Abstract
Background:
Dietary phytochemicals consist of a wide variety of biologically active compounds that are ubiquitous in plants, many of which have been reported to have anti-tumor as well as anti-inflammatory properties.
Objective:
In the present study, we aimed to validate these findings by using docking protocols and explicate the possible mechanism of action for a dataset of nine phytochemicals namely boswellic acid, 1-caffeoylquinic acid, ellagic acid, emodin, genistein, guggulsterone, quercetin, resveratrol, and sylibinin from different plants against the nuclear factor- kappaB (NF-κB) precursor protein p105, an important transcription factor reported to be overexpressed in breast cancer.
Materials and Methods:
2-D structures of all phytochemicals were retrieved from PubChem Compound database and their subsequent conversion into 3-D structures was performed by using online software system CORINA. The X-ray crystallographic structure of the NF-κB precursor p105 was extracted from Brookhaven Protein Data Bank. Molecular docking simulation study was carried out by using AutoDock Tools 4.0.
Results:
Our results showed significant binding affinity of different phytochemicals with the Rel homology domain of the NF-κB precursor protein p105. Quercetin and 1-caffeoylquinic acid were found to be very effective inhibitors against target molecule as they showed binding energy of −12.11 and −11.50 Kcal/mol, respectively. The order of affinity of other ligands with p105 was found as follows: guggulsterone > sylibinin > emodin > resveratrol > genistein > boswellic acid > ellagic acid.
Conclusion:
Our in silico study has explored the possible chemopreventive mechanism of these phytochemicals against the NF-κB precursor protein p105 and deciphered that quercetin, 1-caffeoylquinic acid and guggulsterone were the potent inhibitors against target molecule. In addition, large scale preclinical and clinical trials are needed to explore the role of these chemotherapeutic molecules against the NF-κB precursor protein p105 in cure and prevention of breast cancer.
Keywords: 1-Caffeoylquinic acid, boswellic acid, breast cancer, emodin, guggulsterone, molecular docking, NF-κB, rel homology domain, sylibinin
INTRODUCTION
Breast cancer is the most commonly diagnosed cancer and the primary cause of cancer death in females worldwide. In general, incidence rates are high in Western and Northern Europe, Australia, New Zealand, North America, intermediate in South America, Northern Africa, low in sub-Saharan Africa, and Asia.[1] However, in many African and Asian countries including India, incidence and mortality rates have been rising.[2,3] According to Indian Council of Medical Research (ICMR) 2010 bulletin, breast cancer is the number one cancer in females with 50 000 deaths in the year 2010 and the annual rate of increase is about 2.7%. The factors that contribute to variation in incidence rates largely stem from differences in reproductive and hormonal factors and the availability of early detection services.[4,5]
Previously published reports indicate several molecular mechanisms which are responsible for breast cancer and the NF-κB pathway is one of them which have been reported to be highly activated in breast cancer.[6,7] There is growing evidence implicating all vertebrate Rel/NF-κB transcription factors in human tumors.[8,9] The transcription factors of the Rel/NF-κB family play an essential role in a number of physiological processes including inflammatory, stress, and immune responses, apoptosis and cellular proliferation.[10–12] The members of this family include RelA (p65), RelB, c-Rel, p50/p105 (NF-κB1), and p52/p100 (NF-κB2).[13] These proteins are structurally-related through an approximately 300 amino acid N-terminal domain called the Rel homology (RH) domain, which contains sequences important for DNA binding, dimerization, and inhibitor (IκB) binding. The C-terminal halves of RelA, RelB, and c-Rel contain transcriptional activation domains, whereas the C-terminal halves of p105 and p100 contain inhibitory domains. The mature DNA-binding forms of p105 and p100 are shortened forms called p50 and p52, respectively. Nearly all Rel/NF-κB proteins can form homodimers and heterodimers, which bind to DNA target sites to influence gene expression. The most common dimer is a p50-RelA heterodimer, specifically called NF-κB. In most normal cells, these Rel/NF-κB dimers are retained in the cytoplasm as an inactive complex through the direct binding of an IκB inhibitor. Various signals can lead to degradation of the IκB protein and the resultant translocation of the active Rel/NF-κB complex into the nucleus to differentially regulate the expression of genes involved in various cellular processes.[14,15] Therefore, inhibition of this pathway could provide a new insight in the assessment and management of breast cancer.
With advancement of research, new medications are regularly being developed to treat early and late stages of breast cancer. These chemically synthesized drugs, though being effective, often have some major side effects including nausea, loss of appetite, vomiting, diarrhea, drowsiness, mouth sores, headache, muscle/joint pain, numbness/tingling/burning of the hands/feet, weakness, dizziness, vaginal bleeding, hair thinning, and weight change etc. In a competitive bid to explore new therapeutic agents, research has been focused to find out natural drugs that are cost effective, easily available and have lesser side effects. Several dietary phytochemicals like emodin, guggulsterone, resveratrol, sylibinin, ellagic acid, genistein, boswellic acid, 1-cafeoylquinic acid, and quercetin are natural chemopreventive agents that have been found to be potent inhibitors of NF-κB pathway with anticarcinogenic properties.[6,7,16–22] [Figures 1a–i]. These compounds may block any one or more steps in the NF-κB signaling pathway such as the signals that activate the NF-κB signaling cascade, translocation of NF-κB into the nucleus, and DNA binding of the dimers. Therefore, in present study, we aimed to validate the above findings by using docking simulation studies and elucidate the feasible mechanistic aspects of above mentioned phytochemicals from different plants against the NF-κB precursor protein p105.
Figure 1.
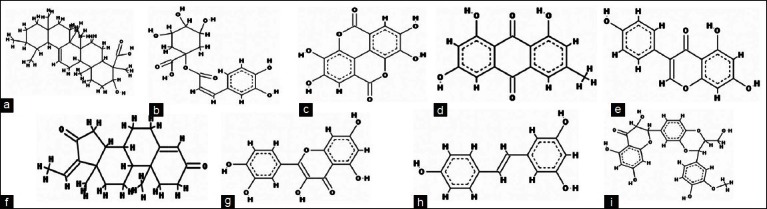
Chemical structure of dietary phytochemicals (a) Boswellic Acid (b) 1-Caffeoylquinic acid (c) Ellagic Acid (d) Emodin (e) Genistein (f) Guggulsterone (g) Quercetin (h) Resveratrol (i) Sylibinin
MATERIALS AND METHODS
Retrieval of protein 3D structure
The crystal structure of NF-κB (PDB: 1SVC) taken in this study was extracted from Brookhaven Protein Data Bank (http://www.rcsb.org/pdb). NF-κB was prepared for docking in such a way that all heteroatoms (i.e., nonreceptor atoms such as water, ions, etc.) were removed. CharMM force field was assigned and further subjected to two steps energy minimization to remove the bad steric clashes using steepest descent and conjugate gradient algorithm for 1000 steps at RMS gradient of 0.01 and 0.05, respectively, during the energy minimization process the backbone was fixed.[23]
Retrieval of ligands 3D structure
Ligands of interest [Table 1] were searched on PubChem database (http://pubchem.ncbi.nlm.nih.gov). The PubChem database contains structural and functional information about different organic compounds. Each compound has its unique compound identification number (CID). The structure of a compound is summarized as its SMILES (Simplified Molecular Input Line Entry Specification) string. This string was taken for each of the ligand of interest and submitted to another online software system CORINA (http://www.molecular-networks.com/products/corina). It takes SMILES string as input and generates 3D structure of the molecule whose structural coordinates file can be downloaded in PDB format suitable for AutoDock 4.0.[24] CharMM force field was applied to them and further subjected to single step energy minimization using steepest descent algorithm for 500 steps at RMS gradient of 0.01.[23] The 2-D structures of different ligands are shown in Figures 1a–i.
Table 1.
Dietary phytochemicals and their sources
Active site identification
Active site of protein was identified by Q-Site finder (http://www.modelling.leeds.ac.uk/qsitefinder) which works by binding hydrophobic probes to the protein, and finding clusters of probes with the most favorable binding energy. These clusters are placed in rank order of the likelihood of being a binding site according to the sum total binding energies for each cluster. It takes PDB files or PDB ID of proteins as input, predicts binding sites and gives residues responsible for making a particular binding site along with a graphic view of that particular binding site as output.[25]
Docking simulation
Docking studies were performed using the AutoDock Tools 4.0 in order to find the preferred binding conformations of the ligands in the receptor.[26] The analysis of the binding conformation uses a scoring function based on the free energy of binding.[27] Among the stochastic search algorithms offered by AutoDock suite, we choose the Lamarckian Genetic Algorithm (LGA) which combines global search (Genetic Algorithm alone) to local search (Solis and Wets algorithm.[28] The grid parameter file of receptor was generated using AutoDock 4.0. A grid-box was generated that was large enough to cover the entire receptor binding site and accommodate ligand to move freely. The number of grid points in x, y, and z-axes were 60 × 60 × 60 Å. The distance between two connecting grid points was 0.375 Å. The center of the ligand in the X-ray crystal structure was used as the center of the grid-box. AutoDock4 and a Lamarckian Genetic Algorithm (LGA) were used for receptor-fixed ligand-flexible docking calculations.[29] Ten search attempts were performed for ligand. The maximum number of energy evaluations before the termination of LGA run was 2500000 and the maximum number of generations of the LGA run before termination was 27000. Other docking parameters were set to the software's default values. During docking process, a maximum of ten different conformations was considered for the ligand. The conformer with the lowest binding free energy was used for further analysis.
Visualization of docked complex
The docked complex of protein and ligand was visualized by Ligplot (http://biochem.ucl.ac.uk/bsm/ligplot/ligplot.html) and ghost script viewer. Ligplot is a command line based program for automated plotting of protein−ligand interactions from the 3-D structure coordinates file of protein−ligand complex and generates schematic diagrams of this interaction, showing interacting residues of protein and ligand, mediated by hydrogen bonds and hydrophobic interactions.[30] H-bonds are indicated by dashed lines between the atoms involved, while an arc represents hydrophobic contacts with spokes radiating toward the ligand atoms they contact. The contacted atoms are shown with spokes radiating back. After successful execution, ligplot generates post script file.
Ghost script viewer (http://www.cs.wisc.edu/~ghost/gsview/) is a GUI based application for viewing the post script files. This program was used to view the script files generated by ligplot for each protein−ligand interaction. After opening the post script file, GSV provides option to convert the image to one of the standard image formats or the portable document format (PDF). Ligplots showing molecular interaction of the NF-κB precursor p105 with different ligands is presented in Figures 2a–i.
Figure 2.
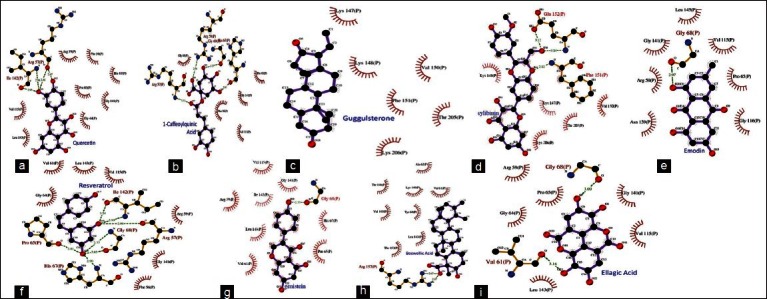
Ligplots showing molecular interaction of the NF-kB precursor p105 with different dietary phytochemicals (a) quercetin (b) 1-caffeoylquinic acid (c) guggulsterone (d) sylibinin (e) emodin (f) resveratrol (g) genistein (h) boswellic acid (i) ellagic acid
Validation of docking protocol
The complex of NF-κB precursor p105 co-crystallized with inhibitors is still unsolved till date. Therefore, a common self-docking procedure to authenticate the accuracy of the docking protocol used in this study was not feasible. In order to conquer this situation, a structurally similar active compound (CID: 21600688) was used as test set and docked in to the binding site of the protein. The top ranking conformational clusters from this dock were evaluated in terms of root mean square deviation between docked position and experimentally determined position for the ligand. The low RMS (1.20 Å) between the experimental and docked co-ordinates of ligand indicated same binding orientation that favored the validation of adopted docking protocol [Figure 3].
Figure 3.
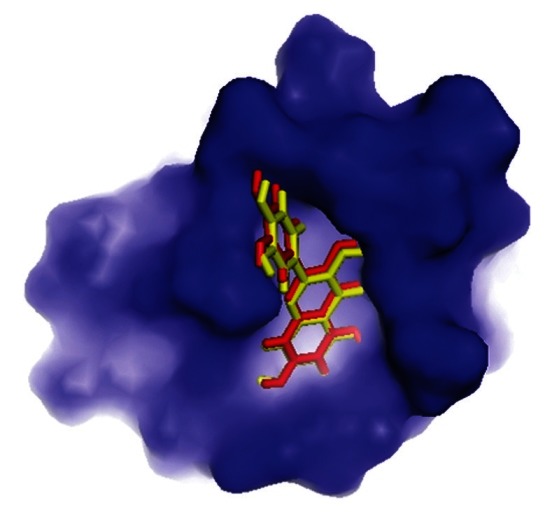
Validation of docking protocol
RESULTS
Among all natural compounds docked with the NF-κB precursor protein p105, quercetin, a compound derived from onion, apple, and citrus fruits was found to bind with the best efficacy in the RH domain of p105 showing binding energy of −12.11 Kcal and inhibition constant (Ki) of 0.002 μM which was followed by 1-caffeoylquinic acid derived from quince and guggulsterone derived from guggulu with binding energies of −11.15 and −9.84 Kcal and inhibition constant (Ki) of 0.007 and 0.06 μM, respectively [Figures 2a–c; Table 2]. ARG57, ILE142, VAL115, LEU143, ARG59, PHE56, HIS67, PRO65, GLY141, and GLY64 residues of p105 were engaged in hydrophobic interactions out of which ARG57, ILE142, and VAL6 residues were involved in hydrogen bond formation with quercetin providing stability of its molecular interactions with p105. ARG59, GLY69, HIS67, ARG57, GLY68, PRO65, GLY141, PHE56, and VAL115 residues showed hydrophobic interactions with 1-caffeoylquinic acid in which HIS67 and ARG59 residues take part in hydrogen bonding. Similarly LYS147, LYS148, VAL150, PHE151, THR205, and LYS206 residues of p105 were also involved in hydrophobic interactions.
Table 2.
Molecular interaction studies of phytochemicals with the NF-κB precursor p105
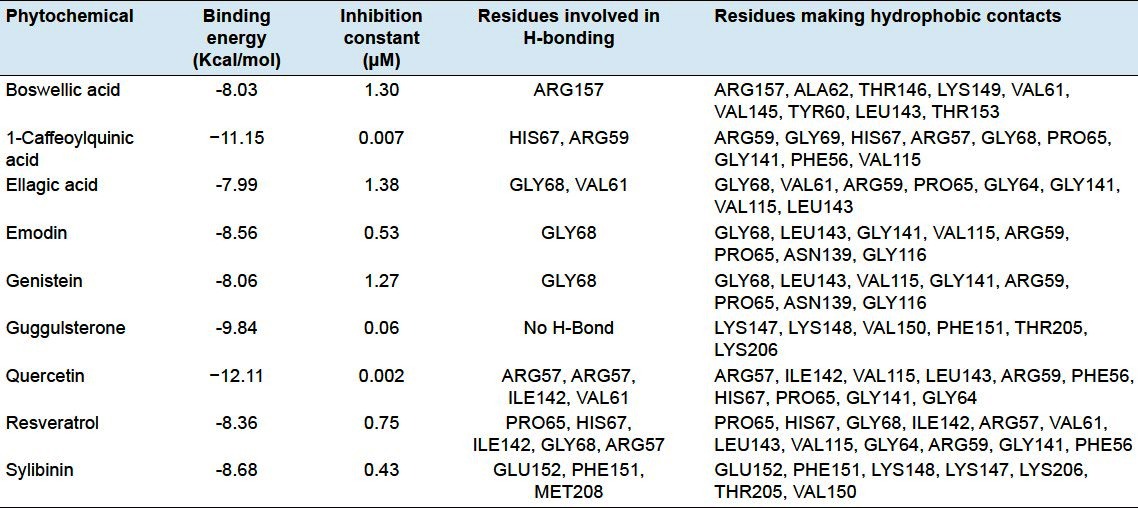
It was found that apart from quercetin, 1-caffeoylquinic acid and guggulsterone other natural compounds namely boswellic acid, ellagic acid, emodin, genistein, resveratrol, and sylibinin also showed significant binding in the RH domain of p105 with binding energies of -8.03, -7.99, -8.56, -8.06, -8.36, and -8.68 Kcal, respectively, with inhibition constants in the range of 0.43−1.38 μM [Figures 2d–i; Table 2]. The current study also revealed some amino acid residues of the RH domain that were playing important role in the proper orientation of natural compounds within this domain. These include ARG59, VAL61, PRO65, HIS67, GLY68, ILE142, PHE151, GLU152, ARG157, and MET208 which were involved in making hydrogen bonds with different ligands. GLY68 was engaged in hydrogen bond formation at four instances and might be considered as a key residue in the binding of these dietary agents with the NF-κB precursor protein p105.
DISCUSSION
The overexpression/activation of NF-κB transcription factor has already been reported in breast cancer cells, where it affects cell proliferation, suppresses apoptosis as well as promotes tumor growth.[31–33] Indeed, a variety of breast cancer cell lines displays increased NF-κB DNA binding activity.[34] The p50/RelA heterodimer has been reported to be activated in 86% of estrogen receptor (ER)-negative, Her2/ErbB2-positive breast tumors.[35] In fact, Her2/ErbB2 activates NF-κB by a mechanism that involves the PI3K/Akt pathway and utilizes calpain to degrade IκBα.[36] ERs have been shown to regulate NF-κB negatively by a variety of mechanisms.[37]
Interestingly, other NF-κB family members have also been implicated in breast cancer. Transgenic mice with mammary cell-specific c-Rel expression have been reported to develop mammary tumors.[38] Moreover, approximately 40% of breast tumors showed an increased binding potential of p50- and p52-containing complexes with an increased expression of Bcl-3.[39] Additionally, it was reported that enhanced DNA-binding activity associated with the p50 NF-κB subunit is associated with ER-positive breast cancers destined for early relapse despite adjuvant endocrine therapy with tamoxifen, suggesting an involvement of this aspect of the NF-κB pathway in endocrine-resistant breast cancer.[40] Recently, Eddy et al. (2005) provided evidence that NF-κB activation in breast cancer cell lines is associated with aberrant expression of the IKKε subunit.[41] Interestingly, it was found that, in certain cancer cells, IKKε can induce the phosphorylation of RelA at Ser-536.[42] These results raise the possibility that breast tumor subtypes exhibit different forms of NF-κB activation, some associated with enhanced p50/RelA DNA binding, some with enhanced p50- or p52-DNA binding activity, some with enhanced c-Rel expression, and some with increased Ser-536 phosphorylation of RelA.
All the phytochemicals mentioned in this study have already been shown to have anticarcinogenic properties and inhibit the NF-κB pathway in different cancer cell lines. Emodin has been shown to inhibit tumor necrosis factor-alpha (TNF-α)-induced NF-κB activation and IκB degradation in human vascular endothelial cells.[16] Guggulsterone has been reported to inhibit NF-κB and IκBα kinase activation in human non-small cell lung carcinoma (H1299) and human lung epithelial cell carcinoma (A549) cells.[17] TNF-induced activation of NF-κB in myeloid (U-937), lymphoid (Jurkat), and epithelial (HeLa and H4) cells has been shown to be suppressed by resveratrol.[18] Sylibinin strongly inhibited the growth and survival of human endothelial cells via cell cycle arrest and downregulation of survivin, Akt and NF-κB.[19] In another study, sylibinin inhibited constitutive and TNF-α-induced activation of NF-κB and sensitized human prostate carcinoma DU145 cells.[20] Similarly, ellagic acid induced apoptosis through inhibition of NF-κB in human pancreatic cancer cells.[21] Inhibition of NF-κB activity by genistein is mediated via Notch-1 signaling pathway in human pancreatic cancer cell line BxPC-3.[6] Gong et al. (2003) have reported that inactivation of NF-κB by genistein is mediated via Akt signaling pathway in human breast cancer cell line MDA-MB-231.[7]
Our docking results illustrated these dietary phytochemicals as potent inhibitors of the NF-κB pathway and initially demonstrated the binding of these natural compounds with the NF-κB precursor protein p105 in its RH domain (amino acid sequence 42-367) which contains sequences important for DNA binding and dimerization. Our results showed that quercetin bind with the best efficacy in the RH domain of NF-κB which was followed by 1-caffeoylquinic acid and guggulsterone. Since these phytochemicals bind p105 in its RH domain, there may be two consequences, first, the dimerization of shortened form p50 could be hampered and second, if somehow p50 forms a functional homo- or a heterodimer with other members of the Rel/NF-κB family, the DNA binding of these functional dimers at kB sites would be hindered and ultimately the genes which are regulated by NF-κB would not be expressed. So, based on above results, quercetin, 1-caffeoylquinic acid, and guggulsterone were found to be more potent inhibitors of NF-κB pathway in comparison to boswellic acid, ellagic acid, emodin, genistein, resveratrol, and sylibinin. Thus, we propose that molecular interaction of these natural compounds with NF-κB DNA binding domain (RH domain) might be the reason for their NF-κB inhibitory activities.
CONCLUSIONS
In summary, the results of the present study have provided a mechanism by which phytochemicals inhibited the NF-κB pathway involved in the regulation of many genes responsible for tumor growth and metastasis. Furthermore, this study would be useful in both understanding the inhibitory mode of dietary phytochemicals as well as in rapidly and accurately predicting the activities of newly designed inhibitors on the basis of docking scores. These models would also provide some valuable clues in structural modification for designing new and more potent chemotherapeutic agents against breast cancer. In addition, further in vitro and in vivo studies are required to explore the role of these chemotherapeutic molecules against the NF-κB precursor protein p105 in cure and prevention of breast cancer.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
REFERENCES
- 1.Althuis MD, Dozier JD, Anderson WF, Devesa SS, Brinton LA. Global trends in breast cancer incidence and mortality 1973-1997. Int J Epidemiol. 2005;34:405–12. doi: 10.1093/ije/dyh414. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Nambooze S, Wabwire-Mangen F, Wabinga HR. Changing cancer incidence in Kampala, Uganda, 1991-2006. Int J Cancer. 2010;126:1187–95. doi: 10.1002/ijc.24838. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Whelan S, Ferlay J, Storm H, editors. Cancer Base No. 7. I-VIII. Lyon: IARC Press; 2005. Cancer incidence in five continents. [Google Scholar]
- 4.Jemal A, Center MM, Desantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 5.Mackay J, Jemal A, Lee NC, Parkin DM. Atlanta, GA: American Cancer Society; 2006. The cancer atlas. [Google Scholar]
- 6.Gilmore TD. The Rel/NF-κB signal transduction pathway: Introduction. Oncogene. 1999;18:6925–37. doi: 10.1038/sj.onc.1203237. [DOI] [PubMed] [Google Scholar]
- 7.Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–47. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 8.Baeuerle PA, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–79. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 9.Barnes PJ, Karin M. Nuclear factor-κB: A pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–71. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 10.Wulczyn FG, Krappmann D, Scheidereit C. The NF-κB/ Rel and IκB gene families: Mediators of immune response and inflammation. J Mol Med. 1996;74:749–69. doi: 10.1007/s001090050078. [DOI] [PubMed] [Google Scholar]
- 11.Chen FE, Ghosh G. Regulation of DNA binding by Rel/ NF-κB transcription factors: Structural views. Oncogene. 1999;18:6845–52. doi: 10.1038/sj.onc.1203224. [DOI] [PubMed] [Google Scholar]
- 12.Karin M. How NF-κB is activated: The role of the IκB kinase (IKK) complex. Oncogene. 1999;18:6867–74. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 13.Pahl HL. Activators and target genes of Rel/NF-κB transcription factors. Oncogene. 1999;18:6853–66. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 14.Kumar A, Dhawan S, Aggarwal BB. Emodin (3-methyl-1,6,8- trihydroxyanthraquinone) inhibits TNF-induced NF-κB activation, IκB degradation, and expression of cell surface adhesion proteins in human vascular endothelial cells. Oncogene. 1998;17:913–8. doi: 10.1038/sj.onc.1201998. [DOI] [PubMed] [Google Scholar]
- 15.Shishodia S, Aggarwal BB. Guggulsterone inhibits NF-κB and IκBα kinase activation, suppresses expression of antiapoptotic gene products, and enhances apoptosis. J Biol Chem. 2004;279:47148–58. doi: 10.1074/jbc.M408093200. [DOI] [PubMed] [Google Scholar]
- 16.Manna SK, Mukhopadhyay A, Aggarwal BB. Resveratrol suppresses TNF-induced activation of nuclear transcription factors NF-κB, activator protein-1, and apoptosis: Potential role of reactive oxygen intermediates and lipid peroxidation. J Immunol. 2000;164:6509–19. doi: 10.4049/jimmunol.164.12.6509. [DOI] [PubMed] [Google Scholar]
- 17.Singh RP, Dhanalakshmi S, Agarwal C, Agarwal R. Silibinin strongly inhibits growth and survival of human endothelial cells via cell cycle arrest and downregulation of survivin, Akt and NF- κB: Implications for angioprevention and antiangiogenic therapy. Oncogene. 2005;24:1188–202. doi: 10.1038/sj.onc.1208276. [DOI] [PubMed] [Google Scholar]
- 18.Dhanalakshmi S, Singh RP, Agarwal C, Agarwal R. Silibinin inhibits constitutive and TNFα-induced activation of NF-κB and sensitizes human prostate carcinoma DU145 cells to TNFα- induced apoptosis. Oncogene. 2002;21:1759–67. doi: 10.1038/sj.onc.1205240. [DOI] [PubMed] [Google Scholar]
- 19.Edderkaoui M, Odinokova I, Ohno I, Gukovsky I, Go VL, Pandol SJ, et al. Ellagic acid induces apoptosis through inhibition of nuclear factor-κB in pancreatic cancer cells. World J Gastroenterol. 2008;14:3672–80. doi: 10.3748/wjg.14.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Inhibition of nuclear factor B activity by genistein is mediated via Notch-1 signaling pathway in pancreatic cancer cells. Int J Cancer. 2006;118:1930–6. doi: 10.1002/ijc.21589. [DOI] [PubMed] [Google Scholar]
- 21.Gong L, Li Y, Nedeljkovic-Kurepa A, Sarkar FH. Inactivation of NF-κB by genistein is mediated via Akt signaling pathway in breast cancer cells. Oncogene. 2003;22:4702–9. doi: 10.1038/sj.onc.1206583. [DOI] [PubMed] [Google Scholar]
- 22.Aggarwal BB, Shishodia S. Molecular targets of dietary agents for prevention and therapy of cancer. Biochem Pharmacol. 2006;71:1397–421. doi: 10.1016/j.bcp.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Brooks BR, Brooks CL, III, Mackerell AD, Nilsson L, Petrella R, Roux B, et al. CHARMM: The biomolecule simulation program. J Comput Chem. 2009;30:1545–615. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–62. [Google Scholar]
- 25.Laurie AT, Jackson RM. Q-SiteFinder: An energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–16. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 26.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: Parallel application of AutoDock 2.4. J Comput Aided Mol Des. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 27.Huey R, Morris GM, Olson AJ, Goodsell DS. Semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–52. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 28.Solis FJ, Wets JB. Minimization by random search techniques. Math Oper Res. 1981;6:19–30. [Google Scholar]
- 29.Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: Applications of AutoDock. J Mol Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protien Eng. 1995;8:127–34. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 31.Nakshatri H, Goulet RJ. NF-κB and breast cancer. Curr Probl Cancer. 2002;26:282–309. doi: 10.1067/mcn.2002.129977. [DOI] [PubMed] [Google Scholar]
- 32.Biswas DK, Dai SC, Cruz A, Weiser B, Graner E, Pardee AB. The nuclear factor kappa B (NF-κB): A potential therapeutic target for estrogen receptor negative breast cancers. Proc Natl Acad Sci U S A. 2001;98:10386–91. doi: 10.1073/pnas.151257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biswas DK, Martin KJ, McAlister C, Cruz AP, Graner E, Dai SC, et al. Apoptosis caused by chemotherapeutic inhibition of nuclear factor-κB activation. Cancer Res. 2003;63:290–5. [PubMed] [Google Scholar]
- 34.Nakshatri H, Bhat-Nakshatri P, Martin DA, Goulet RJ, Jr, Sledge GW., Jr Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol Cell Biol. 1997;17:3629–39. doi: 10.1128/mcb.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biswas DK, Shi Q, Baily S, Strickland I, Ghosh S, Pardee AB, et al. NF-κB activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natl Acad Sci U S A. 2004;101:10137–42. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pianetti S, Arsura M, Romieu-Mourez R, Coffey RJ, Sonenshein GE. Her-2/neu overexpression induces NF-κB via a PI3-kinase/Akt pathway involving calpain-mediated degradation of IκB-α that can be inhibited by the tumor suppressor PTEN. Oncogene. 2001;20:1287–99. doi: 10.1038/sj.onc.1204257. [DOI] [PubMed] [Google Scholar]
- 37.Kalaitzidis D, Gilmore TD. Transcription factor cross-talk: The estrogen receptor and NF-κB. Trends Endocrinol Metab. 2005;16:46–52. doi: 10.1016/j.tem.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Romieu-Mourez F, Kim DW, Shin SM, Demicco EG, Landesman- Bollag E, Seldin DC, et al. Mouse mammary tumor virus c-rel transgenic mice develop mammary tumors. Mol Cell Biol. 2003;23:5738–54. doi: 10.1128/MCB.23.16.5738-5754.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS., Jr Selective activation of NF-κB subunits in human breast cancer: Potential roles for NF-κB2/p52 and for Bcl-3. Oncogene. 2000;19:1123–31. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y, Eppenberger-Castori S, Eppenberger U, Benz CC. The NF-κB pathway and endocrine-resistant breast cancer. Endocr Relat Cancer. 2005;12(Suppl 1):S37–46. doi: 10.1677/erc.1.00977. [DOI] [PubMed] [Google Scholar]
- 41.Eddy SF, Guo S, Demicco EG, Romieu-Mourez R, Landesman- Bollag E, Seldin DC, et al. Inducible IκB kinase/IκB kinase e expression is induced by CK2 and promotes aberrant nuclear factor-κB activation in breast cancer cells. Cancer Res. 2005;65:11375–83. doi: 10.1158/0008-5472.CAN-05-1602. [DOI] [PubMed] [Google Scholar]
- 42.Adli M, Baldwin AS. IKK-i/IKK-epsilon controls constitutive, cancer cell-associated NF-κB activity via regulation of ser-536 p65/RelA phosphorylation. J Biol Chem. 2006;281:26976–84. doi: 10.1074/jbc.M603133200. [DOI] [PubMed] [Google Scholar]


