Abstract
There has been an explosion of interest in studying gene-environment interactions (GxE) as they relate to the development of psychopathol-ogy. In this article, I review different methodologies to study gene-environment interaction, providing an overview of methods from animal and human studies and illustrations of gene-environment interactions detected using these various methodologies. Gene-environment interaction studies that examine genetic influences as modeled latently (e.g., from family, twin, and adoption studies) are covered, as well as studies of measured genotypes. Importantly, the explosion of interest in gene-environment interactions has raised a number of challenges, including difficulties with differentiating various types of interactions, power, and the scaling of environmental measures, which have profound implications for detecting gene-environment interactions. Taking research on gene-environment interactions to the next level will necessitate close collaborations between psychologists and geneticists so that each field can take advantage of the knowledge base of the other.
Keywords: genetics, association, review, GxE, psychopathology
INTRODUCTION
Gene-environment interaction (GxE) has become a hot topic of research, with an exponential increase in interest in this area in the past decade. Consider that PubMed lists only 24 citations for “gene environment interaction” prior to the year 2000, but nearly four times that many in the first half of the year 2010 alone! The projected publications on gene-environment interaction for 2008–2010 are on track to constitute more than 40% of the total number of publications on gene-environment interaction indexed in PubMed. Where does all this interest stem from? It may, in part, reflect a merging of interests from fields that were traditionally at odds with one another. Historically, there was a perception that behavior geneticists focused on genetic influences on behavior at the expense of studying environmental influences and that developmental psychologists focused on environmental influences and largely ignored genetic factors. Although this criticism is not entirely founded on the part of either field, methodological and ideological differences between these respective fields meant that genetic and environmental influences were traditionally studied in isolation. More recently, there has been recognition on the part of both of these fields that both genetic and environmental influences are critical components to developmental outcome and that it is far more fruitful to attempt to understand how these factors come together to impact psychological outcomes than to argue about which one is more important. As Kendler and Eaves argued in their article on the joint effect of genes and environments, published more than two decades ago:
It is our conviction that a complete understanding of the etiology of most psychiatric disorders will require an understanding of the relevant genetic risk factors, the relevant environmental risk factors, and the ways in which these two risk factors interact. Such understanding will only arise from research in which the important environmental variables are measured in a genetically informative design. Such research will require a synthesis of research traditions within psychiatry that have often been at odds with one another in the past. This interaction between the research tradition that has focused on the genetic etiology of psychiatric illness and that which has emphasized environmental causation will undoubtedly be to the benefit of both. (Kendler & Eaves 1986, p. 288)
The PubMed data showing an exponential increase in published papers on gene-environment interaction suggest that that day has arrived. This has been facilitated by the rapid advances that have taken place in the field of genetics, making the incorporation of genetic components into traditional psychological studies a relatively easy and inexpensive endeavor. But with this surge of interest in gene-environment interaction, a number of new complications have emerged, and the study of gene-environment interaction faces new challenges, including a recent backlash against studying gene-environment interaction (Risch et al. 2009). Addressing these challenges will be critical to moving research on gene-environment interaction forward in a productive way.
In this article, I first review different study designs for detecting gene-environment interaction, providing an overview of methods from animal and human studies. I cover gene-environment interaction studies that examine genetic influences as modeled latently as well as studies of measured genotypes. In the study of latent gene-environment interaction, specific genotypes are not measured, but rather genetic influence is inferred based on observed correlations between people who have different degrees of genetic and environmental sharing. Thus, latent gene-environment interaction studies examine the aggregate effects of genes rather than any one specific gene. Molecular genetic studies, in contrast, have generally focused on one specific gene of interest at a time. Relevant examples of gene-environment interaction across these different methodologies are provided, though these are meant to be more illustrative than exhaustive, intended to introduce the reader to relevant studies and findings generated across these various designs. Subsequently I review more conceptual issues surrounding the study of gene-environment interaction, covering the nature of gene-environment interaction effects as well as the challenges facing the study of gene-environment interaction, such as difficulties with differentiating various types of interactions, and how issues such as the scaling of environmental measures can have profound implications for studying gene-environment interaction. I include an overview of epigenet-ics, a relatively new area of study that provides a potential biological mechanism by which the environment can moderate gene expression and affect behavior. Finally, I conclude with recommendations for future directions and how we can take research on gene-environment interaction to the next level.
DEFINING GENE-ENVIRONMENT INTERACTION AND DIFFERENTIATING GENE-ENVIRONMENT CORRELATION
It is important to first address some aspects of terminology surrounding the study of gene-environment interaction. In lay terms, the phrase gene-environment interaction is often used to mean that both genes and environments are important. In statistical terms, this does not necessarily indicate an interaction but could be consistent with an additive model, in which there are main effects of the environment and main effects of genes. But in a statistical sense an interaction is a very specific thing, referring to a situation in which the effect of one variable cannot be understood without taking into account the other variable. Their effects are not independent. When we refer to gene-environment interaction in a statistical sense, we are referring to a situation in which the effect of genes depends on the environment and/or the effect of the environment depends on genotype. We note that these two alternative conceptualizations of gene-environment interaction are indistinguishable statistically. It is this statistical definition of gene-environment interaction that is the primary focus of this review (except where otherwise noted).
It is also important to note that genetic and environmental influences are not necessarily independent factors. That is to say that although some environmental influences may be largely random, such as experiencing a natural disaster, many environmental influences are not entirely random (Kendler et al. 1993). This phenomenon is called gene-environment correlation. Three specific ways by which genes may exert an effect on the environment have been delineated (Plomin et al. 1977, Scarr & McCartney 1983): (a) Passive gene-environment correlation refers to the fact that among biologically related relatives (i.e., nonadoptive families), parents provide not only their children’s genotypes but also their rearing environment. Therefore, the child’s genotype and home environment are correlated. (b) Evocative gene-environment correlation refers to the idea that individuals’ genotypes influence the responses they receive from others. For example, a child who is predisposed to having an outgoing, cheerful disposition might be more likely to receive positive attention from others than a child who is predisposed to timidity and tears. A person with a grumpy, abrasive temperament is more likely to evoke unpleasant responses from coworkers and others with whom he/she interacts than is a cheerful, friendly person. Thus, evocative gene-environment correlation can influence the way an individual experiences the world. (c) Finally, active gene-environment correlation refers to the fact that an individual actively selects certain environments and takes away different things from his/her environment, and these processes are influenced by an individual’s genotype. Therefore, an individual predisposed to high sensation seeking may be more prone to attend parties and meet new people, thereby actively influencing the environments he/she experiences. Evidence exists in the literature for each of these processes. The important point is that many sources of behavioral influence that we might consider “environmental” are actually under a degree of genetic influence (Kendler & Baker 2007), so often genetic and environmental influences do not represent independent sources of influ-ence. This also makes it difficult to determine whether the genes or the environment is the causal agent. If, for example, individuals are genetically predisposed toward sensation seeking, and this makes them more likely to spend time in bars (a gene-environment correlation), and this increases their risk for alcohol problems, are the predisposing sensation-seeking genes or the bar environment the causal agent? In actuality, the question is moot—they both played a role; it is much more informative to try to understand the pathways of risk than to ask whether the genes or the environment was the critical factor. Though this review focuses on gene-environment interaction, it is important for the reader to be aware that this is but one process by which genetic and environmental influences are intertwined. Additionally, gene-environment correlation must be taken into account when studying gene-environment interaction, a point that is mentioned again later in this review. Excellent reviews covering the nature and importance of gene-environment correlation also exist (Kendler 2011).
METHODS FOR STUDYING GENE-ENVIRONMENT INTERACTION
Animal Research
Perhaps the most straightforward method for detecting gene-environment interaction is found in animal experimentation: Different genetic strains of animals can be subjected to different environments to directly test for gene-environment interaction. The key advantage of animal studies is that environmental exposure can be made random to genotype, eliminating gene-environment correlation and associated problems with interpretation. The most widely cited example of this line of research is Cooper and Zubek’s 1958 experiment, in which rats were selectively bred to perform differently in a maze-running experiment (Cooper & Zubek 1958). Under standard environmental conditions, one group of rats consistently performed with few errors (“maze bright”), while a second group committed many errors (“maze dull”). These selectively bred rats were then exposed to various environmental conditions: an enriched condition, in which rats were reared in brightly colored cages with many moveable objects, or a restricted condition, in which there were no colors or toys. The enriched condition had no effect on the maze bright rats, although it substantially improved the performance of the maze dull rats, such that there was no difference between the groups. Conversely, the restrictive environment did not affect the performance of the maze dull rats, but it substantially diminished the performance of the maze bright rats, again yielding no difference between the groups and demonstrating a powerful gene-environment interaction. A series of experiments conducted by Henderson on inbred strains of mice, in which environmental enrichment was manipulated, also provides evidence for gene-environment interaction on several behavioral tasks (Henderson 1970, 1972). These studies laid the foundation for many future studies, which collectively demonstrate that environmental variation can have considerable differential impact on outcome depending on the genetic make-up of the animal (Wahlsten et al. 2003). However, animal studies are not without their limitations. Gene-environment interaction effects detected in animal studies are still subject to the problem of scale (Mather & Jinks 1982), as discussed in greater detail later in this review.
Human Research
Traditional behavior genetic designs
Demonstrating gene-environment interaction in humans has been considerably more difficult where ethical constraints require researchers to make use of natural experiments so environmental exposures are not random. Three traditional study designs have been used to demonstrate genetic influence on behavior: family studies, adoption studies, and twin studies. These designs have been used to detect gene-environment interaction also, and each is discussed in turn.
Family studies
Demonstration that a behavior aggregates in families is the first step in establishing a genetic basis for a disorder (Hewitt & Turner 1995). Decreasing similarity with decreasing degrees of relatedness lends support to genetic influence on a behavior (Gottesman 1991). This is a necessary, but not sufficient, condition for heritability. Similarity among family members is due both to shared genes and shared environment; family studies cannot tease apart these two sources of variance to determine whether familiality is due to genetic or common environmental causes (Sherman et al. 1997). However, family studies provide a powerful method for identifying gene-environment interaction. By comparing high-risk children, identified as such by the presence of psy-chopathology in their parents, with a control group of low-risk individuals, it is possible to test the effects of environmental characteristics on individuals varying in genetic risk (Cannon et al. 1990). In a high-risk study of Danish children with schizophrenic mothers and matched controls, institutional rearing was associated with an elevated risk of schizophrenia only among those children with a genetic predisposition (Cannon et al. 1990). When these subjects were further classified on genetic risk as having one or two affected parents, a significant interaction emerged between degree of genetic risk and birth complications in predicting ventricle enlargement: The relationship between obstetric complications and ventricular enlargement was greater in the group of individuals with one affected parent as compared to controls, and greater still in the group of individuals with two affected parents (Cannon et al. 1993). Another study also found that among individuals at high risk for schizophrenia, experiencing obstetric complications was related to an earlier hospitalization (Malaspina et al. 1999).
Another creative method has made use of the natural experiment of family migration to demonstrate gene-environment interaction: The high rate of schizophrenia among African-Caribbean individuals who emigrated to the United Kingdom is presumed to result from gene-environment interaction. Parents and siblings of first-generation African-Caribbean probands have risks of schizophrenia similar to those for white individuals in the area. However, the siblings of second-generation African-Caribbean probands have markedly elevated rates of schizophrenia, suggesting that the increase in schizophrenia rates is due to an interaction between genetic predispositions and stressful environmental factors encountered by this population (Malaspina et al. 1999, Moldin & Gottesman 1997). Although family studies provide a powerful design for demonstrating gene-environment interaction, there are limitations to their utility. High-risk studies are very expensive to conduct because they require the examination of individuals over a long period of time. Additionally, a large number of high-risk individuals must be studied in order to obtain a sufficient number of individuals who eventually become affected, due to the low base rate of most mental disorders. Because of these limitations, few examples of high-risk studies exist.
Adoption studies
Adoption and twin studies are able to clarify the extent to which similarity among family members is due to shared genes versus shared environment. In their simplest form, adoption studies involve comparing the extent to which adoptees resemble their biological relatives, with whom they share genes but not family environment, with the extent to which adoptees resemble their adoptive relatives, with whom they share family environment but not genes. Adoption studies have been pivotal in advancing our understanding of the etiology of many disorders and drawing attention to the importance of genetic factors. For example, Heston’s historic adoption study was critical in dispelling the myth of schizophrenogenic mothers in favor of a genetic transmission explaining the familiality of schizophrenia (Heston & Denney 1967). Furthermore, adoption studies provide a powerful method of detecting gene-environment interactions and have been called the human analogue of strain-by-treatment animal studies (Plomin & Hershberger 1991). The genotype of adopted children is inferred from their biological parents, and the environment is measured in the adoptive home. Individuals thought to be at genetic risk for a disorder, but reared in adoptive homes with different environments, are compared to each other and to control adoptees. This methodology has been employed by a number of research groups to document gene-environment interactions in a variety of clinical disorders: In a series of Iowa adoption studies, Cadoret and colleagues demonstrated that a genetic predisposition to alcohol abuse predicted major depression in females only among adoptees who also experienced a disturbed environment, as defined by psychopathology, divorce, or legal problems among the adoptive parents (Cadoret et al. 1996). In another study, depression scores and manic symptoms were found to be higher among individuals with a genetic predisposition and a later age of adoption (suggesting a more transient and stressful childhood) than among those with only a genetic predisposition (Cadoret et al. 1990). In an adoption study of Swedish men, mild and severe alcohol abuse were more prevalent only among men who had both a genetic predisposition and more disadvantaged adoptive environments (Cloninger et al. 1981). The Finnish Adoptive Family Study of Schizophrenia found that high genetic risk was associated with increased risk of schizophrenic thought disorder only when combined with communication deviance in the adoptive family (Wahlberg et al. 1997). Additionally, the adoptees had a greater risk of psychological disturbance, defined as neuroticism, personality disorders, and psychoticism, when the adoptive family environment was disturbed (Tienari et al. 1990). These studies have demonstrated that genetic predispositions for a number of psychiatric disorders interact with environmental influences to manifest disorder.
However, adoption studies suffer from a number of methodological limitations. Adoptive parents and biological parents of adoptees are often not representative of the general population. Adoptive parents tend to be socioeconomically advantaged and have lower rates of mental problems, due to the extensive screening procedures conducted by adoption agencies (Kendler 1993). Biological parents of adoptees tend to be atypical, as well, but in the opposite way. Additionally, selective placement by adoption agencies is confounding the clear-cut separation between genetic and environmental effects by matching adoptees and adoptive parents on demographics, such as race and religion. An increasing number of adoptions are also allowing contact between the biological parents and adoptive children, further confounding the traditional genetic and environmental separation that made adoption studies useful for genetically informative research. Finally, greater contraceptive use is making adoption increasingly rare (Martin et al. 1997). Accordingly, this research strategy has become increasingly challenging, though a number of current adoption studies continue to make important contributions to the field (Leve et al. 2010; McGue et al. 1995, 1996).
Twin studies
Twins provide a number of ways to study gene-environment interaction. One such method is to study monozygotic twins reared apart (MZA). MZAs provide a unique opportunity to study the influence of different environments on identical genotypes. In the Swedish Adoption/Twin Study of Aging, data from 99 pairs of MZAs were tested for interactions between childhood rearing and adult personality (Bergeman et al. 1988). Several significant interactions emerged. In some cases, the environment had a stronger impact on individuals genetically predisposed to be low on a given trait (based on the cotwin’s score). For example, individuals high in extraversion expressed the trait regardless of the environment; however, individuals predisposed to low extraversion had even lower scores in the presence of a controlling family. In other traits, the environment had a greater impact on individuals genetically predisposed to be high on the trait: Individuals predisposed to impulsivity were even more impulsive in a conflictual family environment; individuals low on impulsivity were not affected. Finally, some environments influenced both individuals who were high and low on a given trait, but in opposite directions: Families that were more involved masked genetic differences between individuals predisposed toward high or low neuroticism, but greater genetic variation emerged in less controlling families.
The implementation of population-based twin studies, inclusion of measured environments into twin studies, and advances in biometrical modeling techniques for twin data made it possible to study gene-environment interaction within the framework of the classic twin study. Traditional twin studies involve comparisons of monozygotic (MZ) and dizy-gotic (DZ) twins reared together. MZ twins share all of their genetic variation, whereas DZ twins share on average 50% of their genetic make-up; however, both types of twins are age-matched siblings sharing their family environments. This allows heritability, or the proportion of variance attributed to additive genetic effects, to be estimated by (a) doubling the difference between the correlation found between MZ twins and the correlation found between DZ twins, for quantitative traits, or ( b ) comparing concordance rates between MZs and DZs, for qualitative disorders (McGue & Bouchard 1998). Biometrical model-fitting made it possible for researchers to address increasingly sophisticated research questions by allowing one to statistically specify predictions made by various hypotheses and to compare models testing competing hypotheses. By modeling data from subjects who vary on exposure to a specified environment, one could test whether there is differential expression of genetic influences in different environments.
Early examples of gene-environment interaction in twin models necessitated “grouping” environments to fit multiple group models. The basic idea was simple: Fit models to data for people in environment 1 and environment 2 separately and then test whether there were significant differences in the importance of genetic and environmental factors across the groups using basic structural equation modeling techniques. In an early example of gene-environment interaction, data from the Australian twin register were used to test whether the relative importance of genetic effects on alcohol consumption varied as a function of marital status, and in fact they did (Heath et al. 1989). Having a marriage-like relationship reduced the impact of genetic influences on drinking: Among the younger sample of twins, genetic liability accounted for but half as much variance in drinking among married women (31%) as among unmarried women (60%). A parallel effect was found among the adult twins: Genetic effects accounted for less than 60% of the variance in married respondents but more than 76% in unmarried respondents (Heath et al. 1989). In an independent sample of Dutch twins, religiosity was also shown to moderate genetic and environmental influences on alcohol use initiation in females (with nonsignificant trends in the same direction for males): In females without a religious upbringing, genetic influences accounted for 40% of the variance in alcohol use initiation compared to 0% in religiously raised females. Shared environmental influences were far more important in the religious females (Koopmans et al. 1999). In data from our population-based Finnish twin sample, we also found that regional residency moderates the impact of genetic and environmental influences on alcohol use. Genetic effects played a larger role in longitudinal drinking patterns from late adolescence to early adulthood among individuals residing in urban settings, whereas common environmental effects exerted a greater in-fluence across this age range among individuals in rural settings (Rose et al. 2001). When one has pairs discordant for exposure, it is also possible to ask about genetic correlation between traits displayed in different environments.
One obvious limitation of modeling gene-environment interaction in this way was that it constrained investigation to environments that fell into natural groupings (e.g., married/unmarried; urban/rural) or it forced investigators to create groups based on environments that may actually be more continuous in nature (e.g., religiosity). In the first extension of this work to quasi-continuous environmental moderation, we developed a model that allowed genetic and environmental influences to vary as a function of a continuous environmental moderator and used this model to follow-up on the urban/rural interaction reported previously (Dick et al. 2001). We believed it likely that the urban/rural moderation effect reflected a composite of different processes at work. Accordingly, we expanded the analyses to incorporate more specific information about neighborhood environments, using government-collected information about the specific municipalities in which the twins resided (Dick et al. 2001). We found that genetic influences were stronger in environments characterized by higher rates of migration in and out of the municipality; conversely, shared environmental influences predominated in local communities characterized by little migration. We also found that genetic predispositions were stronger in communities composed of a higher percentage of young adults slightly older than our age-18 Finnish twins and in regions where there were higher alcohol sales. Further, the magnitude of genetic moderation observed in these models that allowed for variation as a function of a quasi-continuous environmental moderator was striking, with nearly a fivefold difference in the magnitude of genetic effects between environmental extremes in some cases.
The publication of a paper the following year (Purcell 2002) that provided straightforward scripts for continuous gene-environment interaction models using the most widely used program for twin analyses, Mx (Neale 2000), led to a surge of papers studying gene-environment interaction in the twin literature. These scripts also offered the advantage of being able to take into account gene-environment correlation in the context of gene-environment interaction. This was an important advance because previous examples of gene-environment interaction in twin models had been limited to environments that showed no evidence of genetic effects so as to avoid the confounding of gene-environment interaction with gene-environment correlation. Using these models, we have demonstrated that genetic influences on adolescent substance use are enhanced in environments with lower parental monitoring (Dick et al. 2007c) and in the presence of substance-using friends (Dick et al. 2007b). Similar effects have been demonstrated for more general externalizing behavior: Genetic influences on antisocial behavior were higher in the presence of delinquent peers (Button et al. 2007) and in environments characterized by high parental negativity (Feinberg et al. 2007), low parental warmth (Feinberg et al. 2007), and high paternal punitive discipline (Button et al. 2008). Further, in an extension of the socioregional-moderating effects observed on age-18 alcohol use, we found a parallel moderating role of these socioregional variables on age-14 behavior problems in girls in a younger Finnish twin sample. Genetic influences assumed greater importance in urban settings, communities with greater migration, and communities with a higher percentage of slightly older adolescents.
Other psychological outcomes have also yielded significant evidence of gene-environment interaction effects in the twin literature. For example, a moderating effect, parallel to that reported for alcohol consumption above, has been reported for depression symptoms (Heath et al. 1998) in females. A marriage-like relationship reduced the influence of genetic liability to depression symptoms, paralleling the effect found for alcohol consumption: Genetic factors accounted for 29% of the variance in depression scores among married women, but for 42% of the variance in young unmarried females and 51% of the variance in older unmarried females (Heath et al. 1998). Life events were also found to moderate the impact of factors influencing depression in females (Kendler et al. 1991). Genetic and/or shared environmental influ-ences were significantly more important in influencing depression in high-stress than in low-stress environments, as defined by a median split on a life-event inventory, although there was insufficient power to determine whether the moderating influence was on genetic or environmental effects.
More than simply accumulating examples of moderation of genetic influence by environmental factors, efforts have been made to integrate this work into theoretical frameworks surrounding the etiology of different clinical conditions. This is critical if science is to advance beyond individual observations to testable broad theories. A 2005 review paper by Shanahan and Hofer suggested four processes by which social context may moderate the relative importance of genetic effects (Shanahan & Hofer 2005). The environment may (a) trigger or (b) compensate for a genetic predisposition, (c) control the expression of a genetic predisposition, or (d ) enhance a genetic predisposition (referring to the accentuation of “positive” genetic predispositions). These processes are not mutually exclusive and can represent different ends of a continuum. For example, the interaction between genetic susceptibility and life events may represent a situation whereby the experience of life events triggers a genetic susceptibility to depression. Conversely, “protective” environments, such as marriage-like relationships and low stress levels, can buffer against or reduce the impact of genetic predispositions to depressive problems. Many different processes are likely involved in the gene-environment interactions observed for substance use and antisocial behavior. For example, family environment and peer substance use/delinquency likely constitute a spectrum of risk or protection, and family/friend environments that are at the “poor” extreme may trigger genetic predispositions toward substance use and antisocial behavior, whereas positive family and friend relationships may compensate for genetic predispositions toward substance use and antisocial behavior. Social control also appears to be a particularly relevant process in substance use, as it is likely that being in a marriage-like relationship and/or being raised with a religious upbringing exert social norms that constrain behavior and thereby reduce genetic predispositions toward substance use. Further, the availability of the substance also serves as a level of control over the ability to express genetic predispositions, and accordingly, the degree to which genetic influences will be apparent on an outcome at the population level. In a compelling illustration of this effect, Boardman and colleagues used twin data from the National Survey of Midlife Development in the United States and found a significant reduction in the importance of genetic influences on people who smoke regularly following legislation prohibiting smoking in public places (Boardman et al. 2010).
Molecular analyses
All of the analyses discussed thus far use latent, unmeasured indices of genetic influence to detect the possible presence of gene-environment interaction. This is largely because it was possible to test for the presence of latent genetic influence in humans (via comparisons of correlations between relatives with different degrees of genetic sharing) long before molecular genetics yielded the techniques necessary to identify specific genes influencing complex psychological disorders. However, recent advances have made the collection of deoxyribonucleic acid (DNA) and resultant genotyping relatively cheap and straightforward. Additionally, the publication of high-profile papers brought gene-environment interaction to the forefront of mainstream psychology. In a pair of papers published in Science in 2002 and 2003, respectively, Caspi and colleagues analyzed data from a prospective, longitudinal sample from a birth cohort from New Zealand, followed from birth through adulthood. In the 2002 paper, they reported that a functional polymorphism in the gene encoding the neurotransmitter-metabolizing enzyme monoamine oxidase A (MAOA) moderated the effect of maltreatment: Males who carried the genotype conferring high levels of MAOA expression were less likely to develop antisocial problems when exposed to maltreatment (Caspi et al. 2002). In the 2003 paper, they reported that a functional polymorphism in the promoter region of the serotonin transporter gene (5-HTT) was found to moderate the influence of stressful life events on depression. Individuals carrying the short allele of the 5-HTT promoter polymorphism exhibited more depressive symptoms, diagnosable depression, and suicidality in relation to stressful life events than did individuals homozygous for the long allele (Caspi et al. 2003). Both studies were significant in demonstrating that genetic variation can moderate individuals’ sensitivity to environmental events. These studies sparked a multitude of reports that aimed to replicate, or to further extend and explore, the findings of the original papers, resulting in huge literatures surrounding each reported gene-environment interaction in the years since the original publications (e.g., Edwards et al. 2009, Enoch et al. 2010, Frazzetto et al. 2007, Kim-Cohen et al. 2006, McDermott et al. 2009, Prom-Wormley et al. 2009, Vanyukov et al. 2007, Weder et al. 2009). It is beyond the scope of this review to detail these studies; however, of note was the publication in 2009 of a highly publicized meta-analysis of the interaction between 5-HTT, stressful life events, and risk of depression that concluded there was “no evidence that the serotonin transporter genotype alone or in interaction with stressful life events is associated with an elevated risk of depression in men alone, women alone, or in both sexes combined” (Risch et al. 2009). Further, the authors were critical of the rapid embracing of gene-environment interaction and the substantial resources that have been devoted to this research. The paper stimulated considerable backlash against the study of gene-environment interactions, and the pendulum appeared to be swinging back the other direction. However, a recent review by Caspi and colleagues entitled “Genetic Sensitivity to the Environment: The Case of the Serotonin Transporter Gene and Its Implications for Studying Complex Diseases and Traits” highlighted the fact that evidence for involvement of 5-HTT in stress sensitivity comes from at least four different types of studies, including observational studies in humans, experimental neuroscience studies, studies in nonhuman primates, and studies of 5-HTT mutations in rodents (Caspi et al. 2010). Further, the authors made the distinction between different cultures of evaluating gene-environment interactions: a purely statistical (theory-free) approach that relies wholly on meta-analysis (e.g., such as that taken by Risch et al. 2009) versus a construct-validity (theory-guided) approach that looks for a nomological network of convergent evidence, such as the approach that they took.
It is likely that this distinction also reflects differences in training and emphasis across different fields. The most cutting-edge genetic strategies at any given point, though they have changed drastically and rapidly over the past several decades, have generally involved athe-oretical methods for gene identification (Neale et al. 2008). This was true of early linkage analyses, where ~400 to 1,000 markers were scanned across the genome to search for chromosomal regions that were shared by affected family members, suggesting there may be a gene in that region that harbored risk for the particular outcome under study. This allowed geneticists to search for genes without having to know anything about the underlying biology, with the ideas that the identification of risk genes would be informative as to etiological processes and that our understanding of the biology of most psychiatric conditions is limited. Although it is now recognized that linkage studies were underpowered to detect genes of small effect, such as those now thought to be operating in psychiatric conditions, this atheoretical approach was retained in the next generation of gene-finding methods that replaced linkage—the implementation of genome-wide association studies (GWAS) (Cardon 2006). GWAS also have the general framework of scanning markers located across the entire genome in an effort to detect association between genetic markers and disease status; however, in GWAS over a million markers (or more, on the newest genetic platforms) are analyzed. The next technique on the horizon is sequencing, in which entire stretches of DNA are sequenced to know the exact base pair sequence for a given region (McKenna et al. 2010). From linkage to sequencing, common across all these techniques is an atheoretical framework for finding genes that necessarily involves conducting very large numbers of tests. Accordingly, there has been great emphasis in the field of genetics on correction for multiple testing (van den Oord 2007). In addition, the estimated magnitude of effect size of genetic variants thought to influence complex behavioral outcomes has been continually shifted downward as studies that were sufficiently powered to detect effect sizes previously thought to be reasonable have failed to generate positive findings (Manolio et al. 2009). GWAS have led the field to believe that genes influencing complex behavioral outcomes likely have odds ratios (ORs) on the order of magnitude of 1.1. This has led to a need for incredibly large sample sizes, requiring meta-analytic GWAS efforts with several tens of thousands of subjects (Landi et al. 2009, Lindgren et al. 2009).
It is important to note there has been increasing attention to the topic of gene-environment interaction from geneticists (Engelman et al. 2009). This likely reflects, in part, frustration and difficulty with identifying genes that impact complex psychiatric outcomes. Several hypotheses have been put forth as possible explanations for the failure to robustly detect genes involved in psychiatric outcomes, including a genetic model involving far more genes, each of very small effect, than was previously recognized, and failure to pay adequate attention to rare variants, copy number variants, and gene-environment interaction (Manolio et al. 2009). Accordingly, gene-environment interaction is being discussed far more in the area of gene finding than in years past; however, these discussions often involve atheoretical approaches and center on methods to adequately detect gene-environment interaction in the presence of extensive multiple testing (Gauderman 2002, Gauderman et al. 2010). The papers by Risch et al. (2009) and Caspi et al. (2010) on the interaction between 5-HTT, life stress, and depression highlight the conceptual, theoretical, and practical differences that continue to exist between the fields of genetics and psychology surrounding the identification of gene-environment interaction effects.
THE NATURE OF GENE-ENVIRONMENT INTERACTION
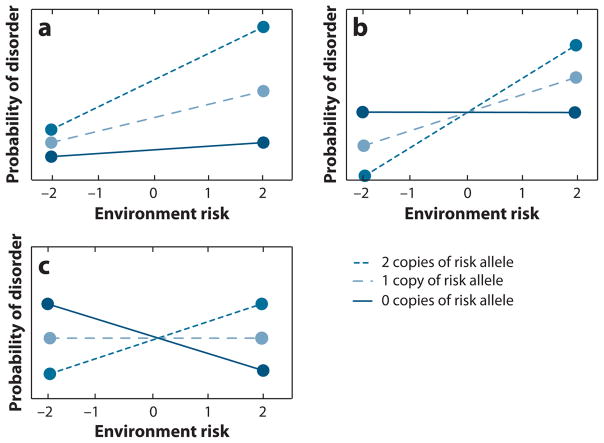
An important consideration in the study of gene-environment interaction is the nature, or shape, of the interaction that one hypothesizes. Figure 1 illustrates the two primary types of interactions. One type of interaction is the fan-shaped interaction (Figure 1a). In this type of interaction, the influence of genotype is greater in one environmental context than in another. This is the kind of interaction that is hypothesized by a diathesis-stress framework, whereby genetic influences become more apparent, i.e., are more strongly related to outcome, in the presence of negative environmental conditions. There is a reduced (or no) association of genotype with outcome in the absence of exposure to particular environmental conditions. The literature surrounding depression and life events would be an example of a hypothesized fan-shaped interaction: When life stressors are encountered, genetically vulnerable individuals are more prone to developing depression, whereas in the absence of life stressors, these individuals may be no more likely to develop depression. In essence, it is only when adverse environmental conditions are experienced that the genes “come on-line.” Gene-environment interactions in the area of adolescent substance use are also hypothesized to be fan-shaped, where some environmental conditions will allow greater opportunity to express genetic predispositions (allowing for more variation by genotype as in the right side of Figure 1a), and other environments will exert social control in such a way as to curb genetic expression (Shanahan & Hofer 2005), leading to reduced genetic variance (as on the left side of Figure 1a). Twin analyses yielding evidence of genetic influences being more or less important in different environmental contexts are generally suggestive of fan-shaped interactions. Changes in the overall heritability do not necessarily dictate that any one specific susceptibility gene will operate in a parallel manner; however, a change in heritability suggests that at least a good portion of the involved genes (assuming many genes of approximately equal and small effect) must be operating in that manner for a difference in heritability by environment to be detectable. The diathesis-stress model has largely been the dominant model in psychiatry. Gene-finding efforts have focused on the search for vulnerability genes, and gene-environment interaction has been discussed in the context of these genetic effects becoming more or less important under particular environmental conditions.
Figure 1.
Different types of gene-environment interactions.
More recently, an alternative framework has been proposed by Belsky and colleagues—the differential susceptibility hypothesis—in which the same individuals who are most adversely affected by negative environments may also be those who are most likely to benefit from positive environments. Rather than searching for “vulnerability genes” influencing psychiatric and behavioral outcomes, they propose the idea of “plasticity genes,” or genes involved in responsivity to environmental conditions (Belsky et al. 2009). Belsky and colleagues reviewed the literatures surrounding gene-environment interactions associated with three widely studied candidate genes, MAOA, 5-HTT, and DRD4, and suggested that the results provide evidence for differential susceptibility associated with these genes (Belsky et al. 2009). Their hypothesis is closely related to the concept of biological sensitivity to context (Ellis & Boyce 2008). The idea of biological sensitivity to context has its roots in evolutionary developmental biology, whereby selection pressures should favor genotypes that support a range of phenotypes in response to environmental conditions because this flexibility would be beneficial from the perspective of survival of the species. However, biological sensitivity to context has the potential for both positive effects under more highly supportive environmental conditions and negative effects in the presence of more negative environmental conditions. This theory has been most fully developed and discussed in the context of stress reactivity (Boyce & Ellis 2005), where it has been demonstrated that highly reactive children show disproportionate rates of morbidity when raised in adverse environments, but particularly low rates when raised in low-stress, highly supportive environments (Ellis et al. 2005). In these studies, high reactivity was defined by response to different laboratory challenges, and the authors noted that the underlying cellular mechanisms that would produce such responses are currently unknown, though genetic factors are likely to play a role (Ellis & Boyce 2008).
Although fan-shaped and crossover interactions are theoretically different, in practice, they can be quite difficult to differentiate. In looking at Figures 1a and b, one can imagine several “variations on the theme” for both fan-shaped and crossover interactions. In general for a fan-shaped interaction, a main effect of genotype will be present as well as a main effect of the environment. In the fan-shaped interaction shown in Figure 1a, there is a main effect of genotype at both environmental extremes; it is simply far stronger in environment 2 (far right side of the graph) as compared to environment–2 (far left side). But one could imagine a fan-shaped interaction where there was no genotypic effect at one extreme (e.g., the lines converge to the same phenotypic mean at environment–2). Further, fan-shaped interactions can differ in the slope of the lines for each genotype, which indicate how much the environment is modifying genetic effects. In the crossover interaction shown in Figure 1b, the lines cross at environment 0 (i.e., in the middle). But crossover interactions can vary in the location of the crossover. It is possible that crossing over only occurs at the environmental extreme. As previously noted, the crossing over of the genotypic groups in the Caspi et al. publications of the interactions between the 5-HTT gene, life events, and depression (Caspi et al. 2003) and between MAOA, maltreatment, and antisocial behavior (Caspi et al. 2002) occurred at the extreme low ends of the environmental measures, and the degree of crossing over was quite modest. Rather, the shape of the interactions (and the way the interactions were conceptualized in the papers) was largely fan-shaped, whereby certain genotypic groups showed stronger associations with outcome as a function of the environmental stressor. Also, in both cases, the genetic variance was far greater under one environmental extreme than the other, rather than being approximately equivalent at both ends of the distribution (but with genotypic effects in opposite directions) as is the case in the crossover effect illustrated in Figure 1b. In general, it is assumed that main effects of genotype will not be detected in crossover interactions, but this will actually depend on the frequency of the different levels of the environment. This is also true of fan-shaped interactions, but to a lesser degree. Note that the crossover depicted in Figure 1b does indicate a main effect of the environment. However, here too substantial variation can be imagined. For example, if the crossover took the shape seen in Figure 1c, assuming approximately equal distributions of environmental levels, you would not find a main effect for genotype or environment. Interactions of this sort are assumed to be relatively rare.
Evaluating the relative importance, or frequency of existence, of each type of interaction is complicated by the fact that there is far more power to detect crossover interactions than fan-shaped interactions. Knowing that most of our genetic studies are likely underpowered, we would expect a preponderance of crossover effects to be detected as compared to fan-shaped effects purely as a statistical artifact. Further, even when a crossover effect is observed, power considerations can make it difficult to determine if it is “real.” For example, an interaction observed in our data between the gene CHRM2, parental monitoring, and adolescent externalizing behavior yielded consistent evidence for a gene-environment interaction, with a crossing of the observed regression lines (as in Figure 1b). However, the mean differences by genotype were not significant at either end of the environmental continuum, so it is unclear whether the crossover reflected true differential susceptibility or simply overfitting of the data across the environmental levels containing the majority of the observations, which contributed to a crossing over of the regression lines at one environmental extreme (Dick et al. 2011). Larger studies would have greater power to make these differentiations; however, there is the unfortunate paradox that the samples with the greatest depth of phenotypic information, allowing for more complex tests about risk associated with particular genes, usually have much smaller sample sizes due to the trade-off necessary to collect the rich phenotypic information. This is an important issue for gene-environment interaction studies in general: Most have been underpowered, and this raises concerns about the likelihood that detected effects are true positives. There are several freely available programs to estimate power (Gauderman 2002, Purcell et al. 2003), and it is critical that papers reporting gene-environment interaction effects (or a lack thereof) include information about the power of their sample in order to interpret the results.
Another widely contested issue is whether gene-environment interactions should be examined only when main effects of genotype are detected. Perhaps not surprisingly, this is the approach most commonly advocated by statistical geneticists (Risch et al. 2009) and that was recommended by the Psychiatric GWAS Consortium (Psychiatr. GWAS Consort. Steer. Comm. 2008). However, this strategy could preclude the detection of crossover interaction effects as well as gene-environment interactions that occur in the presence of relatively low-frequency environments. In addition, if genetic effects are conditional on environmental exposure, main effects of genotype could vary across samples, that is to say, a genetic effect could be detected in one sample and fail to replicate in another if the samples differ on environmental exposure.
Another issue with the detection and interpretation of gene-environment interaction effects involves the range of environments being studied. For example, if we assume that the five levels of the environment shown in Figure 1b represent the true full range of environments that exist, if a particular study only included individuals from environments 0–2, it would conclude that there is a fan-shaped gene-environment interaction. Belsky and colleagues (2009) have suggested this may be particularly problematic in the psychiatric literature because only in rare exceptions (Bakermans-Kranenburg & van Ijzendoorn 2006, Taylor et al. 2006) has the environment included both positive and negative ends of the spectrum. Rather, the absence of environmental stressors has usually constituted the “low” end of the environment, e.g., the absence of life stressors (Caspi et al. 2003) or the absence of maltreatment (Caspi et al. 2002). This could lead individuals to conclude there is a fan-shaped interaction because they are essentially failing to measure, with reference to Figure 1b, environments –2 and –1, which represent the positive end of the environmental continuum. In looking at Figures 1a and b, one can imagine a number of other incorrect conclusions that could be drawn about the nature of gene-environment interaction effects as a result of restricted range of environmental measures. For example, in Figure 1b, measurement of individuals from environments –2 to 0 would lead one to conclude that genetic effects play a stronger role at lower levels of environmental exposure. Measurement of individuals from environments 0 to 2 would lead one to conclude that genetic effects play a stronger role at higher levels of exposure to the same environmental variable. In Figure 1a, if measurement of individuals was limited to environments –2 and –1, depending on sample size, there may be inadequate power to detect deviation from a purely additive genetic model, e.g., the slope of the genotypic lines may not be significantly different.
It is also important to note that not only are there several scenarios that would lead one to make incorrect conclusions about the nature of a gene-environment interaction effect, there are also scenarios that would lead one to conclude that a gene-environment interaction exists when it actually does not. Several of these are detailed in a sobering paper by my colleague Lindon Eaves, in which significant evidence for gene-environment interaction was detected quite frequently using standard regression methods, when the simulated data reflected strictly additive models (Eaves 2006). This was particularly problematic when using logistic regression where a dichotomous diagnosis was the outcome. The problem was further exaggerated when selected samples were analyzed.
An additional complication with evaluating gene-environment interactions in psychology is that often our environmental measures don’t have absolute scales of measurement. For example, what is the “real” metric for measuring a construct like parent-child bonding, or maltreatment, or stress? This becomes critical because fan-shaped interactions are very sensitive to scaling. Often a transformation of the scale scores will make the interaction disappear. What does it mean if the raw variable shows an interaction but the log transformation of the scale scores does not? Is the interaction real? Is one metric for measuring the environment a better reflection of the “real” nature of the environment than another? Many of the environments of interest to psychologists do not have true metrics, such as those that exist for measures such as height, weight, or other physiological variables. This is an issue for the study of gene-environment interaction. It becomes even more problematic when you consider that logistic regression is the method commonly used to test for gene-environment interactions with dichotomous disease status outcomes. Logistic regression involves a logarithmic transformation of the probability of being affected. By definition, this changes the nature of the relationship between the variables being modeled. This compounds problems associated with gene-environment interactions being scale dependent.
EPIGENETICS: A POTENTIAL BIOLOGICAL MECHANISM FOR GENE-ENVIRONMENT INTERACTION
An enduring question remains in the study of gene-environment interaction: how does the environment “get under the skin”? Stated in another way, what are the biological processes by which exposure to environmental events could affect outcome? Epigenetics is one candidate mechanism. Excellent recent reviews on this topic exist (Meaney 2010, Zhang & Meaney 2010), and I provide a brief overview here. It is important to note, however, that although epigenetics is increasingly discussed in the context of gene-environment interaction, it does not relate directly to gene-environment interaction in the statistical sense, as differentiated previously in this review. That is to say that epi-genetic processes likely tell us something about the biological mechanisms by which the environment can affect gene expression and impact behavior, but they are not informative in terms of distinguishing between additive versus interactive environmental effects.
Although variability exists in defining the term, epigenetics generally refers to modifi-cations to the genome that do not involve a change in nucleotide sequence. To understand this concept, let us review a bit about basic genetics. The expression of a gene is influenced by transcription factors, which bind to specific sequences of DNA. It is through the binding of transcription factors that genes can be turned on or off. Epigenetic mechanisms involve changes to how readily transcription factors can access the DNA. Several different types of epigenetic changes are known to exist that involve different types of chemical changes that can regulate DNA transcription. One epigenetic process that affects transcription binding is DNA methylation. DNA methylation involves the addition of a methyl group (CH3) onto a cytosine (one of the four base pairs that make up DNA). This leads to gene silencing because methylated DNA hinders the binding of transcription factors. A second major regulatory mechanism is related to the configuration of DNA. DNA is wrapped around clusters of histone proteins to form nu-cleosomes. Together the nucleosomes of DNA and histone are organized into chromatin. When the chromatin is tightly condensed, it is difficult for transcription factors to reach the DNA, and the gene is silenced. In contrast, when the chromatin is opened, the gene can be activated and expressed. Accordingly, modifications to the histone proteins that form the core of the nucleosome can affect the initiation of transcription by affecting how readily transcription factors can access the DNA and bind to their appropriate sequence.
Epigenetic modifications of the genome have long been known to exist. For example, all cells in the body share the same DNA; accordingly, there must be a mechanism whereby different genes are active in liver cells than, for example, brain cells. The process of cell specialization involves silencing certain portions of the genome in a manner specific to each cell. DNA methylation is a mechanism known to be involved in cell specialization. Another well-known example of DNA methylation involves X-inactivation in females. Because females carry two copies of the X chromosome, one must be inactivated. The silencing of one copy of the X chromosome involves DNA methylation. Genomic imprinting is another long-established principle known to involve DNA methylation. In genomic imprinting the expression of specific genes is determined by the parent of origin. For example, the copy of the gene inherited from the mother is silenced, while the copy inherited from the father is active (or vice versa). The silent copy is inactive through processes involving DNA methylation. These changes all involve epigenetic processes parallel to those currently attracting so much attention. However, the difference is that these known epigenetic modifications (cell specialization, X inactivation, genomic imprinting) all occur early in development and are stable. The discovery that epigenetic modifications continue to occur across development, and can be reversible and more dynamic, has represented a major paradigm shift in our understanding of environmental regulation of gene expression.
Animal studies have yielded compelling evidence that early environmental manipulations can be associated with long-term effects that persist into adulthood. For example, maternal licking and grooming in rats is known to have long-term influences on stress response and cognitive performance in their offspring (Champagne et al. 2008, Meaney 2010). Further, a series of studies conducted in macaque monkeys demonstrates that early rearing conditions can result in long-term increased aggression, more reactive stress response, altered neurotransmitter functioning, and structural brain changes (Stevens et al. 2009). These findings parallel research in humans that suggests that early life experiences can have long-term effects on child development (Loman & Gunnar 2010). Elegant work in animal models suggests that epigenetic changes may be involved in these associations (Meaney 2010, Zhang & Meaney 2010).
Evaluating epigenetic changes in humans is more difficult because epigenetic marks can be tissue specific. Access to human brain tissue is limited to postmortem studies of donated brains, which are generally unique and unrepresentative samples and must be interpreted in the context of those limitations. Nonetheless, a recent study of human brain samples from the Quebec Suicide Brain Bank found evidence of increased DNA methylation of the exon 1F promoter in hippocampal samples from suicide victims compared with controls—but only if suicide was accompanied with a history of childhood maltreatment (McGowan et al. 2009). Importantly, this paralleled epigenetic changes originally observed in rat brain in the ortholog of this locus. Another line of evidence suggesting epigenetic changes that may be relevant in humans is the observation of increasing discordance in epigenetic marks in MZ twins across time. This is significant because MZ twins have identical genotypes, and therefore, differences between them are attributed to environmental influences. In a study by Fraga and colleagues (2005), MZ twins were found to be epigenetically indistinguishable during the early years of life, but older MZ twins exhibited remarkable differences in their epigenetic profiles. These findings suggest that epigenetic changes may be a mechanism by which environmental influences contribute to the differences in outcome observed for a variety of psychological traits of interest between genetically identical individuals.
The above studies complement a growing literature demonstrating differences in gene expression in humans as a function of environmental experience. One of the first studies to analyze the relationship between social factors and human gene expression compared healthy older adults who differed in the extent to which they felt socially connected to others (Cole et al. 2007). Using expression profiles obtained from blood cells, a number of genes were identified that showed systematically different levels of expression in people who reported feeling lonely and distant from others. Interestingly, these effects were concentrated among genes that are involved in immune response. The results provide a biological mechanism that could explain why socially isolated individuals show heightened vulnerability to diseases and illnesses related to immune function. Importantly, they demonstrate that our social worlds can exert biologically significant effects on gene expression in humans (for a more extensive review, see Cole 2009).
CONCLUSIONS
This review has attempted to provide an overview of the study of gene-environment interaction, starting with early animal studies documenting gene-environment interaction, to demonstrations of similar effects in family, adoption, and twin studies. Advances in twin modeling and the relative ease with which gene-environment interaction can now be modeled has led to a significant increase in the number of twin studies documenting changing importance of genetic influence across environmental contexts. There is now widespread documentation of gene-environment interaction effects across many clinical disorders (Thapar et al. 2007). These findings have led to more integrated etiological models of the development of clinical outcomes. Further, since it is now relatively straightforward and inexpensive to collect DNA and conduct genotyping, there has been a surge of studies testing for gene-environment interaction with specific candidate genes. Psychologists have embraced the incorporation of genetic components into their studies, and geneticists who focus on gene finding are now paying attention to the environment in an unprecedented way. However, now that the initial excitement surrounding gene-environment interaction has begun to wear off, a number of challenges involved in the study of gene-environment interaction are being recognized. These include difficulties with interpreting interaction effects (or the lack thereof), due to issues surrounding the measurement and scaling of the environment, and statistical concerns surrounding modeling gene-environment interactions and the nature of their effects.
So where do we go from here? Individuals who jumped on the gene-environment interaction bandwagon are now discovering that studying this process is harder than it first appeared. But there is good reason to believe that gene-environment interaction is a very important process in the development of clinical disorders. So rather than abandon ship, I would suggest that as a field, we just need to proceed with more caution. I close with some thoughts about how we can move forward in this way.
Knowledge Is Power
As every student enrolled in Psychology 101 is taught, simply making an individual aware of something (such as their being in a psychology study) can change behavior. More widespread recognition of the issues surrounding the study of gene-environment interactions, as delineated above, will hopefully lead to more thoughtful and careful evaluations of hypothesized gene-environment interaction effects. Studying gene-environment interactions is not as simple as plugging a genotype and environment into a regression equation and seeing if the interaction term yields p < 0.05. Scientists conducting gene-environment interaction research and reviewers of papers evaluating gene-environment interaction effects need to be keenly aware of these issues so that due diligence can be carried out to evaluate interaction effects that are detected (or not) in a given sample.
Use What We Already Know from the Twin and Developmental Literatures
It has been suggested that twin studies are no longer necessary in the era of molecular genetics. The argument is that there is no longer any need to infer genetic influence now that we can directly measure genotypes. However, studies of latent genetic influence (as inferred from family, adoption, and twin studies) and studies of measured genotypes actually yield very different information and have complementary strengths and weaknesses, as has been nicely reviewed by my colleague Kenneth Kendler in previous papers (Kendler 2005, 2010). Information about aggregate genetic risk, as yielded by twin studies, gives us an idea of the big picture. It is essentially a satellite picture, providing an overview of the general landscape. On the other hand, molecular genetics offers a level of detail about the underlying biology that twin studies cannot. The corresponding metaphor would be the photographer on the ground who is taking pictures of the individual rocks and trees. But lost in that level of detail is information about the overall picture.
Perhaps because of the aggregate nature of genetic influence as studied in twin designs, findings from twin studies have a far better record of replication than findings from specific candidate gene association studies. Heritabilities for clinical disorders have been remarkably consistent across populations, and gene-environment interaction effects have also yielded consistent results (e.g., (Legrand et al. 2007, Rose et al. 2001). This is not the case with gene finding efforts, where replications have been notoriously difficult, both for main effects (e.g., Lind et al. 2009, Wang et al. 2004) and gene-environment interactions (Caspi et al. 2003, Risch et al. 2009).
Parallel to the way that evidence for heritability from twin studies for a given outcome was used to justify searching for specific genes involved in that outcome, evidence for gene-environment interaction from twin studies can also be used to develop hypotheses to test gene-environment interactions associated with specific, identified genes. Change in the overall heritability across environmental contexts does not necessarily dictate that any one specific susceptibility gene will operate in a parallel manner; however, a change in heritability suggests that at least a good portion of the involved genes (assuming many genes of approximately equal and small effect) must be operating in that manner for a difference in heritability by environment to be detected. In this sense, you are loading the dice when you test for specific candidate gene-environment interaction effects with an environment that has already been shown to moderate the overall importance of genetic influences on that outcome. This is the strategy we have used to further characterize the risk related to genes associated with alcohol dependence in the Collaborative Study on the Genetics of Alcoholism. On the basis of our twin studies suggesting that genetic influences on adolescent substance use are moderated by parental monitoring (Dick et al. 2007c) and peer substance use (Dick et al. 2007a), we tested for moderation of the association of GABRA2 (Edenberg et al. 2004) and CHRM2 (Wang et al. 2004) as a function of parental monitoring and peer group antisocial behavior, respectively. We found evidence for gene-environment interaction effects in the direction predicted by the twin studies, namely, genetic effects were enhanced under conditions of lower parental monitoring (Dick et al. 2009) and higher peer group antisocial behavior (Latendresse et al. 2010).
Twin studies are not the only place from which to draw hypotheses about environmental influences that are likely to moderate genetic effects. The developmental literature contains a wealth of studies demonstrating differential effects of the environment across children with differing temperaments and/or who differ on family history. Because temperament and family history both provide information about the child’s genetic predisposition, these kinds of interactions can also serve as starting points for developing hypotheses about gene-environment interaction effects associated with specific genes. In addition to the twin evidence suggesting that parental monitoring moderated the importance of genetic effects, numerous studies in the developmental literature suggest the importance of this construct in moderating associations between early temperament/family history and the subsequent development of child behavior problems. For example, Bates and colleagues found that across two independent samples, a difficult childhood temperament was related to the subsequent development of externalizing behavior, but only in the context of lower parental control (Bates et al. 1998). Further, Molina and colleagues have found that density of family history of alcoholism is related to the development of behavior problems in children, but only in the context of poor parenting (a measure that included reduced parental monitoring) (Molina et al. 2010). These studies both find that associations between predisposing factors (both known to at least partially reflect genetic influ-ence) and child behavior problems are stronger under conditions of lower parental monitoring, paralleling the finding from twin studies that genetic influences were stronger under conditions of lower parental monitoring. They provide a compelling rationale to study parental monitoring as a moderator of the effects associated with specific candidate genes involved in substance use and externalizing behavior, an effect which has now been demonstrated with respect to GABRA2 (Dick et al. 2009).
Statistics Are Not a Substitute for Critical Thinking
One of the take-home messages from this review is that interpreting results from gene-environment interaction studies is not straightforward. There are valid reasons why “real” gene-environment interaction effects might not be detected, why results might vary across studies, or why our statistics might yield “significant evidence” for gene-environment interaction when none really exists. An uncritical tally of whether specific gene-environment interaction effects replicate or not, without paying attention to issues such as the measurement of the outcome, measurement of the environment, statistics employed, sampling strategy, and sample size, across studies will undoubtedly lead to mixed results for any given gene-environment interaction effect in the literature. Statistics such as meta-analyses have become very popular in genetics, largely due to recognition of the incredibly large sample sizes it will take to identify genes of small effect. Meta-analytic techniques are particularly appropriate in this area, where genotypic data can be standardized across studies, and outcomes are often measured using standardized assessments (e.g., DSM diagnoses as assessed in structured clinical interviews). However, in the area of gene-environment interactions, where studies often have very different designs and goals, some of which include the explicit attempt to explore the boundaries of originally reported gene-environment interactions, meta-analyses should not be conducted without careful attention to these issues.
Play Nice in the Sandbox
Research on gene-environment interactions is inherently interdisciplinary; it sits at the intersection between genetics and psychology. However, this perspective has not been embraced to the extent that it could be, and (in my opinion) must be, in order to do really good research in this area. Most of the gene-environment interaction research in psychology has been limited to the “usual suspects”—purportedly functional polymorphisms in MAOA, 5-HTT, DRD4, and a few others (Belsky et al. 2009). However, the evidence for those polymorphisms truly being functional is often ambiguous (Cirulli & Goldstein 2007). In addition, in the field of genetics, we would never test a single marker in a gene in order to make conclusions about the relevance of that gene in a given genotype. Rather, with data from the human genome project and the HapMap project, we now know something about the structure of most genes in the human genome (Manolio et al. 2008). Further, there are many polymorphic markers available across most genes of interest. It is possible that multiple locations in a gene could have various forms that lead to differential function of that gene contributing to differential susceptibility to an outcome (McClellan & King 2010). Today it would be nearly impossible to publish a paper in a genetics journal without saying something about the coverage of the gene provided by the set of genotyped markers (Pettersson et al. 2009).
As one example, the Taq1A allele, originally thought to be in the DRD2 gene, has an extensive literature surrounding it, with reported associations with a number of phenotypes related to substance use, smoking, and a variety of other phenotypes related to impulsivity (Dick et al. 2007d, Noble 2000). Figure 2 shows a screen-shot of output from the program Haploview (Barrett et al. 2005) illustrating the linkage disequilibrium (LD) structure of the chromosomal region surrounding the DRD2 gene. Along the top of the figure is the base pair position of the chromosomal region, as listed in kilobases to give an idea of scale. Directly underneath, the triangles indicate SNPs that were genotyped in the samples on which the Haploview data are based. The SNP highlighted in green is the marker commonly referred to as DRD2 Taq1A. Beneath the SNPs, the genes in the region are listed. The length of the line reflects the length of the gene. One will note that the Taq1A allele is actually located in a small gene next to DRD2 called ANKK1. It is not located in DRD2 despite the large literature making claims about whether DRD2 was involved in many different phenotypes of interest based on genotyping at this marker. Below the genes is the LD plot, where shading indicates the degree of correlation between markers (shown here as the small hash marks at the top of the figure) as measured by D′ (Hedrick & Kumar 2001), with darker red shading indicating higher correlations; blue or white shading indicates the markers are unlinked or uncorrelated. What stands out is the block-like correlational structure, yielding inverted red triangles that indicate groups of SNPs where there is high LD across that group of SNPs and low LD with surrounding SNPs located outside the block. This block-like structure is observed throughout the genome (Gabriel et al. 2002). Knowing the correlation pattern is critical for the selection of markers and the interpretation of genetic association results. For example, DRD2 spans at least two blocks on the figure. If different studies genotyped SNPs from different blocks, they could reach different conclusions about whether DRD2 was “associated” with outcome, depending on the location of the marker they chose and where the actual associated SNP was. Further, also note that the LD block that contains Taq1A spans the genes DRD2, ANKK1, and TTC12. This makes it very difficult to know which gene is actually important for an observed association. More extensive genotyping across these genes and the other gene located very nearby in the region, NCAM1, has suggested that the association with substance-use phenotypes extends to multiple genes in this region (Dick et al. 2007d, Gelernter et al. 2006, Yang et al. 2008). This underscores the necessity of understanding genomic structure in order to evaluate the role of hypothesized genes of interest.
Figure 2.
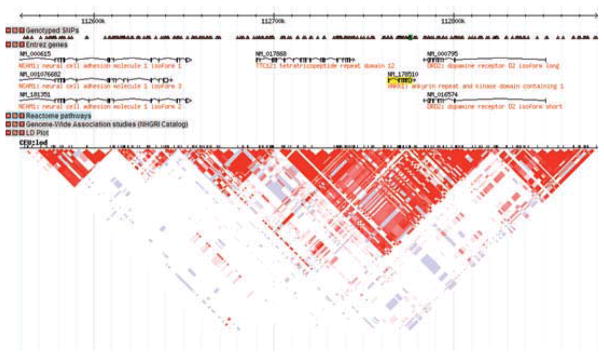
Screenshot from Haploview (Barrett et al. 2005) showing the linkage disequilibrium structure surrounding the DRD2 gene.
In the same way that psychologists pay careful attention to the measurement of their outcomes of interest and potential environmental factors of relevance, care must be taken in characterizing genes of interest. The genetics research being carried out by psychologists should be of the same caliber as that being conducted in other areas of genetics, and it must keep up with the rapid advances going on in that field. Otherwise it will not be taken seriously. This does not mean that all psychologists need to be “gene-finders” or to carry out GWAS. But it does mean that anyone involved in this kind of research should understand the complexities of studying genetics and be connected to the latest developments in genetics. Because of the rapid pace at which the field of genetics moves, this necessitates having collaborators who are tied more centrally to the world of genetics and/or (for the younger generation of psychologists with interest in this area) to obtain focused training in genetics, ideally through a postdoctoral training experience. With ~25,000 genes in the human genome, thousands of genetic association papers published, and GWAS papers being turned out every day, I find it impossible to believe that the handful of usual suspects are the only genes of interest for clinical outcomes. As large-scale gene-finding studies continue to report associations with new and novel genes of interest, these genes too deserve further study by psychologists to delineate how associated risk may be modified by environmental factors.
The bridge between psychology and genetics is not a one-way street. Although psychologists have much to learn from geneticists, they also have much to offer. As geneticists have grown interested in incorporating environmental information into genetic studies, they have been guilty of using environmental measures that would be considered naive by psychologists who are devoted to careful characterization of these constructs. Much of the large-scale gene identification work to date has been dominated by studies of binary diagnostic outcomes. Although the use of binary diagnoses brings the advantage of standardized, reliable assessments across studies and sites, there is reason to believe that these phenotypes are not ideal for gene finding. Psychologists have a long history of careful measurement of phenotype and of studying intermediate phenotypes and mechanistic processes. The application of these skills to the field of genetics holds great promise both to aid in gene identification and to help with the characterization of risk associated with identi-fied genes. However, this promise will only be recognized when psychologists and geneticists work closely together, have patience with one another concerning differences in training and ideology, and respect the relative contributions that each field can bring to the study of gene-environment interaction.
SUMMARY POINTS.
Gene-environment interaction refers to the phenomenon whereby the effect of genes depends on the environment, or the effect of the environment depends on genotype. There is now widespread documentation of gene-environment interaction effects across many clinical disorders, leading to more integrated etiological models of the development of clinical outcomes.
Twin, family, and adoption studies provide methods to study gene-environment interaction with genetic effects modeled latently, meaning that genes are not directly measured, but rather genetic influence is inferred based on correlations across relatives. Advances in genotyping technology have contributed to a proliferation of studies testing for gene-environment interaction with specific measured genes. Each of these designs has its own strengths and limitations.
Two types of gene-environment interaction have been discussed in greatest detail in the literature: fan-shaped interactions, in which the influence of genotype is greater in one environmental context than in another; and crossover interactions, in which the same individuals who are most adversely affected by negative environments may also be those who are most likely to benefit from positive environments. Distinguishing between these types of interactions poses a number of challenges.
The range of environments studied and the lack of a true metric for many environmental measures of interest create difficulties for studying gene-environment interactions. Issues surrounding power, and the use of logistic regression and selected samples, further compound the difficulty of studying gene-environment interactions. These issues have not received adequate attention by many researchers in this field.
Epigenetic processes may tell us something about the biological mechanisms by which the environment can affect gene expression and impact behavior. The growing literature demonstrating differences in gene expression in humans as a function of environmental experience demonstrates that our social worlds can exert biologically significant effects on gene expression in humans.
Much of the current work on gene-environment interactions does not take advantage of the state of the science in genetics or psychology; advancing this area of study will require close collaborations between psychologists and geneticists.
Acknowledgments
This manuscript was prepared with support by AA15416 and K02AA018755 from the National Institute of Alcohol Abuse and Alcoholism. The author also acknowledges helpful comments from Dr. Kenneth Kendler on an earlier version of this manuscript.
Glossary
- Gene
unit of heredity; a stretch of DNA that codes for a protein
- GxE
geneenvironment interaction
- Epigenetics
modifications to the genome that do not involve a change in nucleotide sequence
- Heritability
the proportion of total phenotypic variance that can be accounted for by genetic factors
- MZ
monozygotic
- DZ
dizygotic
- DNA
deoxyribonucleic acid
- Polymorphism
a location in a gene that comes in multiple forms
- Allele
natural variation in the genetic sequence; can be a change in a single nucleotide or longer stretches of DNA
- GWAS
genome-wide association study
- ORs
odds ratios
- Phenotype
the observed outcome under study; can be the manifestation of both genetic and/or environmental factors
- Chromosome
a single piece of coiled DNA containing many genes, regulatory elements, and other nucleotide sequences
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Bakermans-Kranenburg MJ, van Ijzendoorn MH. Gene-environment interaction of the dopamine D4 receptor (DRD4) and observed maternal insensitivity predicting externalizing behavior in preschoolers. Dev Psychobiol. 2006;48(5):406–9. doi: 10.1002/dev.20152. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–65. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bates JE, Pettit GS, Dodge KA, Ridge B. Interaction of temperamental resistance to control and restrictive parenting in the development of externalizing behavior. Dev Psychol. 1998;34:982–95. doi: 10.1037//0012-1649.34.5.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky J, Jonassaint C, Pluess M, Stanton M, Brummett B, Williams R. Vulnerability genes or plasticity genes? Mol Psychiatry. 2009;14(8):746–54. doi: 10.1038/mp.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeman CS, Plomin R, McClearn GE, Pedersen NL, Friberg LT. Genotype-environment interaction in personality development: identical twins reared apart. Psychol Aging. 1988;3:399–406. doi: 10.1037//0882-7974.3.4.399. [DOI] [PubMed] [Google Scholar]
- Boardman JD, Blalock CL, Pampel FC. Trends in genetic influences on smoking. J Health Soc Behav. 2010;51:108–23. doi: 10.1177/0022146509361195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce WT, Ellis BJ. Biological sensitivity to context: I. An evolutionary-developmental theory of the origins and functions of stress reactivity. Dev Psychopathol. 2005;17(2):271–301. doi: 10.1017/s0954579405050145. [DOI] [PubMed] [Google Scholar]
- Button TM, Corley RP, Rhee SH, Hewitt JK, Young SE, Stallings MC. Delinquent peer affiliation and conduct problems: a twin study. J Abnorm Psychol. 2007;116(3):554–64. doi: 10.1037/0021-843X.116.3.554. [DOI] [PubMed] [Google Scholar]
- Button TM, Lau JY, Maughan B, Eley TC. Parental punitive discipline, negative life events and gene-environment interplay in the development of externalizing behavior. Psychol Med. 2008;38(1):29–39. doi: 10.1017/S0033291707001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoret RJ, Troughton E, Merchant LM, Whitters A. Early life psychosocial events and adult affective symptoms. Robbins & Rutter 1990. 1990:300–13. [Google Scholar]
- Cadoret RJ, Winokur G, Langbehn D, Troughton E, Yates WR, Stewart MA. Depression spectrum disease, I: the role of gene-environment interaction. Am J Psychiatry. 1996;153:892–99. doi: 10.1176/ajp.153.7.892. [DOI] [PubMed] [Google Scholar]
- Cannon TD, Mednick SA, Parnas J. Two pathways to schizophrenia in children at risk. Robbins & Rutter. 1990:314–27. [Google Scholar]
- Cannon TD, Mednick SA, Parnas J, Schulsinger F, Praestholm J, Vestergaard A. Developmental brain abnormalities in the offspring of schizophrenic mothers. I Contributions of genetic and perinatal factors. Arch Gen Psychiatry. 1993;50:551–64. doi: 10.1001/archpsyc.1993.01820190053006. [DOI] [PubMed] [Google Scholar]
- Cardon LR. Genetics. Delivering new disease genes. Science. 2006;314(5804):1403–5. doi: 10.1126/science.1136668. [DOI] [PubMed] [Google Scholar]
- Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE. Genetic sensitivity to the environment: the case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am J Psychiatry. 2010;167(5):509–27. doi: 10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, McClay J, Moffitt TE, Mill J, Martin J, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–54. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–89. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Champagne DL, Bagot RC, van HF, Ramakers G, Meaney MJ, et al. Maternal care and hippocam-pal plasticity: evidence for experience-dependent structural plasticity, altered synaptic functioning, and differential responsiveness to glucocorticoids and stress. J Neurosci. 2008;28(23):6037–45. doi: 10.1523/JNEUROSCI.0526-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Goldstein DB. In vitro assays fail to predict in vivo effects of regulatory polymorphisms. Hum Mol Genet. 2007;16(16):1931–39. doi: 10.1093/hmg/ddm140. [DOI] [PubMed] [Google Scholar]
- Cloninger CR, Bohman M, Sigvardsson S. Inheritance of alcohol abuse: cross-fostering analysis of adopted men. Arch Gen Psychiatry. 1981;38:861–68. doi: 10.1001/archpsyc.1981.01780330019001. [DOI] [PubMed] [Google Scholar]
- Cole SW. Social regulation of human gene expression. Curr Dir Psychol Sci. 2009;18:132–37. doi: 10.1111/j.1467-8721.2009.01623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Sung CY, Rose RM, Cacioppo JT. Social regulation of gene expression in human leukocytes. Genome Biol. 2007;8(9):R189. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RM, Zubek JP. Effects of enriched and restricted early environments on the learning ability of bright and dull rats. Can J Psychol. 1958;12:159–64. doi: 10.1037/h0083747. [DOI] [PubMed] [Google Scholar]
- Dick DM, Latendresse SJ, Lansford JE, Budde J, Goate A, et al. The role of GABRA2 in trajectories of externalizing behavior across development and evidence of moderation by parental monitoring. Arch Gen Psychiatry. 2009;66:649–57. doi: 10.1001/archgenpsychiatry.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Meyers JL, Latendresse SJ, Creemers HE, Lansford JE, et al. CHRM2, parental monitoring, and adolescent externalizing behavior: evidence for gene-environment interaction. Psychol Sci. 2011 doi: 10.1177/0956797611403318. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Pagan JL, Holliday C, Viken R, Pulkkinen L, et al. Gender differences in friends’ influences on adolescent drinking: a genetic epidemiological study. Alcohol Clin Exp Res. 2007a;31:2012–19. doi: 10.1111/j.1530-0277.2007.00523.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Pagan JL, Viken R, Purcell S, Kaprio J, et al. Changing environmental influences on substance use across development. Twin Res Hum Genet. 2007b;10(2):315–26. doi: 10.1375/twin.10.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Rose RJ, Viken RJ, Kaprio J, Koskenvuo M. Exploring gene-environment interactions: socioregional moderation of alcohol use. J Abnorm Psychol. 2001;110:625–32. doi: 10.1037//0021-843x.110.4.625. [DOI] [PubMed] [Google Scholar]
- Dick DM, Viken R, Purcell S, Kaprio J, Pulkkinen L, Rose RJ. Parental monitoring moderates the importance of genetic and environmental influences on adolescent smoking. J Abnorm Psychol. 2007c;116(1):213–18. doi: 10.1037/0021-843X.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Wang J, Plunkett J, Aliev F, Hinrichs AL, et al. Family-based association analyses of alcohol dependence phenotypes across DRD2 and neighboring gene ANKK1. Alcohol Clin Exp Res. 2007d;31:1645–53. doi: 10.1111/j.1530-0277.2007.00470.x. [DOI] [PubMed] [Google Scholar]
- Eaves LJ. Genotype x environment interaction in psychopathology: fact or artifact? Twin Res Hum Genet. 2006;9(1):1–8. doi: 10.1375/183242706776403073. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, et al. Variations in GABRA2, encoding the α2 subunit of the GABA-A receptor are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–14. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Dodge KA, Latendresse SJ, Lansford JE, Bates J, et al. MAOA uVNTR and early physical discipline interact to influence delinquent behavior. J Child Psychol Psychiatry. 2010;51(6):679–87. doi: 10.1111/j.1469-7610.2009.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis BJ, Boyce WT. Biological sensitivity to context. Curr Dir Psychol Sci. 2008;17(3):183–87. [Google Scholar]
- Ellis BJ, Essex MJ, Boyce WT. Biological sensitivity to context: II. Empirical explorations of an evolutionary-developmental theory. Dev Psychopathol. 2005;17(2):303–28. doi: 10.1017/s0954579405050157. [DOI] [PubMed] [Google Scholar]
- Engelman CD, Baurley JW, Chiu YF, Joubert BR, Lewinger JP, et al. Detecting gene-environment interactions in genome-wide association data. Genet Epidemiol. 2009;33(Suppl 1):S68–73. doi: 10.1002/gepi.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Steer CD, Newman TK, Gibson N, Goldman D. Early life stress, MAOA, and gene-environment interactions predict behavioral disinhibition in children. Genes Brain Behav. 2010;9(1):65–74. doi: 10.1111/j.1601-183X.2009.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg ME, Button TM, Neiderhiser JM, Reiss D, Hetherington EM. Parenting and adolescent antisocial behavior and depression: evidence of genotype x parenting environment interaction. Arch Gen Psychiatry. 2007;64(4):457–65. doi: 10.1001/archpsyc.64.4.457. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102(30):10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazzetto G, Di LG, Carola V, Proietti L, Sokolowska E, et al. Early trauma and increased risk for physical aggression during adulthood: the moderating role of MAOA genotype. PLoS One. 2007;2(5):e486. doi: 10.1371/journal.pone.0000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–29. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ, Thomas DC, Murcray CE, Conti D, Li D, Lewinger JP. Efficient genome-wide association testing of gene-environment interaction in case-parent trios. Am J Epidemiol. 2010;172(1):116–22. doi: 10.1093/aje/kwq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, et al. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet. 2006;15:3498–507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- Gottesman II. Schizophrenia Genesis: The Origins of Madness. New York: Freeman; 1991. [Google Scholar]
- Heath AC, Eaves LJ, Martin NG. Interaction of marital status and genetic risk for symptoms of depression. Twin Res. 1998;1(3):119–22. doi: 10.1375/136905298320566249. [DOI] [PubMed] [Google Scholar]
- Heath AC, Jardine R, Martin NG. Interactive effects of genotype and social environment on alcohol consumption in female twins. J Stud Alcohol. 1989;50:38–48. doi: 10.15288/jsa.1989.50.38. [DOI] [PubMed] [Google Scholar]
- Hedrick P, Kumar S. Mutation and linkage disequilibrium in human mtDNA. Eur J Hum Genet. 2001;9:969–72. doi: 10.1038/sj.ejhg.5200735. [DOI] [PubMed] [Google Scholar]
- Henderson ND. Genetic influences on the behavior of mice can be obscured by laboratory rearing. J Comp Physiol Psychol. 1970;72:505–11. doi: 10.1037/h0029743. [DOI] [PubMed] [Google Scholar]
- Henderson ND. Relative effects of early rearing environment and genotype on discrimination learning in house mice. J Comp Physiol Psychol. 1972;79:243–53. doi: 10.1037/h0032554. [DOI] [PubMed] [Google Scholar]
- Heston LL, Denney D. Interactions between early life experience and biological factors in schizophrenia. In: Rosenthal D, Kety SS, editors. The Transmission of Schizophrenia. Oxford: Pergamon; 1967. pp. 363–76. [Google Scholar]
- Hewitt JK, Turner JR. Behavior genetic approaches in behavioral medicine: an introduction. In: Turner JR, Cardon LR, Hewitt JK, editors. Behavior Genetic Approaches in Behavioral Medicine. New York: Plenum; 1995. pp. 3–16. [Google Scholar]
- Kendler KS. Twin studies of psychiatric illness: current status and future directions. Arch Gen Psychiatry. 1993;50:905–15. doi: 10.1001/archpsyc.1993.01820230075007. [DOI] [PubMed] [Google Scholar]
- Kendler KS. Psychiatric genetics: a methodologic critique. Am J Psychiatry. 2005;162:3–11. doi: 10.1176/appi.ajp.162.1.3. [DOI] [PubMed] [Google Scholar]
- Kendler KS. A conceptual overview of gene-environment interaction and correlation in a developmental context. In: Kendler KS, Jaffee S, Romer D, editors. The Dynamic Genome and Mental Health. New York: Oxford Univ. Press; 2011. In press. [Google Scholar]
- Kendler KS, Baker JH. Genetic influences on measures of the environment: a systematic review. Psychol Med. 2007;37:615–26. doi: 10.1017/S0033291706009524. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Eaves LJ. Models for the joint effect of genotype and environment on liability to psychiatric illness. Am J Psychiatry. 1986;143:279–89. doi: 10.1176/ajp.143.3.279. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. Life events and depressive symptoms: a twin study perspective. In: McGuffin P, Murray R, editors. The New Genetics of Mental Illness. Oxford: Butterworth-Heinemann; 1991. pp. 146–64. [Google Scholar]
- Kendler KS, Neale MC, Kessler R, Heath A, Eaves L. A twin study of recent life events and difficulties. Arch Gen Psychiatry. 1993;50:789–96. doi: 10.1001/archpsyc.1993.01820220041005. [DOI] [PubMed] [Google Scholar]
- Kim-Cohen J, Caspi A, Taylor A, Williams B, Newcombe R, et al. MAOA, maltreatment, and gene-environment interaction predicting children’s mental health: new evidence and a meta-analysis. Mol Psychiatry. 2006;11(10):903–13. doi: 10.1038/sj.mp.4001851. [DOI] [PubMed] [Google Scholar]
- Koopmans JR, Slutske WS, van Baal GC, Boomsma DI. The influence of religion on alcohol use initiation: evidence for genotype X environment interaction. Behav Genet. 1999;29(6):445–53. doi: 10.1023/a:1021679005623. [DOI] [PubMed] [Google Scholar]
- Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85(5):679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latendresse SJ, Bates J, Goodnight JA, Dodge KA, Lansford JE, et al. Differential susceptibility to adolescent externalizing trajectories: examining the interplay between CHRM2 and peer group antisocial behavior. Child Dev. 2010 doi: 10.1111/j.1467-8624.2010.01517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand LN, Keyes M, McGue M, Iacono WG, Krueger RF. Rural environments reduce the genetic influence on adolescent substance use and rule-breaking behavior. Psychol Med. 2008;38(9):1341–50. doi: 10.1017/S0033291707001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leve LD, Neiderhiser JM, Scaramella LV, Reiss D. The early growth and development study: using the prospective adoption design to examine genotype-environment interplay. Behav Genet. 2010;40:306–14. doi: 10.1007/s10519-010-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Luciano M, Horan MA, Marioni RE, Wright MJ, et al. No association between cholinergic muscarinic receptor 2 (CHRM2) genetic variation and cognitive abilities in three independent samples. Behav Genet. 2009;39(5):513–23. doi: 10.1007/s10519-009-9274-z. [DOI] [PubMed] [Google Scholar]
- Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(6):e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman MM, Gunnar MR. Early experience and the development of stress reactivity and regulation in children. Neurosci Biobehav Rev. 2010;34(6):867–76. doi: 10.1016/j.neubiorev.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspina D, Sohler NL, Susser ES. Interaction of genes and prenatal exposures in schizophrenia. In: Susser ES, Brown AS, Gorman JM, editors. Prenatal Exposures in Schizophrenia. Washington, DC: Am. Psychiatr. Press; 1999. pp. 35–59. [Google Scholar]
- Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118(5):1590–605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Boomsma D, Machin G. A twin-pronged attack on complex traits. Nat Genet. 1997;17:387–92. doi: 10.1038/ng1297-387. [DOI] [PubMed] [Google Scholar]
- Mather K, Jinks J. Biometrical Genetics. London: Chapman & Hall; 1982. [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–17. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- McDermott R, Tingley D, Cowden J, Frazzetto G, Johnson DD. Monoamine oxidase A gene (MAOA) predicts behavioral aggression following provocation. Proc Natl Acad Sci USA. 2009;106(7):2118–23. doi: 10.1073/pnas.0808376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–48. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Bouchard TJ., Jr Genetic and environmental influences on human behavioral differences. Annu Rev Neurosci. 1998;21:1–24. doi: 10.1146/annurev.neuro.21.1.1. [DOI] [PubMed] [Google Scholar]
- McGue M, Sharma A, Benson P. Parent and sibling influences on adolescent alcohol use and misuse: evidence from a US adoption cohort. J Stud Alcohol. 1995;57:8–18. doi: 10.15288/jsa.1996.57.8. [DOI] [PubMed] [Google Scholar]
- McGue M, Sharma A, Benson P. The effect of common rearing on adolescent adjustment: evidence from a U.S. adoption cohort. Dev Psychol. 1996;32(6):604–13. [Google Scholar]
- McKenna AH, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ. Epigenetics and the biological definition of gene x environment interactions. Child Dev. 2010;81(1):41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- Moldin SO, Gottesman II. At issue: genes, experience, and chance in schizophrenia—positioning for the 21st century. Schizophr Bull. 1997;23:547–61. doi: 10.1093/schbul/23.4.547. [DOI] [PubMed] [Google Scholar]
- Molina BSG, Donovan JE, Belendiuk KA. Familial loading for alcoholism and offspring behavior: mediating and moderating influences. Alcohol Clin Exp Res. 2010;34(11):1972–84. doi: 10.1111/j.1530-0277.2010.01287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Ferreira MAR, Medland SE, Posthuma D. Statistical Genetics: Gene Mapping Through Linkage and Association. New York: Taylor & Francis; 2008. [Google Scholar]
- Neale MC. The use of Mx for association and linkage analyses. GeneScreen. 2000;1:107–11. [Google Scholar]
- Noble EP. The DRD2 gene in psychiatric and neurological disorders and its phenotypes. Pharmacoge-netics. 2000;1:309–33. doi: 10.1517/14622416.1.3.309. [DOI] [PubMed] [Google Scholar]
- Pettersson FH, Anderson CA, Clarke GM, Barrett JC, Cardon LR, et al. Marker selection for genetic case-control association studies. Nat Protoc. 2009;4(5):743–52. doi: 10.1038/nprot.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, DeFries JC, Loehlin JC. Genotype-environment interaction and correlation in the analysis of human behavior. Psychol Bull. 1977;84:309–22. [PubMed] [Google Scholar]
- Plomin R, Hershberger S. Genotype-environment interaction. In: Wachs TD, Plomin R, editors. Conceptualization and Measurement of Organism-Environment Interaction. Washington, DC: Am. Psychol. Assoc; 1991. pp. 29–43. [Google Scholar]
- Prom-Wormley EC, Eaves LJ, Foley DL, Gardner CO, Archer KJ, et al. Monoamine oxidase A and childhood adversity as risk factors for conduct disorder in females. Psychol Med. 2009;39(4):579–90. doi: 10.1017/S0033291708004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatr. GWAS Consort. Steer. Comm. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2008;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- Purcell S. Variance components models for gene-environment interaction in twin analysis. Twin Res. 2002;5:554–71. doi: 10.1375/136905202762342026. [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–50. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA. 2009;301(23):2462–71. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins LN, Rutter M, editors. Straight and Devious Pathways from Childhood to Adulthood. London: Cambridge Univ. Press; 1990. [Google Scholar]
- Rose RJ, Dick DM, Viken RJ, Kaprio J. Gene-environment interaction in patterns of adolescent drinking: regional residency moderates longitudinal influences on alcohol use. Alcohol Clin Exp Res. 2001;25:637–43. [PubMed] [Google Scholar]
- Scarr S, McCartney K. How people make their own environments: a theory of genotype-environment effects. Child Dev. 1983;54:424–35. doi: 10.1111/j.1467-8624.1983.tb03884.x. [DOI] [PubMed] [Google Scholar]
- Shanahan MJ, Hofer SM. Social context in gene-environment interactions: retrospect and prospect. J Gerontol B Psychol Sci Soc Sci. 2005;60(Spec 1):65–76. doi: 10.1093/geronb/60.special_issue_1.65. [DOI] [PubMed] [Google Scholar]
- Sherman SL, DeFries JC, Gottesman II, Loehlin JC, Meyer JM, et al. Recent developments in human behavioral genetics: past accomplishments and future directions. Am J Hum Genet. 1997;60:1265–75. doi: 10.1086/515473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens HE, Leckman JF, Coplan JD, Suomi SJ. Risk and resilience: early manipulation of macaque social experience and persistent behavioral and neurophysiological outcomes. J Am Acad Child Adolesc Psychiatry. 2009;48(2):114–27. doi: 10.1097/CHI.0b013e318193064c. [DOI] [PubMed] [Google Scholar]
- Taylor SE, Way BM, Welch WT, Hilmert CJ, Lehman BJ, Eisenberger NI. Early family environment, current adversity, the serotonin transporter promoter polymorphism, and depressive symptomatology. Biol Psychiatry. 2006;60(7):671–76. doi: 10.1016/j.biopsych.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Thapar A, Harold G, Rice F, Langley K, O’Donovan M. The contribution of gene-environment interaction to psychopathology. Dev Psychopathol. 2007;19(4):989–1004. doi: 10.1017/S0954579407000491. [DOI] [PubMed] [Google Scholar]
- Tienari P, Lahti I, Sorri A, Naarala M, Moring J, et al. Adopted-away offspring of schizophrenics and controls: the Finnish Adoptive Family Study of Schizophrenia. Robins & Rutter 1990. 1990:365–79. [Google Scholar]
- Van Den Oord EJ. Controlling false discoveries in genetic studies. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(5):637–44. doi: 10.1002/ajmg.b.30650. [DOI] [PubMed] [Google Scholar]
- Vanyukov MM, Maher BS, Devlin B, Kirillova GP, Kirisci L, et al. The MAOA promoter polymorphism, disruptive behavior disorders, and early onset substance use disorder: gene-environment interaction. Psychiatr Genet. 2007;17(6):323–32. doi: 10.1097/YPG.0b013e32811f6691. [DOI] [PubMed] [Google Scholar]
- Wahlberg K, Wynne LC, Oja H, Keskitalo P, Pykalainen L, et al. Gene-environment interaction in vulnerability to schizophrenia: findings from the Finnish Adoptive Family Study of Schizophrenia. Am J Psychiatry. 1997;154:355–62. doi: 10.1176/ajp.154.3.355. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, Metten P, Phillips TJ, Boehm SL, Burkhart-Kasch S, et al. Different data from different labs: lessons from studies of gene-environment interaction. J Neurobiol. 2003;54(1):283–311. doi: 10.1002/neu.10173. [DOI] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, et al. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum Mol Genet. 2004;13:1903–11. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- Weder N, Yang BZ, Douglas-Palumberi H, Massey J, Krystal JH, et al. MAOA genotype, maltreatment, and aggressive behavior: the changing impact of genotype at varying levels of trauma. Biol Psychiatry. 2009;65(5):417–24. doi: 10.1016/j.biopsych.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Haplotypic variants in DRD2, ANKK1, TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcohol Clin Exp Res. 2008;32(12):2117–27. doi: 10.1111/j.1530-0277.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TY, Meaney MJ. Epigenetics and the environmental regulation of the genome and its function. Annu Rev Psychol. 2010;61:439–66. doi: 10.1146/annurev.psych.60.110707.163625. [DOI] [PubMed] [Google Scholar]