Abstract
Pathogenic Neisseria meningitidis isolates contain a polysaccharide capsule that is the main virulence determinant for this bacterium. Thirteen capsular polysaccharides have been described, and nuclear magnetic resonance spectroscopy has enabled determination of the structure of capsular polysaccharides responsible for serogroup specificity. Molecular mechanisms involved in N. meningitidis capsule biosynthesis have also been identified, and genes involved in this process and in cell surface translocation are clustered at a single chromosomal locus termed cps. The use of multiple names for some of the genes involved in capsule synthesis, combined with the need for rapid diagnosis of serogroups commonly associated with invasive meningococcal disease, prompted a requirement for a consistent approach to the nomenclature of capsule genes. In this report, a comprehensive description of all N. meningitidis serogroups is provided, along with a proposed nomenclature, which was presented at the 2012 XVIIIth International Pathogenic Neisseria Conference.
Keywords : Neisseria meningitidis, capsule, serogroup, bacteria, nomenclature
Thirteen Neisseria meningitidis serogroups have been described on the basis of serologic differences of the capsule; of these 13 serogroups, 6 (A, B, C, W, X, Y) cause invasive meningococcal disease. The polysaccharide capsule is a key virulence determinant, and for serogroups A, C, W, and Y, it forms the basis of polysaccharide conjugate vaccines. In one of the first reports distinguishing N. meningitidis, disease isolates were serologically classified into types I–IV on the basis of agglutination reactions with immune rabbit serum (1). In 1950, the subcommittee on Neisseria of the Nomenclature Committee of the International Association of Microbiologists recommended that types I and III be combined into serogroup A; type II become serogroup B; a type II subgroup, termed type II-α, become serogroup C; and type IV become serogroup D. After the report of a fourth serogroup, Z′ (later shown to be serogroup E), 3 new serogroups (X– Z) were identified by using double agar diffusion (2,3). In 1981, three more serogroups (H, I, K) were proposed, and a fourth (serogroup L) was identified in 1983 (4,5).
Nuclear magnetic resonance spectroscopy enabled determination of the structure of capsular polysaccharides responsible for serogroup specificity, and structures for 12 of the 13 serogroups (all but serogroup D) from N. meningitidis capsular polysaccharides have been reported (6–15). Molecular mechanisms of capsular polysaccharide synthesis have been elucidated; genes involved in polysaccharide biosynthesis and cell surface translocation are clustered at a single chromosomal locus termed cps. Genes within this locus are divided into 6 regions: A–D, D′, and E (16). Genes in region A encode enzymes for biosynthesis of the capsular polysaccharide, and genes in regions B and C are implicated in the translocation of the high molecular weight polysaccharides to the cell surface.
Complete nucleotide sequences of cps loci encoding serogroups A–C, W, and Y have been elucidated. Serogroup-specific capsule biosynthesis genes located in region A have been published for serogroup X, and nucleotide sequences for serogroups E, L, and Z have been submitted to GenBank (accession nos. AJ576117, AF112478, and AJ744766, respectively) (17–19). This study provides a comprehensive description of all N. meningitidis serogroups and presents proposed revisions to the nomenclature.
Materials and Methods
Strain Selection
Serogroup D, H, I, and K isolates were obtained from a collection maintained at the National Institute for Biological Standards and Controls, Potters Bar, UK; the isolates were originally from a collection (1980s) from the People’s Republic of China Committee for Culture Collection of Microorganisms. Serogroup E, W, X, and Y isolates were from the Bavarian N. meningitidis carriage collection (20). Three serogroup L isolates were analyzed with additional serogroup H, I, and K isolates obtained from Paula Kriz (Czech Republic), who had obtained the isolates from Fraser Ashton, who had acquired them from People’s Republic of China (Table 1). The sequenced genomes from serogroup A isolate Z2491, serogroup B isolate H44/76, and serogroup C isolates FAM18 and 053442 were used (Table 1) (21–24). Isolates were grown on Mueller-Hinton agar supplemented with 5% (vol/vol) defibrinated sterile horse blood for 15 h at 37°C in a 5% (vol/vol) CO2 atmosphere. DNA was extracted by using the Wizard Genomic DNA Purification Kit (Promega, Southampton, UK) according to the manufacturer’s instructions.
Table 1. Description of Neisseria meningitidis isolates*.
| Isolate name | Serogroup | Origin | Disease status | Strain designation | GenBank accession no. |
|---|---|---|---|---|---|
| N. meningitidis Z2491 | A | The Gambia | Invasive | A:P1.7, 13–1: F1–5: ST-4 (cc4) | AL157959 |
| N. meningitidis H44/76 | B | Norway | Invasive | B:P1.7,16: F3–3: ST-32 (cc32) | CP002420 |
| N. meningitidis FAM18 | C | United States | Invasive | C:P1.5,2: F1–30: ST-11 (cc11) | AM421808 |
| N. meningitidis 053442 | C | PRC | Invasive | C:P1.7–2,14: F3–3: ST-4821 (cc4821) | CP000381 |
| N. meningitidis 29013 | D | PRC | Unknown | C:P1.22,14–6: F3–16: ST-8723 (cc213) | ERR028660 |
| N. meningitidis α707 | E | Germany | Carrier | E:P1.5–1,2–2: F4–3: ST-254 (cc254) | HF562982 |
| N. meningitidis 29031 | H | PRC | Carrier | H:P1.21,3: F4–21: ST-4959 (-) | ERR028662 |
| N. meningitidis H-ASH/87 | H | PRC | Carrier | H:P1.21,3: F4–21: ST-4959 (-) | ERR036095 |
| N. meningitidis H-ZANE/83 | H | PRC | Carrier | H:P1.21,3: F4–21: ST-4959 (-) | ERR036096 |
| N. meningitidis 29043 | I | PRC | Carrier | I:P1.22,14–6: F5–2: ST-5594 (-) | ERR028663 |
| N. meningitidis I-ZANE/87 | I | PRC | Carrier | I:P1.22,14–6: F5–2: ST-5594 (-) | ERR036097 |
| N. meningitidis 29046 | K | PRC | Carrier | K:P1.7–2,14: F5–14: ST-8724 (-) | ERR028664 |
| N. meningitidis K-ASH/87 | K | PRC | Carrier | K:P1.7–2,14: F5–14: ST-8724 (-) | ERR036087 |
| N. meningitidis WUE3608 | L | Unknown | Carrier | L:P1.18–1,3: F1–5: ST-963 | HF562986 |
| N. meningitidis L-ASHTON/87 | L | PRC | Carrier | L:P1.22,14: F1–38: ST-8902 (-) | ERR036088 |
| N. meningitidis 21033 | L | Dublin, Ireland, UK | Carrier | L:P1.7–2,13–1:F1–5: ST-3258 (-) | ERR063490 |
| N. meningitidis α275 | W | Germany | Carrier | W:P1.18–1,3: F4–1: ST-22 (cc22) | HF562987 |
| N. meningitidis WUE171 | W | Germany | Unknown | W:P1.5–9, 10: F3–6: ST-11 (cc11) | HF562992 |
| N. meningitidis α388 | X | Germany | Carrier | X:P1.5–1,2–2: F5–1: ST-765 (cc254) | HF562988 |
| N. meningitidis α162 | Y | Germany | Carrier | Y:P1.5–2,10–1: F4–1: ST-23 (cc23) | HF562989 |
| N. meningitidis WUE172 | Y | Unknown | Unknown | Y:P1.5,2: F1–1: ST-166 (cc11) | HF562992 |
| N. meningitidis WUE173 | Z | Unknown | Unknown | Z:P1.7–1,1: F1–7: ST-4443 (-) | HF562991 |
*PRC, People’s Republic of China; UK, United Kingdom.
PCR and Nucleotide Sequencing of the Capsule Locus
The genes for the N. meningitidis cps locus are located between a gene encoding a putative inner membrane transport protein (NMC0044 FAM18 genome annotation) and a gene encoding the sodium/glutamate symport carrier protein, gltS (NMC0069). PCR reactions were performed for isolates 29013, α707, 29031, 29043, 29046, 3608, α275, α388, α162, WUE171, WUE172, and WUE173 (Table 1) by using the Expand Long Template PCR System (Roche Applied Science, Burgess Hill, UK) according to the manufacturer’s recommended protocol, with annealing at 55°C and extension at 68°C for 20 min. Initial reactions used 1 of the following primer pairs: gltS (NMC0069) to tex (NMC0059) (primers gltS 5′- CCGACCAAGCCGTATTGC + ATGATACTCGAAGGCGTGGTT-3′ and tex 5′- TGTCGAAGCCGTCCATAATCT + GCCCTGTCCAACAAGTTCGT-3′) and tex to NMC0044 (primers tex 5′- CGCCCGGTTCGTCATCC + TTGCTGCTGGTAGGCGAATCC -3′ and NMC0044 5′- CGGGCGAACACGGTAAT + TATCGTTGGTGCGCTGGTTAT-3′).
Genome Sequencing
Genomic data for serogroups D, H, I, K, and L were obtained by using the Illumina sequencing platform and de novo assembly (Illumina, San Diego, CA, USA), using the shuffle and Velvet Optimization scripts found within Velvet 1.1.03 (25) (Table 1). By using VelvetOptimiser version 2.1.7 (http://bioinformatics.net.au/software.velvetoptimiser.shtml), optimal k-mer lengths ranging from 41 to 65 were selected and contigs were generated. These contigs were deposited in the Bacterial Isolate Genome Sequence Database (BIGSDB) along with the isolate provenance and name, after which contigs were scanned for genes within the cps locus and tagged and alleles were assigned (26). New alleles were manually checked for a correct start and stop codon and aligned with known alleles before assignation. Additional data (e.g., multilocus sequence type [MLST]), and PorA, PorB, and FetA designations were obtained.
Annotation and Bioinformatic Methods
Predicted proteins were clustered into homology groups by using TribeMCL algorithm with a cutoff of 1e-70 (http://doc.bioperl.org/bioperl-run/lib/Bio/Tools/Run/TribeMCL.html#General). The genes within the cps loci that encoded proteins with the same homology group were assigned the same name, exceptions being those genes found in Region A. BLASTp (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) searches were done by using a reference sequence database with default settings.
Annotation was performed by using the genome viewer Artemis (27). The cps nucleotide sequence locus for each serogroup was stored in BIGSDB and linked with the corresponding isolate record.
MEGA5 (http://megasoftware.net/) was used to calculate overall p-distances and G+C content and to obtain the number of polymorphisms observed within each gene. The cps loci were compared by using the Artemis Comparison Tool (www.sanger.ac.uk/resources/software/act/).
Results and Discussion
General Organization
All of the serogroups examined had comparable cps loci, and regions occurred in the following order D-A-C-E-D′-B, revealing a conserved gene synteny and a replication of the genomic rearrangements (Figure). Genes in regions B–D, D′, and E were conserved, whereas genes in region A were diverse; this finding is consistent with the distinct biochemical composition found within each serogroup (Table 2).
Figure.
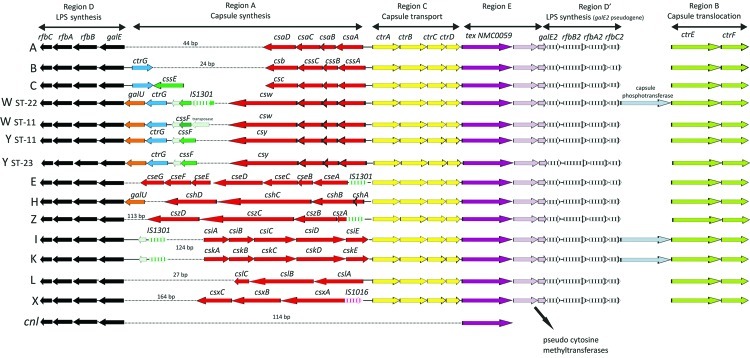
Genetic organization of the cps locus among Neisseria meningitidis serogroups A (N. meningitidis Z2491); B (N. meningitidis H44/76); C (N. meningitidis FAM18, 053442, and 29013); W α275 (clonal complex sequence type [ST] 22); W WUE171 (clonal complex ST-11); Y α162 (clonal complex ST-11); Y WUE172 (ST-23); E (N. meningitidis α707); H (N. meningitidis 29031); I (N. meningitidis 29043); K (N. meningitidis 29046); L (N. meningitidis WUE3608); X (N. meningitidis α388); and Z (N. meningitidis WUE173); and a cnl N. meningitidis isolate. Letters on left represent serogroups. Arrows depict gene orientation.
Table 2. Genetic diversity of Neisseria meningitidis cps genes*.
| Region, locus | Size, bp | No. alleles | No. polymorphisms (%) | p-distance | dN/dS ratio | G+C content |
|---|---|---|---|---|---|---|
| MLST | ||||||
| abcZ | 433 | 11 | 52 (11) | 0.046 | 0.04 | 51 |
| adk | 465 | 7 | 18 (4) | 0.015 | 0.02 | 52 |
| aroE | 490 | 11 | 131 (7) | 0.102 | 0.28 | 56 |
| fumC | 465 | 12 | 27 (6) | 0.02 | 0.02 | 57 |
| gdh | 501 | 8 | 18 (4) | 0.014 | 0.05 | 52 |
| pdhC | 480 | 13 | 61 (13) | 0.041 | 0.08 | 56 |
|
pgm
|
450 |
10 |
62 (14) |
0.055 |
0.11 |
54 |
| Region B | ||||||
| ctrE | 2,115 | 13 | 386 (18) | 0.061 | 0.2 | 51 |
|
ctrF
|
1,260 |
12 |
59 (5) |
0.012 |
0.14 |
49 |
| Region C | ||||||
| ctrA | 1,179 | 11 | 175 (15) | 0.032 | 0.18 | 47 |
| ctrB | 1,164 | 8 | 264 (23) | 0.051 | 0.10 | 46 |
| ctrC | 726 | 8 | 177 (24) | 0.06 | 0.12 | 43 |
|
ctrD
|
651 |
7 |
87 (13) |
0.057 |
0.1 |
46 |
| Region D | ||||||
| rfbA | 867 | 13 | 75 (9) | 0.034 | 0.26 | 53 |
| rfbB | 1,083 | 15 | 312 (28) | 0.067 | 0.12 | 53 |
| rfbC | 558 | 14 | 124 (220 | 0.09 | 0.22 | 55 |
|
galE
|
1,020 |
15 |
382 (37) |
0.119 |
0.12 |
50 |
| Region E | ||||||
| tex | 2,274 | 13 | 173 (8) | 0.019 | 0.09 | 59 |
| NMC0060 | 1,007 | 8 | 24 (2.4) | 0.003 | 0.91 | 40 |
| NMC0061 | 345 | 5 | 24 (7) | 0.013 | 0.19 | 36 |
*Genes in region A were too diverse among serogroups for direct comparison. Region D′ was omitted because galE2 is truncated and rfbA, rfbB, and rfbC were duplicates. MLST, multilocus sequence type.
The lowest GC content was in regions A and C, indicative of genes in these regions resulting from horizontal recombination (Table 2; Table 3, Appendix); regions B, D, and E displayed GC contents of 50%, 52%, and 52%, respectively, similar to those found in the Neisseria genomes (21,22,33). Bacterial genomic GC content varies considerably among species but remains uniform within a bacterial genome, such that genes acquired through horizontal genetic exchange will have a GC content different from the overall GC content found within the genome.
Table 3. Nomenclature of Neisseria meningitidis capsule genes and description*.
| Region, serogroup | Proposed nomenclature (NEIS)† | Previous nomenclature | Protein function | Size, bp | G+C content, % | Ref |
|---|---|---|---|---|---|---|
| Region A | ||||||
| A | csaA (NEIS2157) | mynA/sacA | UDP-N-acetyl-D-glucosamine 2-epimerase | 1,119 | 25 | (18) |
| csaB (NEIS2158) | mynB/sacB | Polymerase linking N-acetyl-D-mannosamine-1-phosphate monomers | 1,638 | 28 | (18) | |
| csaC (NEIS2159) | mynC/sacC | O-acetyltransferase | 744 | 30 | (28) | |
| csaD (NEIS2160) | mynD/sacD | Capsule transport | 864 | 35 | (18) | |
| B, C, W, Y | cssA (NEIS0054) | siaA/synX/synA/neuC | N-acetylglucosamine-6-P 2-epimerase | 1,134 | 31 | (17) |
| cssB (NEIS0053) | siaB/synB/neuA | CMP-N-acetylneuraminic acid synthase | 687 | 41 | ||
| cssC (NEIS0052) | siaC/synC/neuB | N-acetylneuraminic acid synthetase | 1,050 | 40 | ||
| csb (NEIS2161) | siaDB/synD | Polysialyltransferase | 1,488 | 28 | ||
| csc (NEIS0051) | siaDC/synE | Polysialyltransferase | 1,479 | 31 | ||
| csy (NEIS2163) | siaDY/synF | Polymerase linking glucose and N-acetylneuraminic acid | 3,114 | 31 | ||
| csw (NEIS2162) | siaDW/synG | Polymerase linking galactose and N-acetylneuraminic acid | 3,114 | 31 | ||
| cssE (NEIS0050) | oatC | O-acetyltransferase | 1,383 | 29 | (29) | |
| cssF (NEIS2164) | oatWY | O-acetyltransferase | 636 | 33 | (29) | |
| ctrG (NEIS0049) | ctrG/NMB0065 | Putative role in surface expression of sialic capsules | 957 | 33 | (30) | |
| E | cseA (NEIS2165) | cap29eA | Unknown | 1,197 | 33 | |
| cseB (NEIS2166) | cap29eB | Unknown | 639 | 38 | ||
| cseC (NEIS2167) | cap29eC | Glycosyltransferase | 1,461 | 40 | ||
| cseD (NEIS2168) | Fusion of cap29eD and cap29eE | Glycosyltransferase | 2,198 | 39 | ||
| cseE (NEIS2169) | cap29eF | 3-deoxy-D-manno-octulosonic acid 8-phosphate synthase | 852 | 28 | ||
| cseF (NEIS2170) | cap29eG | CMP-2-keto-3-deoxyoctulosonic acid synthetase and 3-deoxy-D-manno-octulosonate 8-phosphatase | 1,290 | 26 | ||
| cseG (NEIS2171) | cap29eH | D-arabinose 5-phosphate isomerase | 947 | 28 | ||
| H | cshA (NEIS2173) | – | Glycerol-3-phosphate cytidylyltransferase | 399 | 29 | |
| cshB (NEIS2174) | – | Putative phosphotransferase with licD domain (involved in phosphorylcholine decoration of teichoic acid) | 1,269 | 33 | ||
| cshC (NEIS2177) | – | Teichoic acid synthase | 3,453 | 34 | ||
| cshD (NEIS2178) | – | Unknown, no putative conserved domains | 1,035 | 38 | ||
| I | csiA (NEIS2179) | – | UDP-N-acetylglucosamine 2-epimerase | 1,125 | 36 | |
| csiB (NEIS2180) | – | UDP-N-acetyl-D-mannosamine dehydrogenase | 1,269 | 45 | ||
| csiC (NEIS2181) | – | Glycosyltransferase group 1 | 2,289 | 36 | ||
| csiD (NEIS2182) | – | Glycosyltransferase group 2 | 2,520 | 34 | ||
| csiE (NEIS2183) | – | Putative glycosyl transferase group 1 | 972 | 42 | ||
| K | cskA (NEIS2179) | – | UDP-N-acetylglucosamine 2-epimerase | 1,125 | 36 | |
| cskB (NEIS2180) | – | UDP-N-acetyl-D-mannosamine dehydrogenase | 1,269 | 45 | ||
| cskC (NEIS2190) | – | Glycosyltransferase group 1 | 2,289 | 36 | ||
| cskD (NEIS2191) | – | Glycosyltransferase group 2 | 2,520 | 34 | ||
| cskE (NEIS2183) | – | Putative glycosyl transferase group 1 | 972 | 42 | ||
| L | cslA (NEIS2184) | lcbA | Capsule phosphotransferase | 1,101 | 30 | |
| cslB (NEIS2185) | lcbB | Capsule polymerase | 2,634 | 28 | ||
| cslC (NEIS2186) | lcbC | Acetyltransferase | 651 | 39 | ||
| X | csxA (NEIS2187) | xcbA | Capsule polymerase | 1,461 | 39 | (19) |
| csxB (NEIS2188) | xcbB | Unknown | 1,053 | 39 | ||
| csxC (NEIS2189) | xcbC | Unknown | 771 | 35 | ||
| Z | cszA (NEIS2173) | capZA | Glycerol-3-phosphate cytidylyltransferase | 399 | 43 | |
| cszB (NEIS2174) | capZB | Putative phosphotransferase with licD domain (involved in phosphorylcholine decoration of teichoic acid) | 1,290 | 45 | ||
| cszC (NEIS2175) | capZC | Teichoic acid synthase | 3,825 | 36 | ||
|
|
cszD (NEIS2176) |
capZD
|
Unknown, no conserved domains detected |
1,626 |
38 |
|
| Region B | ctrE (NEIS0066) | lipA/ctrE | Capsule translocation | 2,115 | 51 | (31) |
|
|
ctrF (NEIS0067) |
lipB/ctrF
|
Capsule translocation |
1,260 |
49 |
(31) |
| Region C | ctrA (NEIS0055) | ctrA | Capsule polysaccharide export outer membrane protein | 1,179 | 47 | (32) |
| ctrB (NEIS0056) | ctrB | Capsule polysaccharide export inner membrane protein | 1,164 | 46 | ||
| ctrC (NEIS0057) | ctrC | Capsule polysaccharide export inner membrane protein | 726 | 43 | ||
| ctrD (NEIS0058) | ctrD | Capsule polysaccharide export ATP binding protein | 651 | 46 |
*Ref, reference; –, no previous nomenclature. Blank cells in last column indicate no reference. †In addition to the common gene name, these loci are allocated a value-free nomenclature consistent with the FAM18 genome annotation but using the prefix NEIS instead of NMC.
Nomenclature
Reports identifying genes and proteins involved in N. meningitidis capsule biosynthesis, combined with the increasing availability of bacterial genomes, have necessitated a more unified approach to the nomenclature of genes within N. meningitidis cps locus (Table 3, Appendix). The capsule locus from each serogroup has been uploaded to the BIGSDB platform, enabling sequences from each gene within the N. meningitidis cps locus to be indexed and multiple cps loci to be typed. However, during the process, it was found that the nomenclature of some genes within this locus posed a problem. For instance, genes within region B were thought to encode lipidation enzymes and, thus, have been known as lipA and lipB. However, within the annotated genomes from N. meningitidis FAM18, MC58, Z2491, and 05342 and from N. lactamica 020–06, two other distinct lipA and lipB genes have been described encoding a lipoic acid synthetase and a lipoate-protein ligase protein, respectively. Furthermore, there are, in some instances, multiple names for the same gene.
Continued surveillance of meningococcal disease combined with the use of multiple names for some genes made it apparent that a consistent approach to the nomenclature of capsule genes was needed. To meet that need, we propose a comprehensive description of all N. meningitidis serogroups and a nomenclature for the cps locus, which was presented at the 2012 XVIIIth International Pathogenic Neisseria Conference (Table 3, Appendix).
We propose that the capsule biosynthesis genes within region A should be termed cs (for capsule synthesis) followed by a letter representing the serogroup and by a capital letter defining each gene according to the Demerec system of genetic nomenclature (34). For example, serogroup A capsule biosynthesis genes would be termed csaA–D (cs for capsule synthesis and a for serogroup A) (Table 3, Appendix). Under this proposal, the sialic acid capsule biosynthesis genes would be termed cssA–C (cs for capsule synthesis and s for sialic acid capsule).
Serogroups B, C, W, and Y are commonly associated with invasive meningococcal disease, and rapid diagnosis of the serogroup is key in monitoring the epidemiology of the disease and in developing prevention strategies. Thus, it is necessary to have nomenclature that identifies the serogroup simply and quickly. The polysialyltransferase genes belonging to serogroups B and C share >70% sequence identity and should be termed csb and csc, respectively. The equivalent gene in serogroups W and Y is also a sialyltransferase, but it also has a glycosyltransferase function and is distinct from the serogroups B and C genes. These distinctions should be reflected in the nomenclature: we propose that the gene for serogroups W and Y be termed csw and csy, respectively. The O-acetyltransferases should be termed cssE for serogroup C and cssF for serogroups W and Y; the nomenclature for ctrG, which has been shown to have a role in surface translocation of sialic capsules, should be retained (30). The remaining serogroups would also follow this scheme (Table 3, Appendix). Genes within region C that encode the capsule transport genes have been termed ctrA–D (after capsule transport), and this nomenclature should be preserved (Table 3, Appendix) (32). Genes within region B that are involved in capsule translocation should be designated ctrE and ctrF (31). Capsule genes are accessible through the PubMLST database (http://pubmlst.org/neisseria). In addition to the common gene name, these loci are allocated a value-free nomenclature consistent with the FAM18 genome annotation but using the prefix NEIS instead of NMC (Table 3, Appendix).
The W-135 and 29E serogroup designations originated at the Walter Reed Army Institute of Research as a result of a paper published by Evans et al. (35). We propose to rename these W and E because the numbers are historic and supply no useful information.
Region A Capsule Biosynthesis Genes
Serogroup A
The serogroup A capsule is composed of repeating units of O-acetylated (α1→6)-linked N-acetyl-D-mannosamine-1-phosphate (13). The capsule biosynthesis region is 4,365 bp long and contains 4 genes: csaA–D (also known as mynA–D or sacA–D) (18,28). The first gene, csaA, encodes the UDP-N-acetyl-D-glucosamine (UDP-GlcNAc) 2-epimerase, which converts UDP-GlcNAc into UDP-N-acetyl-D-mannosamine (UDP-ManNAc); csaB is the polymerase linking ManNAc-phosphate monomers together with csaC, encoding an O-acetyltransferase, which transfers acetyl groups to ManNAc (18,28). The fourth gene, csaD, is predicted to be involved in either capsule transport or in cross-linking of the capsule to the meningococcal cell surface. The serogroup A cps did not contain insertion sequences, and genes within region A contained some of the lowest GC content values among all serogroups (Table 3, Appendix).
Serogroups B, C, W, and Y
Capsular polysaccharides from serogroups B, C, W, and Y are composed of sialic acid derivatives; serogroups B and C express (α2→8)- and (α2→9)-linked sialic acid homopolymers, and alternating sequences of D-galactose or D-glucose and sialic acid are expressed by serogroups W and Y (6,7). Region A is 5,313 bp long in serogroup B, 6,690 bp long in serogroup C, and averages ≈7,581 bp in serogroups W and Y.
All 4 serogroups contain the conserved cssA–C genes for cytidine-5′-monophosphate-N-acetylneuraminic acid synthesis; other designations for cssA–C include siaA–C, synA–C, and neuA–C (17). These are followed by csb, csc, csw, and csy genes (siaD, siaDBCWY, or synD–G), which are the capsule polymerases and determine the functional and nucleotide specificity for the 4 serogroups.
Serogroups C, W, and Y contain O-acetyltransferase genes termed cssE (oatC) and cssF (oatWY) (29). The 2 sequenced serogroup C and serogroup Y isolates harbored functional cssE/cssF genes with an IS1301 transposase adjacent to cssF in serogroup Y. Serogroup W cssF was interrupted by the insertion sequence IS1301. Another gene, ctrG, found in all 4 serogroups, encoded a protein essential in enabling the correct expression of sialic acid polysaccharides (30). The orientation of this gene differed between serogroups such that it was in the same direction as the css genes in serogroups B and C and the opposite in serogroups W and Y (Figure).
Serogroups W and Y contained an additional gene in region A, galU, encoding UTP-glucose-1-phosphate uridylyltransferase, which was also found in region A of serogroup H. BLASTp searches of the derived amino acid sequence of GalU from serogroups W, Y, and H revealed that this protein shared an average of 49% sequence identity with GalU proteins from Streptococcus pneumoniae isolates. GalU has an essential function in capsule formation among S. pneumoniae isolates, catalyzing the reversible formation of UDP-Glc (uridine diphosphate glucose) and inorganic pyrophosphate from UTP (uridine 3-phosphate) and glucose 1-phosphate, and a role in virulence has been recognized for GalU in several bacterial species (36). Additional galU genes were found adjacent to the gene argH encoding argininosuccinate lyase in the sequenced genomes from isolates α275, 29031, H-ASH/87, and H-ZANE/83, and although the genes were conserved, they were not identical to the galU genes found within the cps locus. The genomes belonging to N. meningitidis Z2491, MC58, FAM18, and 053442; N. lactamica 020–06; and N. gonorrhoeae FA1090 also contained galU genes adjacent to argH. These were highly conserved (p-distance = 0.015) but were more distantly related to those found within the cps locus (p-distance = 0.180), indicating a different origin for cps-associated galU genes.
The serogroup D isolate was found to contain serogroup C capsule biosynthesis genes (Figure and Technical Appendix Figure 1, panel A). The cps locus from the prototype serogroup D isolates deposited by Gordon and Murray in 1917 and Branham in 1928 also contained serogroup C-specific capsule genes; however, neither isolate gave precipitins with antiserum to serogroup C, suggesting that these isolates were unencapsulated (C. Frasch. pers. comm.). The presence of internal stop codons in the ctrA and ctrE genes is consistent with this and further confirms that serogroup D does not exist.
Serogroup E
The serogroup E capsule consists of alternating D-galactosamine and 2-keto-3-deoxyoctulosonate (KDO) residues (9). Region A is ≈9,613 bp long, including IS1301, and contains 7 genes, cseA–G (formerly 29eA–H); gene cseE encodes a putative 3-deoxy-phosphooctulonate synthase, cseF encodes a putative 3-deoxy-D-manno-octulosonate cytidylyltransferase, and cseG encodes a D-arabinose 5-phosphate isomerase protein. The deduced amino acid sequence from cseE shared 83% sequence identity with a 3-deoxy-8-phosphooctulonate synthase belonging to N. elongata subsp. glycolytica and 81% identity with N. subflava NJ9703; cseF shared 66% sequence identity with a putative 3-deoxy-D-manno-octulosonate cytidylyltransferase belonging to multiple bacterial species, including Pseudomonas putida and Escherichia coli. The genes cseD and cseE were found to be fused and were predicted to form 1 protein; this prediction is in agreement with the suggestion that the genes for KDO synthesis were dispensable probably because of complementation of lipopolysaccharide–KDO synthesis elsewhere in the genome (37).
Serogroups H and Z
The biochemical structures of serogroups H and Z contain monosaccharide glycerol-3-phosphate repeat units and share similarities with teichoic acid polymers. Region A varied from 6,182 bp in serogroup H to 7,198 bp in serogroup Z, with the latter containing the insertion element IS1301. Four genes were present in both serogroups (Figure and Technical Appendix Figure 3, panel A). BLASTp searches of the deduced amino acid sequences of the first 2 genes, termed cshA/cszA and cshB/cszB, shared 91% sequence identity with each other and also shared 76% and 63% sequence identity with the capsule biosynthesis genes cps2B and cps2C belonging to Actinobacillus pleuropneumoniae serovars 2, 3, 6, 7, 9, 11, and 13 (38). These genes are predicted to encode a glycerol-3-phosphate cytidylyltransferase, and a hypothetical protein containing a LicD domain for a role in phosphorylcholine incorporation into teichoic acid polymers has been suggested (38). The third genes in region A, termed cshC and cszC, each shared 65% sequence identity with a putative teichoic synthase genes (cps2D and cps9D, respectively) belonging to A. pleuropneumoniae serovars 2, 7, 9, and 13. Last, cshD and cszD were 75% homologous to A. pleuropneumoniae capsule synthesis genes (cps7E).
Serogroups L and X
Serogroup L capsule polysaccharides contain 2-acetamido-2-deoxy-D-glucosyl residues and phosphate groups. Region A contained 3 genes, cslA–C (formerly lcbA–C) and was 4,438 bp long. BLASTp searches of the deduced amino acid sequence from cslA predicted a capsule phosphotransferase sharing 73% sequence identity with LcbA proteins from N. mucosa C102 (GenBank accession no. ACRG00000000) and N. subflava NJ9703 (GenBank accession no. ACEO00000000), whereas cslC encoded an acetyltransferase protein. A gene similar to cslA was identified between regions D′ and B among serogroups I and K and a serogroup W isolate belonging to clonal complex sequence type 22 (Figure); BLASTp searches of the deduced amino acid sequence revealed that these genes shared 97% sequence identity. The longest gene, cslB, was 2,633 bp and putatively encoded a capsule polymerase (Figure and Technical Appendix Figure 2).
The serogroup X capsular polysaccharide is composed of (α-1→4)-linked N-acetylglucosamine 1-phosphate (15). In agreement with findings in a previous study, we found that region A in the serogroup X isolate contained 3 genes, csxA–C (xcbA–C) and was 4,467 bp long, including the insertion sequence, IS1016, located upstream of csxA (Figure and Technical Appendix Figure 2) (19). The deduced amino acid sequence from csxA shared 40% sequence identity with the previously mentioned LcbA protein belonging to N. mucosa C102, indicating that csxA encoded a putative capsule phosphotransferase; however, the remaining 2 serogroup X capsule biosynthesis genes did not share substantial sequence identities with any known protein.
Serogroups I and K
Different structural compositions have been described for these serogroups, with serogroup I consisting of O-acetylated alternating N-acetyl-guluronic acid and N-acetylmannosaminuronic acid units and serogroup K composed of O-acetylated disaccharide repeat units containing N-acetylmannosaminuronic acid (10,11). Both serogroups had almost identical capsule biosynthesis genes, csiA–E or cskA–E; region A was ≈9,026 bp long (Figure and Technical Appendix Figure 3, panel B). MLST analysis showed that serogroup I and K isolates investigated in this study did not possess the same sequence types or PorA and FetA variable regions (Table 1). Antiserum is not commercially available to verify these serogroups; however, the immunochemical difference between serogroup I and K polysaccharides may reflect differences in the original methods used to purify them (10,11). In addition, 2 nonsynonymous substitutions were observed in the capsule biosynthesis genes csiC/cskC and csiD/cskD, for which the deduced amino acid sequence encoded putative glycosyltransferases belonging to families 1 and 2, respectively. The capsule polymerases belonging to serogroups W and Y (csw and csy) are closely related; a single amino acid substitution in the N-terminal glycosyltransferase domain of the capsule polymerase produces the glucose or galactose substrate specificity for the enzyme (39) (online Technical Appendix). It is therefore possible that the nonsynonymous changes observed in the glycosyltransferases, csiC/cskC and csiD/cskD, may produce the serogroup I and K capsules. Further investigation of these serogroups is required.
The derived amino acid sequences from csiA, B, and E/cskA, B, and E shared >70% sequence identity with capsule biosynthesis proteins belonging to Mannheimia haemolytica serotype A1. The capsule of M. haemolytica serotype A1 is composed of a disaccharide repeat of N-acetylmannosaminuronic acid linked with N-acetylmannosamine.
Regions B and C
The genes ctrE and ctrF (formerly lipA and lipB) are required for surface expression of a properly anchored capsule polymer. The ABC (ATP binding cassette) transport system is characterized by the hydrophobic outer and inner membrane proteins CtrA and CtrB, respectively; the integral inner membrane–associated protein, CtrC; and the ATP binding protein, CtrD, and homologous genes are found among other group II capsule-expressing bacteria (40) (see Technical Appendix).
Conclusions
We compared nucleotide sequence data from complete cps loci from all described N. meningitidis serogroups, revealing that the genetic organization is similar for all loci and that region A contains the capsule-specific biosynthesis genes. Distinct capsule operons corresponding to serogroups A, B, C, E, H, I, K, L, W, X, Y, and Z have been described herein, with the serogroup D capsule described as being an unencapsulated serogroup C variant. Nucleotide sequence data for each capsule locus have been deposited in BIGSDB, with genes from each region defined and organized into schemes (26). However, it became apparent that a consistent approach to the nomenclature of capsule genes was required. In 2012, the approach detailed in this paper was approved at the XVIIIth International Pathogenic Neisseria Conference in Würzburg, Germany.
Horizontal genetic transfer of cps genes in region A was evident in serogroups H, I, K, and Z isolates, indicating acquisition of genes from external sources, including the bacterial species A. pleuropneumoniae and M. haemolytica. Combined with the low GC content observed in regions A and C, these data are consistent with acquisition of capsular genetic material from other species. Serogroup determination has been unresolved in some isolates. These isolates may contain capsule genes that are not expressed and will not be detectable by using conventional seroagglutination techniques, or they may be serogroup E, H, I, K, L, X, or Z, which are not routinely searched for and for which commercial antiserum is not available. This study provides additional tools to detect all capsule loci and may ultimately permit determination of the distribution of all serogroups among N. meningitidis populations and detection of cnl isolates.
Artemis comparison tool analyses of Neisseria meningitidis.
Acknowledgments
We thank Paula Kriz, Dominique Caugant, and Robert Mulhall for their kind donation of isolates.
M.C.J.M. is a Wellcome Trust Senior Research Fellow.
Biography
Dr Harrison is a postdoctoral research assistant in the Department of Zoology, University of Oxford. Her research interests include the molecular epidemiology and population structure of Neisseria meningitidis.
Footnotes
Suggested citation for this article: Harrison OB, Claus H, Jiang Y, Bennett JS, Bratcher HB, Jolley KA, et al. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg Infect Dis [Internet]. 2013 April [date cited]. http://dx.doi.org/10.3201/eid1904.111799
References
- 1.Gordon MH. Identification of the meningococcus. J Hyg (Lond). 1918;17:290–315 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slaterus KW. Serological typing of meningococci by means of micro-precipitation. Antonie van Leeuwenhoek. 1961;27:305–15 and . 10.1007/BF02538460 [DOI] [PubMed] [Google Scholar]
- 3.Vedros NA, Ng J, Culver G. A new serological group (E) of Neisseria meningitidis. J Bacteriol. 1968;95:1300–4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding SQ, Ye RB, Zhang HC. Three new serogroups of Neisseria meningitidis. J Biol Stand. 1981;9:307–15 and . 10.1016/S0092-1157(81)80056-X [DOI] [PubMed] [Google Scholar]
- 5.Ashton FE, Ryan A, Diena B, Jennings HJ. A new serogroup (L) of Neisseria meningitidis. J Clin Microbiol. 1983;17:722–7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu TY, Gotschlich EC, Dunne FT, Jonssen EK. Studies on the meningococcal polysaccharides. II. Composition and chemical properties of the group B and group C polysaccharide. J Biol Chem. 1971;246:4703–12 . [PubMed] [Google Scholar]
- 7.Bhattacharjee AK, Jennings HJ, Kenny CP, Martin A, Smith IC. Structural determination of the polysaccharide antigens of Neisseria meningitidis serogroups Y, W-135, and BO1. Can J Biochem. 1976;54:1–8 and . 10.1139/o76-001 [DOI] [PubMed] [Google Scholar]
- 8.Jennings HJ, Lugowski CW, Ashton FE, Ryan JA. The structure of the capsular polysaccharide obtained from a new serogroup (L) of Neisseria meningitidis. Carbohydr Res. 1983;112:105–11 and . 10.1016/0008-6215(83)88270-6 [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharjee AK, Jennings HJ, Kenny CP. Structural elucidation of the 3-deoxy-D-manno-octulosonic acid containing meningococcal 29-e capsular polysaccharide antigen using carbon-13 nuclear magnetic resonance. Biochemistry. 1978;17:645–51 and . 10.1021/bi00597a013 [DOI] [PubMed] [Google Scholar]
- 10.Michon F, Brisson JR, Roy R, Ashton FE, Jennings HJ. Structural determination of the capsular polysaccharide of Neisseria meningitidis group I: a two-dimensional NMR analysis. Biochemistry. 1985;24:5592–8 and . 10.1021/bi00341a046 [DOI] [PubMed] [Google Scholar]
- 11.Van der Kaaden A, Gerwig GJ, Kamerling JP, Vliegenthart JF, Tiesjema RH. Structure of the capsular antigen of Neisseria meningitidis serogroup K. Eur J Biochem. 1985;152:663–8 and . 10.1111/j.1432-1033.1985.tb09246.x [DOI] [PubMed] [Google Scholar]
- 12.van der Kaaden A, van Doorn-van Wakeren JI, Kamerling JP, Vliegenthart JF, Tiesjema RH. Structure of the capsular antigen of Neisseria meningitidis serogroup H. Eur J Biochem. 1984;141:513–9 and . 10.1111/j.1432-1033.1984.tb08222.x [DOI] [PubMed] [Google Scholar]
- 13.Liu TY, Gotschlich EC, Jonssen EK, Wysocki JR. Studies on the meningococcal polysaccharides. I. Composition and chemical properties of the group A polysaccharide. J Biol Chem. 1971;246:2849–58 . [PubMed] [Google Scholar]
- 14.Jennings HJ, Rosell K-G, Kenny CP. Structural elucidation of the capsular polysaccharide antigen of Neisseria meningitidis serogroup Z using 13C nuclear magnetic resonance. Can J Chem. 1979;57:2902–7 . 10.1139/v79-474 [DOI] [Google Scholar]
- 15.Bundle DR, Smith IC, Jennings HJ. Determination of the structure and conformation of bacterial polysaccharides by carbon 13 nuclear magnetic resonance. Studies on the group-specific antigens of Neisseria meningitidis serogroups A and X. J Biol Chem. 1974;249:2275–81 . [PubMed] [Google Scholar]
- 16.Frosch M, Weisgerber C, Meyer TF. Molecular characterization and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidis group B. Proc Natl Acad Sci U S A. 1989;86:1669–73 and . 10.1073/pnas.86.5.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claus H, Vogel U, Muhlenhoff M, Gerardy-Schahn R, Frosch M. Molecular divergence of the sia locus in different serogroups of Neisseria meningitidis expressing polysialic acid capsules. Mol Gen Genet. 1997;257:28–34 and . 10.1007/PL00008618 [DOI] [PubMed] [Google Scholar]
- 18.Swartley JS, Liu LJ, Miller YK, Martin LE, Edupuganti S, Stephens DS. Characterization of the gene cassette required for biosynthesis of the (alpha1→6)-linked N-acetyl-D-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J Bacteriol. 1998;180:1533–9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tzeng YL, Noble C, Stephens DS. Genetic basis for biosynthesis of the (alpha 1→4)-linked N-acetyl-D-glucosamine 1-phosphate capsule of Neisseria meningitidis serogroup X. Infect Immun. 2003;71:6712–20 and . 10.1128/IAI.71.12.6712-6720.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claus H, Maiden MC, Wilson DJ, McCarthy ND, Jolley KA, Urwin R, et al. Genetic analysis of meningococci carried by children and young adults. J Infect Dis. 2005;191:1263–71 and . 10.1086/428590 [DOI] [PubMed] [Google Scholar]
- 21.Parkhill J, Achtman M, James KD, Bentley SD, Churcher C, Klee SR, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–6 and . 10.1038/35006655 [DOI] [PubMed] [Google Scholar]
- 22.Bentley SD, Vernikos GS, Snyder LA, Churcher C, Arrowsmith C, Chillingworth T, et al. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 2007;3:e23 and . 10.1371/journal.pgen.0030023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budroni S, Siena E, Hotopp JC, Seib KL, Serruto D, Nofroni C, et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci U S A. 2011;108:4494–9 and . 10.1073/pnas.1019751108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng J, Yang L, Yang F, Yang J, Yan Y, Nie H, et al. Characterization of ST-4821 complex, a unique Neisseria meningitidis clone. Genomics. 2008;91:78–87 and . 10.1016/j.ygeno.2007.10.004 [DOI] [PubMed] [Google Scholar]
- 25.Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics. 2010;Chapter 11:Unit 5. http://10.1002/0471250953.bi1105s31 [DOI] [PMC free article] [PubMed]
- 26.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595 and . 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–5 and . 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 28.Gudlavalleti SK, Datta AK, Tzeng YL, Noble C, Carlson RW, Stephens DS. The Neisseria meningitidis serogroup A capsular polysaccharide O-3 and O-4 acetyltransferase. J Biol Chem. 2004;279:42765–73 and . 10.1074/jbc.M313552200 [DOI] [PubMed] [Google Scholar]
- 29.Claus H, Borrow R, Achtman M, Morelli G, Kantelberg C, Longworth E, et al. Genetics of capsule O-acetylation in serogroup C, W-135 and Y meningococci. Mol Microbiol. 2004;51:227–39 and . 10.1046/j.1365-2958.2003.03819.x [DOI] [PubMed] [Google Scholar]
- 30.Hobb RI, Tzeng YL, Choudhury BP, Carlson RW, Stephens DS. Requirement of NMB0065 for connecting assembly and export of sialic acid capsular polysaccharides in Neisseria meningitidis. Microbes Infect. 2010;12:476–87 and . 10.1016/j.micinf.2010.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzeng YL, Datta AK, Strole CA, Lobritz MA, Carlson RW, Stephens DS. Translocation and surface expression of lipidated serogroup B capsular polysaccharide in Neisseria meningitidis. Infect Immun. 2005;73:1491–505 and . 10.1128/IAI.73.3.1491-1505.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frosch M, Edwards U, Bousset K, Krausse B, Weisgerber C. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol Microbiol. 1991;5:1251–63 and . 10.1111/j.1365-2958.1991.tb01899.x [DOI] [PubMed] [Google Scholar]
- 33.Bennett JS, Bentley SD, Vernikos GS, Quail MA, Cherevach I, White B, et al. Independent evolution of the core and accessory gene sets in the genus Neisseria: insights gained from the genome of Neisseria lactamica isolate 020–06. BMC Genomics. 2010;11:652 and . 10.1186/1471-2164-11-652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demerec M, Adelberg EA, Clark AJ, Hartman PE. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966;54:61–76 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans JR, Artenstein MS, Hunter DH. Prevalence of meningococcal serogroups and description of three new groups. Am J Epidemiol. 1968;87:643–6 . [DOI] [PubMed] [Google Scholar]
- 36.Bonofiglio L, Garcia E, Mollerach M. Biochemical characterization of the pneumococcal glucose 1-phosphate uridylyltransferase (GalU) essential for capsule biosynthesis. Curr Microbiol. 2005;51:217–21 and . 10.1007/s00284-005-4466-0 [DOI] [PubMed] [Google Scholar]
- 37.Frosch M, Vogel U. Structure and genetics of the meningococcal capsule. In: Frosch M, Maiden MCJ, editors. Handbook of meningococcal disease infection, biology, vaccination, clinical management. Weinheim (Germany): Wiley-VCH; 2006. p. 145–62. [Google Scholar]
- 38.Xu Z, Chen X, Li L, Li T, Wang S, Chen H, et al. Comparative genomic characterization of Actinobacillus pleuropneumoniae. J Bacteriol. 2010;192:5625–36 and . 10.1128/JB.00535-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Claus H, Stummeyer K, Batzilla J, Muhlenhoff M, Vogel U. Amino acid 310 determines the donor substrate specificity of serogroup W-135 and Y capsule polymerases of Neisseria meningitidis. Mol Microbiol. 2009;71:960–71 and . 10.1111/j.1365-2958.2008.06580.x [DOI] [PubMed] [Google Scholar]
- 40.Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315 and . 10.1146/annurev.micro.50.1.285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Artemis comparison tool analyses of Neisseria meningitidis.


