Ad libitum fed male mice were injected with intraperitoneally with glucose (1.5 g/kg body weight) at ZT1 or ZT13. Blood glucose were measured from tail-vein blood collected at the designated times using Nova Max Glucometer. Tissues were collected for RNA and protein isolation.
Statistical Analysis
Data are presented as means ± SEM. Statistical analysis was with GraphPad Prism by ANOVA, post-hoc Bonferroni’s test or t test where appropriate. Statistical analysis was accepted as significant if P-value was < 0.05.
Supplementary Material
Highlights.
The hexosamine/O-GlcNAc pathway regulates cellular clock oscillation
OGT promotes expression of BMAL1/CLOCK target genes in cultured cells and liver
Rhythmic O-GlcNAcylation stabilizes BMAL1 and CLOCK by inhibiting their ubiquitination
Hepatic manipulation of OGT perturbs the diurnal rhythm of glucose homeostasis
ACKNOWLEDGEMENTS
We thank P. Sassone-Corsi for Myc-BMAL1 and Myc-CLOCK constructs, K. Lamia for pGL3-Per2, S. Panda for U2OS-B6 cell line. We are grateful to Clifford Slayman and Yao Wu for inspiring discussions. This work was supported by NIH R01 DK089098, American Diabetes Association, and Ellison Medical Foundation to X.Y., NIH R21 NS058330, R01 NS055035, R01 NS056443, and R01 GM098931 to M.N.N., NIH R01 HL083320, R01 HL094419, P01 HL078825, and P20 RR024489 to S.P.J., CSC-Yale World Scholars Program scholarship to M.L., Brown-Coxe fellowship to H.B.R. and NIH F32GM096577 to M.E.H..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION:
Supplemental information includes four figures, two tables, and additional experimental procedures.
REFERENCES
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell metabolism. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science (New York, NY) 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone L, Hirayama J, Giordano F, Tamaru T, Palvimo JJ, Sassone-Corsi P. Circadian clock control by SUMOylation of BMAL1. Science (New York, NY) 2005;309:1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & development. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science (New York, NY) 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, Kirkpatrick DS, Pham VC, Lill JR, Bakalarski CE, Wu J, Phu L, Katavolos P, Lafave LM, Abdel-Wahab O, Modrusan Z, Seshagiri S, Dong K, Lin Z, Balazs M, Suriben R, Newton K, Hymowitz S, Garcia-Manero G, Martin F, Levine RL, Dixit VM. Loss of the Tumor Suppressor BAP1 Causes Myeloid Transformation. Science. 2012 doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan DJ, Pat BM, Laczy B, Bradley JA, Tsai JY, Grenett MH, Ratcliffe WF, Brewer RA, Nagendran J, Villegas-Montoya C, Zou C, Zou L, Johnson RL, Jr, Dyck JR, Bray MS, Gamble KL, Chatham JC, Young ME. O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. The Journal of biological chemistry. 2011;286:44606–44619. doi: 10.1074/jbc.M111.278903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nature reviews.Molecular cell biology. 2012;13:312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between OGlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annual Review of Biochemistry. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. The Journal of biological chemistry. 2002;277:44244–44251. doi: 10.1074/jbc.M206233200. [DOI] [PubMed] [Google Scholar]
- Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, Hunt DF, Hart GW. A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. The Journal of biological chemistry. 2009;284:5148–5157. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of Biological Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hong HK, Chong JL, Indacochea AA, Lee SS, Han M, Takahashi JS, Hogenesch JB. Brain-specific rescue of Clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet. 2012;8:e1002835. doi: 10.1371/journal.pgen.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW. A role for OGlcNAcylation in setting circadian clock speed. Genes & development. 2012 doi: 10.1101/gad.182378.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee Y, Lee MJ, Park E, Kang SH, Chung CH, Lee KH, Kim K. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Molecular and cellular biology. 2008;28:6056–6065. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Ruan HB, Singh JP, Zhao L, Zhao T, Azarhoush S, Wu J, Evans RM, Yang X. O-GlcNAc transferase is involved in glucocorticoid receptor-mediated transrepression. J Biol Chem. 2012;287:12904–12912. doi: 10.1074/jbc.M111.303792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Schroder E, Edelmann SE, Hughes ME, Kornacker K, Balke CW, Esser KA. Age-associated disruption of molecular clock expression in skeletal muscle of the spontaneously hypertensive rat. PloS one. 2011;6:e27168. doi: 10.1371/journal.pone.0027168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annual review of neuroscience. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, Zhao L, Bennett AM, Samuel VT, Wu J, Yates JR, 3rd, Yang X. O-GlcNAc Transferase/Host Cell Factor C1 Complex Regulates Gluconeogenesis by Modulating PGC-1alpha Stability. Cell Metab. 2012;16:226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Zocchi L, Kinoshita C, Borrelli E, Sassone-Corsi P. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PloS one. 2010;5:e8561. doi: 10.1371/journal.pone.0008561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan FK, Davidson AJ. Glucose, but not fat phase shifts the feeding-entrained circadian clock. Physiology & Behavior. 1998;65:277–288. doi: 10.1016/s0031-9384(98)00166-8. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science (New York, NY) 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Stratmann M, Suter DM, Molina N, Naef F, Schibler U. Circadian Dbp Transcription Relies on Highly Dynamic BMAL1-CLOCK Interaction with E Boxes and Requires the Proteasome. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Panda S, DiTacchio L. A high-throughput assay for siRNA-based circadian screens in human U2OS cells. PloS one. 2008;3:e3457. doi: 10.1371/journal.pone.0003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Torii M, Liu H, Hart GW, Hu ZZ. dbOGAP - an integrated bioinformatics resource for protein O-GlcNAcylation. BMC bioinformatics. 2011;12:91. doi: 10.1186/1471-2105-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson LJ, Facundo HT, Ngoh GA, Ameen M, Brainard RE, Lemma KM, Long BW, Prabhu SD, Xuan YT, Jones SP. O-linked beta-N-acetylglucosamine transferase is indispensable in the failing heart. Proc Natl Acad Sci U S A. 2010;107:17797–17802. doi: 10.1073/pnas.1001907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. A wheel of time: the circadian clock, nuclear receptors, and physiology. Genes & development. 2010;24:741–747. doi: 10.1101/gad.1920710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, Evans RM. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


