Abstract
A recent meta-analysis identified seven single-nucleotide polymorphisms (SNPs) with suggestive evidence of association with multiple sclerosis (MS). We report an analysis of these polymorphisms in a replication study that includes 8,085 cases and 7,777 controls. A meta-analysis across the replication collections and a joint analysis with the discovery data set were performed. The possible functional consequences of the validated susceptibility loci were explored using RNA expression data. For all of the tested SNPs, the effect observed in the replication phase involved the same allele and the same direction of effect observed in the discovery phase. Three loci exceeded genome-wide significance in the joint analysis: RGS1 (P value = 3.55 × 10–9), IL12A (P = 3.08 × 10–8) and MPHOSPH9/CDK2AP1 (P = 3.96 × 10–8). The RGS1 risk allele is shared with celiac disease (CD), and the IL12A risk allele seems to be protective for celiac disease. Within the MPHOSPH9/CDK2AP1 locus, the risk allele correlates with diminished RNA expression of the cell cycle regulator CDK2AP1; this effect is seen in both lymphoblastic cell lines (P = 1.18 × 10–5) and in peripheral blood mononuclear cells from subjects with MS (P = 0.01). Thus, we report three new MS susceptibility loci, including a novel inflammatory disease locus that could affect autoreactive cell proliferation.
Keywords: multiple sclerosis, susceptibility loci, eQTL
Introduction
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system associated with demyelination, which is thought to have an underlying autoimmune etiology. Increasing evidence indicates that disease onset is associated with environmental factors interacting with an underlying genetic susceptibility. The past 3 years have seen the discovery of susceptibility loci (such as CD58, CLEC16A, IL2RA, IL7RA) outside of the Major Histocompatibility Complex that exceed the generally accepted threshold of genome-wide significance (P value <5 × 10–8) and have been validated in at least two independent publications.1–4 Several other loci have met this level of significance but await further replication.5,6
Recently, we reported the results of a meta-analysis of genome-wide association studies that included 2624 MS subjects and 7220 control subjects.7 The replication phase of this study was performed in an independent set of 2214 MS subjects and 2116 controls, and the combined analysis led to the validation of three new loci that met genome-wide significance: TNFRSF1A, IRF8 and CD6. Furthermore, this analysis highlighted seven loci with suggestive evidence of association to MS: CXCR4 (rs882300, combined P value = 1.37 × 10–7), IL12A (rs4680534, P value = 5.58 × 10–6), MPHOSPH9/CDK2AP1 (rs1790100, P value = 7.21 × 10–7), OLIG3/TNFAIP3 (rs9321619, P value = 1.71 × 10–5), PTGER4 (rs6896969, P value = 2.40 × 10–7), RGS1 (rs2760524, P value = 9.77 × 10–6 and ZMIZ1 (rs1250540, P value = 1.59 × 10–6).
In this study, we report an attempt to validate these seven putative MS susceptibility loci by genotyping the implicated polymorphisms in new samples and then combining the results with those of the published meta-analysis and its replication effort. Subsequently, we go on to explore the possible functional repercussions of three newly confirmed MS susceptibility loci.
Results
We conducted a replication study typing the single-nucleotide polymorphisms (SNPs) rs882300, rs4680534, rs1790100, rs9321619, rs6896969, rs2760524 and rs1250540 in the sample sets described in Table 1. Detailed clinical information on the included subjects is available in Supplementary Table 1. This replication effort is an extension of the formerly reported effort7 and involved 8085 MS patients and 7777 healthy controls. These numbers therefore include the replication sample collections from the United Kingdom and the United States of America that were used previously.7 The data from each sample collection were merged using a fixed-effect meta-analysis strategy, based on the observed and the expected allele dosage.
Table 1.
Sample data sets used in the replication study
| Data sets | Number of cases | Number of controls | Total | Genotyping platform |
|---|---|---|---|---|
| UK-la | 831 | 1030 | 1861 | Sequenom iPLEX Goldb |
| USA-la | 1383 | 1086 | 2469 | Sequenom iPLEX Goldb |
| Finland | 799 | 1082 | 1881 | Sequenom iPLEX Goldb |
| Germany | 937 | 920 | 1857 | Sequenom iPLEX Goldb |
| Italy | 830 | 642 | 1472 | TaqMan (7900 Sequence Detection System)c |
| Sweden | 2103 | 1733 | 3836 | Sequenom iPLEX Goldb |
| UK-2 | 590 | 749 | 1339 | Sequenom iPLEX Goldb |
| USA-2 | 612 | 535 | 1147 | Sequenom iPLEX Goldb |
| Total | 8085 | 7777 | 15862 |
These collections correspond to the ones used in the replication phase of the published genome-wide meta-analysis study.7
SEQUENOM Inc, San Diego, CA, USA.
Applied Biosystems, Foster City, CA, USA.
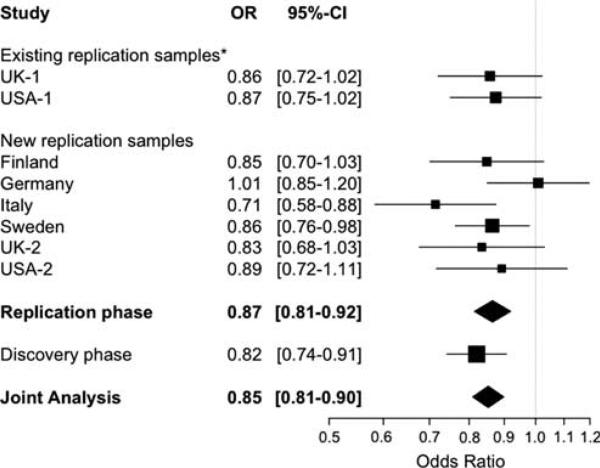
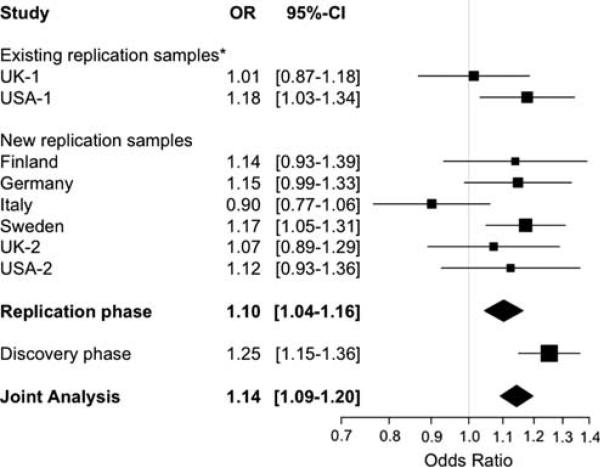
The results of our meta-analysis of the replication samples are described in Table 2. We also conducted a joint analysis, combining the results from the extended replication effort with the results of the discovery phase that was conducted through a genome-wide scan7 (Table 2). For each minor allele, the direction of the association in the replication analysis is consistent with the results of our previous genome-wide meta-analysis.7 Furthermore, six of the seven SNPs exceed a nominal (P value <0.05) threshold of significance in the replication analysis. This observation is unlikely to occur by chance and suggests that most, if not all, of these seven loci will eventually validate as susceptibility loci. At the conclusion of this current replication effort, we have found evidence of association at a genome-wide level of significance (P value <5 × 10–8) in three loci: RGS1 (rs2760524, joint P value = 3.55 × 10–9), IL12A (rs4680534, joint P value = 3.08 × 10–8) and MPHOSPH9/CDK2AP1 (rs1790100, joint P value=3.96 × 10–8). The genetic effect of these three loci in the different sample strata is shown in Figures 1–3. The plots summarizing the meta-analysis results for the remaining four loci are presented in the Supplementary Information (Supplementary Figures 1–4).
Table 2.
Results of the replication and joint analyses
| Replication phase | Discovery phase | Joint analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Locus | BP | A1 | A2 | MAF cases | MAF controls | OR | 95%-CI | Preplication | Pdiscovery | Pjoint |
| 1 | rs2760524 | RGS1 | 190797171 | A | G | 0.150 | 0.169 | 0.87 | (0.81–0.92) | 3.20 × 10–6 | 1.07 × 10–4 | 3.55 × 10–9 |
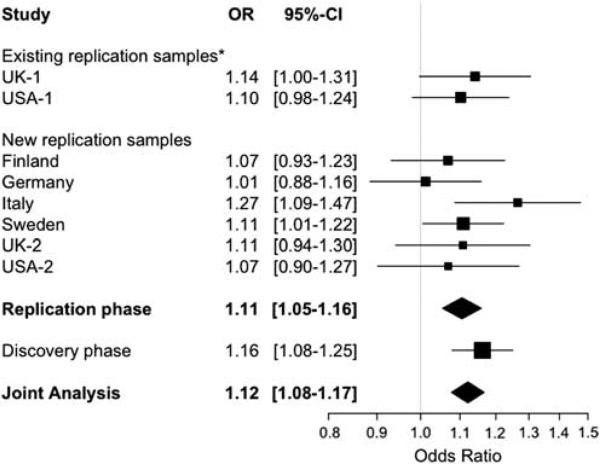
| 3 | rs4680534 | IL12A | 161181639 | C | T | 0.363 | 0.338 | 1.11 | (1.05–1.16) | 2.81 × 10–5 | 6.80 × 10–5 | 3.08 × 10–8 |
| 12 | rs1790100 | MPHOSPH9/CDK2AP1 | 122222678 | G | T | 0.240 | 0.218 | 1.10 | (1.04–1.16) | 3.95 × 10–4 | 2.74 × 10–7 | 3.96 × 10–8 |
| 2 | rs882300 | CXCR4 | 136692725 | T | C | 0.431 | 0.460 | 0.91 | (0.87–0.96) | 9.29 × 10–5 | 5.17 × 10–4 | 4.34 × 10–7 |
| 5 | rs6896969 | PTGER4 | 40460183 | A | C | 0.383 | 0.400 | 0.94 | (0.88–1.00) | 4.11 × 10–2 | 1.44 × 10–7 | 2.80 × 10–6 |
| 10 | rs1250540 | ZMIZ1 | 80706013 | G | A | 0.374 | 0.372 | 1.05 | (0.99–1.11) | 1.18 × 10–1 | 9.89 × 10–6 | 2.31 × 10–4 |
| 6 | rs9321619 | OLIG3/TNFAIP3 | 137916101 | G | A | 0.464 | 0.478 | 0.94 | (0.90–0.99) | 2.29 × 10–2 | 9.34 × 10–4 | 2.85 × 10–4 |
Abbreviations: A1, minor allele; A2, major allele: BP, physical position of the SNP in build 36; chr, chromosome; MAF, minor allele frequency; OR, odds ratio; Pdiscovery P-value from the previous genome-wide meta-analysis study7 (discovery phase); Pjoint, P-value obtained combining the results from the discovery and replication phase; Preplication, P-value obtained pooling together the results from all the replication samples; SNP, single-nucleotide polymorphism.
The MAF in cases and controls was calculated as average allele frequency of the minor allele weighted for the sample size of each replication collection. OR and 95% confidence intervals refer to the meta-analysis performed across all the replication collections. At each locus, the OR is stated relative to the minor allele. 95%-CI refers to the lower and upper bounds of the 95% confidence interval for the OR. All the above results are based on a fixed-effect model. The SNPs are listed in order of decreasing significance at the joint analysis.
Figure 1.
Meta-analysis results within the RGS1 locus. The forest plot summarizes the results obtained for rs2760524 in the RGS1 locus for the replication phase, the discovery phase and the joint analysis. Summary ORs and respective 95% confidence intervals are calculated using the fixed-effect method. *These collections correspond to the ones used previously in the replication arm of the published genome-wide meta-analysis study.7
Figure 3.
Meta-analysis results within the MPHOSPH9/CDK2AP1 locus. The forest plot summarizes the results obtained for rs1790100 in the MPHOSPH9/CDK2AP1 locus for the replication phase, the discovery phase and the joint analysis. Summary ORs and respective 95% confidence intervals are calculated using the fixed-effect method. *These collections correspond to the ones used previously in the replication arm of the published genome-wide meta-analysis study.7
The rs2760524A susceptibility allele (odds ratio (OR) = 0.87, 95% CI=0.81–0.92 in the replication phase) is located on chromosome 1q31 and maps 15 kb distal to the 5′ end of RGS1. This locus has been recently shown to be associated with susceptibility to celiac disease (CD).8 rs2760524 is in strong linkage disequilibrium (LD) with the reported susceptibility SNP for CD (rs2816316, r2 = 0.86 in HapMap 60 unrelated Caucasian individuals of Northern and Western European origin (CEU) samples), and the major allele is associated with susceptibility in both diseases. Further investigations are necessary to understand which one of the two polymorphisms is the better marker and whether the same causal variant affects susceptibility to both diseases.
The rs4680534C allele associated with increased risk of MS (OR = 1.11, 95% CI = 1.05–1.16 in the replication phase) maps ~8 kb distal to the 5′ end of the IL12A gene, which is located on chromosome 3q25.33-q26. For this locus there is evidence of association not only with CD but also with primary biliary cirrhosis.8,9 Our SNP of interest is in modest LD (r2 = 0.48 in HapMap CEU) with rs9811792, one of the susceptibility SNPs for CD; these two SNPs are located in the same LD block immediately 5′ of IL12A. Interestingly, the effect of the rs4680534C allele seems to be different in these two diseases: the risk allele for MS seems to be protective in CD and vice versa. On the other hand, rs4680534 is not in LD with the susceptibility alleles for primary biliary cirrhosis (rs6441286, r2 = 0.02 in HapMap CEU; rs574808, r2<0.001 in HapMap CEU); hence, these signals of association seem to be distinct.
The third locus that met genome-wide significance in the joint analysis contains the rs1790100G susceptibility allele (OR = 1.10, 95% CI = 1.04–1.16 in the replication phase). This polymorphism is located in intron 12 of the MPHOSPH9 gene on chromosome 12q24.31. This region has not been previously identified as a susceptibility locus for inflammatory diseases, and little is known about it.
The remaining four loci have some evidence of replication but do not yet reach the threshold of genome-wide significance. These results could be influenced by the fact that, for three out of these four loci, the implicated polymorphisms were not typed in all of the sample collections because of technical problems (Supplementary Table 2), hence the available sample size was smaller compared with the one used to test the other polymorphisms. Nevertheless, they remain strong candidate MS susceptibility loci, particularly the two SNPs mapping in the PTGER4 and OLIG3/TNFAIP3 loci, which are already known to be associated with other autoimmune diseases.10–12
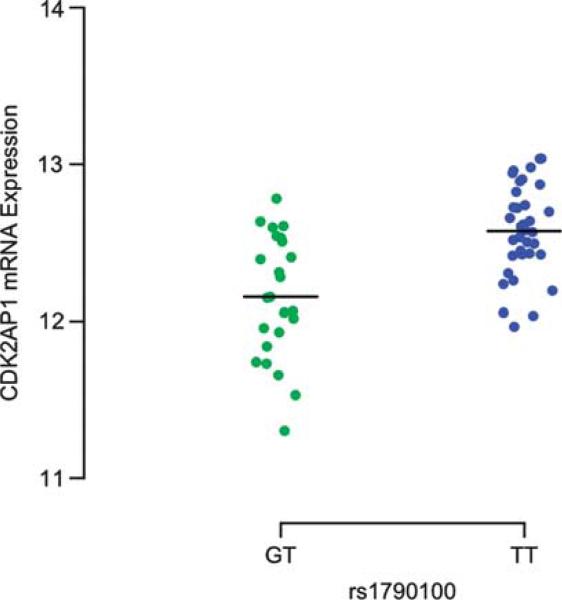
We also explored the possible functional consequences of the seven polymorphisms by performing a quantitative trait analysis that correlates genotyping data with gene expression data. Specifically, we correlated genotype data from the HapMap phase II study13 with publicly available gene-expression data from lymphoblastic cell lines (LCLs) of 60 unrelated subjects of Northern and Western European origin (parents of the CEU trios) using an additive model. Only one of the seven SNPs, rs1790100 in the MPHOSPH9/CDK2AP1 locus, shows evidence of association in ‘cis’: the risk-associated allele correlates with lower CDK2AP1 RNA expression (Figure 4) in the CEU LCL data (linear regression P value = 1.18 × 10–5). The CDK2AP1 gene lies ~40 kb telomeric to the 3′ end of MPHOSPH9 and is within the block of LD containing rs1790100 (Supplementary Figure 5). The correlation of rs1790100G with RNA expression is specific to CDK2AP1: in the available RNA data, there is no significant association with the RNA expression of the other genes located in the same LD block (Supplementary Table 3).
Figure 4.
CDK2AP1 RNA expression relative to rs1790100 in LCLs. The plot illustrates the distribution of CDK2AP1 expression values by genotype classes in LCLs from CEU. A reduced CDK2AP1 expression is observed in the presence of the rs1790100G susceptibility allele. No rs1790100GG homozygotes were observed in this sample of LCLs. A black line denotes the mean value for each category.
Given this result, we analyzed CDK2AP1 gene expression in another publicly available data set, the ‘mRNA-by-SNP-browser’14 (see URL). In this data set generated from 400 LCLs, two of the SNPs associated with the level of CDK2AP1 RNA expression are in strong LD with rs1790100, our SNP of interest (rs1727324, r2 = 1 in HapMap CEU; rs1060105, r2 = 0.87 in HapMap CEU). The minor alleles of these two SNPs correlate with a lower expression of CDK2AP1 (rs1727324, P value = 1.70 × 10–5; rs1060105, P value = 1.60 × 10–4), confirming the results we obtained using the rs1790100 marker and the HapMap LCLs.
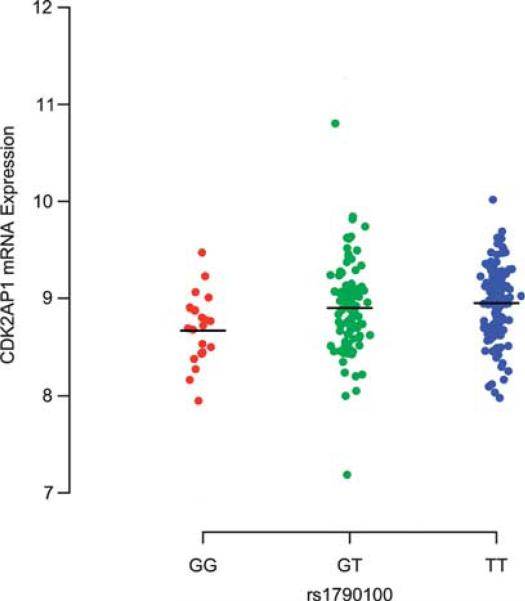
Finally, we further explore these in vitro observations derived from LCLs using ex vivo RNA data obtained from peripheral blood mononuclear cells (PBMCs) in 255 relapsing–remitting MS and clinically isolated demyelinating syndrome (CIS) subjects3 (Supplementary Table 4); as in LCLs, an additive model is used for this quantitative trait analysis. As some of the subjects were treated with an immunomodulatory drug at the time of sampling, we first used the Welch 2 sample t-test to compare the level of CDK2AP1 RNA expression across the treatment regimen categories (Glatiramer acetate/interferon-β/untreated). As seen in Supplementary Figure 6, there is no significant difference in the expression of CDK2AP1 RNA among individuals who were untreated (n = 83), treated with Glatiramer acetate (n = 67) or treated with an interferon-β formulation (n = 105) at the time of sampling. Thus, we merged the RNA data from the three subject categories and correlated the CDK2AP1 expression levels with the rs1790100 genotype. The results of this ex vivo PBMC data analysis are consistent with the results of the LCL analyses: reduced CDK2AP1 expression is observed in the presence of the rs1790100G susceptibility allele (additive model P value = 0.01) (Figure 5). Adding treatment category as a covariate did not change the results (P value = 0.01), giving further evidence to the fact that immunomodulatory drugs do not affect the expression profile of CDK2AP1. Finally, we note that, in our ex vivo data, MS subjects who are homozygous for the risk (‘G’) allele have a substantial reduction of the CDK2AP1 expression profile compared with the other two genotype classes. We therefore tested a recessive model of inheritance, which seems to fit the data better (P value = 0.005). On the other hand, the LCL data are more consistent with an additive model, and additional in vitro and ex vivo data will be useful to dissect the exact nature of the effect of rs1790100G under these different conditions.
Figure 5.
CDK2AP1 RNA expression relative to rs1790100 in PBMCs from subjects with relapsing–remitting MS and CIS. The plot illustrates the correlation between the lower CDK2AP1 RNA expression and the rs1790100G susceptibility allele in RNA data obtained from mononuclear cells of subjects with relapsing–remitting MS and CIS. A black line denotes the location of the mean value for each category.
Discussion
We tested seven putative susceptibility loci for association with MS using an expanded repertoire of independent collections of case–control samples from Finland, Germany, Italy, Sweden, the United Kingdom and the United States of America. The effect observed for each polymorphism in the replication phase is in the same direction as the effect observed in the discovery phase. A joint analysis of replication and discovery data sets also provides robust evidence of association between MS susceptibility and the RGS1 (combined P value = 3.55 × 10–9), IL12A (combined P value = 3.08 × 10–8) and MPHOSPH9/CDK2AP1 (combined P value = 3.96 × 10–8) loci. When comparing the effects of an allele across studies, the IL12A, OLIG3/TNFAIP3, PTGER4 and RGS1 loci were the most consistent: we detected no evidence of between-strata heterogeneity (Supplementary Table 5). Modest heterogeneity was observed in CXCR4 and MPHOSPH9/CDK2AP1 loci; however, the only allele with significant heterogeneity is the one found in the ZMIZ1 locus (I2 = 64.7%, 95% CI = 15.1%–85.4%, P value = 0.01) (Supplementary Table 5). This heterogeneity is most likely due to small sample sizes, as each of the collection remains modest in size and has therefore limited power on its own to provide an accurate estimate of the magnitude of an allele's effect on MS susceptibility. However, we only genotyped seven SNPs across the various sample collections, and therefore we cannot empirically assess and correct for possible population stratification in the different strata of the meta-analysis.
The RGS1 gene encodes a molecule that is a member of the regulators of G-protein signaling (RGS) family. RGS proteins are involved in lymphocyte migration and can influence cell trafficking both during the development of the immune system and during responses to exogenous or infectious agents.15 Specifically, it has been observed that, in RGS1 knockout mice, B cells have a better adhesion to high endothelial venules in lymph nodes; they home better to lymph nodes and move more rapidly within lymph node follicles than do wild-type B cells,16 supporting an involvement of RGS1 in the B-cell mobility into and out of lymph nodes. We can therefore speculate that alterations in RGS1 function mediated by allelic variants could impact the migratory capability of B cells and possibly alter their recruitment to the central nervous system.
The IL12A gene encodes the IL12p35 subunit, which, together with IL12p40, forms the heterodimeric IL-12 cytokine that has a broad range of biological activities. IL-12 is an immunomodulatory cytokine secreted predominantly by monocytes and dendritic cells, and its signaling is thought to be crucial for T-helper 1 (Th1) lymphocyte differentiation; moreover, IL-12 has also been shown to stimulate interferon gamma (IFN-γ) production by T cells and NK cells and to suppress the expansion of T-helper 2 (Th2) cell clones that are thought to have anti-inflammatory properties in MS.17 IL-12, together with IL-23, has been strongly implicated in the pathogenesis of both experimental autoimmune encephalomyelitis (EAE) and MS,18 and antibodies neutralizing IL12p40 prevent clinical EAE in nonhuman primates.19 However, the efficacy of this category of drugs in subjects affected by relapsing–remitting MS has not yet been shown.20
It is interesting to observe that genetic variants in both RGS1 and IL12A have recently been observed to be associated with CD. It is possible that the polymorphisms in the RGS1 locus are tagging SNPs for a common causative inflammatory disease variant that remains to be discovered. This result highlights the evolving story of shared susceptibility loci among different inflammatory disease loci. On the other hand, the IL12A locus illustrates the existence of heterogeneity among the reported associations for a given locus; as seen in other inflammatory disease loci, we report evidence that the allelic variant associated with MS susceptibility is protective for CD.
We refer to the third new MS susceptibility locus as the MPHOSPH9/CDK2AP1 locus, as the best marker, rs1790100, is found within the MPHOSPH9 gene but is strongly correlated with the expression of the neighboring CDK2AP1 gene. Little is known of the function of MPHOSPH9, aside from its homology to M-phase phosphoproteins that are involved in the transition from interphase G2 to mitosis (M) during the cell cycle. The potential biological effect of MPHOSPH9 in MS pathogenesis cannot be readily deduced. However, it is important to remember that all seven of the tested SNPs are most likely to be merely markers for the causal alleles. In fact, it is possible that the causal variant in LD with rs1790100 is physically located within the neighboring CDK2AP1 gene, for which we have evidence that the MS susceptibility allele affects the level of RNA expression in vitro and ex vivo. CDK2AP1 is a highly conserved cellular gene that functions as an S-phase growth suppressor. It encodes p12DOC-1, a negative regulator of DNA replication.21 In addition, it has been observed that p12DOC-1 can induce apoptosis and reduce cell proliferation.22 The MS susceptibility allele rs1790100G is associated with a lower expression of CDK2AP1, suggesting that there may be a reduced inhibition of DNA replication and proliferation mediated by p12DOC-1 in subjects with the risk allele. In the context of MS pathogenesis, we can therefore hypothesize an involvement of this gene in the proliferation and clonal expansion of inflammatory cells. Further functional investigations of this chromosomal region are necessary to refine our understanding of its role in MS and to identify the functional consequence of the causal variant at this locus.
For the remaining loci, we find no evidence of a correlation between the genetic variants and RNA data obtained using the probes contained in the Illumina WG-6 version 1 array of the HapMap phase II experiment. Clearly, more detailed investigations of different cell types and RNA isoforms are indicated to further explore this question.
The list of accepted MS susceptibility loci at this point consists largely of loci with primarily immunological functions: CD6, CD58, CLEC16A, CYP27B1, HLA B, HLA DRB1, IL2RA, IL7R, IRF8, STAT3, TNFRSF1A and TYK2.1–5,7,23–25 We can now add the IL12A, MPHOSPH9/CDK2AP1 and RGS1 loci to this list. Our results are in agreement with recently published genetic studies on MS: the newly confirmed susceptibility alleles are common in the general population, but confer only a modest risk to MS. Thus, individually, these alleles have no predictive power, but aggregate measures of genetic risk seem to be robust in MS and represent a potential strategy to use this information in a clinical setting.26
Several other genes have been put forth as being associated in this and other reports, but they still need to be independently validated. The best available evidence for association is for the 13q31.3, CD40, CD226, CXCR4, GPC5, PTGER4 and TNFAIP3 loci.5,7,27–29KIF1B is an interesting locus that had strong evidence of association in the initial report,6 but this effect has not been observed in additional sample collections tested so far (SJ Sawcer, personal communication). Overall, the list of involved loci is evolving rapidly and will be enriched further by ongoing genome-wide scans in MS.
Recent studies have also highlighted the observation that, in addition to common variants, less common variants can have a role in susceptibility to MS. This is the case in the TNFRSF1A locus,7 which harbors a less common variant (frequency 0.02) with a strong effect (OR = 1.6). It is therefore clear that we need to supplement the current efforts aimed to comprehensively characterize common variants in MS with studies exploring the role of less common and rare variants. The current generation of sequencing platforms is making such studies practical and will allow us to enrich our understanding of the genetic architecture of MS.
Materials and methods
Subjects
This study was conducted within the framework of the International Multiple Sclerosis Genetics Consortium (IMSGC). Samples were collected across different countries, namely, Finland, Germany, Italy, Sweden, United Kingdom and the United States of America. For these last two countries, the subjects were recruited in two different stages: 2214 cases and 2116 controls were initially used as a first replication attempt in our published meta-analysis paper7 (they correspond to the UK-1 and USA-1 data sets in Table 1). Additional individuals were then assessed to confirm the results in the current study (UK-2 and USA-2 data sets in Table 1). Specifically, the latter patient samples were ascertained at two sites within the United States of America (Brigham and Women's Hospital in Boston and the University of California in San Francisco) and from across the United Kingdom. Unrelated controls were obtained from the same US sites and from the British 1958 Birth Cohort Study. In Germany, MS subjects and controls were recruited from the whole country by primary care physicians and neurologists. In the Italian collection, most of the cases and controls were resident of Northern Italy (mainly Northwest Italy). For the majority of them, the place of birth of their grandparents was collected and individuals with Sardinian ancestors were excluded from our analysis, given the unique ancestry of the Sardinian population.30 The Finnish and Swedish sample collections have previously been described.31,32 Summary information on the number of cases and controls recruited in each sample collection is listed in Table 1. The demographic and clinical profiles of the samples are presented in Supplementary Table 1. All subjects were of self-reported European ancestry and met the McDonald criteria for a diagnosis of MS33,34 or for a diagnosis of CIS35, which is often a prelude to a diagnosis of MS and whose genetic architecture is similar to MS.26 We obtained written informed consent from all subjects using documents that were reviewed and approved by the local institutional ethics committee at each center. Genomic DNA was extracted using standard methods.
Genotyping
SNP genotyping was performed at each center using different platforms (Table 1). Because of technical reasons, not all of the SNPs were successfully genotyped in each sample collection (Supplementary Table 2). In particular, SNP rs9321619 failed genotyping in the collections from Finland and Germany; SNP rs6896969 failed genotyping in collections from Finland, Germany and Sweden; and SNP rs1250540 was not genotyped in collections from Germany and Sweden. We took into account the differences in the number of cases and controls across the polymorphisms when we analyzed the data (see the section on statistical analysis). We used PLINK v1.0636 for measuring minor allele frequency, heterozygosity and evidence of deviation from Hardy–Weinberg equilibrium in each data set. The minor allele frequency in cases and controls across all the replication samples was calculated as the average allele frequency of the minor allele weighted for the sample size of each collection. Basic marker performance characteristics for the seven SNPs analyzed are shown in Supplementary Table 6. None of the SNPs significantly deviated from Hardy–Weinberg equilibrium in controls (P value <0.001).
Statistical analysis
We performed a meta-analysis across all of the replication collections assuming a per-allelic model. The minor allele was selected as the reference allele for each polymorphism, and the analyses generated the OR per copy of the reference allele. In our primary analysis, we synthesized data using a fixed-effect meta-analysis approach.37 Secondarily, we also evaluated a random-effects approach38 for our meta-analysis of the replication data. Between-study heterogeneity was assessed with Cochran's Q-test39 and quantified using I2 (and respective 95% confidence intervals).40 We also performed a joint analysis, combining the results from the extended replication effort with the results of the discovery phase that was conducted through a genome-wide scan.7 To combine statistics across different strata, we calculated the respective Z score in each sample collection; in this manner, we summarized both the P value in its magnitude and the direction of the effect in its sign (Z>0 for OR > 1). Thereafter, we obtained an overall Z statistic as a weighted average of each individual Z score, and we calculated the corresponding P value.41 The joint Z score was calculated on the basis of an estimated effective sample size of 2000 cases and 2000 controls for the discovery phase study in subjects genotyped genome wide7 and the number of informative individuals typed across the different SNPs for the replication phase results. All of these statistical analyses were performed in R statistical packages (see URLs).
Lymphoblastic cell line RNA data and quantitative trait analysis
To explore the possible functional consequences of the seven selected genetic variants, we initially used the gene expression data from LCLs of 60 individuals of European ancestry (CEU subjects) genotyped in the International HapMap Project phase II. Transcript levels were obtained from a public database (see URL) in which the RNA expression of each LCL was profiled with an Illumina whole-genome expression array (Sentrix Human-6 Expression BeadChip version 1). Out of the 47 294 transcripts that were interrogated, the normalized values for 14 925 transcripts (14 072 genes) were available for a cis expression QTL (eQTL) analysis.42 We tested for association between SNP and expression variation using a linear regression model. To discover cis effects, only those genes found within the same block of LD as the SNP of interest were tested.
Possible regulatory effects of the seven SNPs were also assessed using the ‘mRNA-by-SNP-browser’ (see URL),14 which contains the results of SNP/RNA expression correlations from 400 LCLs derived from British individuals. This database incorporates association results of ~50 000 transcripts with more than 400 000 SNPs.
Mononuclear cell RNA data from subjects with MS
We used ex vivo data to explore the association of rs1790100 and CDK2AP1 RNA expression. Between July 2002 and October 2007, PBMC samples were collected from relapsing–remitting MS and CIS subjects, as part of the comprehensive longitudinal investigation on MS at the Brigham and Women's Hospital.43 CIS subjects have only one clinical episode of demyelination, whereas MS subjects must have at least two of these episodes or one clinical event and evidence of disease activity in a paraclinical measure such as MRI.33 Nevertheless, these two clinical categories share common pathophysiological events and are also treated in the same manner in the clinical environment. This is why for this analysis we pooled together the RNA data from relapsing–remitting MS and CIS individuals. Some of these subjects were treated with an immunomodulatory drug (Glatiramer acetate or interferon-β) at the time of recruitment. Given the limited number of subjects, we pooled all individuals receiving one of the interferon-β formulations (IFNb1a IM, IFNb1a SC or IFNb1b SC) into a single interferon-β category. None of these subjects were on combination therapy for MS at the time of sampling. PBMC isolation, RNA extraction and quantification and quality control steps are described in detail in a previous publication;3 the expression data based on a Human Genome U133 Plus 2.0 array are available on the Gene Expression Omnibus website.
From our collection, 255 subjects had a CDK2AP1 RNA expression profile and also had a genotype for rs1790100. The quantitative trait analysis module implemented in PLINK v1.0636 was used for the analysis of expression data. A Welch 2 sample t-test was applied to compare the treatment regimen categories (Glatiramer acetate/interferon-β/untreated) across samples for the CDK2AP1 RNA expression levels.
Supplementary Material
Figure 2.
Meta-analysis results within the IL12A locus. The forest plot summarizes the results obtained for rs4680534 in the IL12A locus for the replication phase, the discovery phase and the joint analysis. Summary ORs and respective 95% confidence intervals are calculated using the fixed-effect method. *These collections correspond to the ones used previously in the replication arm of the published genome-wide meta-analysis study.7
Acknowledgements
We are grateful to the patients and the healthy controls for their participation in this study. PLD is a Harry Weaver Neuroscience Scholar Award Recipient of the National MS Society (NMSS). DAH is a Jacob Javits Scholar of NIH. SD is supported by a FISM grant (2008/R/11). BF and IR are authors on behalf of REFGENSEP, a national French clinical and research network funded by INSERM, ARSEP and AFM. The International MS Genetics Consortium is supported by R01NS049477 and by the National Multiple Sclerosis Society. This work was also supported by the Medical Research Council (G0700061) and by the Cambridge NIHR Biomedical Research Centre. The sampling and analysis of the Swedish cohorts have received grant support from The Swedish Research Council, the EU fp6 program neuro-promise (LSHM-CT-2005-018637), as well as from the Bibbi and Niels Jensens Foundation, The Montel Williams Foundation and the Söderberg Foundation.
Appendix
Members of the International Multiple Sclerosis Genetics Consortium
F Esposito1,2, NA Patsopoulos3,4,5, S Cepok6, I Kockum7,8, V Leppä9,10, DR Booth11, RN Heard11, GJ Stewart11, M Cox12, RJ Scott12, J Lechner-Scott12, A Goris13, R Dobosi13, B Dubois13, JD Rioux14,15, AB Oturai16, HB Søndergaard16, F Sellebjerg16, PS Sørensen16, M Reunanen17, K Koivisto18, I Cournu-Rebeix19,20,21, B Fontaine19,20,21, J Winkelmann22,23,24, C Gieger25, C Infante-Duarte26, F Zipp27, L Bergamaschi28, M Leone29, R Bergamaschi30, P Cavalla31, ÅR Lorentzen32,33, I-L Mero33,34, EG Celius34, HF Harbo32,34, A Spurkland35, M Comabella36, B Brynedal7, L Alfredsson37, L Bernardinelli38,39, NP Robertson40, CP Hawkins41,42, LF Barcellos43, G Beecham44, W Bush45, BAC Cree46, MJ Daly4,47,48, AJ Ivinson49, C Aubin48, A Compston50, S D'Alfonso28, JL Haines45, SL Hauser46, B Hemmer6, J Hillert8,37, JL McCauley44, J Oksenberg46, T Olsson7,8, A Palotie9,10, L Peltonen9,10,51, MA Pericak-Vance44, J Saarela9,10, SJ Sawcer50, B Stranger3,4,48, FM Boneschi1,2, G Comi1,2, DA Hafler4,48,52, PIW de Bakker3,4,48 and PL De Jager4,5,48, International Multiple Sclerosis Genetics Consortium members.
1Department of Neurology, Scientific Institute San Raffaele, Milan, Italy; 2Institute of Experimental Neurology, Scientific Institute San Raffaele, Milan, Italy; 3Division of Genetics, Department of Medicine, Brigham & Women's Hospital, Boston, MA, USA; 4Harvard Medical School, Boston, MA, USA; 5Program in Translational NeuroPsychiatric Genomics, Department of Neurology, Brigham & Women's Hospital, Boston, MA, USA; 6Klinikum rechts der Isar, Technische Universität, Münch, Germany; 7Neuroimmunology Unit, Department of clinical neuroscience, Karolinska Institutet at Karolinska University Hospital, Solna, Sweden; 8Center for Molecular Medicine, Karolinska Institutet at Karolinska University Hospital, Solna, Sweden; 9Public Health Genomics Unit, National Institute for Health and Welfare, Helsinki, Finland; 10Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland; 11University of Sydney, Institute for Immunology and Allergy Research, Westmead Millennium Institute, Westmead Hospital, NSW, Australia; 12University of Newcastle, Callaghan, NSW Australia; 13Section for Experimental Neurology, Katholieke Universiteit Leuven, Leuven, Belgium; 14Université de Montréal, Montréal, Québec, Canada; 15Montreal Heart Institute, H1 T 1C8, Montréal, Québec, Canada; 16The Danish Multiple Sclerosis Research Center, Copenhagen University Hospital, Rigshospitalet, DK-2100, Copenhagen, Denmark; 17Department of Neurology, Oulu University Hospital, Oulu, Finland; 18Central Hospital of Seinäjoki, Seinäjoki, Finland; 19INSERM, UMR_S975, Paris, France; 20UPMC Univ Paris 06, UMR_S975, Centre de Recherche Institut du Cerveau et de la Moelle, CNRS 7225, Paris, France; 21Département de Neurologie, Pitié –Salpêtrière Hospital, AP-HP, 75651 Paris, France; 22Klinik für Neurologie, Technische Universität München, München, Deutsch; 23Institut für Humangenetik, Technische Universität München, München, Deutsch; 24Institut für Humangenetik, Helmholtz Zentrum München, München, Deutsch; 25Institute of Epidemiology, Helmholtz Zentrum München-German Research Center for Environmental Health, Munich, Germany; 26Charite-Universitaetsmedizin Berlin, Cecilie Vogt Clinic for Neurology, Berlin, Germany; 27Department of Neurology, University Medicine Mainz, Johannes Gutenberg University, Mainz, Germany; 28Department of Medical Sciences and Interdisciplinary Research Center of Autoimmune Diseases, University of Eastern Piedmont, Novara, Italy; 29Clinica Neurologica, AOU Maggiore della Carità, Novara, Italy; 30Neurological Institute C. Mondino, IRCCS, Pavia, Italy;–31Department of Neurology, Ospedale San Giovanni Battista, Torino, Italy; 32Department of Neurology, Faculty Division Ullevål University Hospital, University of Oslo, Oslo, Norway; 33Institute of Immunology, Rikshospitalet, Oslo University Hospital, Oslo, Norway; 34Department of Neurology, Oslo University Hospital, Ullevål, Oslo, Norway; 35Institute of Basal Medical Sciences, University of Oslo, Blindern, Oslo, Norway; 36Unitat de Neuroimmunologia Clínica, CEM-Cat, Hospital Universitari Vall d'Hebron, Barcelona, Spain; 37Institute of Environmental Medicine, Karolinska Institutet, Stockholm, Sweden; 38University of Pavia, Department of Applied Sciences, Pavia, Italy; 39University of Cambridge, Statistical Laboratory, Centre for Mathematical Sciences, Wilberforce Road, Cambridge, UK; 40Department of Neurology, University Hospital of Wales, Heath Park, Cardiff, UK; 41Human Genomics research group, Keele University, Stoke-on-Trent, UK; 42Department of Neurology, University Hospital North Staffordshire, Stoke-on-Trent, UK; 43Division of Epidemiology, School of Public Health, University of California at Berkeley, Berkeley, CA, USA; 44John P. Hussman Institute for Human Genomics, The University of Miami Miller School of Medicine, Miami, FL, USA; 45Center for Human Genetics Research, Vanderbilt University Medical Center, Nashville, TN, USA; 46Department of Neurology, University of California San Francisco, San Francisco, CA, USA; 47Massachusetts General Hospital, Boston, MA, USA; 48Program in Medical & Population Genetics, Broad Institute of Harvard University and Massachusetts Institute of Technology, Cambridge, MA, USA; 49Harvard Neuro-Discovery Center, Harvard Medical School, Boston, MA, USA; 50University of Cambridge, Department of Clinical Neuroscience, Addenbrooke's Hospital, Cambridge, UK; 51The Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK and 52Department of Neurology, Yale University Medical School, New Haven, CT, USA.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Web Resources
The URLs for data presented herein are as follows:
International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/
GENe Expression VARiation, http://www.sanger.ac.uk/humgen/genevar/
meta: Meta-Analysis, R package http://CRAN.R-project.org/package=meta
rmeta: Meta-Analysis, R package http://CRAN.R-project.org/package=rmeta
‘mRNA-by-SNP-browser’ v1.0.1, http://www.sph.umich.edu/csg/liang/asthma/
The Gene Expression Omnibus accession number for the RNA data obtained from PBMCs and reported in this paper is GSE16214.
Supplementary Information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene)
References
- 1.International Multiple Sclerosis Genetics Consortium (IMSGC) The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun. 2009;10:11–14. doi: 10.1038/gene.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Multiple Sclerosis Genetics Consortium (IMSGC) Refining genetic associations in multiple sclerosis. Lancet Neurol. 2008;7:567–569. doi: 10.1016/S1474-4422(08)70122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Jager P, Baecher-Allan C, Maier LM, Arthur AT, Ottoboni L, Barcellos L, et al. The role of the CD58 locus in multiple sclerosis. Proc Natl Acad Sci USA. 2009;106:5264–5269. doi: 10.1073/pnas.0813310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Multiple Sclerosis Genetics Consortium (IMSGC) Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 5.Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 6.Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J, et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 2008;40:1402–1403. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]
- 7.De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trynka G, Zhernakova A, Romanos J, Franke L, Hunt KA, Turner G, et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut. 2009;58:1078–1083. doi: 10.1136/gut.2008.169052. [DOI] [PubMed] [Google Scholar]
- 13.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 15.Moratz C, Harrison K, Kehrl JH. Regulation of chemokine-induced lymphocyte migration by RGS proteins. Methods Enzymol. 2004;389:15–32. doi: 10.1016/S0076-6879(04)89002-5. [DOI] [PubMed] [Google Scholar]
- 16.Han SB, Moratz C, Huang NN, Kelsall B, Cho H, Shi CS, et al. Rgs1 and Gnai2 regulate the entrance of B lymphocytes into lymph nodes and B cell motility within lymph node follicles. Immunity. 2005;22:343–354. doi: 10.1016/j.immuni.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 18.Segal BM, Dwyer BK, Shevach EM. An interleukin (IL)-10/IL-12 immunoregulatory circuit controls susceptibility to auto-immune disease. J Exp Med. 1998;187:537–546. doi: 10.1084/jem.187.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brok HP, van Meurs M, Blezer E, Schantz A, Peritt D, Treacy G, et al. Prevention of experimental autoimmune encephalomyelitis in common marmosets using an anti-IL-12p40 monoclonal antibody. J Immunol. 2002;169:6554–6563. doi: 10.4049/jimmunol.169.11.6554. [DOI] [PubMed] [Google Scholar]
- 20.Segal BM, Constantinescu CS, Raychaudhuri A, Kim L, Fidelus-Gort R, Kasper LH. Repeated subcutaneous injections of IL12/23 p40 neutralising antibody, ustekinumab, in patients with relapsing-remitting multiple sclerosis: a phase II, double-blind, placebo-controlled, randomised, dose-ranging study. Lancet Neurol. 2008;7:796–804. doi: 10.1016/S1474-4422(08)70173-X. [DOI] [PubMed] [Google Scholar]
- 21.Shintani S, Ohyama H, Zhang X, McBride J, Matsuo K, Tsuji T, et al. p12(DOC-1) is a novel cyclin-dependent kinase 2-associated protein. Mol Cell Biol. 2000;20:6300–6307. doi: 10.1128/mcb.20.17.6300-6307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohno Y, Patel V, Kim Y, Tsuji T, Chin BR, Sun M, et al. Apoptosis, proliferation and p12(doc-1) profiles in normal, dysplastic and malignant squamous epithelium of the Syrian hamster cheek pouch model. Oral Oncol. 2002;38:274–280. doi: 10.1016/s1368-8375(01)00055-0. [DOI] [PubMed] [Google Scholar]
- 23.Jakkula E, Leppa V, Sulonen AM, Varilo T, Kallio S, Kemppinen A, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010;86:285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cenit MC, Alcina A, Marquez A, Mendoza JL, Diaz-Rubio M, de Las Heras V, et al. STAT3 locus in inflammatory bowel disease and multiple sclerosis susceptibility. Genes Immun. 2010;11:264–268. doi: 10.1038/gene.2010.10. [DOI] [PubMed] [Google Scholar]
- 25.Mero IL, Lorentzen AR, Ban M, Smestad C, Celius EG, Aarseth JH, et al. A rare variant of the TYK2 gene is confirmed to be associated with multiple sclerosis. Eur J Hum Genet. 2009;18:502–504. doi: 10.1038/ejhg.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Jager PL, Chibnik LB, Cui J, Reischl J, Lehr S, Simon KC, et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1111–1119. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18:767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comabella M, Craig DW, Camina-Tato M, Morcillo C, Lopez C, Navarro A, et al. Identification of a novel risk locus for multiple sclerosis at 13q31.3 by a pooled genome-wide scan of 500,000 single nucleotide polymorphisms. PLoS One. 2008;3:e3490. doi: 10.1371/journal.pone.0003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hafler JP, Maier LM, Cooper JD, Plagnol V, Hinks A, Simmonds MJ, et al. CD226 Gly307Ser association with multiple autoimmune diseases. Genes Immun. 2009;10:5–10. doi: 10.1038/gene.2008.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Alfonso S, Bolognesi E, Guerini FR, Barizzone N, Bocca S, Ferrante D, et al. A sequence variation in the MOG gene is involved in multiple sclerosis susceptibility in Italy. Genes Immun. 2008;9:7–15. doi: 10.1038/sj.gene.6364437. [DOI] [PubMed] [Google Scholar]
- 31.Kallio SP, Jakkula E, Purcell S, Suvela M, Koivisto K, Tienari PJ, et al. Use of a genetic isolate to identify rare disease variants: C7 on 5p associated with MS. Hum Mol Genet. 2009;18:1670–1683. doi: 10.1093/hmg/ddp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundmark F, Duvefelt K, Iacobaeus E, Kockum I, Wallstrom E, Khademi M, et al. Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat Genet. 2007;39:1108–1113. doi: 10.1038/ng2106. [DOI] [PubMed] [Google Scholar]
- 33.McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50:121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 34.Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the ‘McDonald Criteria’. Ann Neurol. 2005;58:840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 35.Dalton CM, Brex PA, Miszkiel KA, Hickman SJ, MacManus DG, Plant GT, et al. Application of the new McDonald criteria to patients with clinically isolated syndromes suggestive of multiple sclerosis. Ann Neurol. 2002;52:47–53. doi: 10.1002/ana.10240. [DOI] [PubMed] [Google Scholar]
- 36.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 38.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 39.Cochran W. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 40.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 41.de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gauthier SA, Mandel M, Guttmann CR, Glanz BI, Khoury SJ, Betensky RA, et al. Predicting short-term disability in multiple sclerosis. Neurology. 2007;68:2059–2065. doi: 10.1212/01.wnl.0000264890.97479.b1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.